Abstract
The genes for the related human (h) chemokines, PBP (platelet basic protein) and PF4 (platelet factor 4), are within 5.3 kilobases (kb) of each other and form a megakaryocyte-specific gene locus. The hypothesis was considered that the PBP and PF4 genes share a common distal regulatory region(s) that leads to their high-level megakaryocyte-specific expression in vivo. This study examined PBP and PF4 expression in transgenic mice using 4 distinct humanPBP/PF4 gene locus constructs. These studies showed that within the region studied there was sufficient information to regulate tissue-specific expression of both hPBP and hPF4. Indeed this region contained sufficient DNA information to lead to expression levels of PBP and PF4 comparable to the homologous mouse genes in a position-independent, copy number–dependent fashion. These studies also indicated that the DNA domains that led to this expression were distinct for the 2 genes; hPBP expression is regulated by a region that is 1.5 to 4.4 kb upstream of that gene. Expression of hPF4 is regulated by a region that is either intergenic between the 2 genes or immediately downstream of the hPF4 gene. Comparison of the available human and mouse sequences shows conserved flanking region domains containing potential megakaryocyte-related transcriptional factor DNA-binding sites. Further analysis of these regulatory regions may identify enhancer domains involved in megakaryopoiesis that may be useful in the selective expression of other genes in megakaryocytes and platelets as a strategy for regulating hemostasis, thrombosis, and inflammation.
Introduction
Platelet basic protein (PBP) and platelet factor 4 (PF4) are related, platelet-specific chemokines that are expressed at high levels during megakaryopoiesis and stored in platelet α-granules.1 They represent 3% to 5% of total protein releasate from platelets on a molar basis. PBP is N-terminally cleaved to β-thromboglobulin (β-TG) and then to neutrophil-activating peptide-2 (NAP-2).2-4 This final product binds and activates the chemokine receptor CXCR2 on neutrophils.5What role this chemokine has in thrombosis and inflammation is unclear. Unlike PBP and other chemokines, PF4 appears to bind mostly to large, negatively charged molecules such as heparin, and though many biologic functions have been attributed to PF4, its true role in vivo is unknown.6-9
Although the biologic roles of these chemokines need further investigation, their genes offer an opportunity to understand megakaryocyte-specific expression. Like many other genes encoding chemokines, both the PBP and PF4 genes are encoded by 3 exons, and are preceded by a TATA box in the immediate 5′-flanking region.10,11 Transient transcriptional studies with the immediate 5′-flanking region defined a PU.1-binding promoter region upstream of the PBP gene,12 similar to a thrombopoietin-induced enhancer domain upstream of the platelet-specific [alpha]IIbgene.13 Several silencers and promoter domains have been similarly defined in the immediate 5′-flanking region of the PF4 gene, including one GATA-1 binding site.14,15 Reporter gene constructs with 245 bp of the immediate 5′-flanking region of the hPF4 gene showed that this region contained sufficient information to drive tissue-specific expression of a LacZ reporter construct16 and that 1.1 kb of rat PF4 promoter can drive vigorous tissue-specific expression.17 However, neither of these constructs had sufficient information to drive position-independent, copy number–dependent expression.
The genes for PBP and PF4 are closely linked on chromosome 4 with the PBP gene upstream and both transcripts in the same 5′ to 3′ orientation.18-20 Because the 2 genes are both expressed at high levels in developing megakaryocytes, we asked whether the 2 genes are regulated by a common distal regulatory region, similar to those located flanking the α- and β-globin gene loci that are involved in erythroid-specific expression.21 To address these issues, we made a series of transgenic mice with various lengths of the hPBP or the hPF4 genes (or both). Our studies localized distinct regions involved in achieving high level, copy number–dependent expression, at least one for each gene. These potential regulatory regions contain highly conserved domains between human and mouse, which were found to harbor consensus DNA binding sequences of known megakaryocyte-specific transcription factors.
Materials and methods
DNA clones studied
The HindIII fragment used to make theShort-PBP transgenic animals was isolated from a λ phage EMBL3A clone that had been previously characterized by us.10 The Long-PBP and λ-PBP/PF4were also isolated from this same clone as depicted in Figure1. PF4-Only was isolated as anEcoRI fragment from a P1 genomic clone (Genome Systems, St Louis, MO).
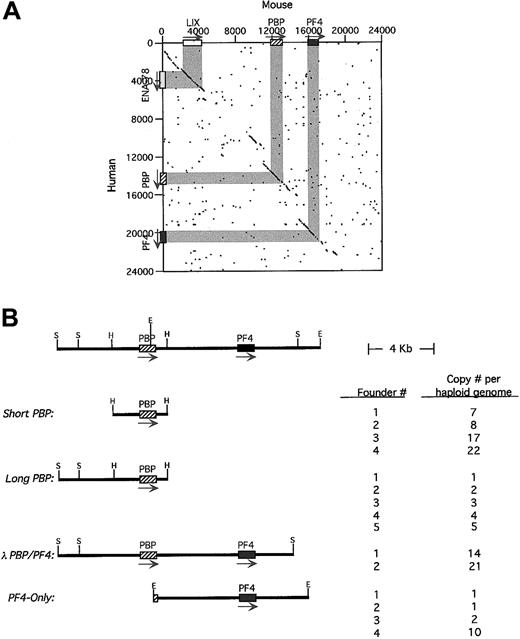
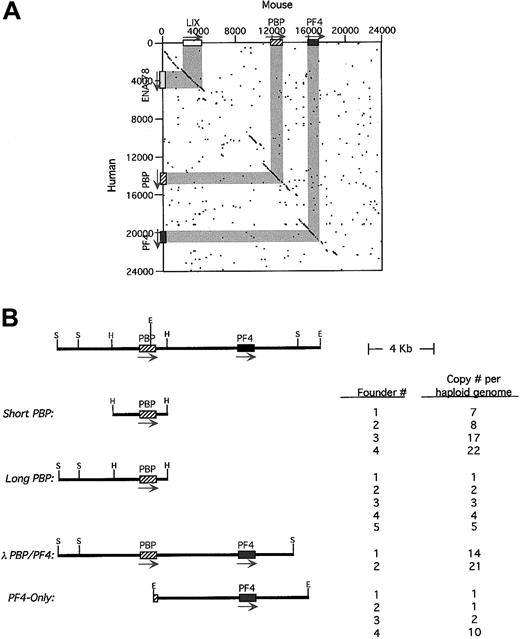
Characterization of the human and mouse
PBP/PF4 gene locus and transgenic constructs. (A) A dot-matrix comparison of the 2 gene loci calculated at 20 bp with 14-bp match. The exons/introns are shown in gray and the arrows indicating the 5′→3′ transcription orientation. (B) A stick figure of the human PBP/PF4 gene locus in humans is shown at the top with a partial restriction map. The constructs used in these studies including the nomenclature are shown. Their copy number per haploid genome is also indicated. H indicatesHindIII; S, SalI; E, EcoRI. The arrow indicates the direction of transcription.
