Granulocyte-macrophage colony-stimulating factor (GM-CSF) is a hematopoietic cytokine that stimulates the production and functional activity of granulocytes and macrophages, properties that have encouraged its clinical use in bone marrow transplantation and in certain infectious diseases. Despite the importance of GM-CSF in regulating myeloid cell numbers and function, little is known about the exact composition and mechanism of assembly of the GM-CSF receptor complex. We have now produced soluble forms of the GM-CSF receptor α chain (sGMRα) and β chain (sβc) and utilized GM-CSF, the GM-CSF antagonist E21R (Glu21Arg), and the βc-blocking monoclonal antibody BION-1 to define the molecular assembly of the GM-CSF receptor complex. We found that GM-CSF and E21R were able to form low-affinity, binary complexes with sGMRα, each having a stoichiometry of 1:1. Importantly, GM-CSF but not E21R formed a ternary complex with sGMRα and sβc, and this complex could be disrupted by E21R. Significantly, size-exclusion chromatography, analytical ultracentrifugation, and radioactive tracer experiments indicated that the ternary complex is composed of one sβc dimer with a single molecule each of sGMRα and of GM-CSF. In addition, a hitherto unrecognized direct interaction between βc and GM-CSF was detected that was absent with E21R and was abolished by BION-1. These results demonstrate a novel mechanism of cytokine receptor assembly likely to apply also to interleukin-3 (IL-3) and IL-5 and have implications for our molecular understanding and potential manipulation of GM-CSF activation of its receptor.
Introduction
Granulocyte-macrophage colony-stimulating factor (GM-CSF) is a cytokine produced by many cells in the body that regulates the production, effector cell function, and survival of myeloid cells.1-4 Macrophages and granulocytes rise in numbers and exhibit a prolonged life span and enhanced effector function in response to GM-CSF,5,6 properties that have encouraged its use in bone marrow transplantation7 and infectious diseases such as those associated with AIDS.8In addition, GM-CSF controls dendritic cell production, differentiation, and function and potentiates responses of CD4+ T cells in vivo.9,10 This dual action of GM-CSF has encouraged its utilization in different vaccination strategies.11 On the other hand, these same properties have implicated GM-CSF in myeloid leukemia and several inflammatory conditions such as asthma12 and rheumatoid arthritis.13
The actions of GM-CSF are mediated by specific receptors composed of 2 different subunits, a receptor α chain (GMRα),14 which provides specificity and the major binding contact, and a β chain (βc),15 which is common with the interleukin-3 (IL-3) and IL-5 receptors, promotes affinity conversion, and acts as the major signal transducer. For this complex to be assembled and to signal, there exist structural and dimerization requirements, some of which have been defined. Extensive structure-function analysis has identified several residues involved in GM-CSF, GMRα, and βc protein interaction and biologic activity. For example, the binding of GM-CSF to GMRα involves an electrostatic interaction between Asp112 in the fourth α helix of GM-CSF and Arg280 in the F-G loop of GMRα.16,17 The biologic activities and high-affinity binding of GM-CSF are exquisitely dependent on Glu21 in the first α helix of GM-CSF, although direct contact with βc has not been demonstrated. Substitution of this amino acid with arginine generates a GM-CSF analog, E21R (Glu21Arg), which exhibits only low-affinity binding and is unable to stimulate cellular proliferation and mature cell functions.18 Importantly, E21R is able to antagonize GM-CSF binding and function19; however, the molecular basis of this antagonism is not fully understood. In βc, residues in the B-C loop (Tyr365, His367, Ile368) and F-G loop (Tyr421) of domain 4 are involved in GM-CSF high-affinity binding and function.20-23 The monoclonal antibody (mAb) BION-1, which binds an area in βc encompassing these loops, blocks GM-CSF binding and biologic activities.24
Dimerization of the α and βc subunits of this family of cytokine receptors is recognized as a crucial step for their activation; however, the exact composition of the assembled complex remains unclear. A number of studies suggest that simple heterodimerization is sufficient to activate the GM-CSF receptor,25 whereas both cross-linking and dominant-negative studies using surface-expressed receptors suggest that the formation of higher-order GM-CSF receptor complexes is required for receptor activation.26,27 Dimerization of βc in particular has also been shown to be an important and necessary step for receptor activation,28,29 probably reflecting the need to bring into close proximity the cytoplasmic domains of 2 βc molecules associated with Janus kinase-2 (JAK-2), resulting in JAK transphosphorylation and receptor phosphorylation. Interestingly, βc has been shown to crystallize as a dimer30 and to exist as a preformed homodimer on the cell surface.26 28Despite these findings, little is known about the full assembly of this family of receptors, the intermediate steps in their formation, and how receptor assembly may be selectively modulated.
In this paper we show for the first time the full assembly of the human GM-CSF receptor in solution. This shows a novel mode of cytokine receptor assembly in which 1 molecule of GM-CSF associates with 1 molecule of GMRα and 2 molecules of βc. In addition, these studies reveal an essential, direct interaction between GM-CSF and βc and provide a molecular understanding of GM-CSF antagonism by E21R or BION-1. This novel mode of receptor assembly may also apply to the IL-5 and IL-3 receptors.
Materials and methods
Human GM-CSF and GM-CSF analogs
Soluble wild-type human GM-CSF was produced in Escherichia coli and recovered from the periplasmic space by osmotic shock as described previously.19 Crude periplasmic extracts were adjusted to 25 mM N-ethylmorpholine HCl (NEM), pH 7.0, loaded onto Q Sepharose Fast Flow (Amersham Biosciences, Sydney, Australia) equilibrated in 25 mM NEM, pH 7.0, and a linear gradient of 0 to 600 mM NaCl in 25 mM NEM, pH 7.0, used to elute the bound proteins. GM-CSF purified by anion exchange was further purified by reversed phase high-performance liquid chromatography (HPLC), lyophilized, dissolved in phosphate-buffered saline (PBS) as previously described,19 and sterile-filtered (0.45 μm). The E21R analog of GM-CSF (BresaGen, Adelaide, South Australia) contains a glutamate to arginine substitution at residue 21 and a modified 12–amino acid leader peptide, MFATSSSTGNDG, to facilitate expression in E coli.31
Radiolabeling of human GM-CSF
To enable phosphorylation of GM-CSF under mild conditions, we made the GM-CSF analog, SGMKIN, in which the amino acids from alanine at position 3 to proline at position 6 were replaced by the peptide sequence RRASV, which is recognized by the catalytic subunit of cyclic adenosine monophosphate (cAMP)–dependent protein kinase from heart muscle.32 Complementary oligonucleotides were used to create a HindIII/NcoI fragment encoding the N-terminal 12 amino acids of SGMKIN. This fragment was ligated with an NcoI/BamHI fragment encoding the C-terminal 116 amino acids of human GM-CSF (hGM-CSF) into HindIII/BamHI–digested pIN-III-OmpH3 expression vector19 to create the plasmid, pSGMKIN. Soluble SGMKIN was expressed in E coli and purified as described for wild-type GM-CSF. The final product was at more than 95% purity by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), and the SGMKIN analog displayed biologic activity indistinguishable from the wild-type GM-CSF (data not shown). Labeling of SGMKIN with 32P used a protocol adapted from Kaelin et al.32 Fifty micrograms of purified SGMKIN was incubated in a 200-μL reaction mix containing 20 mM Tris (tris(hydroxymethyl)aminomethane) HCl, pH 7.5; 100 mM NaCl; 12 mM MgCl2; 10 mM β-mercaptoethanol; 1 μCi/μL (0.037 MBq/μL) [γ-32P]adenosine triphosphate ([γ-32P]ATP) (3000 Ci/mmol [111 000 GBq/mmol]; Geneworks, Adelaide, South Australia), and 1 U/μL of the protein kinase catalytic subunit (Sigma, Castle Hill, Australia). The reaction proceeded at 4°C for 30 minutes, was terminated by the addition of 200 μL of 100 mM EDTA (ethylenediaminetetraacetic acid), adjusted to 0.1% (vol/vol) trifluoroacetic acid (TFA; Auspep, Parkville, Australia), 1% (vol/vol) acetic acid, and loaded onto a Sep-Pak C18 reversed phase cartridge (Waters, Rydalmere, Australia) equilibrated in 0.01% TFA. The cartridge was washed with 0.01% TFA and bound SGMKIN eluted using 10 mL of 50% (vol/vol) acetonitrile in the presence of 0.01% TFA. Ten equal fractions were collected, and those containing the peak of eluted radioactivity were pooled and concentrated using a Speed-vac (Savant Instruments, Farmingdale, NY) to a final volume of approximately 100 μL.
