Abstract
The transcription factor Stat5 mediates the cellular response to activation of multiple cytokine receptors involved in the regulation of proliferation and differentiation of hematopoietic cells. Recently, the human Stat5 gene was found to be translocated to the RARα gene in a patient with acute promyelocytic leukemia indicating that Stat5 might also play a role in cellular transformation. We investigated the mechanism by which Stat5 might exert this function and studied the biochemical and cellular functions of fusion proteins comprising Stat5 and RARα. The expression of Stat5-RARα causes the transcriptional repression of gene transcription, a process that requires the coiled-coil domain of Stat5 (amino acid positions 133-333). Oligomerization of this domain in the Stat5-RARα fusion protein leads to stable binding of the corepressor SMRT independent of all-trans retinoic acid (ATRA) stimulation and is accompanied by an impaired response to differentiation signals in hematopoietic cells. This inhibitory effect on myeloid differentiation cannot be overcome by simultaneous coexpression of RARα. We conclude that Stat5 is capable of interacting with a corepressor complex that alters the pattern of corepressor binding to RARα and its dissociation in response to ATRA stimulation, leading to enhanced repressor activity and a block of hematopoietic differentiation.
Introduction
Stat proteins are signaling components and transcription factors mediating the cellular response to a large number of extracellular growth factors and hormones. On tyrosine phosphorylation through a receptor-associated kinase, they form dimers by intermolecular interactions involving a phosphotyrosine residue and the SH2-domain of the dimerization partner. On nuclear translocation the dimers bind to a DNA motif, TTCNNNGAA, in the promoter region of target genes.1 Transcriptional induction is mediated by a transactivation domain at the C-terminus and a coiled-coil domain at the N-terminus. These domains cooperate in recruiting CBP/p300, a transcriptional activator with histone acetyl transferase activity.2 3
Acetylation of histones is closely linked to transcriptional transactivation in eukaryotic cells. Histone acetyl transferases, such as p300/CBP and P/CAF, acetylate conserved lysine residues in histones, a process that results in chromatin remodeling and supposedly enhanced access of transcription complex components to promoter regions. In contrast, transcription complexes containing corepressors, such as N-CoR and SMRT, can recruit histone deacetylases (HDACs) and cause transcriptional repression.4-6
Altered regulation of histone-modifying enzymes and resulting chromatin modifications have been found to lead to inappropriate repression of genes required for cell differentiation. This has been interpreted as a mechanism in the pathogenesis of various forms of acute myeloid leukemia (AML). Several translocation products have been found in AML that recruit components of the corepressor complex to key transcription factors required for myeloid differentiation.6-8 The molecular mechanisms leading to repression of retinoic acid receptor (RAR)–dependent gene transcription and blocked differentiation have been investigated in detail for the promyelocytic leukemia (PML)–RARα and promyelocytic zinc finger (PLZF)– RARα translocation products in acute promyelocytic leukemia (APL).9-12 In the presence of high doses of all-trans retinoic acid (ATRA) the corepressor complex dissociates only from PML-RARα, but not from PLZF-RARα. This explains why patients with PML-RARα respond with complete remissions to high-dose ATRA treatment, whereas those with PLZF-RARα respond poorly to this therapy.13
Recently, another chromosomal translocation involving the RARα gene has been described. A Stat5-RARα fusion product has been detected in an ATRA-resistant form of APL.14 We have investigated the role of the Stat5 moiety in this fusion protein and find that Stat5 interacts with a corepressor complex containing HDAC activity. In the context of the Stat5-RARα fusion protein, the coiled-coil domain of Stat5 is required for recruitment of the corepressor SMRT. Furthermore, Stat5-RARα blocks differentiation of hematopoietic cells in culture, which is not released by treatment with ATRA.
Materials and methods
Cells, cell culture, and antibodies
293T cells were maintained in Dulbecco modified Eagle medium (Life Technologies, Karlsruhe, Germany) supplemented with 10% fetal calf serum (Biochrom, Berlin, Germany), 50 U/mL penicillin, 50 μg/mL streptomycin, and 2 mM l-glutamine (all from Life Technologies). HL60, HL60R, and U937 cells were cultured in RPMI (Life Technologies) supplemented with 10% fetal calf serum (Biochrom), 50 U/mL penicillin, 50 μg/mL streptomycin, and 2 mMl-glutamine (all from Life Technologies) and differentiated as described previously.9 Briefly, in retrovirally transduced U937 cells, coexpression of enhanced green fluorescent protein (EGFP) and CD14 was detected by fluorescence-activated cell sorting (FACS) 48 hours after induction with vitamin D3 (10−6 M) and transforming growth factor-β (TGF-β; 5 nM). Transduced HL60R or HL60 cells were differentiated with ATRA (10−6 M) with or without trichostatin A (TSA, 120 nM; Upstate Technology, Lake Placid, NY) for 5 days before measuring coexpression of EGFP and CD11b. For flow cytometry, phycoerythrin (PE)–conjugated antihuman CD11b or antihuman CD14 mouse monoclonal IgG1 or mouse PE-IgG1 isotype control antibodies (DPC Biermann, Bad Nauheim, Germany) were used to determine CD11b or CD14 immunofluorescence intensity of HL60, HL60R, or U937 cells before and after differentiation.
Stat5-RARα gene fusions
The expression plasmids were generated by site-directed mutagenesis (Stratagene, La Jolla, CA) at the following positions (amino acid residues indicated): a SmaI restriction site was created at residue 637 in pXM-Stat5a2 and at residue 59 of pC-RARα9 to fuse an EcoRI-SmaI fragment of Stat5a in frame to RARα. An additional SalI site was introduced immediately in front of the coding region of Stat5a and the complementary DNA (cDNA) of the Stat5(1-636)–RARα fusion was then cloned in frame with an amino terminal hemagglutinin (HA) tag into the retroviral vector PINCO,15 or in frame with the Gal4 DNA-binding domain (residues 1-147) of pCMX-Gal4.4 A deletion construct Stat5(1-333)–RARα was generated by introducing aXhoI restriction site at position 334 of Stat5a to fuse anEcoRI-XhoI/blunt-end fragment in frame to RARα. The construct Stat5(1-174)–RARα was cloned by fusing anEcoRI-BamHI/blunt-end fragment of Stat5a in frame to pC-RARα. All constructs were verified by DNA sequencing. Retroviral expression constructs for PML-RARα and PLZF-RARα (PINCO PML–RARα and PINCO PLZF-RARα) have been described previously.16
Cell transfections and transcriptional reporter assays
293T cells were transfected in triplicate with 0.75 μg of the indicated pCMV-Gal4 fusion plasmids, 1.5 μg 2 times UAS-TK-luciferase plasmid, and 1 μg of a promoterless renilla luciferase plasmid by calcium phosphate coprecipitation as described previously.17 Cells were stimulated with ATRA (10−7 M and 10−6 M) for 30 hours before harvesting and luciferase activity was measured using the Dual-Luciferase Reporter Assay system (Promega, Madison, WI) following the protocols provided by the manufacturer. Repression is given relative to the luciferase activity obtained by the DNA-binding domain of Gal4 alone. Experiments were repeated at least 5 times, and results are indicated as the means with SD. Repression and induction are given relative to the luciferase activity obtained by the DNA-binding domain of Gal4 alone. For promoter assays using the β-casein luciferase promoter, 293T cells were transfected in triplicates with the 0.6 μg reporter plasmid together with the expression plasmids for mStat5a, pXM-Stat5a (0.6 μg), prolactin receptor (0.6 μg), pcDNA3-prolactin receptor (0.3 μg), and 1 μg of a promoterless renilla luciferase plasmid as described previously2 and stimulated with 5 μg/mL bovine prolactin or 200 nM TSA (or both) for 30 hours before harvesting. Luciferase activity was measured using the Luciferase Reporter Assay system (Promega).
Protein interaction assays
pCMX-SMRT18 was translated in vitro in the presence of [35S]-methionine using the TNT T7 coupled reticulocyte lysate system (Promega). For coprecipitation experiments using whole cell extracts, 293T cells (5 × 106 cells seeded in 10-mm diameter dishes 24 hours before transfection) were transfected with 20 μg of the diverse Gal4 fusion constructs, or pXM-HA-Stat5a. Forty-eight hours after transfection cells were lysed in NETN buffer (20 mM Tris, pH 8, 100 mM NaCl, 1 mM EDTA, 1 mM DTT, 10% glycerol, 0.5% NP-40) supplemented with 0.5 mM phenylmethylsulfonyl fluoride (Sigma-Aldrich, Taufkirchen, Germany) and a protease inhibitor cocktail (Roche Diagnostics, Mannheim, Germany) as described previously.17 After centrifugation for 5 minutes at 4°C, the supernatants were collected and incubated with radiolabeled SMRT either in the presence or absence of 10−6 M ATRA for 1 hour at 30°C. Then, after addition of the biotinylated 2 times UAS oligo (sense strand: 5′-biotin-GGA TCC TCG GAG GAC TGT CCT CCG CGG ATC CTC GGA GGA ACA GTC CTC CGA GTC GAC) for 30 minutes at 4°C, the DNA-protein complex was captured on streptavidin agarose (Boehringer, Mannheim, Germany) and washed 4 times with NETN buffer. For coimmunoprecipitation experiments, the supernatants containing HA-Stat5a were collected and incubated with radiolabeled SMRT for 1 hour at 30°C. Then, after addition of anti-HA antibody (Boehringer), or anti-FLAG antibody (Sigma-Aldrich) together with protein A/G Sepharose for 60 minutes at 4°C, the immune complexes were washed 4 times with NETN buffer. The precipitated proteins were eluted from complexes by boiling for 5 minutes in Laemmli buffer (ROTH, Karlsruhe, Germany), and 80% of the precipitate was separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and subjected to autoradiography to detect SMRT binding. The remaining 20% was also separated by SDS-PAGE and then transferred to a polyvinylidene difluoride (PVDF) membrane (ROTH). Western blots were blocked for 2 hours with 5% milk and incubated with anti-Gal4 (DNA-binding domain [DBD]) primary antibody (RK5C1; Santa Cruz Biotechnology, Santa Cruz, CA) or anti-HA antibody at 4°C overnight. After extensive washing the blots were incubated with a peroxidase conjugated secondary antibody for 30 minutes. After further washing the proteins were visualized by enhanced chemiluminescence (Pierce, Rockford, IL).
Glutathinone S-transferase (GST) pull-down assays were performed as described elsewhere.17 GST and GST–Stat5(1-331) fusion proteins were expressed in Escherichia coli BL21 codon-positive cells (Stratagene), and equal amounts of each were immobilized on glutathione-Sepharose beads (Sigma-Aldrich). In vitro translated, radiolabeled Gal4-Stat5-RARα and CMX-SMART constructs (TNT T7 coupled reticulocyte lysate system, Promega) were incubated with equal amounts of GST fusion proteins in 100 μL PPI buffer (50 mM Hepes, pH 7.8, 50 mM NaCl, 5 mM EDTA, 1 mM DTT, 0.02% NP-40 containing a protease inhibitor cocktail (Roche Diagnostics) and 0.5 mM phenylmethylsulfonyl flouride (Sigma-Aldrich) for 20 minutes at 25° C. The beads were washed 4 times, and bound proteins eluted by boiling in Laemmli buffer (ROTH). The precipitated proteins were subjected to SDS-PAGE and visualized by autoradiography.
Results
Construction of Stat5-RARα fusion proteins
To investigate whether a chimeric Stat5-RARα protein, found in a patient with ATRA-resistant APL, could contribute to the development of APL, we generated fusion proteins of Stat5a and RARα resembling the translocation product described recently.14 One fusion protein consists of the first 636 amino acids of Stat5 including the N-terminal hooklike domain, a coiled-coil region (amino acids 133-333), and the DBD fused in frame to almost the entire RARα coding sequence. Additionally, we generated deletion mutants lacking either the DBD of Stat5 (Stat5[1-333]–RARα) or the coiled-coil region and the DBD (Stat5[1-174]–RARα) (Figure 1). These constructs as well as RARα were fused in frame with the Gal4 DNA binding domain (residues 1-147) of pCMX-Gal44 for transcriptional repressor assays or cloned into the retroviral vector PINCO15 for transduction of cell lines.
Schematic diagram of Stat5-RARα mutants used.
Structure of full- length Stat5-RARα (Stat5[1-636]–RARα), 2 deletion mutants, and RARα alone are shown. C-C indicates coiled-coil region; DBD, DNA-binding domain; SH2, src-homology region 2; TA, transactivation domain; ZF, zinc finger domain; HBD, hormone-binding domain.
Schematic diagram of Stat5-RARα mutants used.
Structure of full- length Stat5-RARα (Stat5[1-636]–RARα), 2 deletion mutants, and RARα alone are shown. C-C indicates coiled-coil region; DBD, DNA-binding domain; SH2, src-homology region 2; TA, transactivation domain; ZF, zinc finger domain; HBD, hormone-binding domain.
Impaired differentiation of cells expressing Stat5-RARα fusion proteins can be restored by TSA
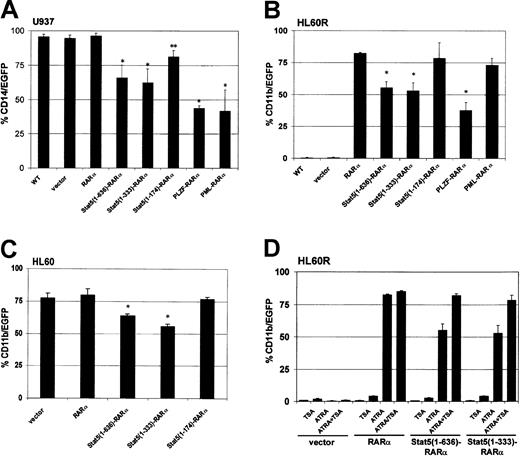
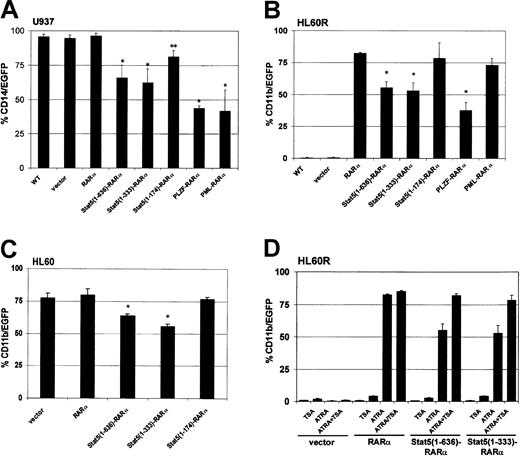
To test the potential of Stat5-RARα fusion proteins to influence myeloid differentiation in cell culture models, hematopoietic cell lines were transduced with retroviral vectors encoding various RARα fusion proteins and green fluorescent protein (GFP). Differentiation was induced in U937 cells with vitamin D3 and TGF-β and the expression of the monocytic surface antigen CD14 was monitored. CD14 expression could be detected in 95% of U937 cells within 48 hours (Figure 2A). CD14 expression was reduced to 60% of the U937 cells expressing the full-length Stat5-RARα fusion protein or Stat5(1-333)–RARα, and to less than 50% of cells expressing PML-RARα or PLZF-RARα. A Stat5(1-174)–RARα fusion protein, which does not contain the coiled-coil region, or the RARα protein alone had little effect on differentiation.
Stat5-RARα fusion proteins containing the coiled-coil domain block hematopoietic differentiation but respond to TSA treatment.
U937 (A), HL60R (B), and HL60 cells (C) were transduced with retroviral vectors coexpressing the indicated Stat5-RARα fusion proteins and EGFP. In U937 cells, coexpression of EGFP and CD14 was detected by FACS 48 hours after induction with vitamin D3 (10−6 M) and TGF-β (5 nM). HL60R and HL60 cells were differentiated with ATRA (10−6 M) for 5 days before measuring coexpression of EGFP and CD11b. (*P < .008, **P < .03). (D) HL60R cells transduced with the indicated vectors were treated with ATRA (10−6 M) for 5 days with or without TSA (120 nM) and then assayed for coexpression of EGFP and CD11b.
Stat5-RARα fusion proteins containing the coiled-coil domain block hematopoietic differentiation but respond to TSA treatment.
U937 (A), HL60R (B), and HL60 cells (C) were transduced with retroviral vectors coexpressing the indicated Stat5-RARα fusion proteins and EGFP. In U937 cells, coexpression of EGFP and CD14 was detected by FACS 48 hours after induction with vitamin D3 (10−6 M) and TGF-β (5 nM). HL60R and HL60 cells were differentiated with ATRA (10−6 M) for 5 days before measuring coexpression of EGFP and CD11b. (*P < .008, **P < .03). (D) HL60R cells transduced with the indicated vectors were treated with ATRA (10−6 M) for 5 days with or without TSA (120 nM) and then assayed for coexpression of EGFP and CD11b.
We also examined whether the Stat5-RARα constructs would interfere with ATRA-induced differentiation of the human promyelocytic cell line, HL60R.9 These cells cannot be induced with ATRA to mature along the granulocytic lineage due to an inactivating mutation in the RARα (Figure 2B). However, transduction of HL60R cells with PINCO-RARα and PINCO–Stat5(1-174)–RARα restored differentiation and induced expression of the surface antigen CD11b in about 80% of the cells after stimulation with ATRA. Expression of the Stat5(1-636)–RARα and Stat5(1-333)–RARα reduced ATRA-induced CD11b expression to only about 55% of the cells.
The effect of the Stat5-RARα on myeloid differentiation was compared to that of PML-RARα and PLZF-RARα. HL60R cells transduced with PML-RARα or PLZF-RARα were treated with ATRA at a concentration of 10−6 M. As expected, PML-RARα–transduced cells showed CD11b expression comparable to that of HL60R cells expressing RARα. In contrast, PLZF-RARα transferred a partial resistance to the effect of ATRA (Figure 2B) similar to Stat5-RARα.