Characterization of the human and mouse
PBP/PF4 gene locus and transgenic constructs. (A) A dot-matrix comparison of the 2 gene loci calculated at 20 bp with 14-bp match. The exons/introns are shown in gray and the arrows indicating the 5′→3′ transcription orientation. (B) A stick figure of the human PBP/PF4 gene locus in humans is shown at the top with a partial restriction map. The constructs used in these studies including the nomenclature are shown. Their copy number per haploid genome is also indicated. H indicatesHindIII; S, SalI; E, EcoRI. The arrow indicates the direction of transcription.
The mouse (m) PF4 gene was isolated from mouse 129 SV λ phage genomic library (Stratagene, La Jolla, CA) using a short region of mPF4 flanking sequence generously provided by Katya Ravid (Boston University, Boston, MA) by standard phage library screening techniques as previously described by us.10 20 Universal T7 primer as well as sequence-specific primer was used to complete the sequence. Additional 5′ and 3′ sequences were obtained from direct sequencing of an mPBP/PF4+ BAC clone isolated by Genome Systems. Dot-matrix homology plot analysis was done by comparing our sequences of the hPBP/PF4 gene locus and supplemental sequences contained within the GenBank public database atwww.ncbi.nih.gov, with our complete sequences of themPBP/PF4 gene locus. Comparative analysis was performed using the DNAsis-Mac version 2.0 (Hitachi Software Engineering, San Bruno, CA) on an Apple Macintosh G4 (Cupertino, CA). Length of comparison was set to 20 bp with a 14-bp match. Analysis of the conserved domains for sequences for consensus GATA, NF-E2, and Ets binding sites was done using Transcription Element Search Software (TESS) developed by the University of Pennsylvania Computational Biology Informatics Laboratory (Philadelphia, PA).
Establishment of transgenic mice
Cesium chloride gradient purified plasmid and λ clone DNAs were cut with appropriate restriction enzymes and run on TAE agarose gel to recover inserts using a Qiaex gel extraction kit (Qiagen, Valencia, CA). The DNAs were then precipitated with ethanol and resuspended in Elutip buffer and passed through Elutip columns (Schleicher & Schuell, Keene, NH). Transgenic mice were generated by pronuclear injections following standard methods at the University of Pennsylvania Transgenic Mice Core Facility. Genomic DNA was isolated from mouse tails using QIAamp DNA Mini Kit (Qiagen). Positive animals were detected by polymerase chain reaction (PCR) analysis of tail DNA using primer pairs for hPF4 or hPBP or both described below and carried out under the following conditions for 30 cycles with Taqpolymerase: denaturing 94°C for 25 seconds, annealing 53°C for 40 seconds, and extension at 72°C for 50 seconds. Amplified products were size fractionated on an agarose gel, ethidium stained, and visualized with UV light.
Positive founder lines were also analyzed by Southern blots as previously described by us.20 Genomic DNA was digested with EcoRI and separated on a 0.8% (wt/vol) agarose gel along with 5 and 10 pg unlabeled probe DNA as controls. Two probes were radiolabeled using a Random Primers Labeling Kit (Boehringer Mannheim, Indianapolis, IN). One was a 1.8-kb hPBP genomic fragment to detect the transgene and the other 1.3 kb fragment was an mαIIb fragment used as a positive mouse genomic control. The filter was washed in 2 × sodium chloride sodium citrate (SSC)/0.1% (wt/vol) (sodium dodecyl sulfate [SDS]) at room temperature, 0.2 × SSC/0.1% (wt/vol) SDS at 65°C, and finally 0.1 × SSC/0.1% (wt/vol) SDS at 65°C, and then exposed on a Phosphorimaging screen (Molecular Dynamics, Sunnyvale, CA). The intensity of bands on film was analyzed by Imagequant PhosphorImager software (Molecular Dynamics). The copy number was determined by comparing the intensity of transgene hPBP band to the genomic control mαIIb band. All studies were approved by the Animal Care and Use Committee of the Children's Hospital of Philadelphia, the University of Pennsylvania, and Thomas Jefferson University.
Reverse transcription–polymerase chain reaction analysis
Tissues examined for RNA expression included the brain, lung, thymus, heart, liver, spleen, small intestine, adrenal, kidney, testes, skeletal muscle, marrow, and purified platelets. Other than the platelets, tissues were obtained after sacrificing the animals by cervical dislocation. The organs were repeatedly rinsed with normal saline to remove residual blood. Tissues were homogenized in RNA Stat-60 (Tel-Test, Friendswood, TX) using a pellet pestle (Kontes Glass, Vineland, NJ). RNA samples were then extracted with chloroform and precipitated in isopropanol. The final RNA pellet was dissolved in water.
Blood was drawn from mice by cardiac puncture and spun at 200gfor 10 minutes to obtain platelet-rich plasma (PRP). The platelets were obtained by centrifugation at 800g for 10 minutes. Platelet RNA were isolated using RNA STAT-60 as described above. Reverse transcription–polymerase chain reaction (RT-PCR) was done using the Superscript II Reverse Transcriptase Kit (Life Technologies, Gaithersburg, MD) following the procedures outlined by the manufacturer (which includes no RT-enzyme control samples). RT-PCR used the following sets of sense/antisense primers:
hPBP: 5′-ATGAGCCTCAGACTTGATAC-3′/5′-ATCAGCAGATTCATGACCTG-3′10
mPBP: 5′-GCCTGCCCACTTCATAACCTC-3′/5′-GGGTCCAGGCACGTTTTTTG-3′
hPF4: 5′-ATCGCACTGAGCACTGAGATC-3′/CTATATAGCAAATGCACACACG-3′11
mPF4: 5′-GTCCAGTGGCACCCTCTTGA-3′/5′-AATTGACATTTAGGCAGCTGA-3′
mαIIb: 5′-GGCTGGAGCACACCTATGAGCT-3′/5′-CTCAACCTTGGGAGATGGGCTG-3′22
mHPRT: 5′-CACAGGACTAGAACACCTGCH-3′/5′-GCTGGTGAAAAGGACCTCT-3′23
The use of RT-PCR to obtain quantitative data regarding the expression of the transgene relative to an endogenous gene requires comparison of products at points when both are in the linear range of amplification. For quantitative studies, the antisense primers were 5′-labeled with fluorescent dye Cy-5 (Integrated DNA Technologies, Coralville, IA). Preliminary experiments showed that the human and mouse PCR products were in the linear range of amplification between cycles 14 to 20. Therefore, PCR reaction mixtures were typically divided into several 15-μL aliquots before initiation of the reaction. PCR was performed at 94°C for 2 minutes, followed by cycles at 94°C for 25 seconds, 50°C for 40 seconds, and 72°C for 40 seconds in a PTC-100 Programmable Thermal Controller (MJ Research, Watertown, MA). At selected cycles aliquots from hPF4 and mPF4 or hPBP and mPBP reactions were removed from the thermocycler and placed on ice. The Cy-5–labeled products were then run on a 10% TBE Ready Gel (Bio-Rad Laboratories, Hercules, CA) and analyzed with a Storm imaging system and Imagequant PhosphorImager software (Molecular Dynamics). The log of the signal intensity of each band was plotted against the cycle number to confirm that the amplification was linear. The signal intensity of the human product was normalized with that of the mouse product to compare the transgene expression levels among the different founder lines. Inclusion of RNase free DNase I (1 U/10 μL reaction, Life Technologies) or DNase free RNase A (1 U/10 μL reaction, Sigma, St Louis, MO) were done in the RT step as controls to verify the RNA nature of the amplified material. Total platelet-derived RNA (1 μg) was added to a PCR tube to which 1 μL 10 × reaction buffer (200 mM Tris-HCl, pH 8.4, 20 mM MgCl2; 500 mM KCl) and 1 μL RNase A or DNase I enzyme was added (total volume = 10 μL). Each tube was incubated at 37°C for 30 minutes. The reaction was stopped by adding 1 μL 25 mM EDTA (pH 8.0) and heating at 70°C for 15 minutes. Oligo dT primer was then added and the RT reaction was followed as described above.
To control for differences in efficiency between the primer pairs, the mPBP and mPF4 and the hPBP and hPF4 complementary DNAs (cDNAs) were all cloned into a single pBluescript SK+ vector (Stratagene). Following the same procedure as described above, the semiquantitative PCR was done using this multigene construct as template. The differences of intensity of the amplified cDNA run on the gel, hPBP versus mPBP and hPF4 versus mPF4 at the same PCR cycles in the range of exponential growth of PCR reaction, were used to determine the differences of the primer pairs amplification efficiency. This information was then used to normalize the expression values determined in the quantitative RT-PCR studies.
Protein detection
Human and mouse platelets were isolated by differential centrifugation of whole blood. Collection of blood from healthy human volunteers was approved by the Institutional Human Review Board at the Children's Hospital of Philadelphia. Samples from both humans and mice were collected in acid-citrate-dextrose (ACD) and centrifuged at 200g for 10 minutes at room temperature as previously described.24 PRP was then collected and prostaglandin E1 (Sigma) was added to a final concentration of 1 μM. Platelets were then obtained by centrifugation at 800g for 10 minutes at room temperature. The pellets were washed twice in 134 mM NaCl, 3 mM KCl, 0.3 mM NaH2PO4, 2 mM MgCl2, 5 mM HEPES, 5 mM glucose, 0.1% NaHCO3, and 1 mM EGTA, pH 6.5, and resuspended in the same buffer except without EGTA. The platelets were then lysed by freezing and thawing the sample twice. The protein concentration of each lysate was determined by the Pierce BCA Protein Assay kit according to the manufacturer's instructions (Pierce, Rockford, IL). Approximately 20 μg total platelet protein was electrophoresed on a 12% SDS-polyacrylamide gel (SDS-PAGE) and stained with Coomassie blue. A duplicate SDS-PAGE gel was transferred to a polyvinylidene difluoride (PVDF) Immobilon-P Transfer Membrane (Millipore, Bedford, MA). The hPF4 protein was detected using RTO, a mouse anti-hPF4 monoclonal antibody generously provided by Gow Arepally (University of New Mexico, Albuquerque, NM),25 and the hPBP protein was detected using a polyclonal rabbit anti–hNAP-2 antibody (PEPRO Tech, Rocky Hills, NJ) that does not cross-react with mouse PBP. Western blot signals were visualized by enhanced chemiluminescence (NEN Life Science Products, Boston, MA) and measured using Imagequant PhosphorImager Software (Molecular Dynamics).
Immunohistochemical staining for hPF4
Tissues from hPF4 transgenic and littermate control mice were immunostained for hPF4 expression using the anti-hPF4 monoclonal antibody RTO. Briefly, formalin-fixed, paraffin-embedded 5-μm sections were deparaffinized in xylene and rehydrated. Endogenous peroxidase activity was quenched with 0.9% peroxide in methanol and unreactive sites blocked with 10% goat serum in 1 × Automation buffer (Biomeda, Foster City, CA) for 20 minutes at 37°C. Slides were then incubated overnight at 4°C with RTO (1 μg/mL), washed in 1 × Automation buffer, and incubated with biotinylated goat-antimouse antibody (Jackson Laboratories, West Grove, PA) diluted 1:200 at 37°C. Slides were washed and incubated with streptavidin–horseradish peroxidase (HRP; Research Genetics, Huntsville, AL) for 30 minutes at 37°C, then were washed and stable DAB chromogen (Research Genetics) was applied for 5 minutes at 20°C. Slides were counterstained with dilute hematoxylin.
Results
Characterization of the PBP/PF4 gene locus
The fact that hPBP and hPF4 form a gene locus has been previously described,20 suggesting that these 2 platelet-specific genes may be coordinately regulated during megakaryopoiesis. We have now completed the characterization of the human PBP/PF4locus and have cloned and characterized the mouse equivalent of thePBP and PF4 genes (GenBank access numbersAF349465 and AF349466). We found that the human genes are 5.3 kb apart. The murine equivalents are also closely linked with both genes oriented in the same 5′→3′ orientation, but with only 3.2 kb in the mice intergenic region. A dot-matrix comparison of the available human and determined mouse sequences shows that homology exists not only within the genes and the immediate 5′- and 3′-flanking regions, but also within several more distal regions as well (Figure 1A). The most 5′ of these conserved regions is actually another CXC chemokine, ENA-78 in human26 and its apparent mouse homologue, LIX.27 This gene is also oriented in the same 5′→3′ orientation as the PBP and PF4 genes. There are 8.9 kb between ENA-78 and hPBP. No other gene was defined up to 40 kb 3′ to the PF4 gene by our analysis. Although the human genome had been shown to contain a nonfunctional duplication of thePBP/PF4 gene complex, ψPBP/PF4alt,10 20 we could not find such a duplication of the mouse genes by analysis of genomic Southern blots (data not shown).
To study the genetic regulation of this region, 4 human transgenic constructs from the hPBP/PF4 gene locus were studied (Figure1B). Short-PBP was a 3.2-kb HindIII fragment that extended 1.4 kb upstream of the hPBP gene's transcriptional start site and 0.8 kb downstream of its poly A signal site, containing only the immediately 5′- and 3′-conserved sequence around PBP.Long-PBP contains an additional 3.0 kb of upstream region, which includes the conserved region upstream of the PBP gene (Figure 1A). The λ-PBP/PF4 construct includes all ofLong-PBP, plus the entire intergenic region, thehPF4 gene and 3.0 kb downstream, including all of the conserved regions flanking both the PBP and PF4genes (Figure 1A). PF4-Only is an EcoRI fragment that begins in the second exon of hPBP, and contains the entire intergenic region, the hPF4 gene and 3.8 kb downstream. This construct is similar to the λ-PBP/PF4 construct, but lacks most of the PBP gene and the conserved region upstream of the PBP gene.
Characterization of transgenic mice: genome copy number and tissue-specific expression
The number of founder animals obtained for each construct is also shown in Figure 1B. Except for the λ-PBP/PF4 construct, at least 4 founders were obtained for each construct. Copy numbers per haploid genome are also indicated in Figure 1B, and except for theλ-PBP/PF4 construct, cover a range of copy numbers.
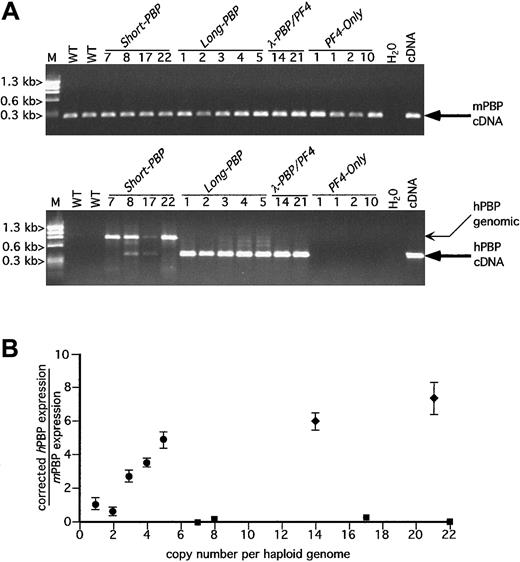
The RT-PCR analysis of brain, lung, thymus, heart, liver, spleen, small intestine, adrenal, kidney, testes, skeletal muscle, marrow, and purified platelets was done using species-specific primers for hPBP, hPF4, mPBP, mPF4, mαIIb, and mHPRT. Expression of the hPBP or hPF4 transgenes and the murine megakaryocyte markers mPBP, mPF4, and mαIIb was limited to platelets, and the 2 hematopoietic tissues, spleen and bone marrow, with no detectable ectopic expression in any of the other tissues examined (data not shown). The ubiquitous mHPRT was readily detectable in these tissues as a positive housekeeping gene control (data not shown). An ethidium bromide gel of the RT-PCR data from platelet RNA, for hPBP and mPBP message, is shown in Figure2A and for hPF4 and mPF4 in Figure3A from transgenic and wild-type mice. In Figures 2A and 3A, the amplifications were done for 30 cycles and are only roughly quantitative. The observed genomic bands seen in several lanes in Figure 2A were only variably present and were not seen at the more limited cycle numbers used for quantitative analysis in Figure 2B. DNase pretreatment prior to RT-PCR also eliminated the genomic bands (data not shown).
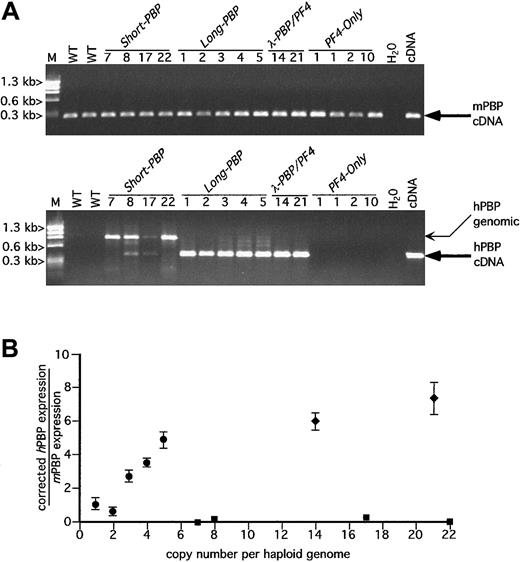
RT-PCR analysis of hPBP versus mPBP.
(A) Ethidium bromide analysis of platelet RT-PCR products for mPBP (top) and for hPBP (bottom) for all of the founder lines studied plus wild-type (WT) mouse platelets and the appropriate cDNA control are included. The founder lines are numbered by transgenic copy. The PCR amplification was carried out for 30 cycles. The anticipated cDNA band is indicated by a large arrow. The genomic DNA band is indicated by a smaller arrow and is only occasionally prominent when there is no or little RNA message. No cDNA bands were seen when the RT step was skipped or when RNase A was included in the RT step (data not shown). WT indicates wild-type litter mates; M, φX HaeIII marker. (B) Relationship between the copy number for the PBP-expression transgenic lines Short-PBP, Long-PBP, andλ-PBP/PF4 and the level of hPBP versus mPBP message are shown. The relative quantity of hPBP to mPBP message was determined using Cy5-labeled primers in the amplification step and measuring final product quantitatively with a Storm imager. Analysis was done of the PCR products within the logarithmically amplifying range of cycles (usually 14-20 cycles) corrected for the relative efficiency of the 2 sets of primer pairs; ▪, Short-PBP; ●,Long-PBP; ♦, λ-PBP/PF4. Data are shown for average ± 1 SD. Experiments were repeated 3 separate times.
RT-PCR analysis of hPBP versus mPBP.
(A) Ethidium bromide analysis of platelet RT-PCR products for mPBP (top) and for hPBP (bottom) for all of the founder lines studied plus wild-type (WT) mouse platelets and the appropriate cDNA control are included. The founder lines are numbered by transgenic copy. The PCR amplification was carried out for 30 cycles. The anticipated cDNA band is indicated by a large arrow. The genomic DNA band is indicated by a smaller arrow and is only occasionally prominent when there is no or little RNA message. No cDNA bands were seen when the RT step was skipped or when RNase A was included in the RT step (data not shown). WT indicates wild-type litter mates; M, φX HaeIII marker. (B) Relationship between the copy number for the PBP-expression transgenic lines Short-PBP, Long-PBP, andλ-PBP/PF4 and the level of hPBP versus mPBP message are shown. The relative quantity of hPBP to mPBP message was determined using Cy5-labeled primers in the amplification step and measuring final product quantitatively with a Storm imager. Analysis was done of the PCR products within the logarithmically amplifying range of cycles (usually 14-20 cycles) corrected for the relative efficiency of the 2 sets of primer pairs; ▪, Short-PBP; ●,Long-PBP; ♦, λ-PBP/PF4. Data are shown for average ± 1 SD. Experiments were repeated 3 separate times.
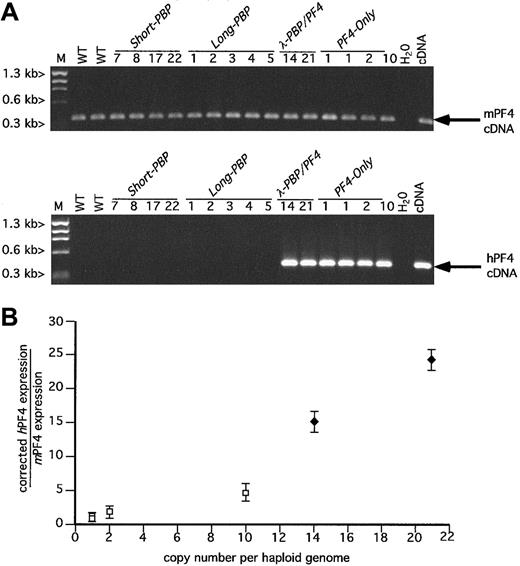
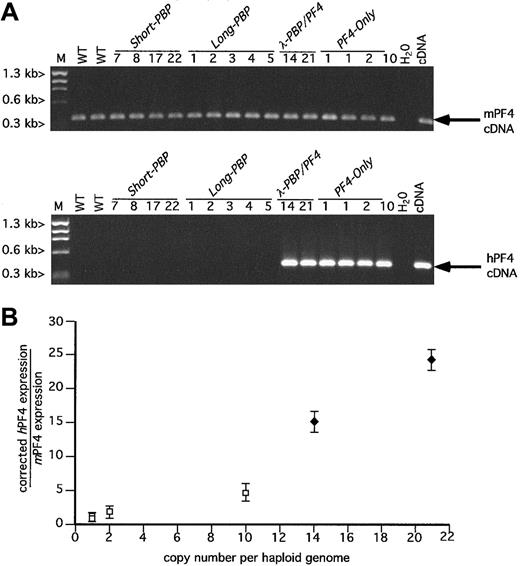
RT-PCR analysis of hPF4 versus mPF4.
(A) Ethidium bromide analysis as in Figure 2A, but for PF4. (B) Relationship between the copy number of the transgene and the platelet level of message of human versus mouse PF4 was determined for the PF4-expression lines λ-PBP/PF4 and PF4-Only. The determination of the relative quantity of hPF4 to mPF4 message was as described in Figure 2B. ■, PF4-Only; ♦,λ-PBP/PF4. Data are shown for average ± 1 SD. Experiments were repeated 3 separate times.
RT-PCR analysis of hPF4 versus mPF4.
(A) Ethidium bromide analysis as in Figure 2A, but for PF4. (B) Relationship between the copy number of the transgene and the platelet level of message of human versus mouse PF4 was determined for the PF4-expression lines λ-PBP/PF4 and PF4-Only. The determination of the relative quantity of hPF4 to mPF4 message was as described in Figure 2B. ■, PF4-Only; ♦,λ-PBP/PF4. Data are shown for average ± 1 SD. Experiments were repeated 3 separate times.
Interestingly, none of the Short-PBP constructs expressed hPBP message well (Figure 2A and solid squares in Figure 2B). Among the 4 founders, only the 8 and 17 copy lines had any detectable hPBP, and this amounted to less than 1% of concurrently expressed mPBP when analyzed over the exponential range of amplification and then corrected for efficiency of amplification of different primer pairs (Figure 2B). Furthermore, 2 other founder lines of the Short-PBPconstruct with similar or higher copy numbers (7 or 22 copies) did not have any detectable expression. Thus, although the Short-PBPconstruct may drive tissue-specific expression, it does so inefficiently and not in a copy number–dependent fashion.
To see if the inclusion of the conserved region further upstream of thehPBP gene was involved in the regulated expression of PBP, the Long-PBP constructs that contained this region were examined (Figure 1A). All of the transgenic lines from this construct expressed high levels of hPBP message (Figure 2A) that on quantitation was approximately equal to endogenous mPBP expression on a per copy basis (Figure 2B). The 2 λ-PBP/PF4 lines, which have the same 3.0 kb of sequence upstream of hPBP, also expressed high levels of hPBP message. Expression per gene copy tends to plateau at high copy numbers in transgenic mice, but it appears that the addition of more 3′ sequence did not further increase PBP message levels detected. Together, these constructs suggest that the distal 5′-flanking region (between 4.4 and 1.4 kb) is necessary and perhaps sufficient to achieve high-level expression of hPBP gene in a tissue-specific and copy number–dependent fashion.
The 4 PF4-Only founder lines expressed high levels of hPF4 message, comparable to endogenous mPF4 levels of message (Figure 3A,B). The level of expression was position independent, copy number dependent (Figure 3B) with the construct with the highest copy number having the highest level of expression. The 2 λ-PBP/PF4 constructs also expressed hPF4 at a high level and seemingly also copy number dependent (Figure 3A,B). Comparison of the 14 copyλ-PBP/PF4 and the 10 copy PF4-Only expression levels suggests that the λ-PBP/PF4 construct drives perhaps an additional 3-fold increase in expression.
PBP and PF4 protein expression by the transgenic lines
Although considerable effort was made to quantitatively analyze mRNA level in the above experiments, we also measured hPBP and hPF4 protein levels as a secondary determination of expression (Figures4 and 5). Human platelet proteins from 4 volunteers were used as a positive control. Levels of PBP and PF4 expression were done on a per milligram basis rather than on a per platelet basis because mouse platelets are much smaller than human platelets.28 Protein concentrations were determined by optical density measurements and confirmed by Coomassie blue stained SDS-PAGE gels (data not shown).
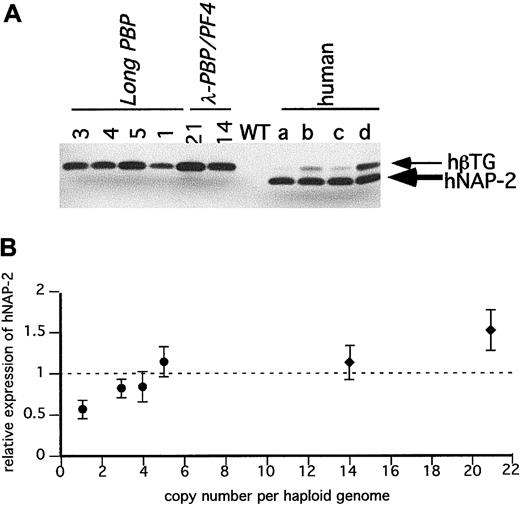
Immunodetection of hPBP in the transgenic mice platelets.
(A) SDS-PAGE gels of equal quantities of platelet protein were immunoblotted with an anti–NAP-2 polyclonal antibody that does not recognize mPBP proteins. Platelet proteins from the transgenic linesLong-PBP and λ-PBP/PF4 were studied. Control lanes include wild-type mice and 4 different normal human volunteers (a through d). (B) The relative expression of hPBP in the transgenic mice to the average value in the 4 human samples per milligram total platelet protein was determined and compared to its copy number. The dotted line is the average human platelet hPBP value. ●,Long-PBP; ♦, λ-PBP/PF4. Data are shown for average ± 1 SD. Experiments were repeated 3 separate times.
Immunodetection of hPBP in the transgenic mice platelets.
(A) SDS-PAGE gels of equal quantities of platelet protein were immunoblotted with an anti–NAP-2 polyclonal antibody that does not recognize mPBP proteins. Platelet proteins from the transgenic linesLong-PBP and λ-PBP/PF4 were studied. Control lanes include wild-type mice and 4 different normal human volunteers (a through d). (B) The relative expression of hPBP in the transgenic mice to the average value in the 4 human samples per milligram total platelet protein was determined and compared to its copy number. The dotted line is the average human platelet hPBP value. ●,Long-PBP; ♦, λ-PBP/PF4. Data are shown for average ± 1 SD. Experiments were repeated 3 separate times.
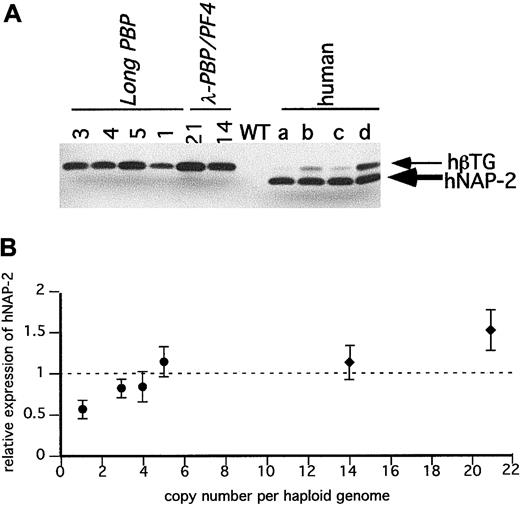
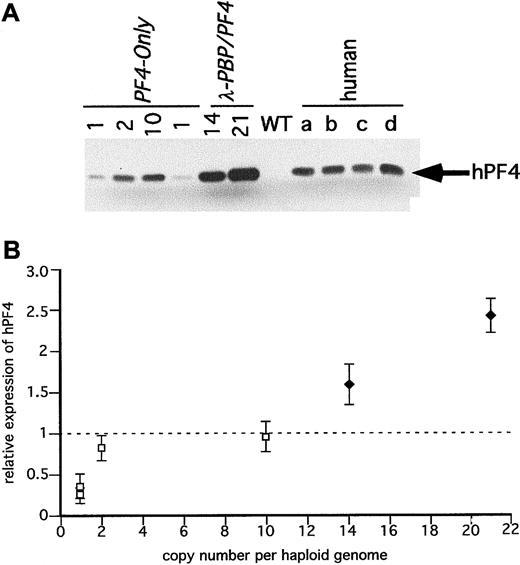
Immunodetection of hPF4 in the transgenic mice platelets.
(A) SDS-PAGE gels of equal quantities of platelet protein were immunoblotted with an anti-hPF4 monoclonal antibody RTO that does not recognize mPF4 proteins. Platelet proteins from the transgenic linesPF4-Only and λ-PBP/PF4 were studied. Control lanes include wild-type mice and 4 different normal human volunteers (a through d). (B) The relative expression of hPF4 in the transgenic mice to the average value in the 4 human samples per milligram total platelet protein was determined and compared to its copy number. The dotted line is that average human platelet hPF4 value. ■PF4-Only; ♦, λ-PBP/PF4. Data are shown for average ± 1 SD.
Immunodetection of hPF4 in the transgenic mice platelets.
(A) SDS-PAGE gels of equal quantities of platelet protein were immunoblotted with an anti-hPF4 monoclonal antibody RTO that does not recognize mPF4 proteins. Platelet proteins from the transgenic linesPF4-Only and λ-PBP/PF4 were studied. Control lanes include wild-type mice and 4 different normal human volunteers (a through d). (B) The relative expression of hPF4 in the transgenic mice to the average value in the 4 human samples per milligram total platelet protein was determined and compared to its copy number. The dotted line is that average human platelet hPF4 value. ■PF4-Only; ♦, λ-PBP/PF4. Data are shown for average ± 1 SD.
Figure 4A is an immunoblot of platelet proteins from 4 human volunteers and from the Long-PBP and λ-PBP/PF4 lines. The double bands seen with human platelets in Figure 4A likely represents differential N-terminal cleavage products of hPBP protein.29 The lower band has the mobility of recombinant NAP-2 (data not shown), and the upper band is likely to be β-TG. In humans, it appears to be variably cleaved to β-TG and NAP-2, and in mice, the predominant product from hPBP appears to be β-TG and not NAP-2. These differences suggest that there is differential cleavage of hPBP between human and mice platelets. Not shown is that platelets from the Short-PBP constructs had no detectable hPBP, consistent with their low levels of hPBP message. The level of hPBP expression increased in the same order as the level of hPBP message seen, although the degree of increase was blunted (Figure 4A,B). This may be due to a limitation in achievable protein expression, or perhaps excess hPBP cannot be efficiently packaged into the platelet α-granules and is lost by the developing megakaryocytes.
Figure 5A is an immunoblot of platelet proteins from the same human volunteers and from the PF4-Only and λ-PBP/PF4lines. Both sets of transgenic lines have detectable hPF4, and the levels increased in the same order as their hPF4 platelet RNA level (Figure 5B). Again, the degree of increase in protein was blunted compared to the degree of increase in message level, perhaps for the same reasons as mentioned above concerning hPBP.
It should be noted that levels of hPBP and hPF4 protein found in a number of lines equal that or exceed that of the proteins found in the tested human samples. For hPBP, the maximum level expressed was 1.5 times the level seen in the human controls, and for hPF4, the maximum level seen was 2.5 times the level seen in the 4 controls. None of the lines studied had decreased viability (being transmitted at the expected mendelian frequency) or had abnormalities in their blood counts (data not shown).
Immunohistochemical staining for hPF4
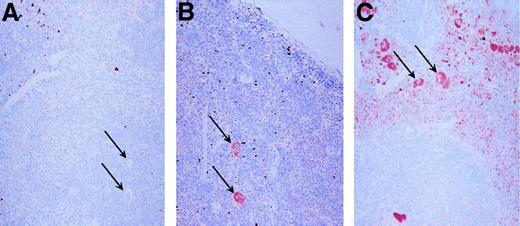
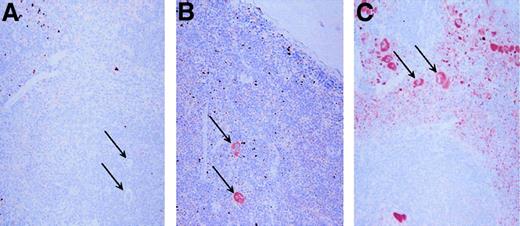
Tissues from PF4 transgenic and wild-type control mice were immunostained for hPF4 expression using RTO. Only the bone marrow and spleen showed positive immunostaining, consistent with the RT-PCR studies mentioned above (data not shown). Figure6 shows that in the spleen, which is a hematopoietic tissue in mice, only mature megakaryocytes show significant staining in the 10-copy PF4-Only and in the 21-copy λ-PBP/PF4 animals, whereas megakaryocytes were unstained in the wild-type control. Of interest is that the platelets in the λ-PBP/PF4 spleen are so intensely stained that their punctate staining allows one to readily distinguish the red pulp, which contains circulating platelets, from the white pulp.
Immunohistochemical localization of PF4.
Spleens were stained with the monoclonal anti-PF4 RTO as the primary antibody. Each spleen was counterstained with hemotoxylin. Panel A represents immunohistochemical studies of the spleen from a wild-type mouse, whereas panel B is from a 10 copy PF4-Only mouse, and panel C is from a 21 copy λ-PBP/PF4 mouse. The arrows point to some, but not all, of the megakaryocytes in each field. Original magnification 20 ×.
Immunohistochemical localization of PF4.
Spleens were stained with the monoclonal anti-PF4 RTO as the primary antibody. Each spleen was counterstained with hemotoxylin. Panel A represents immunohistochemical studies of the spleen from a wild-type mouse, whereas panel B is from a 10 copy PF4-Only mouse, and panel C is from a 21 copy λ-PBP/PF4 mouse. The arrows point to some, but not all, of the megakaryocytes in each field. Original magnification 20 ×.
Discussion
Regulation of megakaryocyte-specific genes has been studied by our laboratory and others to gain insights into the mechanisms underlying megakaryocyte differentiation. Given the paucity of megakaryocytes in bone marrow these studies have previously heavily required the use of cell lines with some, but not complete, megakaryocytic features.30-32 Previous studies of megakaryocyte-specific gene expression in transgenic mice have provided additional insights into this process. However, these studies used reporter gene constructs, which in themselves often contain cryptic regulatory components. Further, these studies have not dealt with the important issues of expression level in megakaryocytes, and copy number dependence and position independence.
Our studies have focused on the PBP/PF4 gene locus because both genes are highly expressed, megakaryocyte-specific chemokines and are physically linked to one another on human chromosome 4.20 The studies presented show this tight linkage to be conserved for the mPBP and mPF4 genes. Further, our studies are the first to show close physical linkage to an additional related CXC chemokine gene ENA-78/LIX, located 8.9 and 6.9 kb upstream of the transcription start site ofhPBP and mPBP genes, respectively. ENA-78 is also expressed in, but not restricted to, megakaryocytes. These data are consistent with our initial supposition that this region could potentially represent a common expression locus for several platelet-specific chemokines.
We also proposed that given their proximity, the PBP andPF4 genes might possess common distal regulatory elements. Phylogenetic footprinting data demonstrated in Figure 1A show that not only are PF4, PBP, and ENA-78 highly conserved across mammalian species, but there are a number of regions outside of the coding regions that are highly conserved. Our data show that thePBP and PF4 genes each contain distinct regulatory regions in their flanking regions that allow for their copy number–dependent and position-independent expression. A region upstream to the PBP gene was necessary for these features for PBP gene expression and may also contribute to an approximate 3-fold enhanced expression of the PF4 gene. Despite this apparent expression augmentation, copy number–contingent high levels of PF4 expression can be achieved independent of this element, being regulated by the flanking regions surrounding thePF4 gene.
Further studies are needed to define the molecular basis by which these 5′-flanking regions regulate the efficient expression of these 2 genes. Such important regulatory elements are likely to be conserved, and indeed, phylogenetic footprinting data analyses for other genes have been useful in localizing important distal regulatory elements,33,34 and may apply to our data as well. For example, our expression data show that the Short-PBP andLong-PBP constructs differ only in that theLong-PBP includes the region 1.4 to 4.4 kb upstream of thehPBP gene. Thus, this region is necessary for high-level tissue-specific and copy number–dependent expression of thehPBP gene. Within this region is a highly conserved approximate 700-bp domain, showing about 60% nucleotide conservation (Figures 1A and 7A). We analyzed this region in the mouse and human sequences for transcription factor consensus binding sequences implicated in the control of megakaryocytic and hematopoietic gene regulation, focusing on GATA-1, NF-E2, Ets, PU.1, and AML-1 binding sites.35-47 Of note is a homologously conserved NF-E2 site beginning 3696 (2151) bp upstream of the PBP start site (with the mouse distance in parenthesis), and a GATA-1 site 3140 (1644) bp upstream (Figure 7A). Whether either site is biologically relevant will now need to be tested. Additionally, several regions contain other GATA-1 and Ets binding sites that are present within about 60 bp of each other in the mouse and human sequences but that may nevertheless be functionally significant. Studies of enhancers have classically demonstrated independence for physical attributes such as distance and orientation. It is possible that there is no evolutionary constraint in this context to keep these binding sites precisely aligned, but rather generally present within the boundaries of the enhancer domain.
Comparison of the homologous domains in the flanking regions of the PBP/PF4 locus.
A comparison of the conserved regions, other than within the coding areas or the immediate 1.0 kb of 5′-flanking region, of the mouse and human PBP and PF4 genes are shown. A “‖” refers to an identical nucleotide. Consensus DNA binding sites for a number of megakaryocyte-specific transcriptional factors are highlighted and the names of the transcriptional factors and their orientations are shown. Panel A represents the conserved domain upstream of the PBP gene, and negative numbering begins at the PBP transcriptional start site.11 Panel B represents the conserved intergenic region, and negative numbering begins at the PF4 transcriptional start site.10 Panel C represents the conserved region downstream of the PF4 gene, and numbering begins at the PF4 transcriptional start site.
Comparison of the homologous domains in the flanking regions of the PBP/PF4 locus.
A comparison of the conserved regions, other than within the coding areas or the immediate 1.0 kb of 5′-flanking region, of the mouse and human PBP and PF4 genes are shown. A “‖” refers to an identical nucleotide. Consensus DNA binding sites for a number of megakaryocyte-specific transcriptional factors are highlighted and the names of the transcriptional factors and their orientations are shown. Panel A represents the conserved domain upstream of the PBP gene, and negative numbering begins at the PBP transcriptional start site.11 Panel B represents the conserved intergenic region, and negative numbering begins at the PF4 transcriptional start site.10 Panel C represents the conserved region downstream of the PF4 gene, and numbering begins at the PF4 transcriptional start site.
Expression of hPF4 from all animals, including the 4PF4-Only founder lines and the 2 λ-PBP/PF4lines, was tissue specific. Previous transgenic studies with 245 bp and 1.1 kb of the immediate 5′-flanking region of the mouse and rat PF4 promoters driving Lac Z reporter genes showed excellent tissue specificity, but the relative degree of expression and copy number dependency and position independence were not well studied.16 17 The present studies with PF4-Onlyshow that the inclusion of the entire PBP and PF4 intergenic region, the intact PF4 gene itself, and 3.8 kb of 3′-flanking sequence was sufficient to achieve megakaryocyte-restricted high level, copy number–dependent and position-independent expression, comparable to the endogenous mPF4 gene. Interestingly, these flanking regions contain several interspecies conserved domains, including an intergenic region beginning 4989 (2728) bp upstream of the hPF4 transcriptional start site (Figure 7B), and a region beginning 1018 (942) bp downstream of the hPF4 gene (Figure 7C). Analysis of these regions again show about 60% nucleotide conservation in these regions. A search for consensus DNA binding sites as described above found multiple consensus sequences for GATA, NF-E2, Ets, PU.1, and AML-1 binding with one homologously conserved site, which was a GATA consensus binding site 1716 (1705) bp downstream of the PF4gene. The functional relevance of these and other sites not readily apparent from the above analysis will require further investigation.
It is intriguing that the λ-PBP/PF4 transgenics have a further increase in hPF4 expression in transgenic animals compared to the PF4-Only transgenics. This could be the result of their having higher copy numbers although the highest PF4-Only had 10 copies and the lowest λ-PBP/PF4 had 14 copies, yet had 3 times the message level. It is possible that the 5′-enhancer region 1.4 to 4.4 upstream of the hPBP gene also regulateshPF4 gene expression. Additional λ-PBP/PF4lines with lower copy numbers and a truncated version ofλ-PBP/PF4, lacking the region from 1.4 to 4.4 kb upstream of hPBP would be needed to see whether the PBP upstream enhancer domain increases PF4 as well as PBP expression. Conversely, additional 3′ sequence of the PBP gene, including the intergenic region and the region downstream of the PF4 gene do not contribute to the level of PBP expression.
Our study clearly shows we have defined important domains for high level, tissue-specific expression within the flanking regions of thePBP and PF4 genes. With the identification ofENA-78 gene immediately upstream of the PBP/PF4gene locus, the question of whether there is a larger megakaryocyte-specific gene locus involved in the regulation of all 3 genes is raised. Given that all known CXC chemokines localize to a single locus on human chromosome 420,48,49 and that our studies show that ENA-78, PBP, and PF4 are clustered and transcribed in the same 5′→3′ direction, this possibility becomes more likely. Further, although ENA-78 was first recognized in epithelial cells, it is highly expressed in platelets,50,51 and nonquantitative RT-PCR suggests that its platelet RNA message is as abundant as PBP.51 Because other CXC chemokines are also expressed in platelets,52 we believe further analysis of the CXC chemokine locus may elucidate other physical and functional links for additional CXC chemokine genes to the PBP/PF4 gene locus.
For over 40 kb downstream of the PF4 gene, we found no other CXC chemokine genes. Whether this means that theENA-78/PBP/PF4 genes are at one end of the CXC chemokine locus is not clear yet. Furthermore, we have yet to determine the precise relative location of the inactive, duplicatedψPBP/PF4alt human gene locus, which is within the same human chromosome 4 locus.20 This duplication, though within the CXC chemokine gene locus, is not closely linked to the functional PBP/PF4 locus.
The protein levels of hPBP and hPF4 in the founder lines are consistent with the semiquantitative RT-PCR data. The 2 λ-PBP/PF4lines have the highest PBP and PF4 RNA and protein levels. The expressions of hPBP and hPF4 in these and a number of other lines are equivalent or higher than PBP and PF4 expressions in human platelets both by immunoblot and tissue immunostaining. Whether these animal lines might be useful to study the biologic and pathobiologic functions of hPBP and hPF4 in mouse models is being pursued. We have already used the highest expressing PF4-Only construct to develop and characterize a mouse model for the pathologic state heparin-induced thrombocytopenia. We found that an accurate mouse model of the disease requires the expression of hPF4 in vivo.53Therefore it is apparent that these constructs are useful not only to better understand the regulation of PBP and PF4gene expression, but also to study the human proteins that are expressed.
We wish to thank Dr Gowthami Arepally at the University of New Mexico, Albuquerque, NM, for providing the mouse anti-hPF4 monoclonal antibody RTO, and Dr Katya Ravid, Boston University, Boston, MA, for sharing unpublished sequences with us. We would also like to thank Zheng Cui, Diana Cassel, and Saul Surrey at the Thomas Jefferson University as well as Jean Richa at the Transgenic Mouse Facility of the University of Pennsylvania for their technical help.
Supported in part by grants from the American Society of Hematology (to C.Z.), the National Institutes of Health HL40387 (to M.P.), HL40387 (to S.E.M.), and HL61865 (to S.E.M. and M.P.R.), and the Nemours Foundation (to S.E.M. and M.P.R.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Chunyan Zhang, The Children's Hospital of Philadelphia, Abramson Research Center, Rm 314, 34th St and Civic Center Blvd, Philadelphia, PA 19104; e-mail: zcy118@yahoo.com.