Production of recombinant soluble GM-CSF receptor subunits
DNA fragments encoding the soluble extracellular domains of GMRα (sGMRα) or βc (sβc) were generated by PCR using the primers 5′-CTGACCGGATCCATGCTTCTCCTGGTGACAAGCC-3′ and 5′-GTACACGGATCCGAATTCTTACCCGTCGTCAGAACCAAATTC-3′ for sGMRα and 5′-CTGACCGGATCCATGGTGCTGGCCCAGGGGCTGC-3′ and 5′-CAGCACGGATCCGAATTCTTACGACTCGGTGTCCCAGGAGCG-3′ for sβc, with EcoRI and BamHI restriction sites underlined. Stop codons were inserted immediately prior to the transmembrane domain for each receptor molecule, following Gly at position 320 for GMRα and Ser at position 438 for βc. The PCR products were digested with BamHI and EcoRI and cloned into the baculovirus transfer vector BacPAK9 (Clontech, Palo Alto, CA) and the sequence of the cloned inserts were verified by cycle sequencing with BigDye chemistry (Applied Biosystems, Foster City, CA). The cDNA encoding sGMRα and sβc was introduced into the genome ofBsu36I-digested BacPAK6 viral DNA (Clontech) by homologous recombination following the manufacturer's instructions. Expression of recombinant protein is under the control of the strong polyhedrin promoter. Large-scale expression of sGMRα or sβc was performed by infection of Sf21 cells, grown in serum-free Ex-Cell 420 medium (JRH Biosciences, Brooklyn, Australia), with recombinant baculovirus at a multiplicity of infection of 0.3. Supernatant containing soluble receptor was harvested following incubation at 27°C for 5 to 7 days.
Purification of soluble GMRα and sβc
Conditioned media containing sGMRα (20 L) or sβc (9 L) were concentrated to less than 1 L using tangential flow filtration cartridges (10 000 molecular weight cutoff, 0.23 m2) (Millipore, Northryde, Australia) operated at 80 kPa and 4°C. Insoluble material in the concentrate was pelleted at 3000gfor 30 minutes and the resulting supernatant filtered (3 μm) prior to affinity chromatography. Affinity matrices were prepared by coupling E21R or the anti-βc mAb, BION-1,24 to cyanogen bromide (CNBr)–activated Sepharose 4B (Amersham Biosciences) following the manufacturer's instructions. Recombinant soluble receptor was bound to the affinity matrix, washed extensively in PBS containing 0.01% (vol/vol) polyoxyethylene 20 sorbitan monolaurate (Tween 20), and bound proteins eluted with 100 mM NaCl, 100 mM sodium acetate (pH 4.0). The eluate fractions were immediately neutralized using 2 M Tris and analyzed for the presence of soluble receptor by SDS-PAGE. Fractions containing purified soluble receptor were pooled and concentrated using a stirred-cell device with a 10 000 molecular weight cutoff, low protein-binding membrane (YM10; Millipore) operated at 300 kPa and 4°C. Concentrated soluble receptor was dialyzed extensively into PBS, sterile-filtered (0.2 μm), and stored at 4°C.
SDS-PAGE
Samples were analyzed on 10% or 12.5% polyacrylamide gels containing 38:1 acrylamide/bisacrylamide under reducing or nonreducing conditions as specified. Bands were visualized by staining with either Coomassie brilliant blue R-250 or silver.33
Mass spectrometry
Electrospray ionization mass spectrometry was performed using a PE/Sciex API100 mass spectrometer (Perkin-Elmer Sciex Instruments, Ontario, Canada). Protein samples were desalted in-line using a 1 × 10 mm reversed phase column eluted with 60% (vol/vol) acetonitrile in the presence of 0.04% (vol/vol) TFA and the primary mass spectrum transformed to give a true-mass profile using instrument software.
Protein analyses by size-exclusion chromatography
Size-exclusion chromatography was initially used to quantify purified soluble receptors and their ligands. Samples were chromatographed on a SMART system with a Superdex 200PC 3.2/30 (3.2 mm × 300 mm) column (Amersham Biosciences) operated at 40 μL/min at 25°C using 150 mM NaCl, 50 mM sodium phosphate, pH 7.0, as running buffer. The area under the protein peak was integrated using the extinction coefficient (absorbance units × mL−1 × mg−1) calculated for each protein: GM-CSF, 0.95; E21R, 0.88; sGMRα, 1.17; sβc, 1.95.
To analyze protein-protein interactions, individual proteins and protein complexes were prepared in a final volume of 50 μL, adjusted with PBS as required, and incubated at 25°C for at least 1 hour. Samples were analyzed by size-exclusion chromatography using the SMART system as described above with data presented from representative experiments (n = 5). The dependence of elution time on the log10 (MW) of protein standards was used to calibrate the column and to generate a trend line for each set of standards. External standards included myoglobin, MW 17 kDa; ovalbumin, MW 44 kDa; γ-globulin, MW 158 kDa; and thyroglobulin, MW 670 kDa (Biorad Laboratories, Hercules, CA). Internal standards were GM-CSF, MW 14.5 kDa; E21R, MW 15.7 kDa; sGMRα, MW 43 kDa; and sβc, MW 101 kDa as determined by mass spectrometry and SDS-PAGE. Soluble βc was found to be a dimer by size-exclusion chromatography consistent with previous reports.30 Calibration curves constructed from the external and internal standards were essentially parallel (see Figure 2A). The calibration curve for the internal standards was extrapolated to higher mass (670 kDa) because this was found to be the limit of the linear range for the external standards.
