Abstract
Knowledge of the composition and interrelationship of the various hematopoietic stem cells (HSCs) that comprise the human HSC pool and the consequence of culture on each class is required for effective therapies based on stem cells. Clonal tracking of retrovirally transduced HSCs in nonobese diabetic/severe combined immunodeficient (NOD/SCID) mice revealed heterogeneity in the repopulation capacity of SCID-repopulating cells (SRCs). However, it is impossible to establish whether HSC heterogeneity is intrinsic or whether the culture conditions required for retroviral transduction induce qualitative and quantitative alterations to SRCs. Here, we report establishment of a clonal tracking method that uses lentivectors to transduce HSCs with minimal manipulation during overnight culture without cytokine stimulation. By serial bone marrow (BM) sampling of mice receiving transplants, short-term SRCs (ST-SRCs) and long-term SRCs (LT-SRCs) were identified on the basis of repopulation dynamics demonstrating that their existence is not an experimental artifact but reflects the state of the HSC pool. However, 4 days of culture in conditions previously used for SRC retroviral transduction significantly reduced SRC number as assessed by clonal analysis. These studies provide a foundation to understand the molecular and cellular determinants of human HSC development and to develop therapies targeted to specific HSC classes.
Introduction
One of the major problems limiting therapies based on stem cells, beyond simple transplantation of mononuclear or enriched CD34+ cells, is the absence of a clear understanding of the composition of the hematopoietic stem cell (HSC) pool in humans. Therapies must be targeted to the right cell. For gene therapy, HSCs capable of long-term repopulation must be transduced, whereas selection and expansion of HSCs that rapidly generate granulocytes, platelets, and erythroid cells might be able to reduce early posttransplantation morbidity that follows high-dose chemotherapy for cancer. Although most therapies under current development involve some form of HSC purification and ex vivo manipulation, it is impossible to rationally devise these procedures without a clear picture of the target cells and their growth requirements. Xenotransplantation of human cells into preimmune sheep1 or, more commonly, immune-deficient mice, provides powerful assay systems to characterize the HSC compartment.2-4 Cells capable of multilineage repopulation, termed severe combined immunodeficiency repopulating cells (SRCs),5-7 have been characterized with respect to frequency, cell surface phenotype, homing and adhesion properties, and cytokine responsiveness.2-4 Cell purification has implied that the human HSC compartment is composed of different classes with distinguishable phenotypic characteristics and proliferative and self-renewal properties.8-10 However, unlike the murine system, it has not yet been possible to obtain homogeneously pure fractions of human HSCs.11 Whereas cell surface markers can be used to prospectively isolate murine stem cell populations with defined developmental properties, single-cell or clonal analysis is still needed to determine whether heterogeneity in repopulation and self-renewal potential still exist in such putatively homogeneous populations due to stochastic elements; a proposition that has recently been observed in single-cell murine HSC transplants.12 Collectively, these studies suggest that a detailed view of the developmental and proliferative properties of the human HSC pool requires long-term dynamic analysis of individual SRCs rather than cell populations.
Currently, viral transduction is the only means of uniquely marking individual HSCs because each random integration site is a distinct clonal marker of the HSC progeny that arise after transplantation.13 We developed an optimized Moloney-based retroviral gene transfer protocol to examine the clonal composition of the transduced cells comprising the human graft during repopulation of nonobese diabetic/severe combined immunodeficient (NOD/SCID) mice.14 The human grafts were typically oligoclonal with substantial heterogeneity in the repopulation capacity and self-renewal potential of SRCs.14 These results implied that the human HSC compartment contained short-term (ST) and long-term (LT) SRCs. Although this study provided insight into the clonal composition of the human HSC compartment, a major unresolved question was whether the ex vivo manipulation required to clonally mark SRCs (eg, 4 days of stimulation with cytokine mixtures that promote cell division) had qualitative or quantitative effects on SRCs. Therefore, the existence of ST-SRCs and LT-SRCs could be an experimental artifact and not indicative of the natural state of the HSC pool. Resolution of this issue will affect our understanding of the human HSC developmental program and the establishment of stem cell–based therapies that involve ex vivo manipulation.
There are a considerable number of experimental and clinical studies on the ex vivo manipulation of HSCs.15-17 Although the conditions leading to significant expansion of hematopoietic progenitors are clear, HSC expansion is more controversial. Quantitative SRC assays have suggested either maintenance or modest expansion during ST culture (< 7 days),18 especially when cultures were initiated with purified Lin–CD34+CD38– cells.19,20 SRC expansion was also reported in longer term cultures using dilute concentrations of Lin–CD34+ cells.21,22 It is possible that low cell concentrations prevent negative feedback from mature cells that arise by differentiation. However, other murine and human studies have suggested that cell culture impairs HSC function via mechanisms that involve differentiation and alterations in cell cycle, homing, and migration.23-30 Human clinical trials represent a definitive test for HSC function; however their results are complicated by the fact that it was impossible to track the fate of expanded cells because autologous unmanipulated cells were coinjected.15 Examination of the engraftment kinetics suggests that there may be expansion of repopulating cells that facilitate early neutrophil recovery, but expanded HSCs were not able to sustain long-term engraftment. Moreover, in several gene therapy trials that use ex vivo culture for retroviral transduction, only low multilineage reconstitution of marked cells occurred unless there was an in vivo selective advantage for the corrected lineage, such as in SCID patients.31,32 The mechanism that underlies the poor maintenance or expansion of LT-HSCs is not clear but may be due to their differentiation into ST-HSCs and committed progenitors or some other impairment of their function.
The resolution of both the nature of the intrinsic properties of the human HSC pool and the consequence of cell culture on HSC expansion requires a method to clonally mark SRCs with minimal ex vivo manipulation and analysis of the repopulation properties of individual HSCs. Lentivectors provide such a solution by efficiently and rapidly transducing SRCs in the absence of cytokine and serum stimulation.26,33,34 We report here the clonal tracking of individual SRCs in NOD/SCID mice before and after culture.
Materials and methods
Sample collection and purification
Samples of cord blood (CB) were obtained according to procedures approved by the institutional review boards of University Health Network and Trillium Hospital. CB samples were collected, purified, and stored as described previously.14 The efficiency of primitive CD34+ cell enrichment, as determined by flow cytometric assessment, was 75% to 95%, except for 1 experiment, which was 38%.
Flow cytometry
Flow cytometric analysis was performed using a FACScalibur (Becton Dickinson, San Jose, CA). Isotype controls were mouse immunoglobulin G conjugated to fluorescein isothiocyanate (FITC), phycoerythrin (PE), both from Becton Dickinson, and PE-cyanine 5 (PC5; Coulter, Fullerton, CA). The CD34 and CD38 expression characteristics of CB cells were assessed using anti–CD34-PC5 with anti–CD38-PE. Human cells from NOD/SCID mice with transplants were assessed using anti–CD45-PC5, anti–CD33-PE or PC5, anti–CD19-PE or PC5 and anti–CD36-PE antibodies. Enhanced green fluorescent protein (EGFP) fluorescence was detected using detector channel FL1 calibrated to the FITC emission profile.
Lentivirus production
Third-generation lentiviral vector containing the central polypurine track (cppt), human phosphoglycerate kinase (huPGK) promoter, EGFP gene, and the woodchuck hepatitis virus postregulatory element (WPRE) was designated CEW.35 Viral supernatants were generated by transient transfection of 293T cells with packaging plasmids and pseudotyped with the vesicular stomatitis virus G protein (VSV-G protein) as previously described.26 High-titer viral stocks were prepared by ultracentrifugation. The functional titers of CEW viral vectors determined by infection of HeLa cells were 3 × 108 to 2 × 109 infectious units (iu)/mL.
Infection and culture protocol
Viral infection was performed as described26 with minor modifications. Briefly, Lin– CB cells were thawed and incubated overnight in serum-free X-VIVO 10 (BioWhittaker, Walkersville, MD) medium supplemented with 1% bovine serum albumin (StemCell Technologies, Vancouver, BC, Canada), 2 mM l-glutamine, and CEW viral supernatants (multiplicity of infection [MOI], 50-150). The viral concentrations were at about 108 iu/mL and cell concentrations at about 106 cells/mL. Half of the transduced cells (referred as minimally cultured cells) were immediately injected into NOD/SCID mice at the dose of 2.19 ± 0.18 × 105 cells (equivalent to 1.55 ± 0.13 × 105 Lin–CD34+ cells; 13 experiments). The other half of the transduced cells were cultured for an additional 3 days with CH-296 fibronectin fragment (Retronectin; Takara Shuzo, Otsu, Japan) under conditions using a cytokine mixture previously described.14 Following culture (viability > 94% and average cell expansion 3.54 ± 0.45-fold), cells were injected into the same number of mice as the minimally cultured cells. The number of expanded cells injected per mouse was 8.24 ± 0.78 × 105 (equivalent to 5.88 ± 0.64 × 105 Lin–CD34+ cells).
Intravenous NOD/SCID mouse repopulation
Transduced cells were transplanted into sublethally irradiated (3.6 Gy) 8- to 12-week old NOD/SCID mice using our standard intravenous protocol for CB cells.14,36 At various intervals after transplantation, BM samples were aspirated from the femur as previously described.14,37 At the end of experiment, mice were killed and peripheral blood (PB), spleen, and cells from the previously aspirated femur or combined BM from both tibia, left femur, and pelvis were analyzed by flow cytometry. The human graft was assessed by flow cytometry for nucleated living cells and myeloid (CD45+CD33+), B lymphoid (CD45+CD19+), and erythroid (CD45–CD36+) cells expressing EGFP.
Intrafemoral transplantation into NOD/SCID mice
The intrafemoral injection procedure was performed as described38,39 with minor modifications.40 Briefly, after mouse anesthesia with a 2.5% avertin solution, the knee was flexed and 25 μL cells were injected with a 28.5-gauge needle through the joint into the right femur (also called injected femur). Lin–CD34+ CB cells were thawed and one half of the cells were transplanted intrafemorally into NOD/SCID mice immediately (day-0 cells); the other aliquot was subjected to 4 additional days (day-4 cells) of ex vivo culture using conditions with cytokines described previously14 and injected into mice. Mice were killed 3 and 6 weeks after transplantation. Cells from the injected right femur, as well as right tibia, left femur, spleen, and PB were analyzed by flow cytometry.
Progenitor assays
Colony-forming cell (CFC) assays of transduced cells and cells derived from engrafted mice were performed as previously described.6,41 Individual granulocytic-monocytic or erythroid colonies from the aspirated femurs at various time points after transplantation were counted at day 12 and collected for subsequent polymerase chain reaction (PCR) or flow cytometry analysis.
PCR analysis
The presence of GFP in individual colonies was determined as previously described14 with modifications. Briefly, the DNA from colonies was extracted by incubating the cells at 50 to 60°C for 1 to 2 hours in 20 μL extraction buffer (50 mM KCl, 10 mM Tris [tris(hydroxymethyl)aminomethane], pH 8.3, 2.5 mM MgCl2, 0.1 mg/mL gelatin, 0.45% Nonidet P40, 0.45% Tween 20, and 120 μg/mL proteinase K). Proteinase K was then inactivated by heating at 95°C for 10 minutes. Ten percent of the reaction mixture was subsequently used for the PCR.
Analysis of the integration sites of retrovirally marked SRCs
The gDNA from BM and spleen of experimental mice was extracted and up to 15 μg was digested with BamHI, which cuts once within the provirus upstream of the EGFP gene. Because the cellularity of BM aspirates was sometimes limiting, in some cases as little as 4 μg was used. Following electrophoresis and transfer to a nylon membrane (Hybond N+, Amersham, Buckinghamshire, England), DNA was hybridized with a GFP probe. Determination of bands, their size, and their kinetics at multiple time points was carried out in a blinded fashion by 4 individuals and was verified by densitometry, which we used to quantify the proliferative capacity of individual clones. To quantify the relative intensity of each band, densitometric analysis was done using ImageQuant software (Molecular Dynamics, Piscataway, NJ). With this method, only clones that produced at least 104 cells when analyzed at 12 weeks could be detected.
Analysis of the presence of the transgene by dot blot
Dot blot analysis of the gDNA extracted from BM and spleen of mice injected with transduced cells was performed as described.14 Different proportions of DNA from nontransduced murine cells and from a clone of the MGIN producer cell line containing a single provirus integration were mixed and used as standards to generate a titration curve. Digestion with EcoR1 and HindIII, which cut outside of the EGFP gene, was used to confirm that the MGIN clone contained only a single proviral insertion. The quantity of total DNA (donor human and recipient murine) immobilized onto individual dots was normalized using a probe for the single-copy human chx10 gene. Human chx10 has more than 92% identity to the murine gene and therefore detects both human and murine DNA. To determine the proportion of transduced human cells within the engrafted NOD/SCID BM, the blots were hybridized with a GFP probe. Densitometric scans followed by ImageQuant (Molecular Dynamics) analysis was used to determine the ratio of proviral DNA to total loaded DNA. Titration curves of these ratios were generated for known standards and used to estimate proviral content of the individual DNA samples.
Statistical analysis
Data are presented as means ± SE. The significance of differences between groups was determined by using the Student t test, Mann-Whitney test, and one-way analysis of variance (ANOVA) (SigmaStat software; Jandel, Chicago, IL).
Results
Efficient lentivector transduction of SRCs
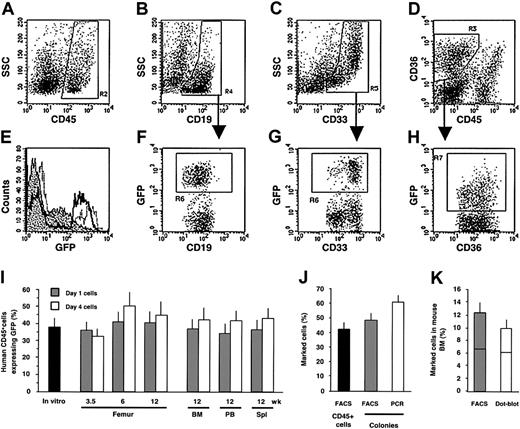
To determine the clonal composition of the minimally manipulated HSC pool and the consequences of ex vivo culture on SRCs, Lin–CD34+ cells were exposed overnight to lentiviral supernatant. Unlike retroviruses, lentivectors efficiently transduce human Lin–CD34+ CB cells in serum-free medium without cytokine stimulation.26 Although cytokines can increase gene transfer efficiency, we avoided their use because cytokine stimulation during transduction increases the probability of multiple integrations per SRC.42 In addition, to minimize the probability of multiple integrations into target cells, we also chose transduction conditions that would achieve approximately 40% gene transfer (Figure 1I). Immediately after transduction (day 1), half of the cells were transplanted into NOD/SCID mice, whereas the balance was subjected to 3 additional days of culture (day 4). The culture conditions were previously optimized for the short-term maintenance and putative expansion of SRCs for our retroviral transduction experiments.14 The efficiency of gene transfer into SRCs was determined for each group of mice at 3.5 and 6 weeks after transplantation by examination of BM aspirates from the same femur, and at the end of the experiment in the aspirated femur, BM (which included cells from nonaspirated femur, tibia, and pelvis), PB, and spleen. Flow cytometry analysis was performed to determine the identities of engrafted human cells including lymphoid (CD45+CD19+; Figure 1B region R4), myeloid (CD45+CD33+; Figure 1C region R5), and erythroid (CD45–CD36+; Figure 1D region R3) lineages, and their expression of GFP (Figures 1E-H). Although a lower level of expression was observed in erythroid cells (Figure 1E, H region R7) compared to myeloid and lymphoid cells (Figure 1E-G region R6), an equivalent proportion of GFP-expressing cells was detected in all lineages (data not shown). Moreover, similar percentages of transduced human cells were observed at all time points in the BM aspirates and in the hematopoietic organs of the mice receiving transplants 12 weeks earlier with either day-1 or day-4 cells, indicating efficient and stable transduction of all repopulating cells (Figure 1I). Comparable frequencies of GFP expression were also observed in human clonogenic progenitors identified at various time points in mouse BM (Figure 1J) confirming efficient transduction of primitive cells that maintain both the progenitor pool and the differentiated cell lineages. Analysis of provirus integration in these colonies by PCR showed that most of the progenitors (79.7%) that contained the provirus also expressed the transgene, indicating that the transgene was not silenced in the progeny of SRCs (Figure 1J), a problem that was observed with older generations of retroviruses.16
Efficiency of lentiviral transduction into SRCs. (A-H) Multilineage engraftment of human cells in the femur 3.5 weeks after intravenous transplantation into the NOD/SCID mice assessed by flow cytometry. The region 2 (R2) for the CD45+ cells (A) was set and used for the assessment of the CD19+ lymphoid (B, R4) and CD33+ myeloid (C, R5) cells. (D) The region R3 was set to define the CD45–CD36+ erythroid population. The proportion of GFP-expressing cells was determined for lymphoid (F, R6), myeloid (G, R6) and erythroid (H, R7) lineages. (E) GFP expression levels for erythroid (dotted area), lymphoid (black line), and myeloid (gray line) cells are compared. (I) Proportion of GFP+ cells estimated by flow cytometry at day 4 of culture (▪). Kinetics of the contribution of gene-marked cells within the human grafts in the aspirated femur and at the end of experiment in BM, blood (PB), and spleen (Spl) following injection of noncultured cells (day 1, ▦) or cultured cells (day 4, □). Each bar represents the mean (± SE) data of 13 experiments and corresponds to 28 (day 1) and 33 (day 4) mice analyzed. (J) Proportion of the GFP+ cells within the human CD45+ grafts (▪) and percentage of GFP+ human colonies (▦) derived from the same grafts were assayed by flow cytometry. Presence of the provirus in the same colonies was determined by PCR (□). Each bar represents the mean (± SE) data of 4 experiments; 217 colonies obtained from 16 mice given transplants of either day 1 or day 4 cells were analyzed. (K) Percentage of GFP+ cells determined by flow cytometry (▦) and proportion of the same cells carrying the provirus analyzed by dot blot (□) in hematopoietic organs at 12 weeks after transplantation. Each bar represents the mean (± SE) data of 80 samples from 32 mice (8 experiments). Horizontal lines are the median value.
Efficiency of lentiviral transduction into SRCs. (A-H) Multilineage engraftment of human cells in the femur 3.5 weeks after intravenous transplantation into the NOD/SCID mice assessed by flow cytometry. The region 2 (R2) for the CD45+ cells (A) was set and used for the assessment of the CD19+ lymphoid (B, R4) and CD33+ myeloid (C, R5) cells. (D) The region R3 was set to define the CD45–CD36+ erythroid population. The proportion of GFP-expressing cells was determined for lymphoid (F, R6), myeloid (G, R6) and erythroid (H, R7) lineages. (E) GFP expression levels for erythroid (dotted area), lymphoid (black line), and myeloid (gray line) cells are compared. (I) Proportion of GFP+ cells estimated by flow cytometry at day 4 of culture (▪). Kinetics of the contribution of gene-marked cells within the human grafts in the aspirated femur and at the end of experiment in BM, blood (PB), and spleen (Spl) following injection of noncultured cells (day 1, ▦) or cultured cells (day 4, □). Each bar represents the mean (± SE) data of 13 experiments and corresponds to 28 (day 1) and 33 (day 4) mice analyzed. (J) Proportion of the GFP+ cells within the human CD45+ grafts (▪) and percentage of GFP+ human colonies (▦) derived from the same grafts were assayed by flow cytometry. Presence of the provirus in the same colonies was determined by PCR (□). Each bar represents the mean (± SE) data of 4 experiments; 217 colonies obtained from 16 mice given transplants of either day 1 or day 4 cells were analyzed. (K) Percentage of GFP+ cells determined by flow cytometry (▦) and proportion of the same cells carrying the provirus analyzed by dot blot (□) in hematopoietic organs at 12 weeks after transplantation. Each bar represents the mean (± SE) data of 80 samples from 32 mice (8 experiments). Horizontal lines are the median value.
In addition, comparison of GFP expression determined by flow cytometry in hematopoietic tissues at 12 weeks and DNA analysis of proviral content by dot blot did not show any significant differences in the proportion of marked cells (Figure 1K). These data confirm that there is no silencing of the transgene delivered by lentivectors into SRCs responsible for long-term repopulation. Moreover, because the same proportion of GFP+ cells was determined by quantitative proviral DNA analysis and by flow cytometry we can conclude that there was on average a single proviral integration per SRC (Figure 1K).
These data indicate that lentivectors, with minimal ex vivo manipulation, are able to rapidly and efficiently transduce SRCs capable of both short-term and long-term engraftment. Moreover, the stability of the contribution that gene marked cells make to the human graft contrasts with the overall decline we observed in most mice given transplants of Moloney-based retrovectors.14 This suggests that the 2 different transduction conditions may result in differences in the number of SRCs that survive or in the transduction of different ratios of ST-SRCs and LT-SRCs.
Ex vivo culture affects SRCs
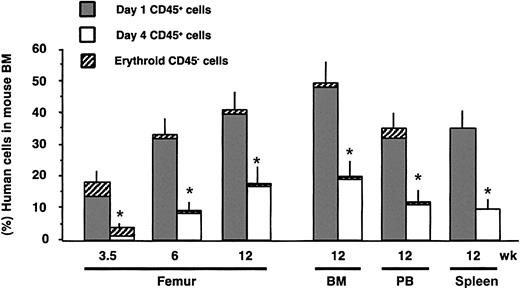
To examine the repopulation dynamics of SRC derived from day-1 and day-4 cells, we evaluated the proportion of engrafted human cells in the murine hematopoietic tissues at different times after transplantation. In contrast to the equivalent proportions of gene-marked cells within the human cell population (Figure 1I), there was a significant difference in the overall levels of human cell engraftment between the 2 groups (Figure 2). The minimally cultured day-1 cells promptly engrafted the murine BM generating graft kinetics previously reported for uncultured CB SRCs.43 By contrast, the cells subjected to ex vivo culture showed delayed engraftment kinetics. Even at 3 months after transplantation, the average level of human cell engraftment for the day-4 cells was only half that obtained with day-1 cells. The reduced level of human cell engraftment observed in the BM aspirates was recapitulated within other hematopoietic tissues including the whole BM, spleen, and PB after 12 weeks (Figure 2), indicating that the reduced engraftment level was not due to increased egress of the engrafted human cells from the BM. These data suggest that culture reduces the number of SRCs and/or impairs their proliferative capacity. The erythroid, myeloid, and lymphoid lineage development in the grafts initiated by day-1 and day-4 cells was similar, indicating that there is no impairment in the differentiation capacities of cultured SRCs, despite the reduced overall engraftment. In addition, gene-marked cells were able to repopulate all 3 hematopoietic lineages in the same manner as unmarked cells (data not shown).
Kinetics of human engraftment. Each bar represents the mean percentage of human engraftment in the periodically sampled BM of mice and in the hematopoietic tissues. Human grafts from the cohort of mice described in Figure 1I were analyzed by flow cytometry and depicted as the percentage of nonerythroid CD45+ (▦, □) plus erythroid CD45–CD36+ (▨) cells. *P < .05 for day 1 versus day 4.
Kinetics of human engraftment. Each bar represents the mean percentage of human engraftment in the periodically sampled BM of mice and in the hematopoietic tissues. Human grafts from the cohort of mice described in Figure 1I were analyzed by flow cytometry and depicted as the percentage of nonerythroid CD45+ (▦, □) plus erythroid CD45–CD36+ (▨) cells. *P < .05 for day 1 versus day 4.
To reduce mouse-to-mouse variability and to test the repopulation capacity of noncultured and cultured cells in the same microenvironment, we developed a dual-engraftment assay in which 2 experimental populations were transduced with lentivectors encoding either the GFP or red fluorescent protein. The results from these studies confirmed the data in Figure 2 and showed that even within the same mouse, engraftment was mostly derived from minimally cultured cells (data not shown).
Clonal contribution and proliferative capacity of individual SRCs
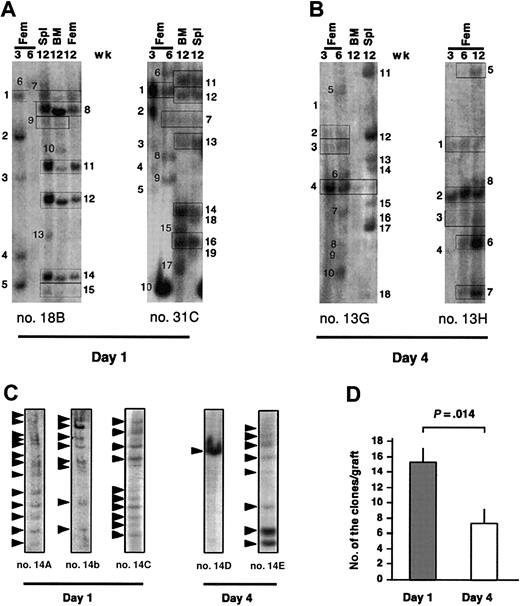
To gain insight into the basis of the decreased repopulation induced by culture, it is necessary to determine the specific fates of individually transduced SRCs from the day-1 and day-4 cells. We performed clonal analysis by Southern blot on cells from serially aspirated femurs and in the hematopoietic tissues at 12 weeks after transplantation from the experiments described in Figures 1 and 2. We have made the assumption that most of the bands represent the clonal descendants of uniquely marked SRCs because each band represents a unique lentiviral integration site (Figure 3A-C) and the dot blot analysis (Figure 1K) indicated that on average there was one insertion per SRC. However, it must be noted that Southern analysis will only detect major clones of transduced cells that generate at least 104 cells.
Southern blot analysis for provirus integration sites within the femur, BM, and spleen of NOD/SCID mice engrafted with noncultured and cultured lentivector-transduced cells. Southern blot analysis of the clonal kinetics in mice engrafted with noncultured (A) and cultured (B) cells. Lanes labeled Fem were loaded with DNA from a femur aspirate at the indicated time points, whereas BM and spleen (Spl) lanes contained DNA from these tissues at 12 weeks. Each unique band has been numbered. The presence of the same-sized bands at multiple time points and different hematopoietic tissues is denoted by rectangles. (C) Comparison of the clonal composition within the BM of mice given transplants of day-1 and day-4 cells from the same CB sample and analyzed at 12 weeks. (D) Total number of clones present in femur, BM, and spleen at 12 weeks after transplantation. Each bar represents the mean data (± SE) from the mice (n = 9 for day 1 and n = 6 for day 4) where the number of clones could be determined in all hematopoietic tissues.
Southern blot analysis for provirus integration sites within the femur, BM, and spleen of NOD/SCID mice engrafted with noncultured and cultured lentivector-transduced cells. Southern blot analysis of the clonal kinetics in mice engrafted with noncultured (A) and cultured (B) cells. Lanes labeled Fem were loaded with DNA from a femur aspirate at the indicated time points, whereas BM and spleen (Spl) lanes contained DNA from these tissues at 12 weeks. Each unique band has been numbered. The presence of the same-sized bands at multiple time points and different hematopoietic tissues is denoted by rectangles. (C) Comparison of the clonal composition within the BM of mice given transplants of day-1 and day-4 cells from the same CB sample and analyzed at 12 weeks. (D) Total number of clones present in femur, BM, and spleen at 12 weeks after transplantation. Each bar represents the mean data (± SE) from the mice (n = 9 for day 1 and n = 6 for day 4) where the number of clones could be determined in all hematopoietic tissues.
Examination of the clonal composition of the BM of mice given transplants of day-1 cells showed that several clones were present at the 3.5-week point, which disappeared or fell below detectable limits at later time points (Figure 3A; eg, mouse 18B, clones 2-5). Other clones appeared at 6 weeks and were gone by 12 weeks (Figure 3A; eg, mouse 18B, clone 6), whereas others did not appear until 12 weeks (Figures 3A; eg, mouse 18B, clones 8-15). Finally, some clones persisted throughout the experiment (Figure 3A; eg, mouse 18B, clone 1). Cultured cells showed a similar pattern of clonal kinetics (Figure 3B). Therefore, we can conclude that SRC heterogeneity exists in the natural state of the stem cell compartment and is not an artifact of cell culture. Our model demonstrates the existence of ST-SRCs that function shortly after transplantation and then disappear, as well as LT-SRCs, which contribute to the graft throughout the experiment or become functional at the later time points. It is not possible to determine the precise ratio of ST-SRC and LT-SRC in the original SRC pool because the BM aspiration method only assesses a limited portion of the total hematopoietic compartment at the early time points.
The Southern blot analysis also revealed a striking difference in the number of clones contributing to long-term human cell engraftment of mice given transplants of day-1 or day-4 cells. The Southern blot analysis of BM from representative mice given transplants of day-1 cells showed the presence of 8 to 13 bands, whereas similar analysis of BM of mice given cultured cells originating from the same sample transduced during the common infection procedure showed 1 to 8 bands (Figure 3C). Of the 15 mice in which the data were available for both cultured and noncultured cells there were on average 15.2 ± 1.8 bands for day-1 versus 7.2 ± 2.0 bands for day-4 cells (Figure 3D). These results provide direct clonal evidence that ex vivo culture markedly reduces the number of LT-SRCs that participate in the repopulation of NOD/SCID mice.
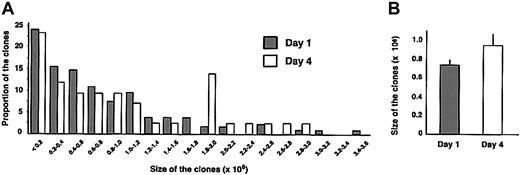
The proliferative capacity of individual SRCs (determined at 12 weeks after transplantation as described in Figure 4) was highly variable (Figure 4A), ranging from less than 2 × 105 cells to more than 3 × 106 cells. The distribution pattern of the clonal size for day-1 and day-4 cells was similar. Although there was a tendency toward an increased proliferative capacity of cultured LT-SRCs, the average size of the clones produced by minimally manipulated or day-4 cultured SRCs was not significantly different (Figure 4B). Thus, in parallel to longevity of repopulation of individual SRCs, these data indicate that significant heterogeneity exists in the proliferative capacity of each SRC. Moreover, the proliferative capacity of the SRCs that are recovered following culture was not impaired compared to the uncultured group.
Analysis of the proliferative capacity of individual clones. The proliferative potential of each unique or common clone detected in femur, BM, and spleen from Figure 3D was estimated by using densitometry analysis of Southern blots. The approximate cellularity of the day 1 (n = 137) and day 4 (n = 43) marked clones was calculated from the intensity of each band, divided by the combined intensities of all bands in a single lane loaded with DNA from the hematopoietic tissue, and multiplied by the number of GFP+ cells collected from this tissue. (A) The proportion of clones corresponding to each clonal size is indicated. (B) Each bar represents the average size (± SE) of the clones from the mice given transplants of day-1 or day-4 cells from 7 experiments.
Analysis of the proliferative capacity of individual clones. The proliferative potential of each unique or common clone detected in femur, BM, and spleen from Figure 3D was estimated by using densitometry analysis of Southern blots. The approximate cellularity of the day 1 (n = 137) and day 4 (n = 43) marked clones was calculated from the intensity of each band, divided by the combined intensities of all bands in a single lane loaded with DNA from the hematopoietic tissue, and multiplied by the number of GFP+ cells collected from this tissue. (A) The proportion of clones corresponding to each clonal size is indicated. (B) Each bar represents the average size (± SE) of the clones from the mice given transplants of day-1 or day-4 cells from 7 experiments.
Repopulation by intrafemoral transplantation
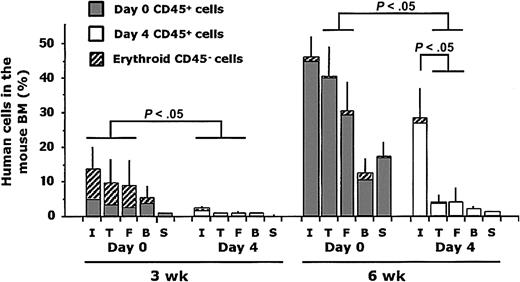
Recently, a novel method of xenotransplantation was developed that relies on the direct delivery of hematopoietic cells into the BM cavity of the femur40 or tibia.38,39 This method results in an increased sensitivity of SRC detection in comparison with the traditional intravenous assay.38 Intrafemoral injection was performed to test whether the detection of SRCs from the day-4 cultured cells could be augmented. Lin– CB cells were injected immediately after thawing (day 0) or after 4 additional days of culture under conditions using cytokines as described.14 At 3 and 6 weeks after transplantation, mice were killed and PB, spleen, and BM from the right tibia, left femur, and injected right femur were analyzed by flow cytometry for the presence of human cells. Examination of the kinetics of human cell engraftment in the different bones or tissues of mice given transplants of noncultured (day 0) and cultured (day 4) cells revealed significant differences between the repopulation patterns (Figure 5). The grafts in the mice given transplants of day-0 cells followed similar kinetics as those in mice undergoing intravenous transplantation with minimally manipulated cells (Figure 2). After intrafemoral injection, human cells engrafted the injected femur and the BM of noninjected bones and cells were found in the circulation indicating spread of cells from the injected femur (Figure 5). By contrast, cultured cells provided low engraftment at 3 weeks after transplantation in all the hematopoietic tissues paralleling intravenous delivery. At 6 weeks, the human grafts in the injected femur derived from cultured SRCs were not significantly smaller than those from day-0 cells. However, the migration of human cells from the injected femur was significantly impaired in mice given cultured cells leading to a decreased proportion of human cells in these recipients. The absence of migration from the injected femur suggests that the repopulating cells themselves sustain an unknown functional impairment of homing and migration as a consequence of culture. Alternatively, culture could induce differentiation, thereby generating a population of somewhat more committed repopulating cells that still retain proliferative and differentiation potential but have lost homing and/or self-renewal properties associated with primitive cells.
Human cell engraftment following intrafemoral delivery of noncultured and cultured cells. Human engraftment was measured at 3 and 6 weeks after transplantation as the percentage of nonerythroid CD45+ (▤, □) plus erythroid CD45–CD36+ (▨) human cells in the injected right femur (I), the right tibia (T), the left femur (F), the blood (B), and the spleen (S). Each bar represents the mean data (± SE) from the mice given transplants of day-0 (n = 24) or day-4 cells (n = 24) from 4 experiments.
Human cell engraftment following intrafemoral delivery of noncultured and cultured cells. Human engraftment was measured at 3 and 6 weeks after transplantation as the percentage of nonerythroid CD45+ (▤, □) plus erythroid CD45–CD36+ (▨) human cells in the injected right femur (I), the right tibia (T), the left femur (F), the blood (B), and the spleen (S). Each bar represents the mean data (± SE) from the mice given transplants of day-0 (n = 24) or day-4 cells (n = 24) from 4 experiments.
Discussion
In this report, we have used lentivector-mediated clonal tracking to establish that the human HSC pool, in its native state, is functionally heterogeneous and comprises individual stem cells with differing lifespan and proliferative potential. Transduced ST-SRCs contributed rapidly to the graft following transplantation, but disappeared at later time points. By contrast, LT-SRCs persisted or expanded over the time of analysis or appeared at the end of the analysis. The designation of LT-SRC is provisional because it is unknown if long-term clones persist beyond 12 weeks or if new clones continuously appear. Future studies using modified strains of NOD/SCID mice that permit human repopulation for 1 year or more will help resolve this issue. A critical element of our analyses is the minimal manipulation of HSCs permitted by lentivector transduction, in contrast to retroviral transduction, which requires several days of culture to force quiescent HSCs into cycle.44 Therefore the functional SRC heterogeneity demonstrated here is likely to be more representative of the native state of the human HSC compartment rather than an experimentally induced artifact of the clonal marking method itself.
A recently reported limitation of lentivectors for clonal tracking is the possibility of more than one proviral integration per SRC.42,45 However, these studies used high vector-to-target cell ratios and cytokine stimulation to increase proviral integration frequency. We chose a strategy predicted by Poisson statistics to achieve approximately 40% transduction (without cytokines) so that on average there would be a single integration per cell. Quantitative analysis for proviral content within the total human cell population detected an average of one integration per SRC, validating the use of lentivectors for clonal tracking. It should be noted that even if there is more than one integration per SRC, the clonal dynamics of SRC function can still be determined because the presence and disappearance of bands were observed. However, this situation could mean that the precise number of SRCs would be overestimated because each band would not correspond to a single SRC.
Although cell culture did not alter the intrinsic heterogeneity of the HSC pool, there was a negative effect on HSC number. Comparison of minimally manipulated cells with those thought to be optimized for stem cell maintenance, expansion, and retroviral transduction revealed significant reductions in SRC number as a consequence of 4-day culture. Clonal tracking of individual SRCs demonstrated that twice as many SRCs participated in long-term engraftment of mice given transplants of minimally cultured cells compared to day-4 cultured cells. Additionally, minimally cultured cells engrafted promptly and with the same kinetics as fresh CB cells, whereas cells cultured for 4 days showed delayed engraftment. Only half the level of engraftment was achieved, despite more than 3-fold expansion of cultured cells in vitro. Delayed and reduced levels of engraftment from the cultured group were also observed in a dual repopulation assay, where differentially marked minimally cultured and cultured cells were injected into the same mouse. These studies provide conclusive evidence that the number of both ST-SRCs and LT-SRCs differs due to short-term ex vivo culture.
Culture conditions similar to those used here were previously shown to increase the number of CD34+CD38– cells and CFCs by 4- and 10-fold, respectively.19 Unfortunately, knowledge of how such conditions affect true HSCs is more limited. We and others have found similar conditions to result in either maintenance or a 2- to 4-fold increase in SRC number after 4 to 7 days of culture as quantified by limiting dilution analysis (LDA).18-20 However, in the context of expansion studies it is important to note that LDA is a statistical method that relies on a detection threshold making it difficult to distinguish between increased SRC proliferative potential following culture (and therefore increased human engraftment detection) or a truly increased SRC number. Our study is the first to address this question at the clonal level using the SRC assay. Also SRC expansion could be affected by subtle culture differences between our data and other studies reporting expansion, including highly purified SRC fractions, low cell concentrations that reduce feedback inhibition from mature cells, and differing cytokine combinations and dosages. Human gene-marking trials provide the only true insight into the consequence of cell culture on HSCs capable of repopulating humans. Although many patients received transplants with retrovirally transduced cells, the overall results were disappointing because only low levels of marked cells were detected during long-term analysis.31 However, it is difficult to distinguish whether HSC transduction efficiency was low or if transduced HSCs were negatively influenced by the requisite cell culture. Recent SCID gene therapy trials used BM and transduction conditions similar to ours. Although lymphoid lineages were highly marked, the corresponding myeloid cells were very low, indicating selective growth of marked lymphoid cells.32,46 In most recent trials, the level of gene transfer in the initial inoculum was high, suggesting that gene-marking efficiency is not the limiting problem. Thus, the reduced SRC numbers we observed using the NOD/SCID model predict that the low level of marking in patients is at least partially attributable to the loss of LT-HSCs during the culture required for retroviral transduction.
Although reduced SRC number appears to be the most direct consequence of short-term culture, the intrafemoral assay revealed a potential qualitative impairment of SRC function. It appears that SRCs are measured more sensitively using the intrafemoral assay due to the direct delivery to the BM cavity.38 In addition, novel SRC classes are detected that do not engraft efficiently if injected by the traditional intravenous route perhaps due to absent homing or migration properties that normally permit circulation after intravenous injection, extravasation through the vascular epithelium, and engraftment of the BM microenvironment.39,40 Whereas injection of cultured cells led to lower engraftment in the injected femur at the 3-week time point compared to noncultured cells (presumably due to reduced numbers of ST-SRCs), by 6 weeks the engraftment levels in the injected femurs were similar. However, the most significant difference was the absence of migration from the injected femur to other bones in the cultured group. These results are difficult to interpret. Using clonal analysis, the same clone can be found in the injected bone and other sites suggesting that the injected SRCs self-renewed soon after transplantation and then migrated to another hematopoietic tissue and began to generate a graft at that site.40 If this suggestion is accurate, in the experiments reported here, it is possible that culture generates a more committed repopulating cell that engrafts poorly if injected intravenously, but it will proliferate extensively if injected directly into the BM cavity. Moreover, these committed repopulating cells would have also lost their ability to self-renew and migrate in vivo following intrafemoral injection. However, we have found that the proliferative potential of the SRCs that survive culture seems to be normal. Thus, an alternate hypothesis would be that cell culture caused impairment of some migration or homing properties of the repopulating cells that continued to manifest itself for several weeks. Overall, these studies indicate that caution must be exercised in extending the duration of HSC expansion cultures prior to transplantation and they demonstrate the importance of a functional assay coupled with clonal analysis in the development of culture conditions that support primitive cells.
Assessment of the biologic function of individual human HSCs is a powerful tool for the characterization of cellular and molecular determinants that govern their developmental program. Moreover, the establishment of highly efficient and reproducible procedures for the transduction of repopulating cells with high proliferative, differentiation, and self-renewal potential will be of substantial benefit to human gene therapy applications. Most current and future therapies based on stem cells will continue to require some form of ex vivo manipulation. For example, expansion of long-term repopulating HSCs would increase the utility of CB samples that currently do not contain enough HSCs to transplant into adult patients. In addition, some cancer therapies would be aided by the expansion and transplantation of HSCs capable of rapidly generating large numbers of myeloid and erythroid cells. Our data provide a foundation to develop procedures for the specific expansion of different classes of human HSCs depending on the clinical need.
Prepublished online as Blood First Edition Paper, September 22, 2003; DOI 10.1182/blood-2003-05-1558.
Supported by grants from the Association pour la Recherche contre le Cancer (F.M.), the Stem Cell Network of National Centres of Excellence (F.M. and J.E.D.), and the National Cancer Institute of Canada (NCIC) with funds from the Canadian Cancer Society, the Canadian Genetic Diseases Network and the Stem Cell Network of the National Centers of Excellence, the Canadian Institutes for Health Research, and a Canada Research Chair (J.E.D.). F.M. and O.I.G. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank P. Savage (Trillium Hospital) for providing CB samples and Ashley R. Cox for her excellent technical assistance.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal