Abstract
Hematopoietic regulation is a complex but dynamic process regulated by intercellular and intracellular interactions within the bone marrow (BM) microenvironment. Through neurokinin-1 (NK-1) and NK-2 receptors, peptides (eg, substance P [SP]) encoded by the preprotachykinin-I gene mediate distinct hematopoietic effects. Cytokines, associated with hematopoietic stimulation, and SP regulate the expression of each other in BM mesenchymal and immune cells. Neutral endopeptidase (NEP) uses SP as a substrate to produce SP(1-4), which inhibits the proliferation of matured myeloid progenitor. This study determines whether the degradation of SP to SP(1-4) by endogenous NEP in BM stroma could be a feedback on hematopoietic stimulation by stem cell factor (SCF). SP(1-4) induced the production of transforming growth factor (TGF)–β and tumor necrosis factor–α in BM stroma. TGF–β production accounted for part of the inhibitory effects by SP(1-4) on the proliferation of early (granulocyte-macrophage colony-forming units) and late (long-term culture-initiating cells) hematopoietic progenitors. Enzyme-linked immunosorbent assays and/or protein-chip arrays indicated a timeline change of SP to SP(1-4) in BM stroma stimulated with SCF, which correlated with increase in NEP messenger RNA. Since SP and its fragment, SP(1-4), interact with the same receptor to mediate opposing hematopoietic effects, 2 interactive studies were done to understand the dual responses of NK-1: (1) a 3-dimensional molecular model of NK-1 and SP and (2) screening of a random dodecapeptide library for SP(1-4) interacting sites. The effects of SP(1-4) on hematopoietic progenitors and the timeline change of SP to SP(1-4), together with the 3-dimensional model, provide a partial explanation for the feedback on the stimulatory effects of SCF and SP on hematopoiesis.
Introduction
In the adult, the bone marrow (BM) is the major site of hematopoiesis. Biological activities in the BM are complexed, albeit controlled, through cell-cell interactions among the hematopoietic stem cell, progenitors, mesenchymal cells, and accessory cells.1 Cellular interactions in the BM could lead to the induction of soluble hematopoietic regulators, such as cytokines, neuropeptides, and neurotrophic factors.2 Other hematopoietic regulators could be derived as neurotransmitters from the innervated nerve fibers in the BM and as hormones from the peripheral circulation.2 Despite the distant source of hormones, they modulate biological responses in the BM through specific receptors on resident BM cells.3
The general consensus is that a finite pool of hematopoietic stem cells produces mature immune and blood cells through a complexed but controlled network that includes cells and soluble factors. Regardless of the pathways that lead to hematopoietic stimulation, the process of feedback is of utmost importance to maintain homeostasis and to protect the pool of hematopoietic stem cells. Understanding this dynamic process in the BM would have an impact on several clinical areas such as gene therapy, chemotherapy, and BM transplantation.4 In this study, we describe a model that might explain the mechanism for negative feedback by the amino terminal of substance P (SP), SP(1-4), on the effects of 2 hematopoietic stimulators: stem cell factor (SCF) and SP. These 2 stimulators regulate the induction of each other in BM cells.2,5SP(1-4) could be derived from degradation of SP by endogenous endopeptidases in the BM.6,7 This study also reports on the functional plasticity of neurokinin-1 (NK-1) receptor to regulate hematopoiesis.8
SP, an undecapeptide, is evolutionarily conserved and is the major peptide derived from the preprotachykinin-I (PPT-I)gene.9,10 PPT-I peptides exert both stimulatory and inhibitory hematopoietic effects,5 which are mediated through G-protein–coupled receptors: NK-1, NK-2, and NK-3.5,11,12 NK-1 is induced in BM cells by cytokines and other stimulatory hematopoietic regulators.2 NK-2 is constitutively expressed in BM cells that are unstimulated or stimulated with suppressive hematopoietic regulators.5NK-1 and NK-2 are not coexpressed in BM cells since NK-1 induction by cytokines is correlated with the down-regulation of NK-2.3 5
Endopeptidases are ubiquitously expressed in hematopoietic cells.7 Dipeptidyl-peptidase IV (CD26), aminopeptidase, angiotensin-converting enzyme, and neutral endopeptidase (NEP)/CD10 use SP as their substrate.6,7,13,14 The carboxyl portion of the truncated peptide can be further digested by aminopeptidases N/CD13,15,16 resulting in Arg-Pro and Arg-Pro-Lys-Pro: SP(1-4). Cytokines, SP, and endopeptidases regulate the expression of each other in BM cells.13,14,17 The proinflammatory properties of SP and cytokines have been linked to the activities of various endopeptidases.14 17
SP fragments could exert immune and hematopoietic regulation.18,19 This study shows that SP(1-4) inhibits the proliferation of late and early hematopoietic progenitors. The parent peptide, SP, and cytokines associated with hematopoietic stimulation regulate the expression of each other, leading to positive hematopoiesis.2 To this end, we investigated whether digestion of SP to SP(1-4) could be a mechanism of hematopoietic feedback on the effects of SCF. This particular cytokine represents a model hematopoietic stimulator, interacting with SP to regulate the expression of each other.20 21 Induction of the following was studied: negative hematopoietic regulators, transforming growth factor (TGF)–β, tumor necrosis factor (TNF)–α, the receptors for SP and its fragments (NK-1 and NK-2), and NEP messenger RNA (mRNA). SP/SP(1-4)–NK-1 interactions were examined in timeline proteomic studies: screening of a random dodecapeptide library, protein-chip array, and 3-dimensional molecular modeling.
Materials and methods
Reagents, cytokines, and antibodies
SP, SP(1-4), thiazolyl blue (dimethyltetrazolium [MTT] assay), spantide, Ficoll-Hypaque, and nonimmune rabbit serum were purchased from Sigma (St Louis, MO). Spantide, SP, and SP(1-4) were dissolved and stored as described.22 Biotin-SP and biotin–NK-A were purchased from Peninsula Laboratories (Belmont, CA). Alkaline phosphatase–conjugated goat antirabbit immunoglobulin (Ig)–G was obtained from Kirkegaard and Perry Laboratories (Gaithersburg, MD). Rabbit anti-SP was purchased from Biogenesis (Brentwood, NH). The Immunology Department of Genetics Institute (Cambridge, MA) provided recombinant human granulocyte-macrophage colony-stimulating factor (rhGM-CSF). Fluorescein isothiocyanate (FITC)–avidin was purchased from Vector Laboratories (Burlingame, CA). SCF, TGF-β1, TNF-α, goat anti–hTNF-α, rabbit anti–hTGF-β, biotinylated goat anti–hSCF receptor (c-kit), nonimmune rabbit IgG, and biotinylated nonimmune goat IgG were purchased from R&D Systems (Minneapolis, MN). Functional studies indicated that anti–hTGF-β exerts specificity by neutralizing the β1 form of TGF proteins.23
Clonogenic assays for granulocyte-macrophage colony-forming units
BM aspirate was obtained from the posterior iliac crest of healthy volunteers. The Institutional Review Board of New Jersey Medical School approved this study. Mononuclear cells were isolated by Ficoll-Hypaque density gradient and then assayed for granulocyte-macrophage colony-forming units (CFU-GMs) in sera-free cultures as described.3 Briefly, 105 BM mononuclear cells per milliliter were plated in methylcellulose with 10 or 1 nM SP(1-4). Each experimental point was assayed in duplicate. Cultures were assayed in parallel in the presence or absence of anti–TGF-β or anti–TNF-α, each ranging between 2 μg/mL and 10 ng/mL. Cultures contained 3 U/mL rhGM-CSF. At day 10, the number of colonies with more than 20 cells were counted. Control cultures, media alone or nonimmune IgG, showed similar numbers of CFU-GMs, ranging between 155 and 80.
Modified long-term culture-initiating cells
Confluent stromal cells were cultured in 25-cm2flasks and then subjected to 150 Gy, delivered by a cesium source. We added 107 bone marrow mononuclear cells to the flask containing the γ-irradiated stroma. Beginning at week 5 of culture, aliquots of cells were assayed for CFU-GMs every week up to 12 weeks.
Preparation and stimulation of BM stroma
Stromal cultures were prepared with BM aspirates from healthy donors as described.24 Briefly, stromal cells were cultured at 33°C, and at day 3, the granulocytes and red blood cells were removed by Ficoll-Hypaque density gradient. Cultures were reincubated with weekly replacement of 50% culture media until confluence.
Cytokine, NK receptor, and c-kit expressions were studied in confluent stroma, stimulated with 10 nM SP(1-4) and/or 10 ng/mL SCF. Stroma was stimulated in 3 mL sera-free α–Eagle minimum essential medium (α-MEM), supplemented with insulin-transferrin-selenium-A (Life Technologies, Grand Island, NY). At 16 hours, stromal monolayers were washed with sera-free α-MEM and then reincubated for 16 hours with media, 10 nM SP(1-4), or 10 ng/mL SCF. Controls included stroma cultured for 16 or 32 hours in media alone. Time-course and dose-response curves established the optimal concentrations of SCF and SP(1-4).
The relative expression of c-kit in BM stroma was determined by immunofluorescence. BM stroma was cultured on coverslips (Fisher Scientific, Springfield, NJ) and then stimulated with 10 ng/mL SCF. At different times after cell stimulation, cells were labeled with 50 ng/mL biotinylated goat anti–c-kit and FITC-avidin. Nonspecific binding was determined with biotinylated nonimmune goat anti-IgG. Cells were immediately examined on an Olympus Probis (New York/New Jersey Scientific, Middlebush, NJ) microscope. The fluorescence intensity at an excitation of 595 nm was dim in unstimulated stroma and increased to bright in cells stimulated for 6 hours to 24 hours.
Quantitation of SP immunoreactivity
Competitive enzyme-linked immunosorbent assay (ELISA) quantitated SP immunoreactivity (SP-IR) as described.3Briefly, Immulon 96-well plates (Dynatech Laboratories, Chantilly, VA) were precoated with streptavidin and then incubated with biotinylated SP (Chiron Mimotopes, Emeryville, CA). Equal volumes (50 μL) of unknown samples and optimum rabbit anti-SP were added to quadruplicate wells. Each sample was assayed as undiluted and in 3 serial dilutions. Complexed anti-SP was detected with alkaline phosphatase–conjugated goat antirabbit IgG and Sigma 104 phosphatase substrate. SP-IR levels were calculated from a standard curve developed with optic density (OD) at 405 nm versus 12 serial dilutions of known SP concentrations, ranging from 100 to 0.08 pg/mL.
Quantitative reverse transcriptase–polymerase chain reaction
Quantitative reverse transcriptase–polymerase chain reaction (RT-PCR) for NK-1, NK-2, and β-PPT-I mRNA was performed as described.3,24 Briefly, 2 μg total RNA from BM stroma was reverse transcribed, and 200 ng complementary DNA (cDNA) was used in PCR reactions with standard DNA at log10-fold dilutions ranging between 10−2 and 10−6 attomole per liter. PCR products (10 μL) were separated on agarose containing ethidium bromide. The DNA was scanned with a Fluorimager (Molecular Dynamics, Sunnyvale, CA), and the densities were analyzed with ImageQuant software version 5.2 (Molecular Dynamics). A standard curve was established for each unknown sample: band densities of unknown/standard DNA versus log10 standard DNA concentration. The concentration of the unknown sample was read at the point where the unknown and standard were equivalent. Construction of DNA standards was previously described.24
Immunofluorescence for NK receptors
BM stroma was stimulated with 10 ng/mL SCF. At 24 hours, cells were washed and then immediately incubated at 4°C for 2 hours with 200 ng/mL biotin-SP or biotin–NK-A. After this, cells were labeled with FITC-avidin for 30 minutes and then examined for fluorescence intensity with an Olympus Provis AX-70 at 480 ± 40 nm and 535 ± 50 nm per emission. Minimal fluorescence was observed in cells colabeled with excess unconjugated SP or NK-A or in cells labeled with FITC-avidin alone. The optimal experimental conditions were determined in dose-response and time-course studies. Specificity of binding by SP or NK-A was studied in competition labeling with the pan-tachykinin antagonist spantide.
TNF-α determination
We seeded 1.5 × 104 L929 fibroblasts in 0.1 mL in 96-well flat-bottomed plates and then incubated them at 37°C. At 16 hours, 4 μg/mL actinomycin D was added to wells. Unknown samples, each with 4 serial dilutions in 50 μL, were added to triplicate wells. A standard curve was established in parallel with 12 serial dilutions of TNF-α beginning at 1.5 nM. Plates were incubated overnight, and the next day, cell viability was determined by means of the MTT assay. Briefly 20 μL MTT at 5 mg/mL was added to each well. The plates were incubated for 2 hours at 37°C and then centrifuged at 800g for 5 minutes. Supernatants were removed and cells washed once with phosphate-buffered saline (PBS) by centrifugation at 2000g for 5 minutes. Thiazolyl blue crystals were dissolved with 100 μL isopropanol, and the OD was determined at 570 to 650 nm with a Kinectic microplate reader (Molecular Devices, Menlo Park, CA).
TGF-β1 quantitation
Growth inhibition of CCL 64 cells forms the basis of the bioassay used to quantitate active TGF-β.25 Each sample was tested in triplicate with 25 μL, 50 μL, or 100 μL supernatants. TGF-β levels were determined from a standard curve established with TGF-β concentrations ranging from 0.001 to 10 ng/mL versus cell concentration. The presence of TGF-β1 was verified by reanalyses of samples containing more than 20 ng/mL TGF-β with neutralizing anti–hTGF-β1.25 Neutralization was determined if the anti–TGF-β1 reversed the growth-inhibitory effect of the supernatants.
Biopanning of phage display library and selection of clones
Peptide-binding sites were determined by screening a random dodecapeptide library, FliTrx, as per manufacturer's instruction (Invitrogen, Carlsbad, CA). Briefly, bacteria were induced with tryptophan and then passaged for 6 hours in 60-mm Petri dishes (Nunclon Delta, Nalge Nunc, Rochester, NY) that were precoated with 20 μg SP(1-4). After the seventh passage, the adherent bacteria were expanded in liquid culture and then subcultured on agar. Forty colonies were expanded, and the pellet from each bacterial culture was boiled in sample buffer and then spotted on methanol-soaked polyvinylidene fluoride (PVDF) transfer membranes (NEN, Boston, MA). The membranes were consecutively incubated with 10 μg/mL SP(1-4) overnight at room temperature and rabbit anti-SP for 2 hours. Anti-SP was detected with alkaline phosphatase–conjugated goat antirabbit IgG followed by incubation for 15 minutes with 5-bromo-4-chloro-3-indolyl-phosphatase/nitroblue tetrazolium phosphate substrate system (Kirkegaard and Perry Laboratories). A single blinded observer scored the relative intensities of the spots, with zero for no detection and 4 for the highest intensity. Twenty clones with assignments between 1 and 4 were selected and further analyzed in Western blots by means of gradient gels of 4% to 20% (Invitrogen).25 Seven clones were finally selected on the basis of the development of a single, dense band at the predicted molecular mass (Mr).
DNA was extracted from each clone (described above) and then sequenced in both orientations at the Molecular Core Facility, UMDNJ–New Jersey Medical School. DNA sequencing was performed with the FliTrx forward and Rsr reverse primers, provided with the peptide library. Each clone was analyzed with the Wisconsin Genetics Computer Group (Madison, WI) package of DNA/protein sequence analysis programs (version 10). Six clones were selected on the basis of greater than 40% identity to the protein sequence of NK-1.26
Computer modeling of SP and NK-1
The transmembrane (TM) portion of NK-1 was modeled by means of WHAT-IF (Inspire Pharmaceuticals, Durham, NC) in conjunction with Swiss-Model (GlaxoSmithKline, Geneva, Switzerland) at the Swiss Institute of Bioinformatics (Geneva) and European Molecular Biology Laboratory (EMBL) (Heidelberg, Germany). The structures of both SP and NK-1 were generated by means of SYBYL 6.6 (Tripos Associates, St Louis, MO). A homolog of the TM region of NK-1 was modeled at the EMBL and Swiss-Model repository by means of WHAT-IF, an algorithm that maps the sequence onto a template structure. WHAT-IF optimizes the structure while adjusting for side-chain collisions and interactions. The template used was the alpha-carbon chain from the crystallized structure of the G-protein– coupled receptor, bacteriorhodopsin.27 The arrangement of the TM helices in bacteriorhodopsin is the conventional model for all members of the G-protein–coupled receptor. The intracellular and extracellular loops were constructed by means of a loop search. SP was constructed ab initio on the basis of its sequence. A molecular dynamics run was performed with the use of a time step of 5, carried out through 500 iterations.
Energy minimization was performed. Docking of SP into NK-1 was performed with SYBYL's Docking engine and Flexidock. All energetics calculations were performed by means of Kollman-United charges with the Tripos force-field engine.
Profiling for SP and SP(1-4) by protein-chip analyses
SP and SP(1-4) were profiled in stromal cell extracts by means of Ciphergen's ProteinChip Technology (Ciphergen Biosystems, Fremont, CA). Weak Cation Exchanger (WCX2) ProteinChip array (for profiling) and the preactivated chip surface, PS1 (for affinity studies) were used for the identification of SP(1-4). The basic method, which was provided by Ciphergen, was adjusted to optimize the analyses that follow. Stromal cells were washed with PBS and then subjected to repeated freeze-thaw. Cell-free lysates were prepared by centrifuging at 4°C for 30 minutes/15 000g. WCX2 was pretreated with 10 mM HCl, and 500 μL each sample was spotted on the array by means of a bioprocessor. The chips were incubated at room temperature for 30 minutes with vigorous shaking to ensure binding of sample to array. Subsequently, chips were washed with 5% Triton/PBS (2×) and with PBS (1×). A saturated solution of α-cyano-4-hydroxy cinnamic acid (CHCA) (Ciphergen Biosystems) was diluted at 1:50 in 50% acetonitrile and 0.5% trifluoroacetic acid. Diluted CHCA (0.5 μL) was added to the spots of the arrays and the chips were dried at room temperature. After this, chips were read on Ciphergen's surface-enhanced laser desorption ionization–time of flight mass spectrometer (SELDI TOF-MS). Accurate mass was determined by collecting 150 averaged laser shots. Stromal extracts from 48-hour–stimulated cultures were selected for further identification of immunoreactive SP(1-4). PS1 chips were pretreated with 50% acetonitrile and then incubated for 45 minutes with 10 μL anti-SP at 1:500 dilution in PBS. Control spots contained nonimmune rabbit serum. The arrays were blocked for 25 minutes with 1M ethanolamine and washed with PBS + 0.5% Triton X (2×) and a final PBS wash step. Five hundred μL of cell extracts was incubated with the use of the bioprocessor. The chips were washed with PBS + Triton X, PBS, rinsed with 5 mM HEPES, and dried. CHCA was applied and SP(1-4) bound to the PS1 chip was analyzed as described above for the profiling studies.
Northern analysis
Northern analyses for steady-state NEP and NK-1 mRNA were performed as described.28 For NEP mRNA, stromal cells were stimulated with SCF for 16 hours in sera-free α-MEM supplemented with insulin-transferrin-selenium-A. At different times afterwards, total RNA was extracted and 10 μg was used for Northern analyses. Membranes were consecutively hybridized with [α-32P]-d-adenosine 5′-triphosphate–labeled cDNA probes for NEP and 18S ribosomal RNA (rRNA) as described.3 NK-1 mRNA was determined with total RNA from stromal cultures that were sequentially stimulated with SCF and SP(1-4), described above. The cDNA for NK-1 was previously described.29 NK-2 cDNA was prepared by RT-PCR with total RNA from BM stroma as template and the same primers as were used for quantitative RT-PCR (described above). The NK-2 fragment, which was equivalent to 274 base pairs, was subcloned into pNoTA/T7 (5 Prime → 3 Prime, Boulder, CO). The cDNA for human NEP and 18S rRNA were purchased from American Type Culture Collection (Manassas, VA).
Statistical analysis
Data were analyzed by means of the Student t test to determine the significance (P value) between experimental values.
Results
Interactions between SCF and SP(1-4) on the induction of TGF-β and TNF-α in BM stroma
The inhibitory effect of SP(1-4) on hematopoiesis is similar to NK-A, which is also a PPT-I peptide.5 The suppressive effect of NK-A was partly mediated through the induction of TGF-β.5 We therefore determined whether SP(1-4) could induce 2 hematopoietic suppressors in BM stroma: TGF-β and TNF-α.28,30 31 Table 1shows that SP(1-4) significantly (P < .05) increases the induction of both cytokines in BM stroma: unstimulated, less than 5 pg/mL TGF-β and less than 10−4 pM TNF-α; stimulated, 90 ± 15 pg/mL TGF-β and 40 ± 4 pM TNF-α.
The next set of experiments determined whether SP(1-4) and SCF could affect the role of each other on the induction of TGF-β and TNF-α in BM stroma. Cells stimulated with SCF showed baseline TNF-α and TGF-β (Table 1). However, when SP(1-4) was added to the SCF-stimulated stroma, the levels of both cytokines were significantly (P < .05) increased: TGF-β, less than 5 pg/mL in the first stimulation with SCF, which was changed to 80 ± 8 pg/mL after a second stimulation with SP(1-4); TNF-α, 0.028 ± 10−4 in the first stimulation to 31 ± 2 pM in the second stimulation (Table 1).
To determine if SCF could affect the induction of TNF-α and TGF-β by SP(1-4), BM stroma was stimulated with SP(1-4), and 16 hours later, cultures were subjected to a second stimulation with SCF. The results, shown in Table 1, indicated that TGF-β levels were reduced from 64 ± 4 pg/mL to less than 5 pg/mL and that TNF-α decreased from 36 ± 6 to 0.025 ± 10−4 pM. The results indicate that SP(1-4) and SCF, negative and positive regulators of hematopoiesis, respectively,8,20 32 showed different effects on the induction of TGF-β and TNF-α in BM stroma.
Effects of SP(1-4) on early and late hematopoietic progenitors
SP(1-4) inhibited the proliferation of hematopoietic progenitors (CFU-GMs) in sera-containing cultures.8 To verify that the sera did not contribute to this inhibitory effect of SP(1-4), clonogenic assays were repeated for CFU-GMs (n = 7) in sera-free cultures. The percentage of inhibition by SP(1-4) on CFU-GMs in the sera-free cultures was significant (P < .05): 55% ± 4% for 1 nM SP(1-4) and 65% ± 6% for 10 nM SP(1-4). This indicates that the inhibitory effect of SP(1-4) on CFU-GMs was independent of the sera in the experimental assay. Since SP(1-4) induces the production of TGF-β and TNF-α (Table 1), which are inhibitors of hematopoiesis,30 studies were performed to determine if these cytokines could mediate the suppression of SP(1-4) on CFU-GMs. Clonogenic assays were performed with 10 nM SP(1-4) and/or various concentrations of anti–TGF-β1 or anti–TNF-α. The data, shown in Figure 1A, represent the point of maximal effect by the anti–TGF-β (0.5 μg/mL). Anti–TGF-β showed significant (P < .05) change in reversing the suppression of SP(1-4) on CFU-GMs, whereas anti–TNF-α showed no change (Figure 1A). Equivalent concentrations of nonimmune rabbit and goat IgG showed no change in CFU-GMs cultured with 10 nM SP(1-4) (data not shown). These results indicate that part of the suppressive effect of SP(1-4) on CFU-GMs could be indirectly mediated by the production of TGF-β1 in BM stroma.
Role of TGF-β and TNF-α in the suppression of CFU-GMs and long-term culture-initiating cell (LTC-IC) proliferation by SP(1-4).
Clonogenic (panel A) and LTC-IC (panel B) assays were performed in the presence of 10 nM SP(1-4). Parallel cultures contained anti–TGF-β or anti–TNF-α. The results are expressed as the mean ± SD of 6 experiments, each performed with a different healthy BM donor. *P < .05 versus SP(1-4) alone; **P < .05 versus SP(1-4) plus anti–TGF-β.
Role of TGF-β and TNF-α in the suppression of CFU-GMs and long-term culture-initiating cell (LTC-IC) proliferation by SP(1-4).
Clonogenic (panel A) and LTC-IC (panel B) assays were performed in the presence of 10 nM SP(1-4). Parallel cultures contained anti–TGF-β or anti–TNF-α. The results are expressed as the mean ± SD of 6 experiments, each performed with a different healthy BM donor. *P < .05 versus SP(1-4) alone; **P < .05 versus SP(1-4) plus anti–TGF-β.
TGF-β suppresses the proliferation of the more primitive hematopoietic progenitors.28 We therefore studied the effects of SP(1-4) on the proliferation of long-term culture-initiating cells (LTC-ICs). The results are shown for CFU-GMs cultured between weeks 10 and 12 (Figure 1B). The pattern of inhibition by SP(1-4) on these relatively primitive hematopoietic progenitors was similar to that of CFU-GMs. In the presence of anti–TGF-β1, there was partial but significant (P < .05) reversal in the inhibitory effect of SP(1-4) on the proliferation of the LTC-ICs (Figure 1B), 48 ± 3 CFU-GM colonies for media control and 37 ± 4 CFU-GM colonies for SP(1-4) plus anti–TGF-β, ± SD. Anti–TNF-α showed no effect on the proliferation of LTC-ICs (Figure 1B). The results showed that SP(1-4) mediated the induction of TGF-β, which suppressed CFU-GMs and partly inhibited the proliferation of LTC-ICs. The secondary production of TNF-α by SP(1-4) in BM stroma showed no evidence of being a mediator in the hematopoietic effect of SP(1-4).
Effects of SP(1-4) on NK-1 and NK-2 expression
Since NK-1 and NK-2 receptors mediated the hematopoietic effects of the tachykinins,5 this section describes the results of studies designed to further determine the interacting receptor for SP(1-4). Owing to the lack of specific antibodies to the human NK-1 and NK-2 receptors, their detection by immunofluorescence was performed with the preferred ligand that was conjugated to biotin: SP for NK-1, and NK-A for NK-2.12 Spantide, a pan-tachykinin antagonist that competes with the ligand for NK receptor binding, was used to determine the specificity of binding. Representative results of 5 different labeling experiments are shown in Figure2. Consistent with published reports,5 the results showed that SP binds to stromal cells in which NK-1 receptor was induced (SCF stimulation, Figure 2B). NK-A did not bind to the SCF-stimulated stroma (Figure 2C). These results are internal controls that show that the induction of NK-1 by cytokines correlated with reduced expression of NK-2.2 5Interestingly, stromal cells stimulated with SCF for 16 hours and then a second time with SP(1-4) showed a reduction in NK-1 expression (SP binding, Figure 2E) and relatively brighter staining for NK-2 (NK-A binding, Figure 2F). These results show that SP(1-4) reduced the expression of NK-1 in SCF-stimulated stroma with concomitant increase in the expression of NK-2.
Immunofluorescence for SP- and NK-A–binding sites in BM stroma.
Cells were stimulated with 10 ng/mL SCF and/or 10 nM SP(1-4) for 24 hours. After this, cells were labeled with biotin-SP or biotin–NK-A and then developed with FITC-avidin. Unstimulated (A,C); SCF-stimulated (B,D) consecutive stimulation with SCF and SP(1-4) (E-F).
Immunofluorescence for SP- and NK-A–binding sites in BM stroma.
Cells were stimulated with 10 ng/mL SCF and/or 10 nM SP(1-4) for 24 hours. After this, cells were labeled with biotin-SP or biotin–NK-A and then developed with FITC-avidin. Unstimulated (A,C); SCF-stimulated (B,D) consecutive stimulation with SCF and SP(1-4) (E-F).
Quantitative RT-PCR was used to determine if the phenotypic expression of NK receptors (Figure 2) correlated to the respective mRNA levels. The levels of NK-2 mRNA in unstimulated and SP(1-4)–stimulated stroma were similar (Table 2). The level of NK-2 mRNA was, however, reduced following a first stimulation with SCF (Table 2). This reduction in NK-2 mRNA was reversed following a second stimulation by SP(1-4) (Table 2). NK-1 mRNA levels were quantitated in BM stroma stimulated with SCF (first stimulation). After 16 hours, the point at which NK-1 mRNA levels were increased (Table 2), stromal cells were restimulated with SP(1-4) (second stimulation). Compared with unstimulated stroma, the second stimulation showed a significant (P < .05) increase in NK-1 mRNA levels (Table 2): 275 ± 14 molecule per microgram total RNA in stimulation with SCF plus SP(1-4) versus less than 1 molecule per microgram total RNA in unstimulated cells. The increase in NK-1 mRNA level by SCF plus SP(1-4) was significantly reduced (P < .05) compared with stroma stimulated with SCF alone: 275 ± 14 versus 1480 ± 34 molecules per microgram total RNA (Table 2). Northern blots with total RNA from selected experimental points supported the difference in NK-1 and NK-2 expression in unstimulated cultures (Figure3, lane 1); SCF stimulation (Figure 3, lane 2); and cultures stimulated consecutively with SCF and SP(1-4) (Figure 3, lane 3). The results showed that SP(1-4) reduced the expression of NK-1 induced by SCF, with a concomitant increase in NK-2 expression.
NK-1 and NK-2 mRNA in BM stroma stimulated with 10 ng/mL SCF and/or 10 nM SP(1-4).
Stroma was stimulated with SCF; at 16 hours, cultures were terminated or washed and then restimulated with SP(1-4) for 16 hours. Total RNA from each culture was studied for NK-1 and NK-2 mRNA by Northern analyses. Lane 1, media alone; lane 2, SCF; lane 3, SCF-SP(1-4).
NK-1 and NK-2 mRNA in BM stroma stimulated with 10 ng/mL SCF and/or 10 nM SP(1-4).
Stroma was stimulated with SCF; at 16 hours, cultures were terminated or washed and then restimulated with SP(1-4) for 16 hours. Total RNA from each culture was studied for NK-1 and NK-2 mRNA by Northern analyses. Lane 1, media alone; lane 2, SCF; lane 3, SCF-SP(1-4).
Induction of NEP and correlation with detectable SP and SP(1-4)
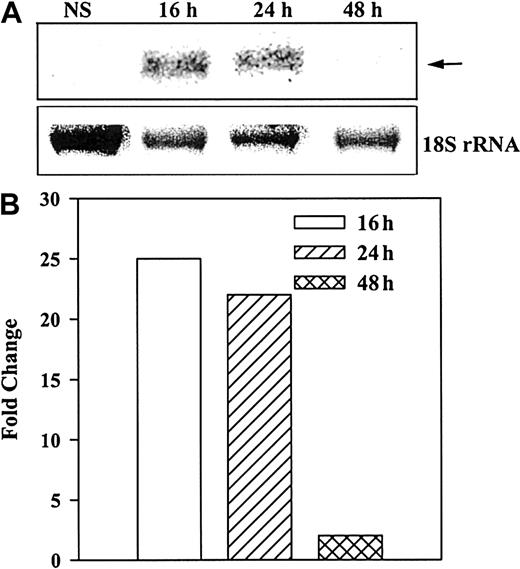
Total RNA from unstimulated and stimulated (10 ng/mL SCF) BM stroma was analyzed for steady-state NEP mRNA levels by Northern analyses. The results (Figure 4A) indicate single bands at the predicted size of 1.8 kilobases. Strong bands were observed at 16 and 24 hours, with no detectable band at 48 hours. Figure 4B shows the fold change of normalized (18S rRNA) mRNA in stimulated over unstimulated stroma. The data indicate that the steady-state mRNA for NEP is optimally increased by SCF after 16 hours and remained detectable up to 24 hours.
Induction of NEP in BM stroma stimulated with SCF.
A representative blot from 3 healthy donors is shown. (A) Northern analyses determined steady-state levels of mRNA for NEP at different times after stimulation of BM stroma with 10 ng/mL SCF. (B) Arrow represents NEP mRNA and lower row represents 18S rRNA. NEP levels were normalized with 18 sRNA and represented as the fold change over unstimulated stroma.
Induction of NEP in BM stroma stimulated with SCF.
A representative blot from 3 healthy donors is shown. (A) Northern analyses determined steady-state levels of mRNA for NEP at different times after stimulation of BM stroma with 10 ng/mL SCF. (B) Arrow represents NEP mRNA and lower row represents 18S rRNA. NEP levels were normalized with 18 sRNA and represented as the fold change over unstimulated stroma.
To determine if the induction of NEP correlated with the degradation of SP, timeline profiling studies were used to determine the change of SP to SP(1-4) in stromal cell extracts stimulated with 10 ng/mL SCF. Analyses for SP and SP(1-4) were performed at different times by means of Ciphergen's ProteinChip arrays. ELISA determined the levels of SP-IR in the supernatant and cell extracts of the stimulated stroma (5 × 106 stroma). An affinity protein chip with mobilized anti-SP determined the presence of SP(1-4) in extracts from 48-hour–stimulated stroma. The results showed fewer than 2 ng SP-IR in unstimulated stroma (n = 5). At 24 hours, the total levels of SP-IR (supernatants and cell extracts) in stimulated stroma were 185 ± 8 ng. SP levels were reduced to 12 ± 5 ng at 48 hours and to baseline levels (less than 2 ng) at 72 hours (Table3).
Since the small size of SP(1-4) limits its detection by methods similar to SP, ProteinChip arrays were determined to be sensitive for the detection of SP(1-4). Stromal extracts were analyzed at 24, 48, and 72 hours of SCF stimulation for peaks that are equivalent to SP and SP(1-4). A peak corresponding to the size of SP (Figure5A) was detected in 5 different samples, which disappeared at 48 and 72 hours. During these same periods (48 and 72 hours), peaks corresponding to SP(1-4) were detected (Figure 4B). To ascertain that the peak corresponding to 495 d in the 48-hour extract was indeed SP(1-4), the analyses was further studied with an affinity chip in which anti-SP was mobilized. Previous studies indicated that this antibody binds to SP(1-4), which explained a single peak at 495 d (Figure 5C) with no detection at approximately 1395 d. Binding of nonimmune rabbit sera to similar chips showed no detectable protein (data not shown). Figure 5D summarizes the dynamics of SP and detectable SP(1-4) in SCF-stimulated stroma. The time spans of reduced SP correlated with up-regulation of NEP mRNA and the detection of molecules corresponding to 495 d, the size of SP(1-4).
Chromatogram for SP profiling studies.
Standard chromatogram for profiling studies for SP (panel A) or SP(1-4) (panel B) and for affinity detection of SP(1-4) (panel C). A scheme showing optimal level of SP at 24 hours, which was reduced at 48 hours. Between 16 and 24 hours, NEP mRNA is induced, which is followed by the appearance of SP(1-4) at 48 hours (panel D).
Chromatogram for SP profiling studies.
Standard chromatogram for profiling studies for SP (panel A) or SP(1-4) (panel B) and for affinity detection of SP(1-4) (panel C). A scheme showing optimal level of SP at 24 hours, which was reduced at 48 hours. Between 16 and 24 hours, NEP mRNA is induced, which is followed by the appearance of SP(1-4) at 48 hours (panel D).
Three-dimensional molecular model of SP/SP(1-4)–NK-1 interactions
Functional studies with specific NK-1 and NK-2 receptor antagonists indicated that the NK-1 subtype mediated the suppressive effects on hematopoiesis by SP(1-4).8 Studies described in this report, however, showed low levels of NK-1 mRNA in BM stroma stimulated with SP(1-4) compared with SCF (Table 2). Relatively reduced fluorescence was also observed for NK-1 labeling in stroma costimulated with SCF and SP(1-4) (Figure 2). Owing to the small size of SP(1-4), the use of standard binding assays poses technical problems for the study of its binding kinetics. We therefore used other approaches to understand SP(1-4)–NK-1 interactions. The first set of studies screened a random dodecapeptide library for SP(1-4) interacting sequences, which resulted in 6 clones that shared significant homology to NK-1 protein residues: clones 7, 23, and 28 aligned with exon 1, and clones 12, 19, and 26 with exon 5 (Figure6). Alignment of the residues from the clones retrieved from the peptide library with NK-1 is shown at the bottom of Figure 6.
Selected clones from a random dodecapeptide library, screened with SP(1-4).
Shown are the amino acid residues of the DNA sequence for each clone. Bold and underlined residues indicate regions that align with NK-1 (top). Alignment of the combined amino acid sequences from the peptide library with NK-1, Accession No. m76675 (bottom).
Selected clones from a random dodecapeptide library, screened with SP(1-4).
Shown are the amino acid residues of the DNA sequence for each clone. Bold and underlined residues indicate regions that align with NK-1 (top). Alignment of the combined amino acid sequences from the peptide library with NK-1, Accession No. m76675 (bottom).
To determine how the residues shown in Figure 6 interact with SP(1-4), we used computer-assisted molecular modeling to generate a 3-dimensional structure of SP and NK-1. This model was also needed to understand the paradigm of NK-1 mediating both positive and negative hematopoietic effects.2,8 We are unaware of a crystal or nuclear magnetic resonance structure for the extracellular and intracellular loops of NK-1, which is a 7-TM, 407-residue, G-protein–coupled receptor.10 The spatial arrangement of its secondary structure is shown in Figure7,33 and the sequences shown for the TM regions were used to model the intracellular and extracellular loops, by means of a loop search (SYBYL). Given a certain root mean square distance between 2 end points, the loop search algorithm searched a database of loops and the highest scoring homolog was selected between 24.3% and 46%.
Spatial assignment of NK-1 residues.
27 Segments of amino acid residues representing the TM regions of NK-1 and the related intracellular and extracellular areas are shown in the 2-dimensional diagram.
Spatial assignment of NK-1 residues.
27 Segments of amino acid residues representing the TM regions of NK-1 and the related intracellular and extracellular areas are shown in the 2-dimensional diagram.
A crystallized structure of SP was not available in any public database. There are interesting aspects of the sequence of SP, which has an amide group at the carboxyl terminal (Figure8A). The position of the 2 phenyl rings of the adjacent Phe7 8 is optimal when they aretrans to each other, and their spatial arrangement lowers the probability for folding of the amino and carboxyl tails toward each other. Pro2 and Pro4 create a kink in the head structure and the electrostatic interaction between positive and negative residues helps to stabilize the kink.
Steric and overhead views of SP and SP binding.
(A) Steric view of SP. Yellow dashed lines represent hydrogen bonds. (B) Overhead view of SP binding to NK-1 represented as solvent accessible surfaces. Colors represent electrostatic potential, with red indicating positive and blue, negative. (C) Overhead view of SP binding to NK-1 with the binding domains shown for SP(1-4).
Steric and overhead views of SP and SP binding.
(A) Steric view of SP. Yellow dashed lines represent hydrogen bonds. (B) Overhead view of SP binding to NK-1 represented as solvent accessible surfaces. Colors represent electrostatic potential, with red indicating positive and blue, negative. (C) Overhead view of SP binding to NK-1 with the binding domains shown for SP(1-4).
After the 3-dimensional structures of SP and NK-1 were generated, they were docked by liphophilic, electrostatic, and H-bond interactions. The 3 types of interactions were performed to ensure that the docking was proper. Docking was done with and without the solvent-accessible surfaces, which were generated in SYBYL. MOLCAD (Tripos Associates) was used to map the lipophilic, electrostatic, and H-bond potentials on the surfaces. DOCK and Flexidock were used to prevent collision while docking the solvent accessible surfaces of SP and NK-1. The results (Figure 8B-C) are consistent with a previous report,33which indicated that the contact sites between SP and NK-1 are mainly on the extracellular loops and a few residues in the extracellular matrix–TM interface. The results are also consistent with the interacting sites retrieved from the peptide library, which indicated that exons 1 and 5 of NK-1 are important for contact with SP(1-4) (Figures 6, 8C). Exon 1 encodes TM 1-3, shown as α-helices 1-3, and exon 7 encodes TM 7, shown as α-helix 7 (Figure 8C).34Indeed, the model, (Figure 8B-C) indicates that the regions of SP(1-4) bind in the pocket within the domain formed by exons 1 and 5. The results show that SP and SP(1-4) can bind into the same pocket within NK-1 and that they could compete for the same interacting site so that eventually SP cannot dock, which could lead to down-regulation of NK-1.
Discussion
A diagram representing the findings of this study is shown in Figure 9. We showed the potential for an additional mechanism in which the PPT-I gene could be involved in modulating hematopoiesis. In the BM, SP(1-4) could be derived through enzymatic digestion by endogenous endopeptidases.6,7 Since SP(4-11) exerts stimulatory effects similar to the parent peptide,8 the effects of the amino terminal fragment raises the question of whether SP(1-4) is effective as a hematopoietic feedback while the carboxyl fragment might be in the vicinity of the receptor. Although this question has to be addressed in more detailed study, the experimental evidence suggests that SP(5-11) might be quickly degraded to a nonfunctional form by other endopeptidases that can use the longer carboxyl fragment as a substrate.15 16 Unpublished and ongoing studies using Ciphergen's ProteinChip technique showed no evidence of SP(5-11). This might be caused by rapid degradation of the carboxyl fragment by other endopeptidases, which would result in SP(1-4) alone.
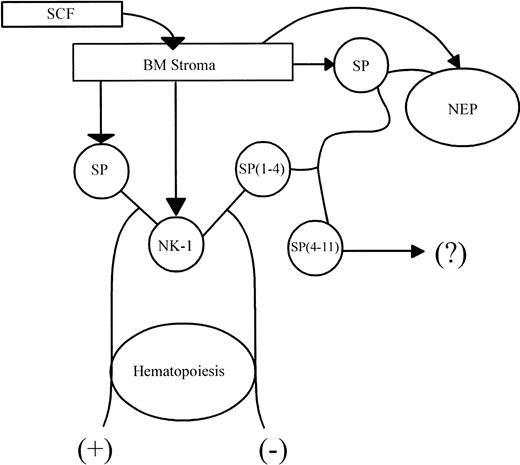
Model of hematopoietic feedback by SP(1-4).
SCF mediated the release of SP from BM stroma with concomitant expression of NK-1. SP–NK-1 interaction leads to hematopoietic stimulation (+). NEP, which is also induced in the SCF-stimulated stroma, uses SP as its substrate to produce SP(1-4). Interaction between SP(1-4) and NK-1 negatively regulates the stimulatory effect of SP on hematopoiesis (−). The fate of SP(4-11) is yet to be determined (?).
Model of hematopoietic feedback by SP(1-4).
SCF mediated the release of SP from BM stroma with concomitant expression of NK-1. SP–NK-1 interaction leads to hematopoietic stimulation (+). NEP, which is also induced in the SCF-stimulated stroma, uses SP as its substrate to produce SP(1-4). Interaction between SP(1-4) and NK-1 negatively regulates the stimulatory effect of SP on hematopoiesis (−). The fate of SP(4-11) is yet to be determined (?).
We reported that the negative hematopoietic effects of SP(1-4) were mediated through NK-1.8 This was a paradox since NK-1 receptor mediates positive hematopoietic responses by interacting with SP.2 Although it was expected that a negative hematopoietic response would be mediated through NK-2 receptor,5 screening of the peptide library did not show a potential interacting site for SP(1-4) within NK-2 (Figure 6). The lack of evidence for NK-2 as a mediator in the effect of SP(1-4) was puzzling since SP(1-4) did not induce the other NK subtype (NK-1) in stromal cells (Table 2). An understanding of the role of NK-1 in the SP(1-4) effect was provided by the appearance of SP(1-4) in SCF-stimulated cells (Figure 5) while NK-1 expression was still up-regulated (Figure 2, Table 2). Analyses of these results suggest that the induction of NK-1 by SCF was blunted after the change of SP to SP(1-4) and that, with time, membrane expression of NK-1 was reduced (Figure 2). The significance of this reduction is the provision of a feedback mechanism on the effects of SCF. Further studies that are beyond the scope of this report are needed to determine if this feedback mechanism is common to other stimulatory hematopoietic growth factors. This question would be pertinent for further understanding of the data shown in this report since several cytokines, associated with inflammatory responses and hematopoietic stimulation, can induce the production of SP.2
Figure 5C summarizes the dynamics between SP and SP(1-4). SP is first detected in stromal cells stimulated with SCF. This is followed by the appearance of NEP and then the degradation of SP to SP(1-4). Further elucidation of these findings and quantitation of SP, SP fragments, and various endopeptidases are the subject of ongoing studies in the laboratory. This model does not explain the signaling pathway mediated by SP(1-4) to induce TGF-β and TNF-α (Table 1). Explanation of such a mechanism could only be speculative. Since NK-1 is a G-protein–coupled receptor that uses the β-arrestins as scaffolds for signaling molecules,35 a possible mechanism that is related to the β-arrestins is being studied. There were significant differences in the levels of TGF-β and TNF-α in stroma stimulated with SP(1-4) during the first and second stimulation (Table 1). Since the cytokine quantitations for each stimulation set were performed separately, the differences suggest that the optimal times for the induction of TNF-α and TGF-β were 16 hours, which was followed by reduced gene expression. The increased levels of TGF-β and TNF-α in stroma stimulated with SP(1-4) could not be due to the initial stimulation with SCF since the levels were similar in stroma stimulated with only SP(1-4) (Table 1).
The 3-dimensional structure of NK-1 and SP allows assumptions regarding interactions between NK-1 and SP and provides a possible explanation for the function of SP(1-4) in hematopoietic feedback. Both SP and its fragment, SP(1-4), can occupy the same pocket within NK-1 (Figure 8B-C). A common pocket for both SP and SP(1-4) indicated that in the presence of SP(1-4), the fit for SP within NK-1 would be hindered. In the absence of SP–NK-1 interactions, the production of stimulatory hematopoietic regulators such as cytokines could be down-regulated.2 Since these cytokines induce the expression of NK-1, the loss of SP–NK-1 could result in down-regulation of NK-1 and a switch to alternative functions by SP(1-4)–NK-1 (Figure 8C). A possibility for this is shown by the production of TGF-β and TNF-α by SP(1-4) in BM stroma (Table 1).
Previous studies5 as well as this report show that a feedback effect on hematopoiesis by the PPT-I gene is not a one-way process, but reversible. The induction of TGF-β and TNF-α, hematopoietic inhibitors28,30 could be blunted by SCF (Table 1), which would promote a switch from negative to positive hematopoiesis. Also, NK-A, a PPT-I gene–derived peptide that exerts hematopoietic suppression,5 could be a substrate of endogenous endopeptidases.36 Degradation of NK-A could make the BM more amenable to hematopoietic stimulation. Another example is SP's ability to protect itself from degradation. This protection could occur by SP's being able to down-regulate the expression of particular endopeptidases, such as CD13, and its complexing to a fibronectin, which is a relatively large molecule.29,37 CD13 is the second enzyme following neutral endopeptidase that could degrade the carboxyl fragment of SP.38 Reduced expression of CD13 would leave SP intact for hematopoietic stimulation. Together, these results show that the positive and negative effects of peptides derived from thePPT-I gene are complex and include other genes that might be regulated by the tachykinin family of peptides.39
When the conventions of head (residues 1-4) and tail (residues 5-11) are used for SP, certain assumptions can be made about the structure of SP. The model, shown in Figure 8A, indicates no helical structure of SP but a curve in the head region. However, Cowsik et al40reported a structure of SP that is partly helical in the tail region when bound to lipids. Although we used a different 3-dimensional structure of SP, the structure of Cowsik et al40 could also dock to the NK-1 model, shown in Figures 8B and 8C. Using molecular dynamics, the model shown in this study is the lowest in energy (−245.7 kJ), although it is possible that the bioactive peptide could be in a higher energy confirmation.
SP binds to NK-1 within an interface between the cell membrane and the extracellular matrix.33,41 The confirmation of SP–NK-1, shown in the 3-dimensional model (Figure 8B-C) allows easy access for endopeptidases to cleave SP. The model accounts for a structural difference between SP(1-4) and SP(5-11) and also for electrostatic (Figure 8B) and lipophilic (not shown) complementarities between SP and NK-1. We hypothesize that the message sequence of SP, located within the carboxyl terminal,42 cannot activate NK-1 if the address sequence, which includes SP(1-4), occupies the pocket (Figure 8C). Without proper configuration of the message, the peptide would not be able to activate the G-protein through a classical mechanism. However the SP(1-4) address alone mediates signaling through the NK-1 receptor. This is demonstrated by the biological functions mediated by SP(1-4) (Table 1, Figure 1).
The negative effect of SP(1-4) on hematopoiesis is not unusual for a small peptide since similar effects were reported for other small peptides.43,44 The biology of PPT-I peptides and their fragments is applicable not only to the BM but also to other lymphoid organs2 and would unravel the finely tuned interplay between NK-A, endopeptidases, SP, and other hematopoietic/inflammatory factors. The inhibitory hematopoietic effects of SP(1-4) suggest that this peptide can influence the self-renewal capabilities of the hematopoietic stem cells. Further research in this area is necessary since such information would be important for application in malignancies that are phenotypically late-stage progenitors with the adapted self-renewal capability, such as leukemia and lymphoma.4
Supported by National Institutes of Health grants HL-54973, HL-57675, and CA89868.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Pranela Rameshwar, PhD, UMDNJ–New Jersey Medical School, 185 South Orange Ave, MSB, Rm E-579, Newark, NJ 07103; e-mail:rameshwa@umdnj.edu.