Analytical ultracentrifugation
The molecular weights of GM-CSF, E21R, sGMRα, sβc, and the binary and ternary complexes were determined by sedimentation equilibrium. Individual proteins and protein complexes were isolated by size-exclusion chromatography using a fast protein liquid chromatography (FPLC) system with a Superdex 200 10/30 (10 mm × 300 mm) column (Amersham Biosciences) operated at 0.5 mL/min at 25°C using 150 mM NaCl, 50 mM sodium phosphate, pH 7.0, as running buffer. Pooled fractions were concentrated using Centricon 10 microconcentrators (Amicon, Beverly, MA). Sedimentation equilibrium experiments were performed using a Beckman XL-A analytical ultracentrifuge equipped with a Ti60 rotor (Beckman, Palo Alto, CA) and filled epon centerpieces (12-mm path length). Sedimentation equilibrium profiles were obtained at 20°C using the rotor speeds indicated. Equilibrium distributions were fitted by nonlinear regression analysis to obtain best-fit values for the M (1-νρ), where M is the molecular weight and ν the partial specific volume of the sedimenting species and ρ the solution density. The compositional molecular weights of the proteins and the partial specific volumes of GM-CSF and E21R were calculated from their amino acid sequences. Partial specific volumes for the glycosylated forms of sGMRα and sβc were calculated assuming these proteins were monomer and dimmer, respectively. A value of 0.622 mL/g was assumed for the partial specific volume of carbohydrate. The experimental value of M (1-νρ) and the molecular weight (Mp) and partial specific volume of the protein component were then used to solve for the weight fraction of bound carbohydrate and hence the partial specific volume of the carbohydrate-bound protein. Values for the partial specific volumes of the GM-CSF/sGMRα and E21R/sGMRα complexes were calculated assuming a 1:1 complex and no volume change on association. A value of 0.72 mL/g was assumed for the GM-CSF/sGMRα/sβc complex.
Cross-linking experiments
Stable cross-linking of sβc or soluble complexes of ligand with sβc was performed by incubation of 2.6 μg sβc with either 2.4 μg GM-CSF or E21R for 1 hour at 25°C followed by addition of BS3 cross-linker (Pierce, Rockford, IL) at a final concentration of 0.1 mg/mL for 10 minutes. The reaction was then stopped by addition of ethanolamine HCl, pH 8.0, to a final concentration of 100 mM. Cross-linked proteins were subjected to reducing SDS-PAGE and compared with non–cross-linked material. Antibody Fab fragments of BION-1 (anti-βc fourth domain blocking mAb) and 2H1 (anti-βc fourth domain control mAb) used in cross-linking experiments were generated by digestion with ficin using the Immunopure IgG1 Fab Preparation Kit (Pierce) following the manufacturer's instructions. Fab fragment (18 μg) was preincubated with 2.6 μg sβc for 30 minutes at 25°C prior to the addition of 2.4 μg GM-CSF in a final volume of 20 μL for a further hour. Cross-linking was then performed as above followed by SDS-PAGE analysis.
Results
Production, purification, and analysis of GM-CSF soluble receptor components
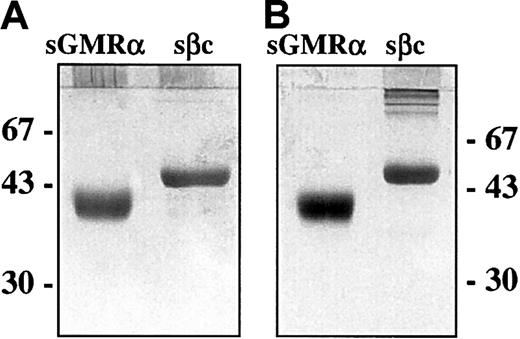
Complementary DNA fragments encoding the extracellular domains of GMRα and βc (sGMRα and sβc) were generated by PCR and cloned into a baculovirus transfer vector. Following introduction into a baculovirus expression system by homologous recombination, the soluble receptor components were generated by infection of Sf21 cells. Purification of the soluble receptors was achieved by affinity chromatography using immobilized ligand for sGMRα and immobilized mAb BION-124 for sβc. Purified soluble receptors were recovered at more than 95% purity as assessed by silver-stained SDS-PAGE under reducing conditions (Figure1A) with an apparent molecular weight (MW) of approximately 43 kDa for sGMRα and 55 kDa for sβc. Importantly, these MWs determined for sGMRα and for sβc did not alter when analyzed under nonreducing conditions (Figure 1B), indicating the absence of disulfide-linked dimers. A small amount of disulfide-aggregated sβc was visible by nonreducing SDS-PAGE (Figure1B) and as an early, minor peak during size-exclusion chromatography (Figure 2C). The absence of detectable disulfide-linked dimers in sβc was confirmed by ion-spray mass spectrometry, which demonstrated that the protein preparation had a major species of 50.623 kDa with several minor species representing glycosylation variants.
SDS-PAGE analysis of purified sGMRα and sβc.
Soluble GMRα and sβc were produced by Sf21 cells infected with recombinant baculovirus encoding appropriate cDNA and affinity purified from the supernatant as described in “Materials and methods.” Soluble GMRα (1 μg) and sβc (0.5 μg) were fractionated by 10% SDS-PAGE under reducing (A) and nonreducing (B) conditions and silver stained. The positions of molecular weight markers are shown in kilodaltons.
SDS-PAGE analysis of purified sGMRα and sβc.
Soluble GMRα and sβc were produced by Sf21 cells infected with recombinant baculovirus encoding appropriate cDNA and affinity purified from the supernatant as described in “Materials and methods.” Soluble GMRα (1 μg) and sβc (0.5 μg) were fractionated by 10% SDS-PAGE under reducing (A) and nonreducing (B) conditions and silver stained. The positions of molecular weight markers are shown in kilodaltons.
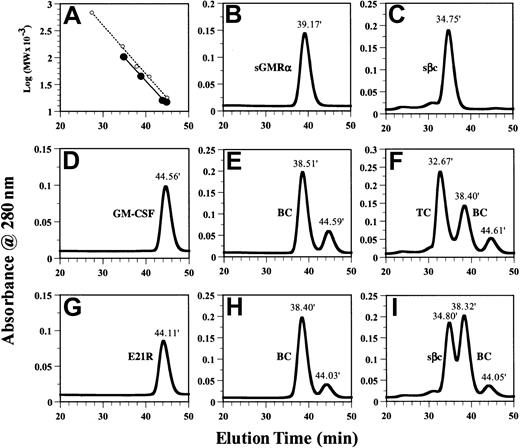
GM-CSF but not the GM-CSF analog E21R induces the assembly of the ternary GM-CSF receptor complex in solution.
The presence and molecular weight of individual proteins and protein complexes were determined using size-exclusion chromatography as described in “Materials and methods.” (A) Linear regression of log10 (MW × 10−3) versus elution times using external (○) and internal (●) standards for calibration of the column. (B-I) Individual proteins sGMRα (B), sβc (C), GM-CSF (D), and E21R (G) were applied separately. Mixtures of sGMRα (6 μM) and GM-CSF (12 μM) (E); sβc (3 μM), sGMRα (6 μM), and GM-CSF (12 μM) (F); sGMRα (6 μM) and E21R (12 μM) (H); sβc (3 μM), sGMRα (6 μM), and E21R (12 μM) (I) were incubated for 1 hour before being applied to the column. The number above each peak represents elution time. Peaks containing binary (BC) or ternary (TC) complexes are indicated.
GM-CSF but not the GM-CSF analog E21R induces the assembly of the ternary GM-CSF receptor complex in solution.
The presence and molecular weight of individual proteins and protein complexes were determined using size-exclusion chromatography as described in “Materials and methods.” (A) Linear regression of log10 (MW × 10−3) versus elution times using external (○) and internal (●) standards for calibration of the column. (B-I) Individual proteins sGMRα (B), sβc (C), GM-CSF (D), and E21R (G) were applied separately. Mixtures of sGMRα (6 μM) and GM-CSF (12 μM) (E); sβc (3 μM), sGMRα (6 μM), and GM-CSF (12 μM) (F); sGMRα (6 μM) and E21R (12 μM) (H); sβc (3 μM), sGMRα (6 μM), and E21R (12 μM) (I) were incubated for 1 hour before being applied to the column. The number above each peak represents elution time. Peaks containing binary (BC) or ternary (TC) complexes are indicated.
The physical properties of sGMRα and sβc were further characterized by size-exclusion chromatography. We initially determined the retention times of sGMRα (Figure 2B), sβc (Figure 2C), GM-CSF (Figure 2D), and E21R (Figure 2G). The individual proteins eluted at 39.17 minutes for sGMRα, 34.75 minutes for sβc, 44.56 minutes for GM-CSF, and 44.11 minutes for E21R. External MW standards for calibration of the size-exclusion chromatography (Figure 2A) indicated that sGMRα, GM-CSF, and E21R were monomeric but that sβc was dimeric. The dimeric nature of sβc was confirmed by cross-linking experiments with purified sβc, which produced a covalent dimer with a MW of 100 kDa as determined by SDS-PAGE (see Figure 8B). The observation that sβc exists as a dimer is consistent with a recent report describing the structure of the extracellular domain of βc expressed in insect cells.30 34 We observed that both the ligands and the receptor components eluted from size-exclusion chromatography earlier than expected from the elution times of the external MW standards (Figure 2A). We chose to use the proteins of interest as internal MW standards and constructed a calibration curve for the internal MW standards that is parallel to that constructed from the external MW standards (Figure 2A). This is expected to provide a superior estimate of the masses of the receptor complexes.
Soluble GMRα interactions with GM-CSF and E21R
Purified sGMRα (6 μM) was incubated with GM-CSF (12 μM) and fractionated on a Superdex 200 column, producing a modest shift (from 39.17 minutes to 38.51 minutes) in the elution time of sGMRα (Figure2E). The shifted peak, with an apparent MW of 48 kDa, contained both GM-CSF and sGMRα as determined by SDS-PAGE analysis of fractions (data not shown). The MW of the GM-CSF/sGMRα binary complex is consistent with a stoichiometry of 1:1 as has previously been described.35 The complete peak shifts observed when sGMRα binds GM-CSF suggest that all of this soluble receptor is competent to bind ligand. Saturation binding experiments revealed that GM-CSF bound to sGMRα with a dissociation constant (Kd) of 1.5 to 9 nM, similar to that seen with cell surface–expressed GMRα (data not shown).
Purified sGMRα (6 μM) was incubated with E21R (12 μM) and fractionated on a Superdex 200 column, producing a modest shift (from 39.17 minutes to 38.40 minutes) in the elution time of sGMRα (Figure 2H). The shifted peak, with an apparent MW of 49 kDa, contained both E21R and sGMRα as determined by SDS-PAGE analysis of fractions (data not shown). The MW of the E21R:sGMRα binary complex is consistent with a stoichiometry of 1:1.
The sβc induces the formation of a GM-CSF ternary complex
Purified sβc (3 μM) was incubated with sGMRα (6 μM) plus GM-CSF (12 μM) and fractionated on a Superdex 200 column, producing a complete shift in the elution time of sβc (from 34.75 minutes to 32.67 minutes) as well as peaks corresponding to the binary complex at 38.40 minutes and free ligand at 44.61 minutes (Figure 2F). The peak eluting at 32.67 minutes had an apparent MW of 155 kDa and contained GM-CSF, sGMRα, and sβc as determined by SDS-PAGE analysis of fractions (Figure 3A). The MW of this ternary GM-CSF receptor complex is consistent with a stoichiometry of 1 GM-CSF:1 sGMRα:2 sβc.
SDS-PAGE analysis of the ternary GM-CSF receptor complex.
Mixtures of sβc, sGMRα, and either GM-CSF (A) or E21R (B) were analyzed by size-exclusion chromatography as described for Figure 2F and I. Fractions were collected at 1-minute intervals, fractionated by 12.5% SDS-PAGE under reducing conditions, and silver stained.33 The positions of individual components are indicated.
SDS-PAGE analysis of the ternary GM-CSF receptor complex.
Mixtures of sβc, sGMRα, and either GM-CSF (A) or E21R (B) were analyzed by size-exclusion chromatography as described for Figure 2F and I. Fractions were collected at 1-minute intervals, fractionated by 12.5% SDS-PAGE under reducing conditions, and silver stained.33 The positions of individual components are indicated.
In contrast, no ternary complex was observed when purified sβc (3 μM) was incubated with sGMRα (6 μM) plus E21R (12 μM) and fractionated on a Superdex 200 column (Figure 2I). Whereas the binary complex eluting at 38.32 minutes contained E21R and sGMRα, the peak at 34.80 minutes contained sβc but no sGMRα or E21R as determined by SDS-PAGE analysis of fractions (Figure 3B).
E21R disrupts the formation of the ternary GM-CSF receptor complex
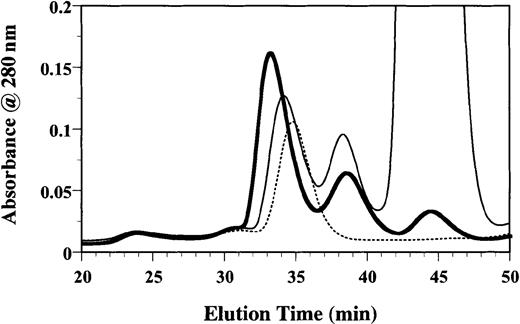
To investigate whether the formation of a binary complex was an intermediate step in the formation of the ternary GM-CSF receptor complex, we tested the effect of E21R in this process. Purified sβc (3 μM) was incubated with sGMRα (6 μM) and GM-CSF (12 μM) for 1 hour. A 100-fold molar excess of E21R was then added, and after a further 1-hour incubation the mixture was fractionated on a Superdex 200 column. In the absence of E21R the ternary GM-CSF receptor complex eluted at 33.12 minutes (Figure 4). Significantly, in the presence of a 100-fold molar excess of E21R (Figure 4) there was a reduction in the amount of ternary GM-CSF receptor complex and an increase in its elution time (34.10 minutes), more comparable with the elution time of free sβc (34.75 minutes). The reduction in the amount of ternary complex along with an increased amount of binary complex (38.40 minutes) and free ligand (44.00 minutes) is consistent with sGMRα preferentially forming a binary complex with E21R, which is unable to recruit sβc into a ternary complex.
E21R prevents formation of the ternary GM-CSF receptor complex.
Following formation of the GM-CSF/sGMRα/sβc ternary complex using a 1:2:4 molar ratio, a 100-fold molar excess of E21R over GM-CSF was added for a further hour at 25°C before size-exclusion chromatography. The chromatogram shows the A280 profile of a GM-CSF/sGMRα/sβc mixture in the absence (thick line) or presence of E21R (thin line) or in sβc alone (dashed line).
E21R prevents formation of the ternary GM-CSF receptor complex.
Following formation of the GM-CSF/sGMRα/sβc ternary complex using a 1:2:4 molar ratio, a 100-fold molar excess of E21R over GM-CSF was added for a further hour at 25°C before size-exclusion chromatography. The chromatogram shows the A280 profile of a GM-CSF/sGMRα/sβc mixture in the absence (thick line) or presence of E21R (thin line) or in sβc alone (dashed line).
Stoichiometry of the ternary GM-CSF receptor complex
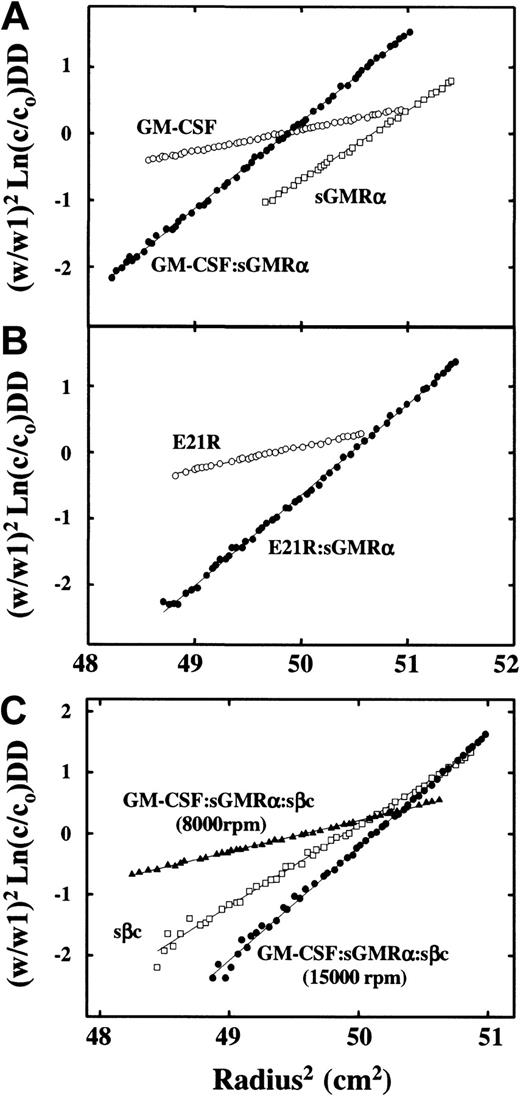
To confirm the 1 GM-CSF:1 sGMRα:2 sβc stoichiometry of the ternary GM-CSF receptor complex obtained by size-exclusion chromatography, we utilized 2 other complementary and independent methods. In one of these the molecular weights of the individual proteins and of the binary and ternary complexes were determined by sedimentation equilibrium. The results showed (Figure5; Table1) values similar to those obtained by gel filtration. The estimates of the molecular weight of the binary complexes GM-CSF/sGMRα, 52.7 kDa (Figure 5A; Table 1), and E21R/sGMRα, 54.8 kDa, (Figure 5B; Table 1), are consistent with a 1:1 stoichiometry. The molecular weight of the ternary GM-CSF/sGMRα/sβc complex was determined to be 135 kDa (Figure 5C and Table 1). This value is consistent with a model where one sβc dimer (97.4 kDa) associates with one GM-CSF/sGMRα binary complex (52.7 kDa) (Table 1) with a theoretical molecular weight of 150.1 kDa.
Analyses of the ternary GM-CSF receptor complex by sedimentation equilibrium.
The individual proteins or protein complexes in 150 mM NaCl/50 mM sodium phosphate, pH 7.0, were centrifuged at 20°C at angular velocity, W rpm, for 16 hours. The equilibrium profiles are presented as (W/W1)2 Ln(c/co) versus the square of the radial distance, where c/co is the optical density at 280 nm divided by the initial optical density and W1 is 20 000 rpm. For a single species, this plot is linear with a slope proportional to the molecular weight of the sedimenting species. The initial concentrations were in the range 0.40 to 0.47 mg/mL, and samples were centrifuged at 20 000 rpm except for E21R, where the initial concentration was 0.2 mg/mL and the angular velocity 15 000 rpm. Panel A samples: GM-CSF (○), sGMRα (■), GM-CSF/sGMRα complex (●). Panel B samples: E21R (○), E21R/sGMRα complex (●). Panel C samples: purified sβc (■) was centrifuged at 15 000 rpm with an initial concentration of 0.45 mg/mL, whereas the GM-CSF/sGMRα/sβc ternary complex at an initial concentration of 0.47 mg/mL was centrifuged at either 8000 rpm (▴) or 15 000 rpm (●) for 16 hours at 20°C.
Analyses of the ternary GM-CSF receptor complex by sedimentation equilibrium.
The individual proteins or protein complexes in 150 mM NaCl/50 mM sodium phosphate, pH 7.0, were centrifuged at 20°C at angular velocity, W rpm, for 16 hours. The equilibrium profiles are presented as (W/W1)2 Ln(c/co) versus the square of the radial distance, where c/co is the optical density at 280 nm divided by the initial optical density and W1 is 20 000 rpm. For a single species, this plot is linear with a slope proportional to the molecular weight of the sedimenting species. The initial concentrations were in the range 0.40 to 0.47 mg/mL, and samples were centrifuged at 20 000 rpm except for E21R, where the initial concentration was 0.2 mg/mL and the angular velocity 15 000 rpm. Panel A samples: GM-CSF (○), sGMRα (■), GM-CSF/sGMRα complex (●). Panel B samples: E21R (○), E21R/sGMRα complex (●). Panel C samples: purified sβc (■) was centrifuged at 15 000 rpm with an initial concentration of 0.45 mg/mL, whereas the GM-CSF/sGMRα/sβc ternary complex at an initial concentration of 0.47 mg/mL was centrifuged at either 8000 rpm (▴) or 15 000 rpm (●) for 16 hours at 20°C.
Sedimentation equilibrium analysis of the molecular weights of GM-CSF, E21R, sGMRα, sβc, and their complexes
| Species . | M (1-vρ) . | v . | MW . | Predicted MW . | Predicted stoichiometry . |
|---|---|---|---|---|---|
| GM-CSF | 3 590 | 0.734 | 13 700 | — | — |
| E21R | 4 050 | 0.734 | 14 300 | — | — |
| sGMRα | 11 800 | 0.706 | 40 700 | — | — |
| sβc | 26 600 | 0.723 | 97 400 | — | — |
| sGMRα plus GM-CSF | 14 600 | 0.718 | 52 700 | 54 400 | 1:1 |
| sGMRα plus E21R | 15 400 | 0.714 | 54 800 | 55 000 | 1:1 |
| sβc plus sGMRα plus GM-CSF | 37 300 | 0.72 | 135 300 | 151 800 | 2:1:1 |
| Species . | M (1-vρ) . | v . | MW . | Predicted MW . | Predicted stoichiometry . |
|---|---|---|---|---|---|
| GM-CSF | 3 590 | 0.734 | 13 700 | — | — |
| E21R | 4 050 | 0.734 | 14 300 | — | — |
| sGMRα | 11 800 | 0.706 | 40 700 | — | — |
| sβc | 26 600 | 0.723 | 97 400 | — | — |
| sGMRα plus GM-CSF | 14 600 | 0.718 | 52 700 | 54 400 | 1:1 |
| sGMRα plus E21R | 15 400 | 0.714 | 54 800 | 55 000 | 1:1 |
| sβc plus sGMRα plus GM-CSF | 37 300 | 0.72 | 135 300 | 151 800 | 2:1:1 |
The buffer used was 150 mM NaCl/50 mM sodium phosphate, pH 7.0, and the temperature was 20°C. The complexes formed between GM-CSF and E21R with sGMRα and between GM-CSF, sGMRα, and sβc were isolated by gel filtration. The initial concentration used for all samples was between 0.40 and 0.47 mg/mL except for E21R, where the starting concentration was 0.2 mg/mL. The reduced molecular weights of the samples, M (1-vρ), were determined by direct fitting of the sedimentation data presented in Figure 5. These values were used to calculate the molecular weight (MW) of the sedimenting species using the partial specific volumes (v) indicated.
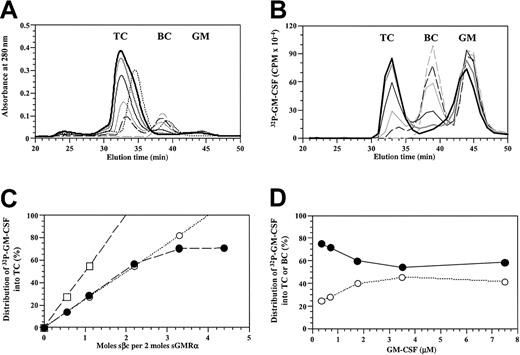
In a separate approach, we used radiolabeled GM-CSF as a tracer molecule. Purified sβc (0 to 7 μM) was titrated against a mixture of sGMRα (3.2 μM) and cold GM-CSF (7.3 μM) spiked with the GM-CSF analog 32P-SGMKIN and subjected to size-exclusion chromatography as above. Addition of sβc to the GM-CSF/sGMRα mixture led to the dose-dependent formation of the ternary complex and depletion of the binary complex (Figure6A). Once the concentration of sβc saturated the available binary complex, a shoulder appeared on the trailing edge of the ternary complex peak, presumably reflecting the presence of free sβc. Formation of the ternary complex was associated with a dose-dependent accumulation of radioactivity at the appropriate elution time of the ternary complex and was accompanied by a reduction of radioactivity at the elution time of the binary complex (Figure 6B). Titration of sβc did not lead to a reduction of radioactivity at the elution time of free ligand, although a modest shift at the leading edge of the free ligand peak was observed. We then determined the distribution of 32P-SGMKIN into the ternary complex and expressed it as a percentage of total label in the ternary and binary complexes versus proportion of sβc present (Figure 6C). When compared with the theoretical distribution predicted for a ternary complex with a GM-CSF/sGMRα/sβc ratio of 1:1:2 or 2:2:2, the observed distribution was consistent with a 1:1:2 stoichiometry. The observed distribution only departed from the modeled linear distribution as the concentration of binary complex became limiting.
Radiolabeled GM-CSF differentially partitions to the ternary GM-CSF receptor complex.
(A-C) A titration of purified sβc (0 to 7.03 μM) against a mixture of 3.2 μM sGMRα and 7.3 μM GM-CSF spiked with32P-labeled SGMKIN. Reaction mixes were set up with 0 (dashed gray), 0.88 μM (dashed black), 1.76 μM (thin gray), 3.52 μM (thin black), 5.27 μM (thick gray), or 7.03 μM (thick black) sβc and incubated at 25°C for 1 hour before size-exclusion chromatography. Fractions were collected at 1-minute intervals. A control reaction was also prepared with 5.27 μM sβc and 3.2 μM sGMRα but no GM-CSF (dashed black). (A) Chromatogram of A280 profiles for each sample with the location of the ternary complex (TC), binary complex (BC), and free ligand (GM) indicated. (B) Distribution of radioactivity among the ternary complex, binary complex, and free ligand for the reactions described in panel A. (C) Radioactive GM-CSF distributed into the ternary complex, expressed as a percentage of the total radioactive GM-CSF in ternary and binary complexes; comparing experimentally observed values for the reactions described in panel A (●) with a theoretical distribution based on 1GM:1α:2β (○) and 2GM:2α:2β (■) models. (D) Titration of GM-CSF (0 to 7 μM) spiked with32P-labeled SGMKIN against a mixture of 3.5 μM sGMRα and 3.5 μM sβc. Reaction mixes were allowed to reach equilibrium at 25°C for at least 2 hours before being fractionated by size-exclusion chromatography. The distribution of radioactivity among ternary (●) and binary (○) complexes was determined and the radioactivity in each complex was expressed as a percentage of total bound counts where counts in TC plus counts in BC is 100%.
Radiolabeled GM-CSF differentially partitions to the ternary GM-CSF receptor complex.
(A-C) A titration of purified sβc (0 to 7.03 μM) against a mixture of 3.2 μM sGMRα and 7.3 μM GM-CSF spiked with32P-labeled SGMKIN. Reaction mixes were set up with 0 (dashed gray), 0.88 μM (dashed black), 1.76 μM (thin gray), 3.52 μM (thin black), 5.27 μM (thick gray), or 7.03 μM (thick black) sβc and incubated at 25°C for 1 hour before size-exclusion chromatography. Fractions were collected at 1-minute intervals. A control reaction was also prepared with 5.27 μM sβc and 3.2 μM sGMRα but no GM-CSF (dashed black). (A) Chromatogram of A280 profiles for each sample with the location of the ternary complex (TC), binary complex (BC), and free ligand (GM) indicated. (B) Distribution of radioactivity among the ternary complex, binary complex, and free ligand for the reactions described in panel A. (C) Radioactive GM-CSF distributed into the ternary complex, expressed as a percentage of the total radioactive GM-CSF in ternary and binary complexes; comparing experimentally observed values for the reactions described in panel A (●) with a theoretical distribution based on 1GM:1α:2β (○) and 2GM:2α:2β (■) models. (D) Titration of GM-CSF (0 to 7 μM) spiked with32P-labeled SGMKIN against a mixture of 3.5 μM sGMRα and 3.5 μM sβc. Reaction mixes were allowed to reach equilibrium at 25°C for at least 2 hours before being fractionated by size-exclusion chromatography. The distribution of radioactivity among ternary (●) and binary (○) complexes was determined and the radioactivity in each complex was expressed as a percentage of total bound counts where counts in TC plus counts in BC is 100%.
The use of radiolabeled GM-CSF also allowed us to investigate whether the presence of sβc in the ternary complex led to affinity conversion. GM-CSF spiked with the GM-CSF analog 32P-SGMKIN was titrated against an equimolar mixture of sGMRα and sβc, allowed to equilibrate, and fractionated by size-exclusion chromatography. For each GM-CSF concentration point, radioactivity bound in the binary and ternary complexes was determined and the proportion in each complex was expressed as a percentage of total bound counts. We found (Figure 6D) a 4-fold preferential distribution of32P-SGMKIN into ternary complexes at subsaturating concentrations of ligand, indicating that the presence of sβc in the ternary complex induces a measurable degree of affinity conversion.
GM-CSF binds sβc in the absence of sGMRα
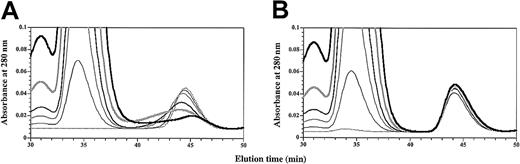
Initial chromatography experiments at approximately equimolar concentrations indicated that GM-CSF was unable to form a complex with sβc in the absence of sGMRα. However, close inspection of the elution profile of radiolabeled GM-CSF in the presence of free sβc (Figure 6B) revealed a modest decrease in the elution time of GM-CSF suggestive of a weak interaction between sβc and GM-CSF. To investigate this further and to determine the specificity of this interaction, we titrated sβc against GM-CSF or the E21R analog (Figure 7). Titration of sβc against GM-CSF had a dose-dependent effect on GM-CSF peak height with a concomitant spreading of the GM-CSF profile to earlier elution times (Figure 7A). Titration of sβc against E21R had no effect on E21R elution time or profile (Figure 7B). These results show that GM-CSF directly interacts with sβc through the functionally important Glu21 residue and that substitution of this residue makes a qualitative difference to the GMRα-independent recognition of βc by GM-CSF.
GM-CSF binds directly to βc.
Purified sβc was titrated (0 to 20 μM) against 5 μM GM-CSF (A) or 5 μM E21R (B). Reaction mixes were set up with 0 (dashed black), 1 μM (thin black), 2.5 μM (medium gray), 5 μM (medium black), 10 μM (thick gray), or 20 μM (thick black) sβc, incubated at 25°C for 2 hours, and fractionated by size-exclusion chromatography.
GM-CSF binds directly to βc.
Purified sβc was titrated (0 to 20 μM) against 5 μM GM-CSF (A) or 5 μM E21R (B). Reaction mixes were set up with 0 (dashed black), 1 μM (thin black), 2.5 μM (medium gray), 5 μM (medium black), 10 μM (thick gray), or 20 μM (thick black) sβc, incubated at 25°C for 2 hours, and fractionated by size-exclusion chromatography.
A second approach confirmed the direct interaction of GM-CSF with βc and extended these findings to the identification of the reciprocal region in βc. We incubated purified sβc with a 3-fold molar excess of GM-CSF or E21R, treated with the BS3cross-linker and analyzed the mixture by SDS-PAGE under reducing conditions (Figure 8). No covalent interactions between sβc and GM-CSF or E21R were observed in the absence of cross-linker (Figure 8A). GM-CSF and E21R were not dimerized by cross-linker under the conditions used, whereas the sβc dimer was partially cross-linked, yielding a band of MW 100 kDa (Figure 8B). Significantly, when GM-CSF was incubated with sβc and cross-linked, a unique band of MW 70 kDa was observed (Figure 8B). Western blotting with anti-βc or anti–GM-CSF antibodies showed that this band contains both sβc and GM-CSF (data not shown). E21R could not be cross-linked directly to sβc as seen by the absence of the 70 kDa band (Figure 8B), thus confirming that Glu21 of GM-CSF is necessary for direct contact with βc. Neither the structurally related cytokine human growth hormone nor sGMRα was able to be cross-linked to sβc under these conditions (data not shown).
Discrete regions in GM-CSF and βc mediate their direct interaction.
Purified sβc was incubated at 25°C for 1 hour alone or in the presence of either GM-CSF or E21R. Samples were left untreated (A) or were treated for 10 minutes with BS3 cross-linker (B). To determine if GM-CSF was interacting with the cytokine-binding site in the fourth domain of βc, purified sβc was preincubated with a Fab fragment of the neutralizing anti-βc mAb, BION-1, or the nonneutralizing control anti-βc mAb, 2H1. GM-CSF was then allowed to bind and the mixture and individual mAb treated with BS3cross-linker (C). Samples were analyzed on 12.5% (A) or 10% (B-C) SDS-PAGE gels and stained with Coomassie. The positions of molecular weight markers are shown in kilodaltons, and the position of sβc cross-linked to GM-CSF is indicated by ◂.
Discrete regions in GM-CSF and βc mediate their direct interaction.
Purified sβc was incubated at 25°C for 1 hour alone or in the presence of either GM-CSF or E21R. Samples were left untreated (A) or were treated for 10 minutes with BS3 cross-linker (B). To determine if GM-CSF was interacting with the cytokine-binding site in the fourth domain of βc, purified sβc was preincubated with a Fab fragment of the neutralizing anti-βc mAb, BION-1, or the nonneutralizing control anti-βc mAb, 2H1. GM-CSF was then allowed to bind and the mixture and individual mAb treated with BS3cross-linker (C). Samples were analyzed on 12.5% (A) or 10% (B-C) SDS-PAGE gels and stained with Coomassie. The positions of molecular weight markers are shown in kilodaltons, and the position of sβc cross-linked to GM-CSF is indicated by ◂.
To determine if the interaction between sβc and GM-CSF was occurring through a functionally relevant region of βc, we used BION-1, a mAb that blocks GM-CSF, IL-3, and IL-5 binding and signaling through βc.24 BION-1 recognizes a discrete region in the fourth domain of βc associated with high-affinity GM-CSF binding and function.20,22 36 Preincubation of sβc with BION-1 Fab fragment prevented GM-CSF from being cross-linked to sβc, as seen by the absence of the 70-kDa band (Figure 8C). The Fab fragment of a mAb that binds to the fourth domain of βc but does not block cytokine binding was unable to perturb the cross-linking of sβc to GM-CSF (Figure 8C).
Discussion
We report here the first demonstration of a fully assembled GM-CSF/GM-CSF receptor ternary complex in solution and describe the molecular interactions required for its formation. It is shown that the ternary complex exhibits a novel mode of cytokine receptor assembly that comprises 1 molecule of GM-CSF and 1 molecule of GM-CSF receptor α chain interacting monovalently with a noncovalently linked dimer of βc. In addition, a direct interaction between GM-CSF and βc in the absence of the receptor α chain could be demonstrated. The recruitment of βc as a preformed dimer may facilitate receptor activation and may also represent a mechanism utilized by the related IL-3 and IL-5 receptors. The GM-CSF ternary complex was demonstrated by gel filtration and sedimentation equilibrium analyses to have a molecular weight of between 135 kDa and 156 kDa, consistent with a GM-CSF/sGMRα/sβc stoichiometry of 1:1:2. In addition, the relative distribution of radiolabeled GM-CSF fitted a ternary complex with a 1:1:2 stoichiometry. The preferential distribution of radiolabeled GM-CSF into the ternary complex is indicative of sβc-mediated, affinity conversion. No disulfide linkages between receptor subunits were observed; there were no differences seen when the ternary complex was analyzed by SDS-PAGE under either reducing or nonreducing conditions or when the free cysteine groups in sβc were blocked with iodoacetamide (data not shown). These results are consistent with previous reports suggesting that GM-CSF receptor heterodimerization is required to activate the GM-CSF receptor,25 the dimeric nature of βc observed both on the cell surface and in solution,26,28,30 the affinity conversion afforded by βc,15,20 and the requirement of at least a βc dimer for function and activation of downstream signaling molecules.28,29 The intermediate binding affinity for GM-CSF in the ternary complex is consistent with a report describing the low-affinity binding of murine GM-CSF to detergent-solubilized GM-CSF receptors extracted from a murine cell line.37 In addition, these results do not rule out the formation of higher-order complexes on the cell surface,27,38 which may lead to further affinity conversion and disulfide linkage required for receptor stabilization, activation, or internalization purposes. The assembly of the human GM-CSF receptor shown here is different from that seen for the IL-639 and LIF40 receptors, which exhibit a stoichiometry of 2:2:2 and 1:1:1, respectively. Interestingly, the dynamics of the GM-CSF receptor assembly are analogous to the IL-6 receptor in that following the binding of ligand to the major binding subunit (α chain) there is recruitment of the signaling subunit (βc or gp130). However, although dimerization of gp130 requires a second IL-6/IL-6Rα chain binary complex, this is not the case with βc, which is recruited to a single GM-CSF/sGMRα binary complex as a preformed dimer. Despite the dimeric nature of sβc and even in the presence of a 2-fold molar excess of the GM-CSF/sGMRα binary complex, we saw no evidence for the formation of a ternary complex with a stoichiometry of 2:2:2. The functional monovalency of sβc may be due to conformational changes within the sβc dimer, induced by the binding of one GM-CSF/sGMRα binary complex that prevents the binding of a second binary complex.
The recruitment of sβc to the GM-CSF/sGMRα binary complex occurs through functionally relevant sites in GM-CSF and βc itself. This is demonstrated by the inability of the GM-CSF analog E21R to form the ternary complex and by the inhibition of sβc cross-linking to GM-CSF by the anti-βc mAb BION-1, which blocks the high-affinity binding of GM-CSF.24 Given that there is an homologous glutamic acid in IL-3 (position 22) and in IL-5 (position 13) and the fact that BION-1 also blocks high-affinity binding of IL-3 and IL-5, it is possible that this mode of receptor assembly will also apply to the IL-3 and IL-5 receptors. The recruitment of dimerized βc and associated JAK-2 molecules may facilitate receptor phosphorylation and activation in this subfamily of receptors. Using a soluble receptor system we could detect for the first time a direct interaction between GM-CSF and βc in the absence of the GM-CSF receptor α subunit. We observed this by gel filtration (Figure 7) and cross-linking studies (Figure 8). The interaction was sensitive to the E21R substitution and the mAb BION-1, indicating that the direct interaction observed between GM-CSF and sβc is chemically and spatially equivalent to the interaction that occurs with the cell membrane–anchored receptor. Considering that all βc-interacting cytokines do so through a chemically and structurally conserved mechanism,36 it is likely that a direct interaction between βc and IL-3 or IL-5 will also exist. The relative affinity of the direct βc interaction for each cytokine may help to explain differences in βc-mediated affinity conversion in the high-affinity binding of IL-3, GM-CSF, or IL-5. Despite the direct interaction of βc with GM-CSF seen in the soluble system, this may not be sufficient to activate the receptor in vivo given the very high concentrations of both receptor and ligand needed to detect this weak interaction (in the micromolar range) and the fact that GMRα intracellular domain has been previously shown to be crucial for GM-CSF signaling.41
In the IL-4 system, a high-affinity (Kd= 0.15 nM) interaction between IL-4 and the IL-4 receptor α chain42 utilizes a chemically and structurally homologous mechanism, suggesting that the type of direct interaction we observed between GM-CSF and βc may be conserved among other cytokines. The direct interaction we detected between GM-CSF and sβc also suggests that conformational changes in the GM-CSF/sGMRα binary complex may not be necessary for the recruitment of βc. However, the monovalent binding of the GM-CSF/sGMRα binary complex to sβc suggests the possibility of an induced conformational change within the extracellular domain of sβc. Conformational changes in the cytoplasmic region of βc may be induced by the assembly of the ternary complex to promote βc/JAK-2 proximity and receptor activation as shown for the erythropoietin receptor.43 The assembly of the human GM-CSF receptor system in solution described herein also provides a useful tool for investigating its dynamics and structural requirements. The initial event in activation of the GM-CSF receptor is the binding of ligand to the GMRα with low affinity prior to recruitment of βc. The soluble system used here revealed a 1:1 stoichiometry of binding between the sGMRα chain and GM-CSF with aKd equivalent to that seen with the full-length GMRα on the cell surface. We were able to show that E21R, a GM-CSF analog defective in high-affinity binding and a specific GM-CSF antagonist currently in phase 2 clinical trials, also binds sGMRα with a 1:1 stoichiometry. Importantly, E21R is incapable of forming a ternary receptor complex and when present in excess is able to prevent the formation of the ternary GM-CSF receptor complex, thus explaining its antagonistic activity. This set of experiments also demonstrates that the assembly of the GM-CSF receptor is a sequential process that involves first the formation of a binary complex. In structural terms it will be interesting to use single point mutants of βc to examine the residues that participate in direct contact with GM-CSF or the GM-CSF receptor α chain. This may be also a useful system for the identification of small molecules that prevent the formation of the ternary complex. Finally, the assembly of the human GM-CSF ternary complex in solution should aid in its crystallization and ultimately in the solving of its structure.
Prepublished online as Blood First Edition Paper, October 10, 2002; DOI 10.1182/blood-2002-06-1903.
Supported by grants from the National Health and Medical Research Council of Australia.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Angel Lopez, Cytokine Receptor Laboratory, Division of Human Immunology, IMVS, Frome Road, Adelaide, South Australia, 5000, Australia; e-mail:angel.lopez@imvs.sa.gov.au.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal