Abstract
Osteoclasts and dendritic cells are derived from monocyte/macrophage precursor cells; however, how their lineage commitment is regulated is unknown. This study investigated the differentiation pathways of osteoclasts and dendritic cells from common precursor cells at the single-cell level. Osteoclastogenesis induced by macrophage colony-stimulating factor (M-CSF) and receptor activator of nuclear factor–κB ligand (RANKL) or tumor necrosis factor-α (TNF-α) is completely inhibited by addition of granulocyte-macrophage colony-stimulating factor (GM-CSF) or interleukin-3 at early stages of differentiation. GM-CSF–treated cells express both c-Fms and RANK and also low levels of CD11c and DEC205, which are detected on dendritic cells. Addition of GM-CSF also reduces expression of both c-Fos and Fra-1, which is an important event for inhibition of osteoclastogenesis. Overexpression of c-Fos by retroviral infection or induction in transgenic mice can rescue a failure in osteoclast differentiation even in the presence of GM-CSF. By contrast, differentiation into dendritic cells is inhibited by M-CSF, indicating that M-CSF and GM-CSF reciprocally regulate the differentiation of both lineages. Dendritic cell maturation is also inhibited when c-Fos is expressed at an early stage of differentiation. Taken together, these findings suggest that c-Fos is a key mediator of the lineage commitment between osteoclasts and dendritic cells. The lineage determination of osteoclast progenitors seen following GM-CSF treatment functions through the regulation of c-Fos expression.
Introduction
Osteoclasts and dendritic cells are the monocyte/macrophage lineage cells derived from hematopoietic stem cells.1-5 However, how commitment to a given lineage is established among macrophages, osteoclasts, and dendritic cells is unclear. Macrophage colony-stimulating factor (M-CSF), granulocyte-macrophage colony-stimulating factor (GM-CSF), and interleukin-3 (IL-3) are macrophage-inducing cytokines with different specificities of action. GM-CSF and IL-3 can induce lineages of hematopoietic cells other than macrophages. The effect of M-CSF is restricted to induction of macrophages, although it is also important for osteoclastogenesis.6-8 We have established an in vitro osteoclast differentiation system using isolated precursor cells and a stromal cell–free culture media containing M-CSF and a soluble form of receptor activator of nuclear factor–κB ligand (sRANKL).9,10 This system enables us to observe directly the effect of various cytokines and transcription factors on osteoclasts. RANK and RANKL were shown to be essential for osteoclast development11,12 and dendritic cell activation.13 In addition to M-CSF and RANKL, osteoclastogenesis is stimulated by IL-1 and tumor necrosis factor-α (TNF-α)14-17 and inhibited by interferon γ (IFN-γ), IL-18, and GM-CSF.18-20 However, it is not known how osteoclastogenesis is inhibited or whether cells differentiate into another lineage following inhibition.
Several transcription factors are known to be essential for osteoclast formation. They include PU.1,21 c-Fos,22,23nuclear factor κB1 (NF-κB1), and NF-κB2.24 PU.1-deficient mice show a macrophage differentiation failure, and osteoclastogenesis is inhibited at an early stage of differentiation. The c-Fos is a component of the dimeric transcription factor AP-1, which includes not only c-Fos but FosB, Fra-1, Fra-2, and Jun proteins such as c-Jun, JunB, and JunD. Mice deficient in c-Fos exhibit osteopetrosis due to an osteoclast differentiation defect, while the number of macrophages increases,23 indicating that inhibition of differentiation occurs later than in PU.1-deficient mice. Fra-1 is thought to be a target of c-Fos,25,26 which is supported by the observation that Fra-1 expression can rescue the osteoclastogenesis defect seen in c-Fos–deficient mice.27 NF-κB is downstream of RANK28 and TNF receptor–associated factor 6 (TRAF6),29 which are also essential for osteoclast differentiation,12,30 and the phenotype of mice doubly mutant in NF-κB1 and NF-κB2 includes an osteoclast differentiation failure.24 Dendritic cells are derived from hematopoietic stem cells4,5,31 and monocyte/macrophage progenitor cells.1 Dendritic cell differentiation is stimulated by GM-CSF, TNF-α, Flt-3 ligand, IL-4, or RANKL.4,13,31-34 Furthermore, RelB, PU.1, and Ikaros are reported to be required for dendritic cell development,35-37 as evidenced by the absence of dendritic cells in both RelB−/− and Ikaros−/−mice35,37 and the increase of monocytes/macrophages seen in Ikaros mutants.37
In this study, we characterized a lineage bifurcation of monocyte/macrophage precursor cells using an in vitro differentiation system. Osteoclast differentiation was inhibited and dendritic cell differentiation was reciprocally stimulated by GM-CSF through suppression of c-Fos. On the other hand, dendritic cell development was inhibited by expression of c-Fos, indicating that early c-Fos expression is involved in the lineage determination of osteoclasts and dendritic cells.
Materials and methods
Fluorescence-activated cell-sorter analysis and cell sorting
Two-month-old C57/BL6 (female) mice were purchased from SCL (Shizuoka, Japan). Transgenic mice in which c-Fos expression is under control of the IFN-α/β–inducible Mx promoter (Mx–c-fos)38,39 were maintained as heterozygotes. Mouse bone marrow cells were harvested from femurs and tibias, and osteoclast precursor cells (c-Kit+c-Fms+RANK− or c-Fms+RANK− cells) were prepared as previously described.10 Briefly, mononuclear cells were stained with biotinylated anti-RANK monoclonal antibody (muRANK-M395) (a gift from Immunex, Seattle, WA) followed by allophycocyanin-conjugated streptavidin (Caltag Laboratories, San Francisco, CA), fluorescein isothiocyanate–conjugated c-Fms (AFS98) (a gift from Toray Industries, Kamakura, Japan), and R-phycoerythrin (R-PE)–conjugated c-Kit (2B8) (Pharmingen, San Diego, CA). Cell sorting was performed using fluorescence-activated cell-sorter (FACS) Vantage or FACS Calibur (Becton Dickinson Immunocytometry Systems, San Jose, CA). Data were analyzed using Lysys II software on a Consort 32 system or Cellquest software (Becton Dickinson).
The c-Kit+c-Fms+RANK− cells were clone-sorted, and each single cell was plated in a well of 96-well culture plates (Primaria; Falcon3872, Becton Dickinson) using the FACS Vantage single-cell sort system and cultured in the presence of M-CSF and sRANKL; GM-CSF and sRANKL; or M-CSF, sRANKL, and GM-CSF; as described below. After 6 days of cultivation, cells were identified, and the frequency of the wells containing osteoclasts or dendritic cells was determined.
Osteoclast differentiation in vitro
A total of 1000 sorted cells were plated on 96-well culture plates (Primaria) and cultured in α–minimal essential medium (Gibco, Gaithersburg, MD) containing 10% fetal bovine serum (JRH Bioscience, Lenexa, KS), 100 ng/mL M-CSF (R&D Systems, Minneapolis, MN), and 25 ng/mL sRANKL (prepared as previously described9) with or without GM-CSF, IL-3, or IL-5 (purchased from R&D Systems) in the presence or absence of IFN-α/β (Sigma Chemical, St Louis, MO) for 6 days. For the GM-CSF blocking assay, a neutralizing antibody against GM-CSF (R&D Systems) was added to the culture medium containing 100 ng/mL M-CSF, 25 ng/mL sRANKL, and 1 ng/mL GM-CSF. In some experiments, various concentrations of TNF-α (R&D Systems) were added to the cultures instead of sRANKL. To block sRANKL and TNF-α, RANK-Fc (R&D Systems) was added to the culture medium containing 100 ng/mL M-CSF and 25 ng/mL sRANKL or 25 ng/mL TNF-α. Cultured cells were subjected to tartrate-resistant acid phosphatase (TRAP) staining (Sigma) or TRAP solution assays as previously described.9
Analysis of c-Fms, RANK, and GM-CSF receptor expression
Osteoclast precursor cells (c-Fms+RANK−cells) were cultured in the presence of M-CSF, GM-CSF, or both for 24 or 72 hours on fibronectin-coated plates that were preincubated with 2% bovine serum albumin/phosphate-buffered saline for 30 minutes at 37°C. Cells were then harvested using a cell scraper (Becton Dickinson) and either analyzed by FACS or used to prepare total RNA for reverse transcriptase–polymerase chain reaction (RT-PCR). For FACS analysis, cells were incubated with CD16/CD32 antibody (2.4G2) Fc block (1:100) on ice for 20 minutes and stained with biotinylated anti–c-Fms or RANK monoclonal antibody for 30 minutes on ice and washed twice. Subsequently, cells were incubated with allophycocyanin-conjugated streptavidin for 30 minutes on ice. For isotype-matched controls, biotinylated rat immunoglobulin G2a (IgG2a) was used. After the final wash, the expression of c-Fms and RANK was analyzed by FACS Calibur. For analysis of GM-CSF receptor expression, RT-PCR analysis was performed as described below.
Colony assay
A colony assay was performed as described previously.10 After 6 days of cultivation, the numbers of colonies were scored under a microscope, and single colonies were aliquotted and placed on culture plates or fibronectin-coated dishes. Cells in the culture plates were cultured with M-CSF and sRANKL or with GM-CSF and sRANKL for 6 days and then stained with TRAP and DEC205 (NLDC-145) (Serotec, Oxford, England). Immunohistochemical staining of DEC205 was performed as described elsewhere.40 Cells on fibronectin-coated plates were cultured in the presence of GM-CSF and sRANKL for 6 days, and the expression of Mac-1 and CD11c was analyzed as described below.
Dendritic cell differentiation in vitro
The c-Kit+c-Fms+RANK− cells were cultured in liquid culture containing M-CSF alone; GM-CSF alone; GM-CSF and sRANKL; M-CSF, sRANKL, and GM-CSF; or in methylcellulose culture containing M-CSF and sRANKL. After 6 days of cultivation, cells were collected and incubated with Fc block (1:100) on ice for 20 minutes. Subsequently, cells were aliquotted for staining with Mac-1 and CD11c or DEC205. For Mac-1 and CD11c staining, cells were stained with biotinylated anti–Mac-1 (Pharmingen) for 30 minutes on ice and washed twice. Cells were then incubated with PE-conjugated anti-CD11c antibody (Pharmingen) and allophycocyanin-conjugated streptavidin (Caltag) for 30 minutes on ice and washed twice. For DEC205 staining, cells were incubated with DEC205 (1:50) for 30 minutes on ice and washed twice and were then stained with B–PE-conjugated anti–rat IgG antibody (Jackson ImmunoResearch Laboratories, West Grove, PA) for 30 minutes on ice. For isotype-matched controls, biotinylated (Pharmingen) and PE-conjugated rat IgG2a (Pharmingen) were used. After the final wash, cells were suspended in washing buffer (5% fetal bovine serum/phosphate-buffered saline), and the expression of Mac-1 and CD11c or DEC205 was analyzed by FACS Calibur.
RT-PCR analysis
Total RNA was isolated from freshly isolated osteoclast precursor cells (c-Fms+RANK− cells) or cultured cells (Rneasy mini kit, Qiagen, Hilden, Germany). Single-strand complementary DNAs (cDNAs) were synthesized with reverse transcription (first-strand cDNA synthesis kit, Clontech Laboratories, Palo Alto, CA). The cDNAs were amplified using the Advantage2 PCR System (PerkinElmer, Norwalk, CT) in a GeneAmp PCR system model 9700 (PerkinElmer) for 30 cycles. Each cycle consisted of 30 seconds of denaturation at 94°C and 4 minutes of annealing/extension at 70°C. Glyceraldehyde-3-phosphate dehydrogenase (G3PDH) was used as an internal control. Primers used for RT-PCR were as follows: CTR (calcitonin receptor)–5′, 5′-TGGTTGAGGTTGTGCCCAATGGAGA-3′; CTR-3′, 5′-CTCGTGGGTTTGCCTCATCTTGGTC-3′; c-Src–5′, 5′-TGATGTTATGAAGA ACTGCTCGCACCTG-3′; c-Src–3′, 5′-TGCCCATTTGCTGGGTACTTTCTTTCTC-3′; c-Fms–5′, 5′-CTGTGAATGGCTCTGATGTCCTGTTCTG-3′; c-Fms–3′, 5′-CTCCCACTT CTCATTGTAGGGCAACTGA-3′; c-Fos–5′, 5′-ATGATGTTCTCGGGTTTCAACG-3′; c-Fos–3′, 5′-CAGTCTGCTGCATAGAAGGAACCG-3′; RANK−5′, 5′-CTTCGACTGGTT CACTGCTCCTAAT-3′; RANK−3′, 5′-TTACTGTTTCCAGTCACGTTCCCAGAGG-3′; PU.1-5′, 5′-CTGAGAACCACTTCACAGAGCTGCAGAG-3′; PU.1-3′, 5′-GCGCCATC TTCTGGTAGGTCATCTTCTT-3′; Fra-1–5′, 5′-GTGCAAGTGGTTCAGCCCAAGAAC TTTT-3′; Fra-1–3′, 5′-GGGTCCTTCTTGTCTCCTTCTGGGATTT-3′; GM-CSF R-3′, 5′-CACGATGACGTCTATCAGCATCGCTTC-3′; G3PDH-5′, 5′-TGAAGGTCGGTGTGA ACGGATTTGGC-3′; G3PDH-3′, 5′-CATGTAGGCCATGAGGTCCACCAC-3′.
Preparation of retroviral vectors expressing full-length or truncated forms of c-Fos and Fra-1
Full-length and truncated (δ 262) forms of c-Fos and Fra-1 (full-length and δ 185) expression retroviral vectors were generated by ligating the RT-PCR products into the pMY-IRES-GFP vector provided by Dr T. Kitamura (University of Tokyo, Institute of Medical Science). The primer sets for RT-PCR are as follows: c-Fos–5′, 5′-GGAATTCATGATGTTCTCGGGTTTCAACG-3′; c-Fos-full–3′, 5′-ACGCGTCGACTCACAGGGCCAGCAGCGTGGGTGA-3′; c-Fos–δ 262–3′, 5′-ACGCGTCGACTCACACGTTGCTGATGCTCTCTTGACTGGCT-3′; Fra-1–5′, 5′GGAATT CATGTACCGAGACTACGGGGAAC-3′; Fra-1- full–3′, ACGCGTCGACTCACAAAGCC AGGAGTGTAGGAGAG-3′; Fra-1–δ 185–3′, 5′-ACGCGTCGACTCAAGAACCACCTGGGTCCTTCTTGT-3′. Single-stranded cDNA was generated from total RNA isolated from cultured osteoclast precursor cells in the presence of M-CSF and sRANKL for 6 days.
Retroviral vectors were transfected into Phoenix-ecotropic cells by Lipofectamine Plus (Gibco) in Dulbecco modified essential medium (Gibco) containing 10% fetal bovine serum. The medium was collected after 24 and 48 hours of incubation as a virus solution and stored at −80°C. For infection of osteoclast precursor cells, retronectin-coated plates (Takara, Otsu, Japan) were used. A total of 1000 sorted c-Kit+c-Fms+RANK−cells were cultured on these plates in the presence of 100 ng/mL M-CSF, 25 ng/mL sRANKL, and 1 ng/mL GM-CSF with 10 multiplicities of infection of various retroviral constructs. After 24 hours of infection, the culture medium was changed to fresh medium containing M-CSF, sRANKL, and GM-CSF. After 5 days of cultivation, cells were stained with TRAP and the number of TRAP-positive cells were scored.
Results
GM-CSF inhibits osteoclast differentiation
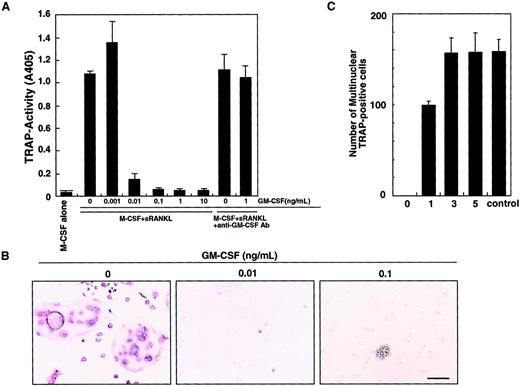
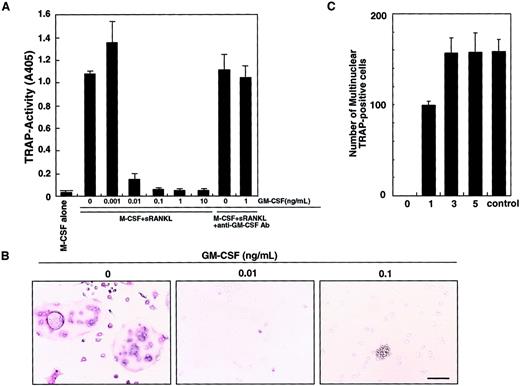
We previously showed that osteoclasts differentiate from isolated osteoclast precursor cells (c-Fms+RANK− cells) in the presence of M-CSF and sRANKL in a stromal cell–free culture system.10 Such osteoclastogenesis is inhibited by GM-CSF in a dose-dependent manner (Figure 1A). This inhibitory effect can be seen at concentrations of GM-CSF as low as 0.01 ng/mL. A small number of mononuclear TRAP-positive cells are observed at 0.01 ng/mL GM-CSF, and the inhibitory effect is completely abolished by addition of a neutralizing antibody against GM-CSF. At 0.1 ng/mL GM-CSF, TRAP-positive cells were not detected and cell clusters were observed (Figure 1B). Because GM-CSF and RANKL are known to induce dendritic cell differentiation and clustering,13 these clusters were interpreted to be dendritic cells (see below). Interestingly, the inhibitory effect of GM-CSF was observed only when GM-CSF was added on day 0, and inhibition decreased as GM-CSF addition was delayed (Figure 1C). These results suggest that after the differentiation signal by M-CSF and RANKL is transduced, cells are no longer competent to respond to GM-CSF.
Inhibition of osteoclastogenesis by GM-CSF.
The c-Fms+RANK− cells were cultured in the presence of M-CSF and sRANKL with or without GM-CSF. After 6 days of cultivation, cells were subjected to a TRAP activity assay (A) and TRAP staining (B). The inhibitory effect of GM-CSF on osteoclast differentiation was dose dependent and abolished by addition of neutralizing antibody against GM-CSF (A). In the absence of GM-CSF, multinuclear TRAP-positive cells were formed (B). Only mononuclear TRAP-positive cells and TRAP-negative cells were observed in the presence of 0.01 ng/mL GM-CSF, and cell clusters that were TRAP-negative were detected in the presence of 0.1 ng/mL GM-CSF (B). Bar = 25 μm. (C) The c-Fms+RANK− cells were cultured in the presence of M-CSF and sRANKL, and 1 ng/mL GM-CSF was added to the culture on days 0, 1, 3, and 5. Inhibition of osteoclastogenesis was observed only when GM-CSF was added on days 0 and 1. Control = no addition of GM-CSF.
Inhibition of osteoclastogenesis by GM-CSF.
The c-Fms+RANK− cells were cultured in the presence of M-CSF and sRANKL with or without GM-CSF. After 6 days of cultivation, cells were subjected to a TRAP activity assay (A) and TRAP staining (B). The inhibitory effect of GM-CSF on osteoclast differentiation was dose dependent and abolished by addition of neutralizing antibody against GM-CSF (A). In the absence of GM-CSF, multinuclear TRAP-positive cells were formed (B). Only mononuclear TRAP-positive cells and TRAP-negative cells were observed in the presence of 0.01 ng/mL GM-CSF, and cell clusters that were TRAP-negative were detected in the presence of 0.1 ng/mL GM-CSF (B). Bar = 25 μm. (C) The c-Fms+RANK− cells were cultured in the presence of M-CSF and sRANKL, and 1 ng/mL GM-CSF was added to the culture on days 0, 1, 3, and 5. Inhibition of osteoclastogenesis was observed only when GM-CSF was added on days 0 and 1. Control = no addition of GM-CSF.
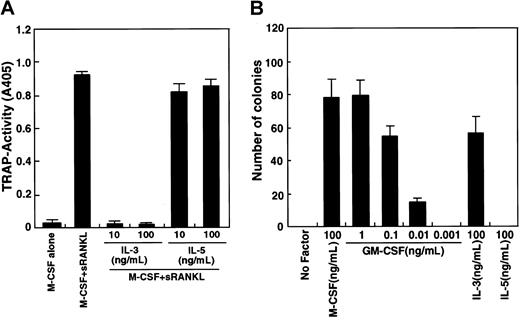
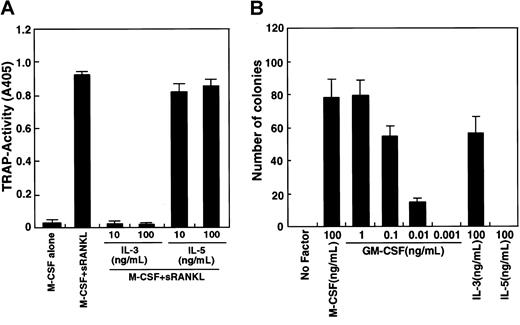
The GM-CSF receptor shares a common β chain with IL-3 and IL-5 receptors.41 To assess the involvement of a potential common β-chain signal in osteoclastogenesis, we determined whether IL-3 and IL-5 inhibit osteoclastogenesis (Figure2A). IL-3 effectively inhibited osteoclast differentiation, while IL-5 did not. It is possible that osteoclast precursor cells (c-Fms+RANK− cells) cultured in this assay are not competent to respond to IL-5. To examine this possibility, a methylcellulose colony assay was performed to analyze whether osteoclast precursor cells respond to IL-3 and IL-5 (Figure 2B). IL-3 and GM-CSF stimulated colony formation by osteoclast precursors, but IL-5 did not, while IL-5 at this dose induced eosinophil colonies from whole bone marrow cells. Colony cells cultured with GM-CSF or IL-3 differentiated into multinuclear TRAP-positive cells when cells were transferred to liquid medium and cultured with M-CSF and sRANKL (data not shown).
Inhibition of osteoclastogenesis by IL-3.
(A) The c-Fms+RANK− cells were cultured in the presence of M-CSF and sRANKL with or without IL-3 and IL-5. Osteoclast differentiation was inhibited by addition of IL-3 but not IL-5. (B) The c-Fms+RANK− cells were cultured in 1.2% methylcellulose medium containing 100 ng/mL M-CSF, IL-5, IL-3, or various concentrations of GM-CSF. Colony formation was observed in the presence of M-CSF, IL-3, and GM-CSF but not IL-5.
Inhibition of osteoclastogenesis by IL-3.
(A) The c-Fms+RANK− cells were cultured in the presence of M-CSF and sRANKL with or without IL-3 and IL-5. Osteoclast differentiation was inhibited by addition of IL-3 but not IL-5. (B) The c-Fms+RANK− cells were cultured in 1.2% methylcellulose medium containing 100 ng/mL M-CSF, IL-5, IL-3, or various concentrations of GM-CSF. Colony formation was observed in the presence of M-CSF, IL-3, and GM-CSF but not IL-5.
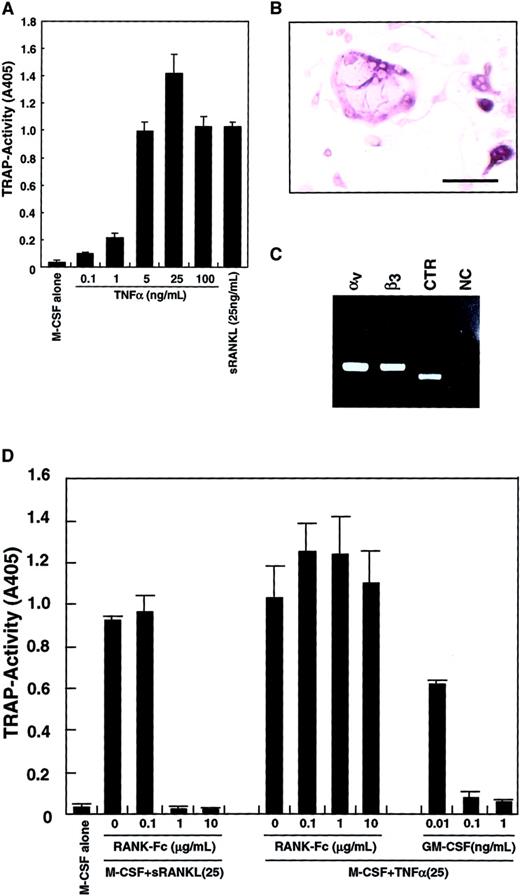
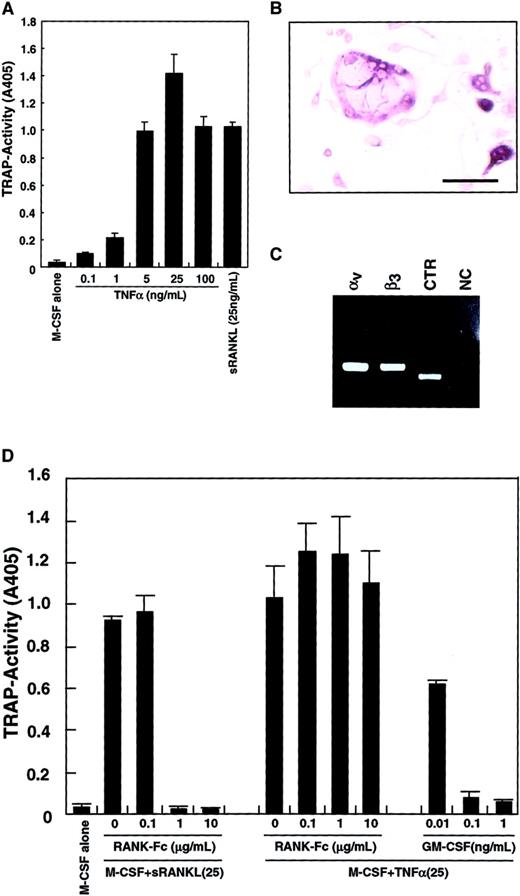
Because it was reported that TNF-α induced osteoclastogenesis,42 43 TNF-α was added to the culture system instead of sRANKL. As shown in Figure3A, differentiation into TRAP-positive cells from osteoclast precursors was observed in the presence of TNF-α and M-CSF. TRAP activity peaked at 25 ng/mL TNF-α, and multinuclear TRAP-positive cells were seen (Figure 3B). The expression of vitronectin receptors (integrin αv and β3) and a CTR was also detected by RT-PCR (Figure 3C), suggesting that these cells were committed to an osteoclast lineage. The TRAP activity induced by M-CSF and TNF-α was not reduced by addition of RANK−-Fc, which completely inhibited osteoclastogenesis stimulated by RANKL (Figure 3D). GM-CSF also strongly inhibited osteoclast differentiation induced by TNF-α in a dose-dependent manner.
Inhibition of TNF-α–induced osteoclastogenesis by GM-CSF.
The c-Fms+RANK− cells were cultured in the presence of 100 ng/mL M-CSF with various concentrations of TNF-α or 25 ng/mL sRANKL. After 6 days of culture, cells were subjected to a TRAP activity assay (A) or TRAP staining (bar = 25 μm) (B). Osteoclastogenesis reached a plateau at 25 ng/mL TNF-α (A), and multinuclear TRAP-positive cells were also formed (B). Expression of vitronectin receptors (integrin αv and β3) or a CTR were detected by RT-PCR in cells cultured with M-CSF and TNF-α for 6 days (C). NC indicates no template control. Osteoclast differentiation was induced by 2 cytokine combinations: M-CSF and sRANKL and, also, M-CSF and TNF-α. Differentiation induced by the former was completely inhibited by the addition of RANK−Fc in a dose-dependent manner, but differentiation induced by the latter was not. By contrast, GM-CSF inhibited both in a dose-dependent manner (D).
Inhibition of TNF-α–induced osteoclastogenesis by GM-CSF.
The c-Fms+RANK− cells were cultured in the presence of 100 ng/mL M-CSF with various concentrations of TNF-α or 25 ng/mL sRANKL. After 6 days of culture, cells were subjected to a TRAP activity assay (A) or TRAP staining (bar = 25 μm) (B). Osteoclastogenesis reached a plateau at 25 ng/mL TNF-α (A), and multinuclear TRAP-positive cells were also formed (B). Expression of vitronectin receptors (integrin αv and β3) or a CTR were detected by RT-PCR in cells cultured with M-CSF and TNF-α for 6 days (C). NC indicates no template control. Osteoclast differentiation was induced by 2 cytokine combinations: M-CSF and sRANKL and, also, M-CSF and TNF-α. Differentiation induced by the former was completely inhibited by the addition of RANK−Fc in a dose-dependent manner, but differentiation induced by the latter was not. By contrast, GM-CSF inhibited both in a dose-dependent manner (D).
RANK and c-Fms are expressed in the presence of GM-CSF
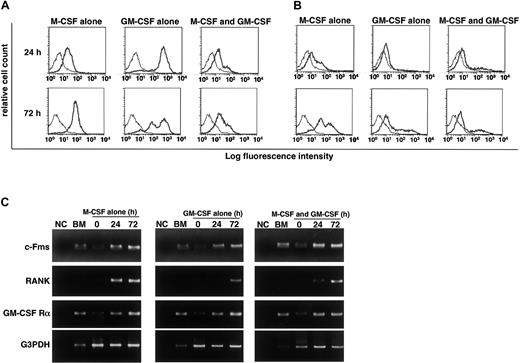
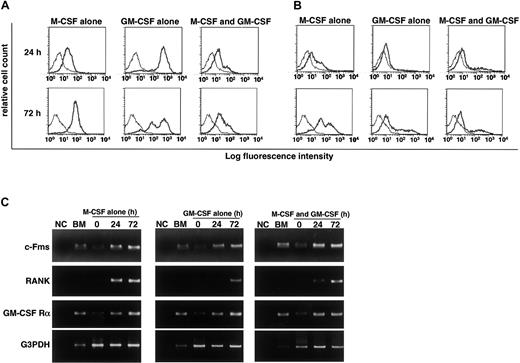
We previously reported that RANK expression is induced by M-CSF and subsequent stimulation by RANKL induces osteoclastogenesis.9 Gliniak and Rohrschneider have shown that expression of c-Fms is reduced in the presence of GM-CSF or IL-3.44 Thus, it is likely that GM-CSF suppresses expression of c-Fms or RANK and that subsequently, the effect of M-CSF or RANKL is inhibited. Because inhibition of osteoclastogenesis occurs at early stages of differentiation, we examined expression of c-Fms and RANK by FACS after 24 and 72 hours of cultivation. Expression of c-Fms, however, was observed in cultures with M-CSF and/or GM-CSF (Figure4A). Because the monoclonal antibody against c-Fms recognizes the ligand binding site, c-Fms expression may be relatively low in the presence of M-CSF due to the occupation of the antibody binding site by M-CSF. Similarly, RANK expression was induced even in the presence of GM-CSF, and the expression was detected in cultures at 24 and 72 hours (Figure 4B). These results were confirmed by RT-PCR analysis (Figure 4C). Our findings indicate that the inhibitory effect of GM-CSF on osteoclastogenesis is not mediated by a blockade in receptor expression. RT-PCR analysis revealed that GM-CSF receptor α chain was expressed in freshly isolated osteoclast precursor cells and was up-regulated in the presence of GM-CSF and/or M-CSF (Figure 4C). Overall, our results indicate that osteoclast precursor cells express both c-Fms and GM-CSF receptor α chain and that RANK is induced by both M-CSF and GM-CSF. The expression of c-Fms and RANK was also observed in the culture with IL-3 (data not shown).
RANK and c-Fms expression is induced in the presence of GM-CSF.
The c-Fms+RANK− cells were cultured in the presence of 100 ng/mL M-CSF or 50 ng/mL GM-CSF alone or both for 24 and 72 hours, and the expression of c-Fms (A) and RANK (B) was examined by FACS. The expression of c-Fms, RANK, and GM-CSF receptor (C) was analyzed by RT-PCR. Expression of c-Fms and RANK was observed in the presence of GM-CSF alone or GM-CSF and M-CSF, and the expression of GM-CSF receptor α chain (GM-CSF Rα) was detected in the presence of M-CSF or M-CSF and GM-CSF. NC indicates no template control.
RANK and c-Fms expression is induced in the presence of GM-CSF.
The c-Fms+RANK− cells were cultured in the presence of 100 ng/mL M-CSF or 50 ng/mL GM-CSF alone or both for 24 and 72 hours, and the expression of c-Fms (A) and RANK (B) was examined by FACS. The expression of c-Fms, RANK, and GM-CSF receptor (C) was analyzed by RT-PCR. Expression of c-Fms and RANK was observed in the presence of GM-CSF alone or GM-CSF and M-CSF, and the expression of GM-CSF receptor α chain (GM-CSF Rα) was detected in the presence of M-CSF or M-CSF and GM-CSF. NC indicates no template control.
Down-regulation of c-Fos by GM-CSF is a key event for the inhibition of osteoclastogenesis
Osteoclastogenesis was completely inhibited by addition of GM-CSF even though cells expressed both c-Fms and RANK. We therefore asked which transcription factors could be involved in this inhibition. First we investigated the expression of the factors such as PU.1, c-Fos, NF-κB1, and NF-κB2 by RT-PCR. The expression of PU.1 and NF-κB1 was not altered by addition of GM-CSF (data not shown); however, the expression of c-Fos was strongly suppressed (Figure 5A), suggesting that inhibition of c-Fos expression might be related to inhibition of osteoclastogenesis.
Down-regulation of c-Fos is a key event for the inhibition of osteoclastogenesis in the presence of GM-CSF.
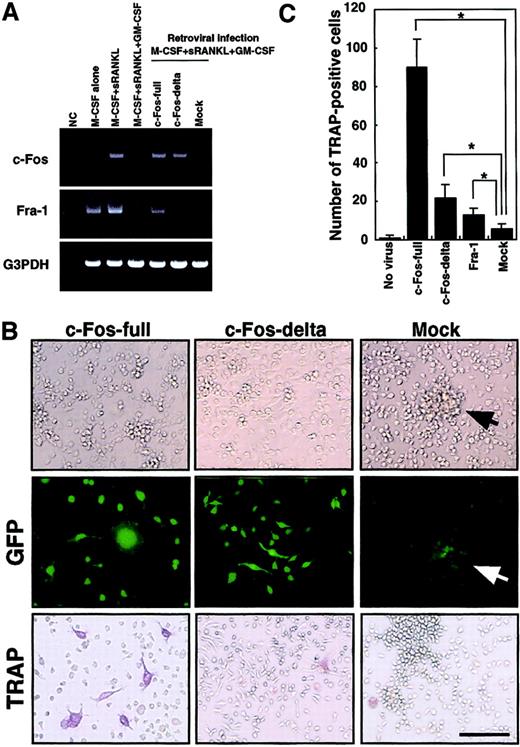
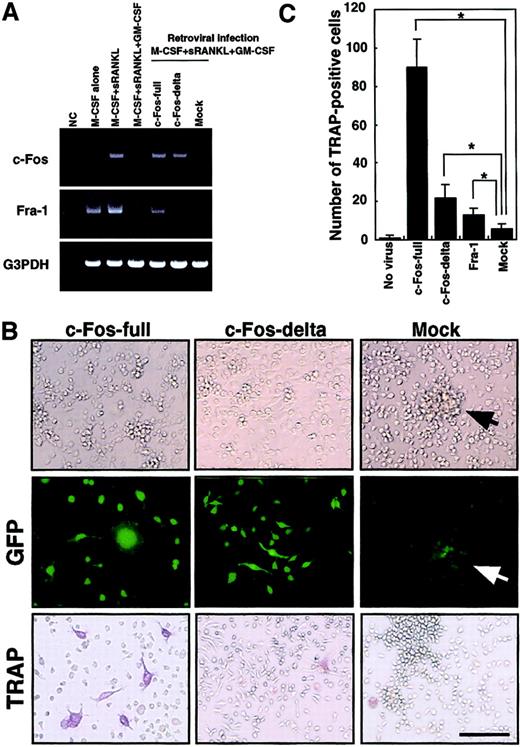
The c-Kit+c-Fms+RANK− cells were cultured in the presence of M-CSF (100 ng/mL) alone; M-CSF and sRANKL (25 ng/mL); and M-CSF, sRANKL, and GM-CSF (1 ng/mL) for 6 days. The effect of retroviral infection with full-length c-Fos (c-Fos–full), truncated c-Fos (c-Fos–δ) or mock-infected cells was examined in cultures containing M-CSF and sRANKL and GM-CSF. Expression of c-Fos, Fra-1, and G3PDH was examined by RT-PCR on day 6 (A). NC indicates no template control. Expression of Fra-1 was induced by c-Fos–full but not by c-Fos–δ. Retroviral infection could be detected by GFP expression visualized with a fluorescence microscope (B). Cell spreading and disappearance of cell clusters were observed following infection with c-Fos–full or c-Fos–δ but not in mock-infected cells (arrow indicates cell cluster). Significant increases in TRAP-positive cells were detected in the c-Fos–full infection (B,C). Bar = 25 μm.
Down-regulation of c-Fos is a key event for the inhibition of osteoclastogenesis in the presence of GM-CSF.
The c-Kit+c-Fms+RANK− cells were cultured in the presence of M-CSF (100 ng/mL) alone; M-CSF and sRANKL (25 ng/mL); and M-CSF, sRANKL, and GM-CSF (1 ng/mL) for 6 days. The effect of retroviral infection with full-length c-Fos (c-Fos–full), truncated c-Fos (c-Fos–δ) or mock-infected cells was examined in cultures containing M-CSF and sRANKL and GM-CSF. Expression of c-Fos, Fra-1, and G3PDH was examined by RT-PCR on day 6 (A). NC indicates no template control. Expression of Fra-1 was induced by c-Fos–full but not by c-Fos–δ. Retroviral infection could be detected by GFP expression visualized with a fluorescence microscope (B). Cell spreading and disappearance of cell clusters were observed following infection with c-Fos–full or c-Fos–δ but not in mock-infected cells (arrow indicates cell cluster). Significant increases in TRAP-positive cells were detected in the c-Fos–full infection (B,C). Bar = 25 μm.
To rescue suppression of c-Fos, we expressed c-Fos in osteoclast precursor cells by retroviral expression. Because cell proliferation is critical for retroviral infection, c-Kit+c-Fms+RANK− cells were used as osteoclast precursors in the following experiments. Because the truncated form of c-Fos lacking its carboxy (C)–terminus is reported to overcome the osteoclastogenesis defect in c-Fos–deficient mice,27 we tested both full-length (c-Fos–full) and truncated forms lacking the C-terminal domain (c-Fos–δ). Retroviral infection of osteoclast precursors was detected by expression of green fluorescent protein (GFP) inserted downstream of an internal ribosome entry site (IRES) sequence in the retroviral vector (Figure 5B). Osteoclast precursors (c-Kit+c-Fms+RANK− cells) were cultured in the presence of M-CSF and sRANKL with 1 ng/mL GM-CSF and 10 multiplicities of infection of various retroviral preparations. GFP expression was observed from day 3 under fluorescence microscopy, and the frequency of GFP-positive cells was 21.1% to 33.3%. Expression of c-Fos was observed by RT-PCR in cells infected by c-Fos–expressing retrovirus but not in mock-infected cells on day 6 (Figure 5A).
We also examined the expression of the closely related transcription factor Fra-1, because Fra-1 was shown to rescue the osteoclast differentiation defect seen in c-Fos–deficient mice.27Interestingly, Fra-1 expression was strongly suppressed by GM-CSF; however, Fra-1 was induced when osteoclast precursors were infected by c-Fos–full but not by retrovirus expressing c-Fos–δ (Figure 5A). Why c-Fos–δ fails to induce Fra-1 is not clear, but this finding correlates with previous reports that Fra-1 is a target of c-Fos.25 26 After 6 days of cultivation, GFP-positive cells infected by both full-length c-Fos and c-Fos–δ retrovirus showed obvious morphologic changes, such as spreading and loss of cell clustering (Figure 5B). In Fra-1–induced cells, cell spreading was not observed and cell clustering was still detected (data not shown), suggesting that the amino (N)–terminus of c-Fos might be required for regulation of cell spreading and clustering. TRAP-positive cells were observed when full length of c-Fos was expressed (Figure 5B,C), although TRAP-positive cells were not seen following c-Fos–δ expression. Because c-Fos–δ failed to induce Fra-1, it is likely that Fra-1 alone is sufficient to induce osteoclastogenesis in the presence of GM-CSF. However, the expression of Fra-1 did not induce osteoclastogenesis as efficiently as did full-length c-Fos (Figure 5C).
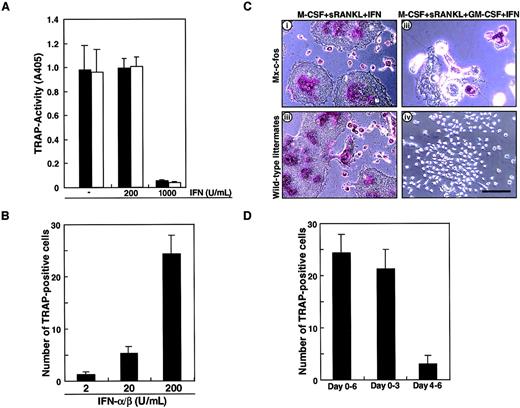
To confirm the role of c-Fos in osteoclast differentiation in the presence of GM-CSF, inducible c-Fos transgenic mice known as Mx–c-fos mice were used to prepare osteoclast precursor cells (c-Kit+c-Fms+RANK− cells). The Mx–c-fos mouse was constructed to induce c-Fos expression by addition of IFN-α/β; however, IFN-α/β is known to inhibit osteoclastogenesis. The frequency of osteoclast precursor cells in bone marrow mononuclear cells was not different in Mx–c-fos mice from that seen in wild-type littermates (data not shown). First, we examined whether osteoclast differentiation was affected by the addition of the inducing agent, IFN-α/β. Osteoclast precursor cells were cultured in the presence of M-CSF and sRANKL with or without various concentrations of IFN-α/β. Osteoclastogenesis was strongly inhibited by 1000 U/mL IFN-α/β in both Mx–c-fos and wild-type littermates even in the absence of GM-CSF (Figure6A). c-Fos was induced within 24 hours by addition of 200 U/mL IFN-α/β (data not shown), and such treatment had no inhibitory effect on osteoclast formation (Figure 6A) even though the c-Fos–inducible gene, 2′-5′ oligoadenylate synthetase, was induced in a dose-dependent manner (data not shown). Subsequently, we examined the effect of c-Fos induction by IFN-α/β on osteoclast formation in the presence of GM-CSF. The effect of IFN-α/β on rescue of the osteoclast differentiation defect was dose-dependent and peaked at 200 U/mL (Figure 6B). Induction of c-Fos in cells prepared from Mx–c-fos mice resulted in formation of multinuclear TRAP-positive cells. Such cells were not observed in similar assays of cells from wild-type littermates in the presence of GM-CSF (Figure 6C). These findings are consistent with the analysis using retroviral infection, which demonstrates a rescue of osteoclast differentiation defect by c-Fos expression in the presence of GM-CSF. The c-Fos induction rescued osteoclastogenesis especially in multinucleation rather than TRAP activity (Figure 6C). Induction of c-Fos at early stages of culture was more efficient at promoting osteoclastogenesis than induction at later stages (Figure 6D), similar to the findings in Figure 1C, showing that cell lineage was determined at early stages of differentiation. The expression of c-Fos increased as GM-CSF addition was delayed (data not shown).
Induction of c-Fos can rescue the osteoclastogenesis failure by GM-CSF.
(A) The c-Kit+c-Fms+RANK− cells prepared from wild-type littermates (■) or Mx–c-fos mice (▪) were cultured in the presence of 100 ng/mL M-CSF and 25 ng/mL sRANKL with (200 or 1000 U/mL) or without IFN-α/β for 6 days, and a TRAP activity assay was performed. Osteoclastogenesis was strongly inhibited by 1000 U/mL IFN-α/β in both Mx–c-fos and wild-type littermates. (B) The c-Kit+c-Fms+RANK− cells prepared from Mx–c-fos mice were cultured in the presence of 100 ng/mL M-CSF, 25 ng/mL sRANKL, and 1 ng/mL GM-CSF with IFN-α/β (2, 20, or 200 U/mL) for 6 days, and a TRAP staining was performed. The formation of TRAP-positive cells was observed in a dose-dependent manner. (C) Osteoclast differentiation was not affected by addition of 200 U/mL IFN-α/β in both wild-type littermates and Mx–c-fos mice, and multinuclear TRAP-positive cells were observed in the absence of GM-CSF (i,iii). The multinuclear TRAP-positive cells were formed when c-Fos was induced by the addition of IFN-α/β in Mx–c-fos mice in the presence of GM-CSF (ii), while no TRAP-positive cells were detected in wild-type littermates (iv). Bar = 100 μm. (D) The c-Kit+c-Fms+RANK− cells were cultured in the presence of M-CSF, sRANKL, and GM-CSF. IFN-α/β was present over the culture period (days 0-6), during the first 3 days (days 0-3), and the last 3 days (day 4-6). After 6 days of culture, the number of TRAP-positive cells were counted. The early expression of c-Fos is more effective than that of late expression in the rescue of osteoclastogenesis.
Induction of c-Fos can rescue the osteoclastogenesis failure by GM-CSF.
(A) The c-Kit+c-Fms+RANK− cells prepared from wild-type littermates (■) or Mx–c-fos mice (▪) were cultured in the presence of 100 ng/mL M-CSF and 25 ng/mL sRANKL with (200 or 1000 U/mL) or without IFN-α/β for 6 days, and a TRAP activity assay was performed. Osteoclastogenesis was strongly inhibited by 1000 U/mL IFN-α/β in both Mx–c-fos and wild-type littermates. (B) The c-Kit+c-Fms+RANK− cells prepared from Mx–c-fos mice were cultured in the presence of 100 ng/mL M-CSF, 25 ng/mL sRANKL, and 1 ng/mL GM-CSF with IFN-α/β (2, 20, or 200 U/mL) for 6 days, and a TRAP staining was performed. The formation of TRAP-positive cells was observed in a dose-dependent manner. (C) Osteoclast differentiation was not affected by addition of 200 U/mL IFN-α/β in both wild-type littermates and Mx–c-fos mice, and multinuclear TRAP-positive cells were observed in the absence of GM-CSF (i,iii). The multinuclear TRAP-positive cells were formed when c-Fos was induced by the addition of IFN-α/β in Mx–c-fos mice in the presence of GM-CSF (ii), while no TRAP-positive cells were detected in wild-type littermates (iv). Bar = 100 μm. (D) The c-Kit+c-Fms+RANK− cells were cultured in the presence of M-CSF, sRANKL, and GM-CSF. IFN-α/β was present over the culture period (days 0-6), during the first 3 days (days 0-3), and the last 3 days (day 4-6). After 6 days of culture, the number of TRAP-positive cells were counted. The early expression of c-Fos is more effective than that of late expression in the rescue of osteoclastogenesis.
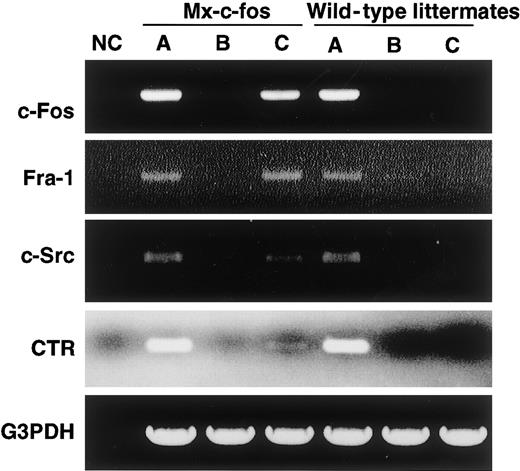
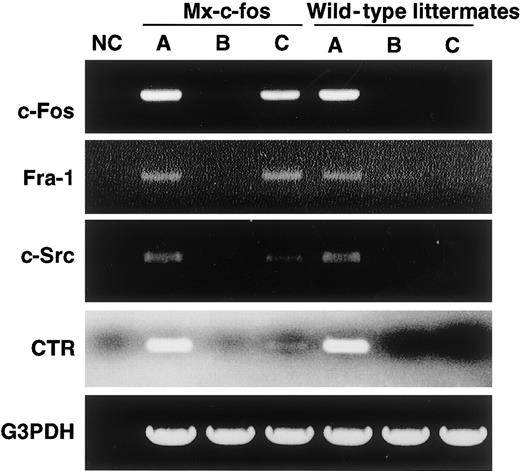
RT-PCR analysis showed that c-Fos was induced by addition of IFN-α/β in cells prepared from the Mx–c-fos mice as was Fra-1, as observed when c-Fos was induced by retroviral infection (Figure7). The expression of CTR and c-Src, which are osteoclast markers, was also induced, indicating that cells were committed to an osteoclast lineage even in the presence of GM-CSF.
Cells are committed to an osteoclast lineage by c-Fos induction, but Jun proteins are not involved in the rescue of osteoclastogenesis.
The expression of c-Fos, Fra-1, c-Src, CTR, and G3PDH was examined in c-Fms+RANK− cells cultured for 6 days with M-CSF and sRANKL (A); M-CSF, sRANKL, and GM-CSF (B); and M-CSF, sRANKL, GM-CSF, and IFN-α/β (C). NC indicates no template control. Expression of Fra-1, c-Src, and CTR was observed following induction of c-Fos.
Cells are committed to an osteoclast lineage by c-Fos induction, but Jun proteins are not involved in the rescue of osteoclastogenesis.
The expression of c-Fos, Fra-1, c-Src, CTR, and G3PDH was examined in c-Fms+RANK− cells cultured for 6 days with M-CSF and sRANKL (A); M-CSF, sRANKL, and GM-CSF (B); and M-CSF, sRANKL, GM-CSF, and IFN-α/β (C). NC indicates no template control. Expression of Fra-1, c-Src, and CTR was observed following induction of c-Fos.
Analysis of dendritic cell differentiation
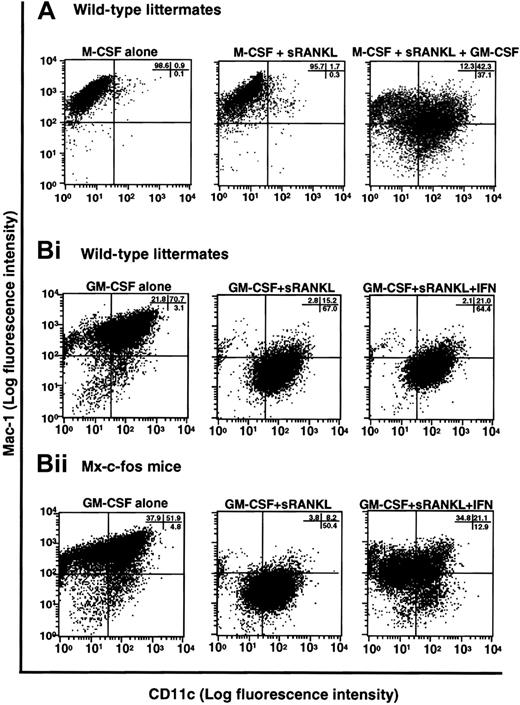
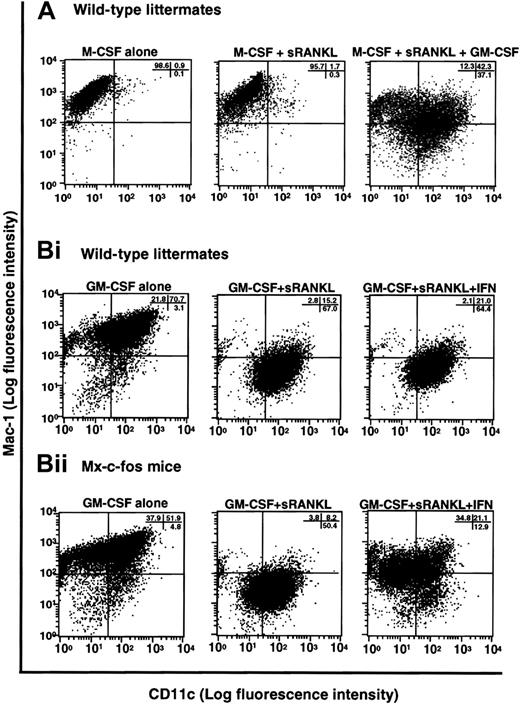
In the presence of M-CSF, sRANKL, and GM-CSF, osteoclastogenesis was completely inhibited, and cell clusters, which are indicative of dendritic cell activation, were observed (Figure 1B). Because GM-CSF is known to be involved in dendritic cell differentiation,32 it is likely that the cells are committed to a dendritic cell lineage. To test this possibility, we examined the expression of CD11c, which is a marker of dendritic cells,45 and Mac-1, which is expressed on macrophages (Figure 8A). Cells cultured with M-CSF alone or with M-CSF and sRANKL (or LZ-RANKL) for 6 days expressed Mac-1 but not CD11c. However, in the presence of M-CSF, sRANKL, and GM-CSF, CD11c expression was induced together with that of Mac-1, indicating a shift from an osteoclast to a dendritic cell lineage. To confirm a bifurcated differentiation of osteoclast precursors to osteoclasts and dendritic cells, clonal analysis was performed by methylcellulose culture. At first, single colonies derived from osteoclast precursors in the presence of M-CSF or GM-CSF were exposed to M-CSF and sRANKL or GM-CSF and sRANKL on day 6 of culture. After 6 more days, cells were stained with TRAP and DEC205, a marker of mature dendritic cells. Both TRAP- and DEC205-positive cells were seen in cells derived from the same colony, indicating that osteoclast precursors can differentiate into osteoclasts or dendritic cells other than macrophages (data not shown). Next, single-cell sorting of c-Kit+c-Fms+RANK− cells was performed, and sorted cells were cultured in the presence of M-CSF and sRANKL; GM-CSF and sRANKL; and M-CSF, sRANKL, and GM-CSF. After 6 days of culture, cells were stained with TRAP and DEC205 to determine the frequency of osteoclast and dendritic cells. In the 2 experiments, the frequency of TRAP-positive cells was 58.3% and 39.6%, while DEC205-positive cells were 54.5% and 33.3%, respectively (Table1). Judging from high plating efficiency, it was suggested that osteoclasts and dendritic cells develop from the same precursors. This result was confirmed by the culture with M-CSF, sRANKL, and GM-CSF. Mixed wells (8.3% and 4.2%) contained both TRAP-positive and -negative cells, and the other wells contained only TRAP-negative cells in 32.3% (Table 2). No well contained only TRAP-positive cells, as observed in the culture with M-CSF and sRANKL, indicating the differentiation into osteoclasts was inhibited by GM-CSF. The c-Kit+c-Fms+RANK− cells have bipotency to respond to both M-CSF and GM-CSF.
Induction of dendritic cell differentiation in the presence of GM-CSF and its inhibition by c-Fos expression.
(A) The c-Kit+c-Fms+RANK− cells were cultured in liquid medium containing M-CSF alone; or M-CSF, sRANKL, and GM-CSF; or 1.2% methylcellulose medium containing M-CSF and sRANKL for 6 days, and the expression of CD11c and Mac-1 was examined by FACS. Expression of CD11c was induced by addition of GM-CSF. (B) The c-Kit+c-Fms+RANK−cells were prepared from wild-type littermates (i) and Mx–c-fos mice (ii) and cultured in the presence of GM-CSF alone; GM-CSF and sRANKL; and GM-CSF, sRANKL, and IFN-α/β. After 6 days of cultivation, cells were harvested and stained with CD11c and Mac-1. The CD11c+Mac-1+ cells were induced by GM-CSF, and the CD11c+Mac-1− cells appeared in the presence of GM-CSF and sRANKL in both wild-type littermates and Mx–c-fos mice. However, the expression of CD11c was reduced by induction of c-Fos in Mx–c-fos mice.
Induction of dendritic cell differentiation in the presence of GM-CSF and its inhibition by c-Fos expression.
(A) The c-Kit+c-Fms+RANK− cells were cultured in liquid medium containing M-CSF alone; or M-CSF, sRANKL, and GM-CSF; or 1.2% methylcellulose medium containing M-CSF and sRANKL for 6 days, and the expression of CD11c and Mac-1 was examined by FACS. Expression of CD11c was induced by addition of GM-CSF. (B) The c-Kit+c-Fms+RANK−cells were prepared from wild-type littermates (i) and Mx–c-fos mice (ii) and cultured in the presence of GM-CSF alone; GM-CSF and sRANKL; and GM-CSF, sRANKL, and IFN-α/β. After 6 days of cultivation, cells were harvested and stained with CD11c and Mac-1. The CD11c+Mac-1+ cells were induced by GM-CSF, and the CD11c+Mac-1− cells appeared in the presence of GM-CSF and sRANKL in both wild-type littermates and Mx–c-fos mice. However, the expression of CD11c was reduced by induction of c-Fos in Mx–c-fos mice.
Finally, we examined the effect of c-Fos on dendritic cell differentiation using Mx–c-fos mice. Cells expressing CD11c and Mac-1 were observed in cultures of cells with GM-CSF, and dendritic cell activation was indicated by reduced expression of Mac-1 (CD11c+Mac-1−)46 in cultures of cells from Mx–c-fos mice with GM-CSF and sRANKL (Figure 8B). Induction of DEC205 was observed in the presence of sRANKL with GM-CSF (data not shown). However, the frequency of CD11c+Mac-1−cells was reduced following c-Fos induction (Figure 8B), and DEC205 expression was suppressed by c-Fos induction even in the presence of GM-CSF and sRANKL (data not shown), indicating that c-Fos negatively affects dendritic cell differentiation. Inhibition of dendritic cell maturation was also observed following addition of M-CSF to cultures containing GM-CSF and sRANKL (Figure 8A), suggesting that M-CSF inhibits dendritic cell differentiation. The inhibitory effect of c-Fos and M-CSF on dendritic cell differentiation was more effective when they were induced or added early (data not shown). Our results suggest that M-CSF and GM-CSF antagonize each other in the differentiation of osteoclasts and dendritic cells. Osteoclast differentiation is inhibited due to the lineage switching promoted by GM-CSF through suppression of c-Fos. Therefore, c-Fos plays a pivotal role at an early switch in differentiation of precursors into osteoclasts or dendritic cells.
Discussion
Bifurcated differentiation of osteoclasts and dendritic cells
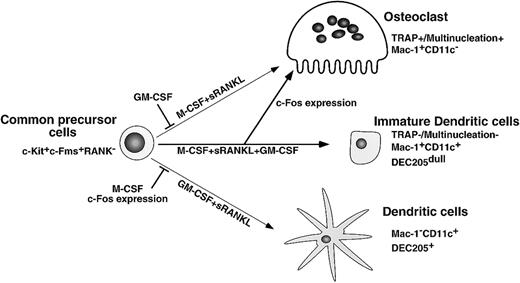
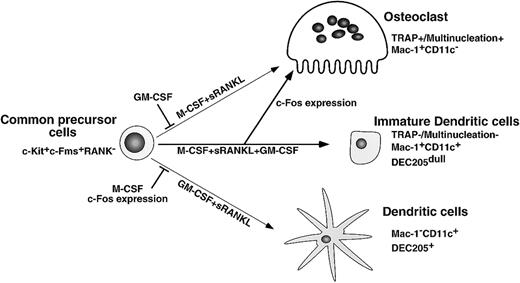
Here we present findings supporting a differentiation and lineage determination model between osteoclasts and dendritic cells (Figure9). We have used a stromal cell–free culture system that allows us to manipulate osteoclast differentiation. M-CSF and sRANKL induce osteoclastogenesis, while GM-CSF and sRANKL induce dendritic cell differentiation from single common precursors (c-Kit+c-Fms+RANK− cells). Bone resorptive multinuclear osteoclasts differentiate from monocyte/macrophage progenitor cells in the presence of M-CSF and sRANKL47 48 and are identified as TRAP-positive cells. Because the inhibition of osteoclastogenesis induced by GM-CSF is observed in all wells, which receive single cells, it is likely that the differentiation pathway of common precursor cells is influenced by extrinsic factors. Reduction of c-Fos is critical for inhibition of osteoclastogenesis by GM-CSF, while expression of c-Fos inhibits dendritic cell maturation. Therefore, c-Fos is a critical regulator of both osteoclast and dendritic cell differentiation.
Differentiation and lineage bifurcation model between osteoclasts and dendritic cells from common precursor cells (c-Kit+c-Fms+RANK− cells).
The differentiation of osteoclasts is inhibited by GM-CSF through reduction of c-Fos expression. By contrast, dendritic cell maturation induced by GM-CSF and sRANKL is inhibited by expression of c-Fos or M-CSF.
Differentiation and lineage bifurcation model between osteoclasts and dendritic cells from common precursor cells (c-Kit+c-Fms+RANK− cells).
The differentiation of osteoclasts is inhibited by GM-CSF through reduction of c-Fos expression. By contrast, dendritic cell maturation induced by GM-CSF and sRANKL is inhibited by expression of c-Fos or M-CSF.
Dendritic cells also differentiate from macrophage precursor cells,1 and GM-CSF plays an important role in their differentiation.49 CD11c+/Mac-1−cells are reported to be more mature dendritic cells than are CD11c+/Mac-1+ cells,46 and this maturation step occurs following addition of sRANKL to cultures with GM-CSF. RANKL is an activating factor of dendritic cells,13 and the activation was confirmed in our study by observation of up-regulation of the maturation marker DEC205 in the presence of both GM-CSF and sRANKL. This maturational step was inhibited by addition of M-CSF. Taken together, our findings indicate that differentiation into an osteoclast or a dendritic cell lineage is reciprocally inhibited by M-CSF and GM-CSF, respectively. To understand how lineages of osteoclasts and dendritic cells bifurcate in the presence of GM-CSF, we examined expression and function of cytokine receptors and transcription factors.
Expression of cytokine receptors in differentiation
M-CSF, GM-CSF, and IL-3 are macrophage-inducible cytokines. The RANK and c-Fms receptors are indispensable for osteoclast differentiation, and we have previously reported that pretreatment of precursor cells with M-CSF is required for osteoclastogenesis as well as RANK induction.9 Although it is reported that expression of c-Fms is inhibited in the presence of GM-CSF or IL-3,44 expression of c-Fms, RANK, and GM-CSF receptors was detected in the presence of M-CSF, GM-CSF, or IL-3 in our culture system, suggesting that inhibition of osteoclastogenesis is not due to down-regulation of receptor expression. Common β-chain signaling shared with GM-CSF, IL-3, and IL-5 receptors could be involved in inhibition of osteoclast differentiation, although in our experiments osteoclast precursors did not respond to IL-5. This inhibition of osteoclastogenesis is considered to have species variability because human GM-CSF did not inhibit human osteoclast differentiation (data not shown). Because cells in the colonies formed in the presence of GM-CSF or IL-3 are able to differentiate into multinuclear osteoclasts, when they are transfered to the liquid culture with M-CSF and RANKL the inhibitory effect of GM-CSF and IL-3 on osteoclastogenesis is reversible. It has been reported that GM-CSF or IL-3 can rescue the osteoclastogenesis defect in op/op mice.50 Our findings indicate that GM-CSF may increase the number of osteoclast precursor cells rather than directly induce the osteoclast differentiation. Recently Niida et al reported that VEGFR-1 is expressed in osteoclasts and VEGF can rescue the osteopetrosis inop/op mice by substituting in part for M-CSF in osteoclastogenesis.51
Critical function of c-Fos in the differentiation switch
Osteopetrosis due to an osteoclast differentiation failure is a phenotype observed following targeted mutation of several genes, including RANK, RANKL, TRAF6, and the transcription factors PU.1, c-Fos, NF-κB1, and NF-κB2.11,12,21-24,30 Among these proteins, we showed that c-Fos expression was specifically suppressed in the presence of GM-CSF. The c-Fos is a component of the AP-1 family of transcriptional regulators composed of c-Fos proteins (c-Fos, FosB, Fra-1, Fra-2) and Jun proteins (c-Jun, JunB, JunD), and c-Fos transgenic mice exhibit osteosarcoma.52,53 Although c-Fos contains a transactivation domain in its C-terminus, a truncated form of c-Fos (c-Fos–δ) lacking the C-terminus effectively rescues the osteoclastogenesis defect seen in c-Fos–deficient mice.27However, c-Fos–δ did not rescue an osteoclastogenesis defect induced by GM-CSF as full length of c-Fos, suggesting that the C-terminal of c-Fos is required to overcome the effect of GM-CSF. Fra-1 is thought to be a target of c-Fos,25,26 and overexpression of Fra-1 rescues the osteoclast defect seen in c-Fos–deficient mice.27 A truncated form of Fra-1 lacking the C-terminus can also rescue osteoclastogenesis in c-Fos–deficient mice,27 but osteoclast induction by full-length and truncated forms of Fra-1 or c-Fos–δ is less effective compared with full length of c-Fos. Interestingly, Fra-1 was induced by expression of full-length c-Fos but not by c-Fos–δ. Taken together, because expression of Fra-1 did not rescue the osteoclastogenesis as full-length c-Fos, the C-terminal of c-Fos is likely to be important for osteoclast differentiation as well as induction of Fra-1 in the presence of GM-CSF. In contrast, morphologic changes, such as cell spreading and loss of clustering, were induced by both the full-length and truncated forms of c-Fos. Cell aggregation, which is an indicator of dendritic cell activation,13 was inhibited by both full-length and truncated c-Fos. Recently, Fra-1 and ΔFosB transgenic mice have been shown to exhibit osteosclerosis due to increased bone formation54,55; however, no bone abnormalities have been observed in FosB knockout mice56 or in mice expressing c-Jun, JunB, FosB, or Fra-2.52
The effect of c-Fos on osteoclastogenesis was confirmed in experiments with cells derived from the Mx–c-fos mice, in which c-Fos expression was induced in osteoclast precursor cells. Using this system, we demonstrated that suppression of c-Fos is crucial for the inhibition of osteoclastogenesis by GM-CSF at bifurcation of osteoclasts and dendritic cells. Because Fra-1 was induced by c-Fos expression as observed in our retroviral experiments, Fra-1 may cooperate with c-Fos to overcome the GM-CSF effect. In addition to its positive effect on osteoclastogenesis, c-Fos negatively affected dendritic maturation, indicating that c-Fos has a reciprocal effect on the differentiation pathway of common precursors.
Our findings indicate that the inhibition of osteoclastogenesis by addition of GM-CSF is not a selection but rather a lineage bifurcation. The expression of CD11c, which is expressed on dendritic cells, was not observed in cells cultured with M-CSF alone or with M-CSF and sRANKL; however, CD11c was induced by addition of GM-CSF in the presence of M-CSF and sRANKL. Also, cell clusters observed in dendritic cells were induced by GM-CSF. These results indicate that differentiation had shifted from an osteoclast to a dendritic lineage. By contrast, early expression of c-Fos or early addition of M-CSF inhibited the maturation of dendritic cells. Taken together, our results indicate that osteoclasts, dendritic cells, and macrophages develop from the same precursor cells and that M-CSF, GM-CSF, and IL-3 antagonize each other in the course of differentiation. Furthermore, early expression of c-Fos is crucial for the differentiation of osteoclasts and the maturation of dendritic cells.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Toshio Suda, Dept of Cell Differentiation, Institute of Molecular Embryology and Genetics (IMEG), Kumamoto University, 2-2-1 Honjo, Kumamoto 860-0811, Japan; e-mail:sudato@gpo.kumamoto-u.ac.jp.