To test whether Stat5-RARα would be able to block ATRA-induced differentiation in the presence of functional RARα, we transduced HL60 cells, which express functional RARα, with PINCO-RARα and PINCO Stat5-RARα constructs (Figure 2C). Although CD11b expression after ATRA stimulation was high in cells transduced with the empty vector (PINCO: 76.2%) or the Δcoiled-coil form of Stat5-RARα (PINCO–Stat5[1-174]–RARα: 78.4%), it was significantly reduced in cells transduced with Stat5(1-636)–RARα or Stat5(1-333)–RARα constructs (62.1% and 55.7%, respectively). These experiments confirm a dominant effect of Stat5-RARα over coexpressed RARα in respect to retinoic acid–induced differentiation. Furthermore, our data indicate that under the experimental conditions used here, Stat5-RARα fusion proteins interfere with myeloid differentiation, by blocking ATRA-dependent and nondependent differentiation pathways in a similar manner as already described for PLZF-RARα.
Addition of TSA to HL60R cells treated with ATRA increased CD11b expression in cells transduced with PINCO–Stat5(1-636)–RARα (55.3% without TSA versus 82% with TSA) and PINCO–Stat5(1-333)–RARα (53.1% without TSA versus 78.2% with TSA) to normal levels but had almost no influence on CD11b expression in HL60R cells transduced with the empty vector (PINCO, 1.1% without TSA versus 3.2% with TSA) or RARα alone (PINCO-RARα, 82.5% without TSA versus 85% with TSA; Figure 2D). These data suggested that Stat5-RARα impaired differentiation by aberrant recruitment of a corepressor complex containing HDAC activity.
The coiled-coil domain of Stat5 enhances transcriptional repression in Stat5-RARα fusion proteins
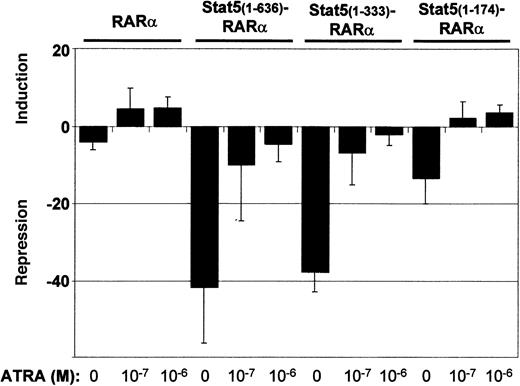
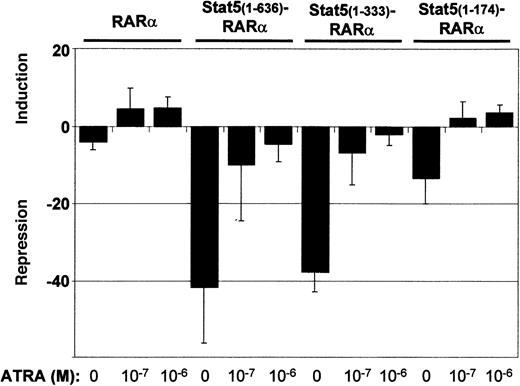
To further analyze Stat5-RARα function, we assessed transcriptional repressor activity in cotransfection experiments of Gal4-Stat5-RARα fusion proteins and a 2 times UAS-TK-luciferase reporter gene (Figure 3). A fusion protein of Gal4 and RARα (Gal4-RARα) caused a 5-fold reduction in luciferase activity when compared to a plasmid expressing only the Gal4 DBD. Stimulation of the cells with ATRA resulted in a 5-fold induction over basal activity. Gal4–Stat5(1-636)–RARα and Stat5(1-333)–RARα exhibited a much stronger repressor activity (40-fold). This repression could only be partially alleviated by ATRA. A Gal4-fusion with Stat5(1-174)–RARα showed lower repressor activity (12-fold) without ATRA and positive induction over basal levels (3-fold) after addition of ATRA to the cells. These experiments show that repressor activities of Stat5-RARα constructs are 8-fold higher compared to those exerted by constructs comprising RARα only and cannot be switched from transcriptional repression to transcriptional activation by ATRA stimulation.
Stat5-RARα fusion proteins enhance transcriptional repression.
293T cells were transfected in triplicate with 0.75 μg of the indicated pCMV-Gal4 fusion plasmids, 1.5 μg of 2 times UAS-TK-luciferase plasmid, and 1 μg of a promoterless renilla luciferase plasmid by calcium phosphate coprecipitation. Cells were stimulated with ATRA (10−7 M and 10−6 M) and repression or induction is given relative to the luciferase activity obtained by the DNA-binding domain of Gal4 alone. Transcriptional repression is given as the mean and SD of 5 experiments.
Stat5-RARα fusion proteins enhance transcriptional repression.
293T cells were transfected in triplicate with 0.75 μg of the indicated pCMV-Gal4 fusion plasmids, 1.5 μg of 2 times UAS-TK-luciferase plasmid, and 1 μg of a promoterless renilla luciferase plasmid by calcium phosphate coprecipitation. Cells were stimulated with ATRA (10−7 M and 10−6 M) and repression or induction is given relative to the luciferase activity obtained by the DNA-binding domain of Gal4 alone. Transcriptional repression is given as the mean and SD of 5 experiments.
Binding of Stat5-RARα to SMRT is not sensitive to ATRA treatment
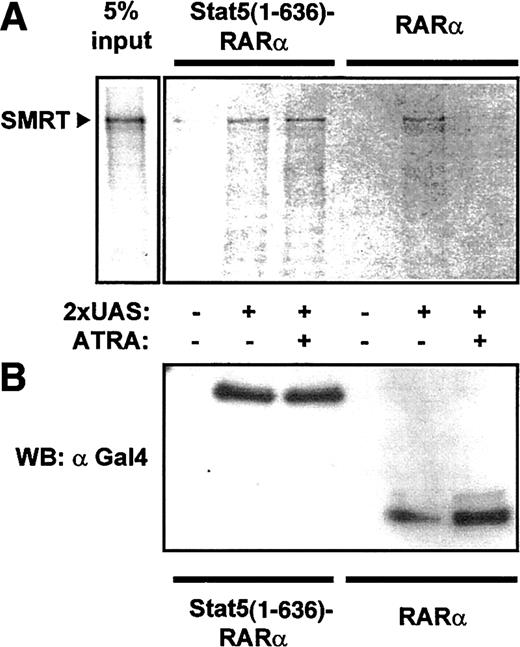
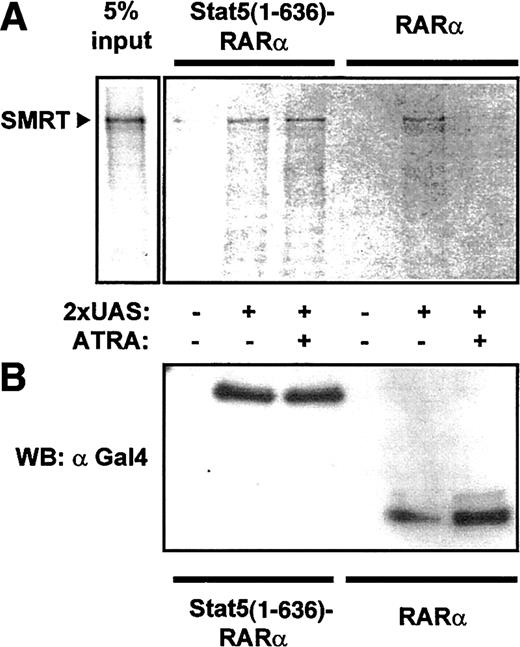
To confirm interaction of Stat5-RARα with a corepressor complex, we performed coprecipitation experiments with cellular lysates from 293T cells expressing Gal4–Stat5(1-636)–RARα and Gal4-RARα. Association of SMRT with Stat5-RARα and RARα was detected after precipitation with an oligonucleotide containing the Gal4 DNA binding sequence (UAS) in the absence of ATRA (Figure4). Because this association was lost in the presence of ATRA for Gal4-RARα but was not altered in cells expressing Gal4–Stat5(1-636)–RARα, we suggest that the Stat5 moiety in the fusion protein prevents dissociation of the corepressor complex even in the presence of hormone.
Stat5-RARα fusion proteins induce ATRA-insensitive interaction with SMRT.
(A) Cellular extracts from 293T cells expressing Gal4 fusions of Stat5(1-636)–RARα and RARα were incubated with radiolabeled SMRT and precipitated with a 2 × UAS oligo. SMRT could be detected in precipitates with Stat5(1-636)–RARα in the absence or presence of ATRA (10−5 M). In contrast, no SMRT binding was seen in the presence of ATRA with extracts containing RARα. (B) Western blot performed with 30% of precipitated proteins using an anti-Gal4 antibody showed equal binding of both Gal4-fusion proteins to the oligo.
Stat5-RARα fusion proteins induce ATRA-insensitive interaction with SMRT.
(A) Cellular extracts from 293T cells expressing Gal4 fusions of Stat5(1-636)–RARα and RARα were incubated with radiolabeled SMRT and precipitated with a 2 × UAS oligo. SMRT could be detected in precipitates with Stat5(1-636)–RARα in the absence or presence of ATRA (10−5 M). In contrast, no SMRT binding was seen in the presence of ATRA with extracts containing RARα. (B) Western blot performed with 30% of precipitated proteins using an anti-Gal4 antibody showed equal binding of both Gal4-fusion proteins to the oligo.
Stat5-dependent induction of the β-casein promoter is strongly enhanced in the presence of TSA
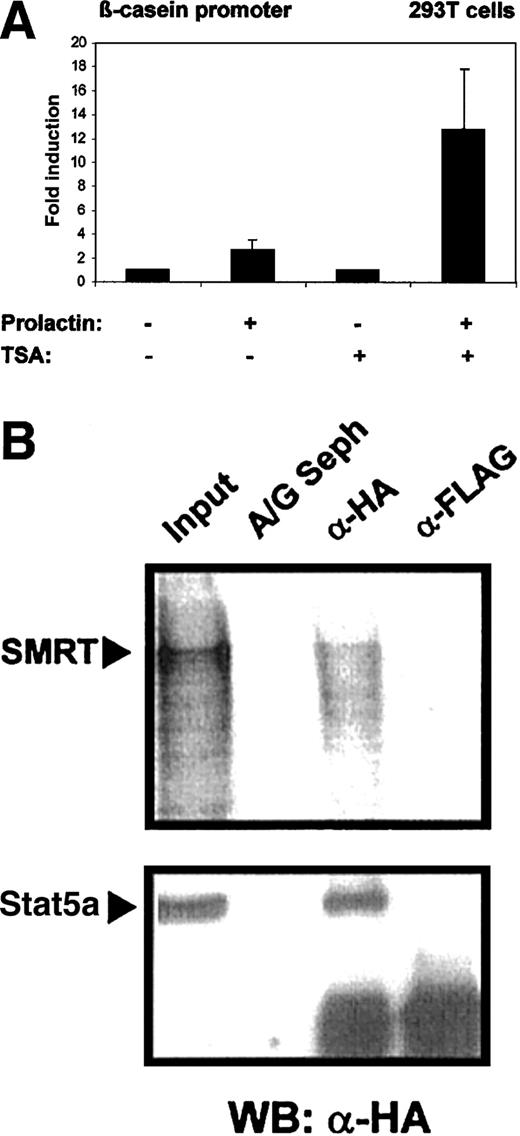
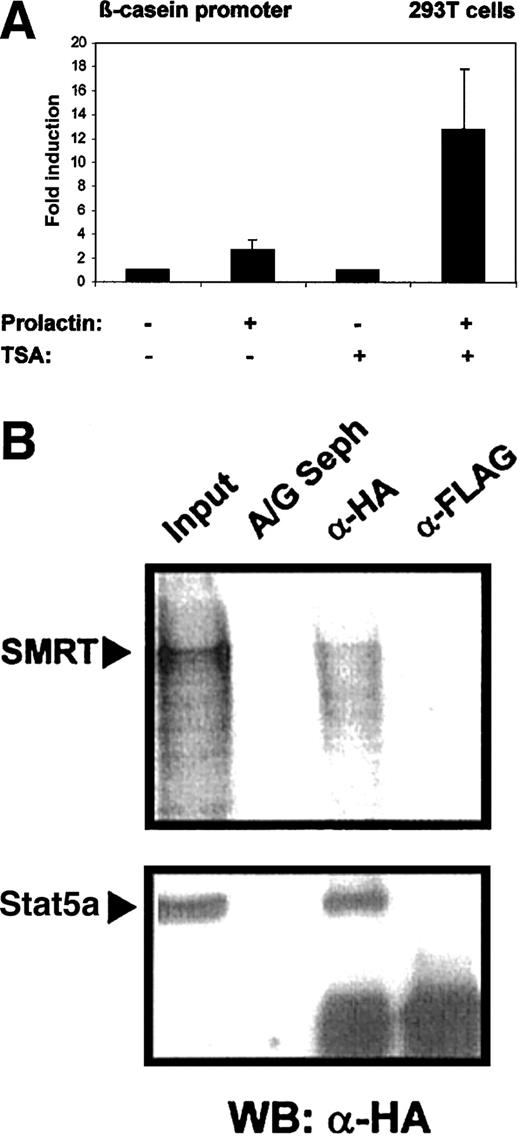
Stat5 was initially discovered as a mediator of prolactin-dependent gene induction. It has been found that histone acetyl transferases are recruited into Stat5-initiated transcription complexes and are crucial for the induction process.2 It is conceivable that HDACs might counteract Stat5-induced transcription. To assess the influence of HDACs on Stat5-dependent gene expression, we measured transcriptional induction by Stat5 in the absence and presence of the HDAC inhibitor TSA. 293T cells were transfected with a β-casein gene promoter-luciferase reporter construct and plasmids encoding Stat5a and the prolactin receptor. Cells were induced by prolactin treatment (Figure 5A). Prolactin stimulation caused a modest induction β-casein promoter (3.5-fold) in the absence of TSA. Promoter activity was increased to 18-fold in the presence of TSA. This indicates that Stat5-dependent transcription is limited in its extent by histone deacetylase activity.
Stat5-dependent β-casein promoter activity is enhanced in the presence of TSA.
(A) 293T cells were transfected in triplicates with 0.6 μg reporter plasmid (β-casein promoter luciferase) together with the expression plasmids for mStat5a, (pXM-Stat5a; 0.6 μg), and prolactin receptor (pcDNA3-prolactin receptor; 0.3 μg) using a calcium phosphate method. Cells were stimulated with or without 5 μg/mL prolactin and 200 nM TSA and equal amounts of the cell lysates were assayed for luciferase activity. Luciferase activity assayed in cells treated with prolactin was given as fold induction over values obtained without prolactin, which were set as 1. Transcriptional induction is given as the mean and SD of 5 experiments. The corepressor SMRT binds to wild-type Stat5a. (B) Cellular extracts from 293T cells transfected with 5 μg HA-tagged Stat5a expression plasmid (pXM-HA-Stat5a) were incubated with radiolabeled SMRT. Subsequent immunoprecipitations were performed with anti-HA monoclonal antibody (α-HA), anti-FLAG antibody (α-Flag), or protein A/G Sepharose alone (A/G Seph). Radiolabeled SMRT protein was detected on a dried gel in extracts precipitated with α-HA antibody, whereas Stat5a was detected by Western blot using anti-HA antibody and both were compared to 10% input.
Stat5-dependent β-casein promoter activity is enhanced in the presence of TSA.
(A) 293T cells were transfected in triplicates with 0.6 μg reporter plasmid (β-casein promoter luciferase) together with the expression plasmids for mStat5a, (pXM-Stat5a; 0.6 μg), and prolactin receptor (pcDNA3-prolactin receptor; 0.3 μg) using a calcium phosphate method. Cells were stimulated with or without 5 μg/mL prolactin and 200 nM TSA and equal amounts of the cell lysates were assayed for luciferase activity. Luciferase activity assayed in cells treated with prolactin was given as fold induction over values obtained without prolactin, which were set as 1. Transcriptional induction is given as the mean and SD of 5 experiments. The corepressor SMRT binds to wild-type Stat5a. (B) Cellular extracts from 293T cells transfected with 5 μg HA-tagged Stat5a expression plasmid (pXM-HA-Stat5a) were incubated with radiolabeled SMRT. Subsequent immunoprecipitations were performed with anti-HA monoclonal antibody (α-HA), anti-FLAG antibody (α-Flag), or protein A/G Sepharose alone (A/G Seph). Radiolabeled SMRT protein was detected on a dried gel in extracts precipitated with α-HA antibody, whereas Stat5a was detected by Western blot using anti-HA antibody and both were compared to 10% input.
Stat5 interacts with a corepressor SMRT
The effect of TSA on Stat5-dependent gene induction might be explained through the direct association of the Stat5 transcription complex with a corepressor molecule. We performed coimmunoprecipitation experiments to investigate the interaction of Stat5 and the corepressor SMRT. Cell lysates were obtained from 293T cells expressing Stat5a containing an HA tag and incubated with in vitro translated radiolabeled SMRT. Immunoprecipitations were performed with FLAG-tag– or HA-tag–specific antibodies and the precipitates were investigated for the presence of SMRT. Coprecipitation of Stat5 and SMRT was detected (Figure 5B).
The coiled-coil domain of Stat5 induces dimerization of Stat5-RARα fusion proteins and mediates binding to SMRT
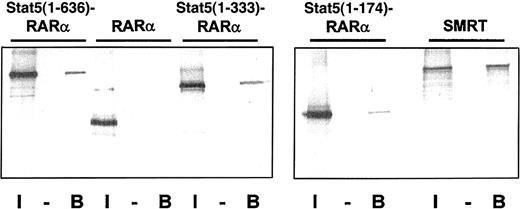
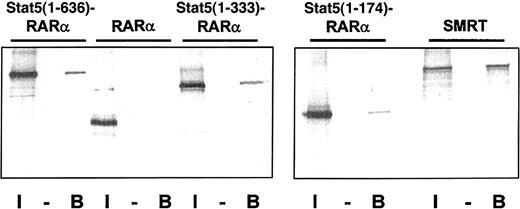
Oligomerization of fusion proteins such as PML-RARα, PLZF-RARα, and AML1-ETO16 19 have been shown to be essential for their leukemogenic potential. We hypothesized that the coiled-coil domain of Stat5 may induce oligomerization of the Stat5-RARα fusions. To test this model we generated a GST fusion protein containing the coiled-coil domain of Stat5 (amino acids 1-331) and investigated if this protein can interact with radiolabeled Gal4-Stat5-RARα fusion proteins (Figure6). Interaction of GST–Stat5(1-331) was shown with Gal4–Stat5(1-636)–RARα and Stat5(1-333)–RARα. Only a weak interaction was observed with Stat5(1-174)–RARα. No interaction was found with RARα. This indicates that the coiled-coil domain mediates homodimerization. We also observed that radiolabeled SMRT interacts with a GST fusion of the Stat5 coiled-coil region indicating that this domain of Stat5 is responsible for the corepressor recruitment in the transcription complex.
The coiled-coil domain of Stat5 induces homodimerization of Stat5-RARα fusion proteins and binding to SMRT.
(A) Radiolabeled Gal4-Stat5-RARα proteins and SMRT were precipitated with GST (−) or GST–Stat5(1-331) (B) fusion proteins and the bound radiolabeled fraction was compared to 10% input (I). Binding of Stat5(1-636)–RARα and Stat5(1-333)–RARα, but not RARα, to GST–Stat5(1-331) is seen, indicating that the coiled-coil domain mediates homodimerization. Furthermore, stable interaction of radiolabeled SMRT and GST–Stat5(1-331) can also be observed.
The coiled-coil domain of Stat5 induces homodimerization of Stat5-RARα fusion proteins and binding to SMRT.
(A) Radiolabeled Gal4-Stat5-RARα proteins and SMRT were precipitated with GST (−) or GST–Stat5(1-331) (B) fusion proteins and the bound radiolabeled fraction was compared to 10% input (I). Binding of Stat5(1-636)–RARα and Stat5(1-333)–RARα, but not RARα, to GST–Stat5(1-331) is seen, indicating that the coiled-coil domain mediates homodimerization. Furthermore, stable interaction of radiolabeled SMRT and GST–Stat5(1-331) can also be observed.
The strongly reduced sensitivity to ATRA, both in the transcriptional repression assay and in differentiation assays, provides evidence that a Stat5-RARα fusion protein can contribute to the pathogenesis of AML. It may also explain why ATRA treatment was not successful in a patient described with such a translocation. Furthermore, it provides an additional example for an AML-associated translocation interfering with myeloid differentiation through the aberrant recruitment of a corepressor complex. This bodes well for the use of HDAC inhibitors in the treatment of leukemias.
Discussion
Typical APL is associated with expression of the PML-RARα fusion gene, which leads to reduced sensitivity to ATRA for the dissociation of the corepressor complex. However, in a rare form of APL that does not respond to ATRA treatment, a variant translocation product, PLZF-RARα, interacts directly in an ATRA-insensitive manner with the HDAC corepressor complex mediated by the BTB/POZ domain of the PLZF protein. Here, we present evidence that in a newly found translocation associated with ATRA-resistant APL, the translocation product, Stat5-RARα,14 interacts with the corepressor SMRT and induces impaired differentiation in hematopoietic cell lines similar to PLZF-RARα. We demonstrate that interaction of Stat5-RARα and SMRT is mediated by the coiled-coil region of Stat5 and is not released by ATRA, explaining the resistance to ATRA treatment observed in the patient described with this translocation. Furthermore, we show interaction of wild-type Stat5a with the corepressor SMRT and enhanced Stat5-induced transcription of target genes in the presence of HDAC inhibitors. These findings indicate that Stat5-regulated gene expression is controlled through association with coactivators and corepressors. Interestingly, hyperactive forms of Stat5 containing single point mutations within the coiled-coil domain of the protein are also impaired in their ability to bind to SMRT,20suggesting a strong correlation between binding of SMRT and the level of Stat5-induced transactivation.
Retinoid receptors belong to the family of nuclear hormone receptors (NHRs), which are ligand-dependent transcriptional regulators. These NHRs induce transcriptional repression in the absence of ligand through binding to the corepressors SMRT and N-CoRs, whereas in the presence of ligand they recruit coactivators with histone acetyl transferase activity inducing chromatin modifications that allow active transcription.21 Chromosomal translocations involving the RARα receptor, such as PML- RARα or PLZF- RARα do not change the affinity to the ligand or binding to retinoic acid response elements, as compared to the wild-type RARα. However, these chimeric receptors cause leukemias by interfering with the function and nuclear localization of RARs under low or physiologic concentrations of retinoic acid (10−9-10−8 M). Presumably, they block activation of retinoic acid–dependent promoters and hematopoietic differentiation by constitutively recruiting corepressor complexes to the partner proteins in the RARα translocation product.22
Interaction of Stat5 with coactivators associated with histone acetyl transferase activity has been shown to be an essential step in the ability of Stat5 to induce transcription of target genes.2,23 Down-regulation of Stat5 activity has also been observed. It has been mainly attributed to an autoregulatory mechanism involving Stat5 dephosphorylation through tyrosine phosphatases or Stat5-induced expression of members of SOCS family, which terminate Stat5 activity by interfering with binding of Stat5 to activated cytokine receptors.24 However, lack of coactivator binding may also shift the function of the molecule from a transcriptional activator to a transcriptional repressor. This could be the case in C-terminally truncated Stat5 molecules (Stat5ΔC), which lack the transactivation domain for p300/CBP coactivator interaction.2 Binding of Stat5ΔC-Stat5wt heterodimers to the Stat5 binding site in target gene promoters negatively regulates expression,25-27 and may result from 2 combined effects: (1) the truncated molecule has a longer residence time on the DNA binding site and thereby blocks the access of the wild-type molecule,25 and (2) in addition it may recruit only corepressors to the Stat5-DNA complex. Similarly, in the translocation product Stat5-RARα, the RARα receptor is fused to a truncated Stat5b, which is C-terminally deleted and lacks part of its SH2 domain and its transactivation domain.14
However, there is also evidence from promoter studies for a direct role of wild-type Stat5 in transcriptional repression that appears to be tissue and promoter specific and requires modulation by interacting molecules.28,29 This has been shown for the activated glucocorticoid receptor, which cooperates with Stat5 to increase β-casein gene expression. It is conceivable that in certain cell types expressing particular cofactors, Stat5 target genes are repressed by the interaction of Stat5 with a corepressor complex containing HDAC activity. Such a mechanism has been suggested recently for the modulation of Stat-dependent transcription, where members of a novel protein family, named PIAS1 and PIAS3, bind to phosphorylated Stat1 and Stat3. To repress Stat1- and Stat3-dependent gene expression, these molecules essentially require an α-helical LXXLL signature motive, which has also been found in cofactors of nuclear receptors and may be involved in the assembly of PIAS-containing corepressor complexes.30
In the case of Stat5, interaction with the corepressor complex appears to be direct, because we present evidence that the coiled-coil region of Stat5 is able to directly recruit SMRT and contributes actively to transcriptional repression in the Stat5-RARα fusion protein. The coiled-coil region of Stat proteins has been implicated in protein-protein interaction and contribute to such diverse functions as transcriptional activation,3 receptor binding, and nuclear translocation.31 Furthermore, recent evidence indicates that a coiled-coil domain or similar helical structures are required for fusion proteins resulting from leukemia-related translocations, to form high-molecular-weight complexes and induce transcriptional repression.16,19 Interestingly, a recent publication shows interaction of the coiled-coil domain of Stat5 with Nmi augmenting Stat5-dependent transcription by enhanced association of p300/CBP coactivator proteins.3 It will be interesting to analyze how the interaction of the coiled-coil domain with coactivators or corepressors is regulated. Further experiments will reveal whether simultaneous binding of both corepressor and coactivator molecules to Stat5 can be observed. Such a situation has been found for the pituitary transcription factor, Pit-1. Disruption of the N-CoR interaction increased Pit-1–dependent target gene expression, mutations that enhance N-CoR binding resulted in reduced transcriptional activity.32
We propose that in a fusion with RARα, the coiled-coil domain of Stat5 is responsible for impaired differentiation in hematopoietic cells and ATRA-insensitive interaction with a corepressor complex, contributing to the development of an ATRA-resistant form of APL. Because stimulation with ATRA and the HDAC inhibitor TSA normalizes differentiation in hematopoietic cells expressing Stat5-RARα, this raises hopes that a combination treatment of ATRA together with an HDAC inhibitor would improve the therapeutic outcome in ATRA-resistant APLs.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Alexander B. Maurer, Georg-Speyer-Haus, Institute for Biomedical Research, Paul-Ehrlich-Str 42-44, 60596 Frankfurt, Germany; e-mail: a.maurer@em.uni-frankfurt.de.

![Fig. 1. Schematic diagram of Stat5-RARα mutants used. / Structure of full- length Stat5-RARα (Stat5[1-636]–RARα), 2 deletion mutants, and RARα alone are shown. C-C indicates coiled-coil region; DBD, DNA-binding domain; SH2, src-homology region 2; TA, transactivation domain; ZF, zinc finger domain; HBD, hormone-binding domain.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/99/8/10.1182_blood.v99.8.2647/5/m_h80822380001.jpeg?Expires=1766168641&Signature=T9~rWKSwYg1rqNZoNbFrkTHBFXEh8FHNk7fMGa1ov7mp7VQlb5Xauc4yICMbIp4Q2QqfUIJqDme5LKI6gMpiGaoCDAy9KFPw0M2wXzzBcnTkoYcpt0Tvrtryr0JHQQu3~iI0cn8qHvOPz7wbP3-ElOCLrodBeFnI0t-0huR7VLrR5xfyF5Nf~njn1oGPckoZFONrrTFtaX9ytCMob~txzusn5jgovHmpcJ0cU9bKd7vSxI~8OJiHb2IySc6Z1ijjzVJS7bgeeymN8phDf6LPdvFwyBDp7u6UmuT6I~XiiTe5DFQixIwfDglPb7GYllaPdaY5KLI5CmhYMc90ocMMnQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Fig. 1. Schematic diagram of Stat5-RARα mutants used. / Structure of full- length Stat5-RARα (Stat5[1-636]–RARα), 2 deletion mutants, and RARα alone are shown. C-C indicates coiled-coil region; DBD, DNA-binding domain; SH2, src-homology region 2; TA, transactivation domain; ZF, zinc finger domain; HBD, hormone-binding domain.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/99/8/10.1182_blood.v99.8.2647/5/m_h80822380001.jpeg?Expires=1767041871&Signature=fa4IEBTWTQshE67uxZLixDMmpZBUfN67o9aM-tbe7KpHOgufaDAUqZevE9b18CNTgf80126gw3tiZ-fzR1HTi4QOiEM2JZmwaSSzgXuKqaD79QEsWsgj4fp3rWNPiLDZus3GQd0dwvSneB0~XAbFxspMkr3qo26Mwc9XWGRhXEB-qbT~XkzWozaXkTPe-nb52zhvg0G4B2L5MiGr5oNL3CNZcarec3e3zR20RqiTzcP2ZJwhr1IDbixsnmJv-8MeL09sz~SBCBPLofqegbVFEDHW13VaYa~SnHWLX~oaMcl7TahaJnfUiZ0~2llH4zwGQJZVoDHeybSPPJoQtLPujQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)