Abstract
The physiologic function of BUBR1, a key component of the spindle checkpoint, was examined by generating BUBR1-mutant mice. BUBR1–/– embryos failed to survive beyond day 8.5 in utero as a result of extensive apoptosis. Whereas BUBR1+/– blastocysts grew relatively normally in vitro, BUBR1–/– blastocysts exhibited impaired proliferation and atrophied. Adult BUBR1+/– mice manifested splenomegaly and abnormal megakaryopoiesis. BUBR1 haploinsufficiency resulted in an increase in the number of splenic megakaryocytes, which was correlated with an increase in megakaryocytic, but a decrease in erythroid, progenitors in bone marrow cells. RNA interference–mediated down-regulation of BUBR1 also caused an increase in polyploidy formation in murine embryonic fibroblast cells and enhanced megakaryopoiesis in bone marrow progenitor cells. However, enhanced megakaryopoiesis in BUBR1+/– mice was not correlated with a significant increase in platelets in peripheral blood, which was at least partly due to a defect in the formation of proplatelet-producing megakaryocytes. Together, these results indicate that BUBR1 is essential for early embryonic development and normal hematopoiesis.
Introduction
The spindle checkpoint (also known as the spindle assembly or mitotic checkpoint) ensures that cells with a defective spindle or with defective spindle-kinetochore interactions do not exit from mitosis. Genetic screening approaches have identified at least 7 genes (BUB1, BUB2, BUB3, MAD1, MAD2, MAD3, and MPS1) that are required for spindle checkpoint control in Saccharomyces cerevisiae.1-3 The spindle checkpoint appears to be highly conserved in mammals.4,5 In addition to homologs of BUB and MAD family members, several additional genes, such as those for CENP-E and Hec1, contribute to spindle checkpoint function in mammalian cells.6-8 We and others have cloned and characterized human BUB1 and BUBR1 (also known as hMAD3; GenBank accession no. AF068760),9-13 which are homologous to yeast BUB1 and MAD3, respectively. Despite its marked sequence homology to yeast MAD3, human BUBR1 possesses a unique COOH-terminal extension that contains a putative serine-threonine kinase domain.
Polyploidy refers to the presence of multiple copies of the haploid chromosome set within a cell. It occurs in specialized types of cells in plants, insects, and mammals. Megakaryocytes, the precursors of platelets, achieve a high level of polyploidy by a process known as endomitosis, which is thought to result from the uncoupling of genome replication from mitosis during the cell cycle (see reviews).14-17 Perturbation of various cell cycle processes, including a block at prophase without dissolution of the nuclear membrane, failure to enter anaphase, or skipping of anaphase and cytokinesis, results in the development of polyploidy.18 The high ploidy of mature megakaryocytes is thought to be attributable predominantly to the skipping of anaphase (anaphase B) and cytokinesis before entry into the next cell cycle.16
Although the occurrence of endomitosis in maturing megakaryocyte (MKs) has been known for decades, the molecular mechanisms that underlie this process remain unclear.16,17 Studies of the effects of small molecules that alter either cytoskeletal structures or the activities of protein kinases have provided some insight into the complex process of polyploidization. Nocodazole, a microtubule-depolymerizing agent capable of triggering mitotic arrest, induces polyploidy formation in K562 cells19,20 and other somatic cells,21 suggesting that endogenous regulators of microtubule dynamics might play a role in endomitosis. Cytochalasin B, an inhibitor of actin polymerization, induces increases in both cell size and ploidy as well as the expression of MK-specific markers in DAMI, HEL, and K562 cells.22 These effects of cytochalasin B might be due in part to prevention of the formation of the actin-based cleavage furrow required for cytokinesis, underscoring the importance of mitotic exit in polyploidy formation.
The disruption of spindle checkpoint components often results in the development of polyploidy. The Tax protein of human T-cell leukemia virus–type I (HTLV-I) interacts with and inactivates human MAD1, and its expression results in the formation of multinucleated cells, a phenotype also characteristic of cells deficient in MAD1 function.23 Deficiency of MAD2 induces premature onset of anaphase, and MAD2+/– cells achieve a higher level of ploidy than do wild-type cells in response to nocodazole treatment.24 To determine the physiologic function of BUBR1, we have now generated BUBR1-mutant mice. We demonstrate that BUBR1-knockout mice die during early embryonic development as a result of an increased prevalence of apoptosis. BUBR1 haploinsufficiency resulted in splenomegaly and extramedullary megakaryopoiesis that was coupled with a decrease erythropoiesis. However, enhanced megakaryopoiesis in BUBR1 mice did not result in an increase in platelets in peripheral blood owing to a defect in the formation of proplatelet-producing megakaryocytes.
Materials and methods
Generation of BUBR1+/– mice
The nucleotide sequence of the BUBR1 genomic locus was obtained by a basic local alignment search tool (BLAST) search of mouse genomic databases. Exon-intron boundaries were determined on the basis both of pairwise sequence comparisons after the BLAST search and of the conserved boundary sequences of splice donor and splice acceptor sites. The BUBR1-mutant mice were generated in collaboration with Lexicon Genetics (The Woodlands, TX) by a gene-trapping method.25 The trapping vector contained both a mutagenic component, consisting of a splice acceptor (SA) sequence fused to a selectable marker (neomycin-resistance) gene with a polyadenylation signal sequence, and a sequence-acquisition component, consisting of an embryonic stem (ES) cell–active mouse gene promoter (from the phosphoglycerate kinase-1, or PGK, gene) fused to a coding sequence of Bruton tyrosine kinase (BTK) gene with a splice donor (SD) site. Two independent ES cell clones in which the trapping vector was integrated at the BUBR1 genomic locus were obtained, and DNA sequencing analysis confirmed that the vector was inserted in intron 1. The nucleotide sequence surrounding the insertion site (*) was determined as 5′ TCTGGGAGGATTCAAGATGTTTGT*AAAGTACCTGCTTTCAACTAATAA 3′.
Genotyping
Pieces of mouse tail were digested for 2 to 3 hours at 55°C with proteinase K (300 μg/mL) in a solution containing 50 mM Tris-Hcl (tris(hydroxymethyl)aminomethane-HCl) (pH 7.5), 100 mM NaCl, 50 mM EDTA (ethylenediaminetetraacetic acid), 5 mM dithiothreitol, 0.05 mM spermidine, and 15% sarkosyl. DNA was extracted once with a volume of phenol equal to the sample volume and once with phenol and chloroform (1:1, vol/vol). After the addition of 0.1 vol 3 M sodium acetate to the extract, the supernatant was mixed with 2 vol ethanol, and the resulting DNA precipitate was dissolved in Tris-EDTA (TE) buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA). Genomic DNA from mouse tail, blastocysts, or mouse embryo fibroblasts (MEFs) was genotyped with a nested polymerase chain reaction (PCR) approach. The nucleotide sequences of the PCR primers were as follows: forward primer A, 5′-GGGAGGATCGAGGAGGTCG-3′; forward primer long-terminal repeat 2 (LTR2), 5′-AAATGGCGTTACTTAAGCTAGCTTGC-3′; reverse primer B, 5′-CTGTTCGCCTTCAGTGCTCAAAATGGTAGTCG-3′; and reverse primer C, 5′-TACACTTGTTAATACTGATTCTATCTGATGT-3′. Genomic DNA was subjected to a first round of PCR amplification with primers LTR2, A, and B, and the resulting products were then subjected to a second round of PCR with primers LTR2 and C. The amplification protocol comprised an initial denaturation at 94°C for 8 minutes; 35 cycles of 94°C for 45 seconds, 55°C for 30 seconds, and 72°C for 1 minute; and a final extension at 72°C for 10 minutes. PCR products were fractionated by electrophoresis on a 1% agarose gel and detected by ethidium bromide staining.
Cell culture and immunoblotting
MEFs were derived from embryonic day 14.5 (E14.5) embryos (produced from BUBR1+/– intercrosses). MEF cells were cultured under 5% CO2 in dishes containing Dulbecco minimum essential medium (DMEM) supplemented with 0.1 mM β-mercaptoethanol, 15% fetal bovine serum (FBS), and antibiotics (100 μg/mL penicillin, 50 μg/mL streptomycin sulfate). HeLa and K562 cells were cultured in DMEM and RPMI-1640, respectively, and the media were supplemented with 10% FBS and antibiotics as for MEF cells. K562 cells treated with phorbol 12-myristate 13-acetate (PMA) (10 nM) for various times were collected for Western blotting or flow cytometric analysis. Equal amounts of proteins were then subjected to sodium dodecyl sulfate (SDS)–polyacrylamide gel electrophoresis and immunoblot analysis with antibodies to BUBR1, Plk3, or α-tubulin (Sigma, St Louis, MO). Immune complexes were detected with an appropriate second antibody conjugated with horseradish peroxidase (Sigma) and enhanced chemiluminescence reagents (Amersham Pharmacia Biotech, Piscataway, NJ).
Analysis of embryos
E3.5 blastocysts were harvested from pregnant females of BUBR1+/– intercrosses and were cultured individually, essentially as described,26 for various times (up to the equivalent of E8.5) in 35-mm culture dishes or chamber slides with DMEM containing 15% FBS. They were examined by phase-contrast microscopy and, at the end of the culture period, were subjected to genotype analysis. Decidua of postimplantation embryos (E5.5 to E8.5) were fixed in 10% formalin, embedded in paraffin blocks, and sectioned. The sections were stained with hematoxylin-eosin for histologic analysis or subjected to the transferase-mediated deoxyuridine triphosphate nick end labeling (TUNEL) technique with a Klenow-FlagEL DNA fragmentation kit (Oncogene Research Products, Boston, MA).
RNA interference
Double-stranded RNAs of 21 nucleotides were synthesized by Dharmacon Research (Lafayette, CO). The targeting sequence was 5′-AAGGGAAGCCGAGCUGUUGAC-3′, corresponding to the coding region of 1281 to 1301 in human BUBR1 (accession no., AF068760) and 1259 to 1279 in mouse BUBR1 (accession no., NM009773) relative to the first nucleotide of the ATG start codon. The control small interfering (si) RNA targets luciferase mRNA (accession no., X65324) of the firefly (Photinus pyralis), and the targeting sequence was 5′-UUCCTACGCTGAGTACTTCGA-3′ (GL-3; Dharmacon Research). Double nucleotides (2 deoxythymidines [dTdTs]) were added at the 3′ end of each strand. RNA duplexes were transfected into MEFs or bone marrow progenitor cells via the Oligofectamine approach (Invitrogen, Carlsbad, CA). Cells transfected with siRNAs for 3 or 5 days (unless otherwise specified) were assayed for BUBR1 expression via immunoblotting. MEF cells transfected with siRNAs for 2 days were also split and were incubated for additional 4 days before fixing and staining with DAPI (4,6-diamidino-2-phenylindole) for ploidy analysis.
CFU-Mk and CFU-E assays
Bone marrow cells were collected by flushing the tibia of BUBR1+/– or wild-type mice (4 pairs per group) with phosphate-buffered saline (PBS), and mononuclear cells were isolated after centrifugation through a Ficoll-Paque cushion. Bone marrow mononuclear cells were cultured for 2 days in StemSpan SF expansion medium (Stem Cell Technologies, La Jolla, CA) containing Flt-3 ligand (100 ng/mL), murine stem cell factor (100 ng/mL), interleukin-3 (10 ng/mL), interleukin-6 (20 ng/mL), and thrombopoietin (TPO) (50 ng/mL) before being used for colony assays. Each MK colony-forming unit (CFU-Mk) colony assay was performed with at least 3 duplicates in the MegaCult-C medium according to the protocol provided by the supplier (Stem Cell Technologies). After 12 days' culture, colonies were fixed, stained for megakaryocytic marker acetylcholine esterase (AChE) with acetylthiocholiniodine (Sigma), and scored. An erythrocyte CFU (CFU-E) assay was performed with a methylcellulose-based medium (Stemgenix, Amherst, NY). Each CFU-E assay was performed with 3 duplicates. Each experiment was repeated at least 2 times.
Isolation of bone marrow progenitor cells
Bone marrow progenitor cells were enriched through negative selection by means of a “SpinSepTM Cell Enrichment” kit (Stem Cell Technologies) according to the instruction provided by the supplier. Enriched progenitor cells were first cultured for 1 day in StemSpan SF medium containing a cytokine cocktail as described in “CFU-Mk and CFU-E assays.” Cells were then transfected with siRNAs targeting BUBR1 or luciferase or left untransfected as control. At 2 days after trasnfection, progenitor cells were used for assays for CFU-Mk's. A majority of transfected progenitor cells were lysed, and equal amounts of cell lysates were analyzed for BUBR1 levels via Western blotting.
Proplatelet-producing megakaryocyte assay
Murine proplatelet assays were essentially as described.27,28 Briefly, bone marrow cells were resuspended at 1 × 106/mL and incubated in serum-free Iscove modified Dulbecco medium (IMDM) supplemented with 1% Nutridoma (Roche, Indianopolis, IN) plus recombinant human TPO (50 ng/mL) for 1 week. Mature megakaryocytes were harvested by low-speed centrifugation (50g for 10 minutes) and then overlayered on a discontinuous bovine serum albumin (BSA) (Sigma) density gradient (0%/1.5%/3.0% BSA in phosphate-buffered saline). The final preparation of megakaryocytes was collected by the low-speed centrifugation and then resuspended in IMDM containing 1% Nutridoma and 50 ng/mL TPO. About 3000 to 5000 cells were seeded in each well in a 96-well plate. At least 3 replicates were seeded for each sample. At 3 days later, the time of maximal proplatelet formation, the number of proplatelet-bearing megakaryocytes was counted by means of an inverted microscope.
Flow cytometry
Splenocytes were isolated by grinding of spleen fragments and filtration of the resulting cell suspension through a 200-mesh stainless steel sieve. Peripheral blood cells or splenocytes were incubated for 5 minutes at room temperature with buffer (150 mM NH4Cl, 10 mM KHCO3, 0.01 mM EDTA [pH 7.2]), after which cell lysis was terminated by the addition of Dulbecco PBS. Flow cytometric analysis of cell surface antigens was performed essentially as described.29 In brief, cells were suspended in PBS containing 0.05% bovine serum albumin and incubated for 30 minutes at room temperature with monoclonal antibodies to CD14, CD41, or Ter119 (BD Biosciences, San Jose, CA). The antibody to CD41 was conjugated with fluorescein isothiocyanate, and the one to Ter119 was conjugated with phycoerythrin. Mouse isotype-matched immunoglobulin G (IgG) conjugated with the appropriate label was used for determination of nonspecific binding. The cells were then washed with PBS before injection into a flow cytometer. Analysis of DNA content by flow cytometry was performed after staining of cells with propidium iodide as described.30
Results
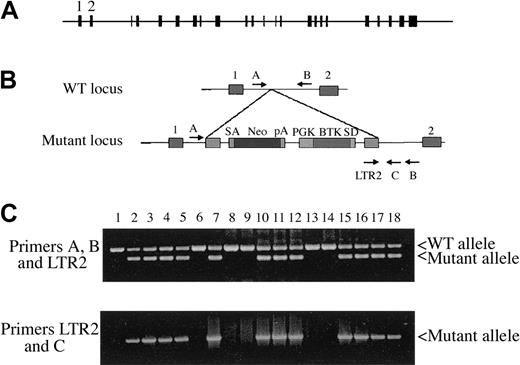
To generate BUBR1-knockout mice, we first identified the mouse gene through a BLAST search of mouse genomic databases. The coding region of BUBR1 was found to span more than 20 kilobase pair (kbp), including 22 introns ranging in size from 130 bp (intron 3) to 6116 bp (intron 2) (Figure 1A). Mouse ES cells with a targeted disruption of the BUBR1 locus were obtained by a gene-trapping method.25 The targeting retroviral vector, which contained a neomycin resistance cassette, was inserted between exons 1 and 2 of BUBR1 (Figure 1B). Two independent 129/Sv-derived ES cell lines were injected into C57/BL6 blastocysts, and the resulting chimeric mice were backcrossed to wild-type C57/BL6 animals. Disruption of BUBR1 and the genotypes of the offspring of various crosses were determined by a nested PCR strategy. Animals positive for the mutant allele amplified by primers LTR2 and B were also positive for the product of primers LTR2 and C (Figure 1C).
Disruption of the mouse BUBR1 locus. (A) Schematic representation of the structure of mouse BUBR1 showing the relative positions and sizes of the 23 exons (filled boxes). (B) Structures of the wild-type (WT) and mutant BUBR1 loci. SA-Neo-pA denotes a splice acceptor sequence fused to the neomycin resistance gene followed by a polyadenylation signal sequence. PGK-BTK-SD denotes PGK gene promoter fused to BTK gene that is flanked by an SD sequence at its 3′ terminus. The positions and relative orientations of 4 PCR primers (A, B, LTR2, and C) used for genotyping are shown. (C) Genotyping of mouse tail DNA by nested PCR. Genomic DNA isolated from the tails of offspring of crosses between wild-type and BUBR1+/– mice was analyzed by PCR with primers A, B, and LTR2 (top panel), and the resulting products were subjected to a second round of PCR with primers LTR2 and C.
Disruption of the mouse BUBR1 locus. (A) Schematic representation of the structure of mouse BUBR1 showing the relative positions and sizes of the 23 exons (filled boxes). (B) Structures of the wild-type (WT) and mutant BUBR1 loci. SA-Neo-pA denotes a splice acceptor sequence fused to the neomycin resistance gene followed by a polyadenylation signal sequence. PGK-BTK-SD denotes PGK gene promoter fused to BTK gene that is flanked by an SD sequence at its 3′ terminus. The positions and relative orientations of 4 PCR primers (A, B, LTR2, and C) used for genotyping are shown. (C) Genotyping of mouse tail DNA by nested PCR. Genomic DNA isolated from the tails of offspring of crosses between wild-type and BUBR1+/– mice was analyzed by PCR with primers A, B, and LTR2 (top panel), and the resulting products were subjected to a second round of PCR with primers LTR2 and C.
During the first year of breeding, we obtained more than 300 live animals from crosses of BUBR1+/– mice. Genotype analysis of live births from BUBR1+/– intercrosses indeed revealed the absence of homozygous mutants and about twice (1.8 ×) the number of heterozygotes as wild-type animals (Table 1), consistent with expected Mendelian segregation. Moreover, analysis of 22 blastocysts randomly obtained from 5 BUBR1+/– intercrosses at E3.5 showed that 23% of the preimplantation embryos were BUBR1–/–, consistent with the expected Mendelian percentage of 25%. Microscopic examination of hematoxylin-eosin (H&E)–stained sections of 58 E6.5 embryos obtained from BUBR1+/– intercrosses revealed that 16 embryos (28%) either had developed abnormally or were resorbed, also consistent with the notion that BUBR1–/– embryos die during early embryonic development.
Analysis of the viability of BUBR1–/– mice
. | BUBR1+/+ × BUBR1+/-, no. (%) . | BUBR1+/- × BUBR1+/-, no. (%) . |
|---|---|---|
| Genotype analysis of live births | ||
| BUBR1+/+ | 33 (49) | 92 (36) |
| BUBR1+/- | 34 (51) | 163 (64) |
| Genotype analysis of preimplantation (E3.5) embryos from 5 pregnant females | ||
| BUBR1+/+ | NA | 5 (23) |
| BUBR1+/- | NA | 12 (55) |
| BUBR-/- | NA | 5 (23) |
| Microscopic examination of postimplantation (E6.5) embryos from 7 pregnant females | ||
| Developed normally | NA | 42 (72) |
| Developed abnormally or resorbed | NA | 16 (28) |
. | BUBR1+/+ × BUBR1+/-, no. (%) . | BUBR1+/- × BUBR1+/-, no. (%) . |
|---|---|---|
| Genotype analysis of live births | ||
| BUBR1+/+ | 33 (49) | 92 (36) |
| BUBR1+/- | 34 (51) | 163 (64) |
| Genotype analysis of preimplantation (E3.5) embryos from 5 pregnant females | ||
| BUBR1+/+ | NA | 5 (23) |
| BUBR1+/- | NA | 12 (55) |
| BUBR-/- | NA | 5 (23) |
| Microscopic examination of postimplantation (E6.5) embryos from 7 pregnant females | ||
| Developed normally | NA | 42 (72) |
| Developed abnormally or resorbed | NA | 16 (28) |
NA indicates not applicable.
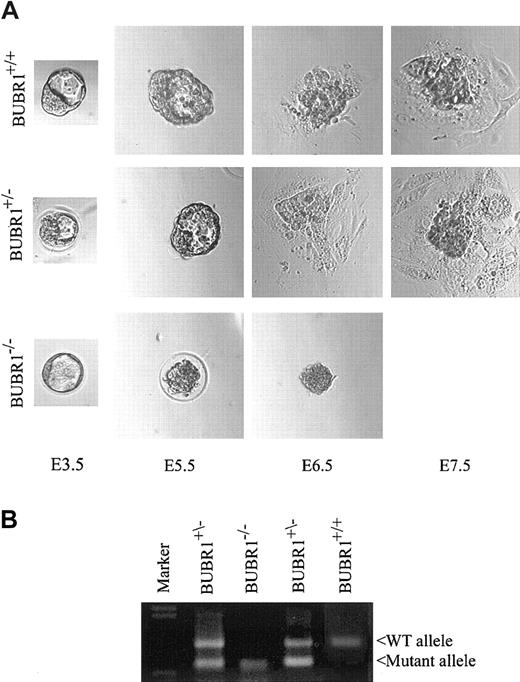
To further examine when BUBR1–/– embryos die, we cultured E3.5 blastocysts in vitro and examined the morphology of the blastocyst-derived cells at various times. The morphology of BUBR1+/– blastocysts did not appear to differ from that of the wild type (Figure 2A). Most wild-type and BUBR1+/– embryos hatched from the zona pellucida and attached to the culture dish, and the trophoblast cells had spread out efficiently by the equivalent of E6.5. The inner cell mass derived from BUBR1+/– embryos maintained a size and shape similar to those apparent for wild-type embryos. In contrast, BUBR1–/– blastocysts did not exhibit robust proliferation, and they became atrophic by the equivalent of E6.5. The genotype of the embryonic cells was determined at the end of the culture period (Figure 2B).
Early embryonic death of BUBR1-null embryos. (A) E3.5 blastocysts of the indicated genotypes were harvested, cultured in vitro, and examined at various times by phase-contrast microscopy. Data are representative of at least 3 independent experiments. Original magnification × 200. (B) At the end of the culture period, cells from the embryos shown in panel A were collected and subjected to genotyping by PCR. The leftmost lane contains DNA size markers.
Early embryonic death of BUBR1-null embryos. (A) E3.5 blastocysts of the indicated genotypes were harvested, cultured in vitro, and examined at various times by phase-contrast microscopy. Data are representative of at least 3 independent experiments. Original magnification × 200. (B) At the end of the culture period, cells from the embryos shown in panel A were collected and subjected to genotyping by PCR. The leftmost lane contains DNA size markers.
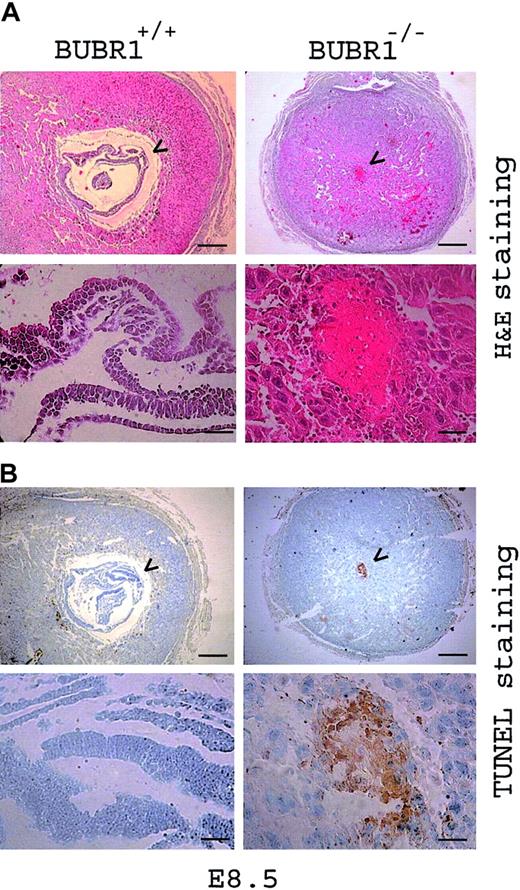
The nature of the death of BUBR1–/– embryos was investigated by subjecting embryos at stages up to E8.5 to fixation in formalin, after which they were embedded in paraffin, sectioned, and either stained with H&E or subjected to the TUNEL technique for the detection of apoptotic cells. As mentioned in the discussion of Table 1, microscopic examination of gross morphology revealed that a substantial proportion of BUBR1–/– embryos at E6.5 were either abnormal or resorbed. Whereas wild-type embryos appeared to develop normally by E8.5, only remnants of BUBR1–/– embryos remained (Figure 3A). TUNEL staining of neighboring sections of the same embryos revealed a large proportion of TUNEL-positive (apoptotic) cells in the atrophic BUBR1–/– embryos but not in the wild-type embryos (Figure 3B).
Cause of death for BUBR1-null embryos.BUBR1-null embryos die of apoptosis. Postimplantation mouse embryos at E8.5 were fixed, embedded in paraffin, and sectioned transversely. Neighboring sections were either stained with hematoxylin-eosin (panel A) or subjected to the TUNEL technique (panel B), and were then examined at low (scale bar = 0.2 μm) and high (scale bar = 5 nm) magnification (upper and lower panels, respectively). Arrowheads indicate embryos or embryonic remnants. Attempts to confirm the genotypes of the abnormally developed or resorbed embryos were rendered difficult by contamination of tissue from the mother. The BUBR1–/– designation of the embryo shown was thus presumed. Gross morphologic examination, however, demonstrated that about one fourth of the embryos resulting from BUBR1+/– intercrosses either developed abnormally or were resorbed (Table 1) and that atrophic embryos were rarely observed to result from intercrosses of wild-type animals, thus supporting the presumptive genotyping.
Cause of death for BUBR1-null embryos.BUBR1-null embryos die of apoptosis. Postimplantation mouse embryos at E8.5 were fixed, embedded in paraffin, and sectioned transversely. Neighboring sections were either stained with hematoxylin-eosin (panel A) or subjected to the TUNEL technique (panel B), and were then examined at low (scale bar = 0.2 μm) and high (scale bar = 5 nm) magnification (upper and lower panels, respectively). Arrowheads indicate embryos or embryonic remnants. Attempts to confirm the genotypes of the abnormally developed or resorbed embryos were rendered difficult by contamination of tissue from the mother. The BUBR1–/– designation of the embryo shown was thus presumed. Gross morphologic examination, however, demonstrated that about one fourth of the embryos resulting from BUBR1+/– intercrosses either developed abnormally or were resorbed (Table 1) and that atrophic embryos were rarely observed to result from intercrosses of wild-type animals, thus supporting the presumptive genotyping.
We noticed that the spleen of adult BUBR1+/– mice was enlarged, with an average mass of approximately 0.15 g, which was more than twice that of the spleen of wild-type mice (Figure 4A). The average body mass of BUBR1+/– mice was 32.9 g, which was less than that of wild-type animals (37.4 g). Thus, the increase in the spleen–body mass ratio in BUBR1+/– mice compared with wild-type mice was even more pronounced than the increase in spleen mass alone. Splenomegaly in BUBR1+/– mice was often accompanied by disruption of the normal architecture of spleen pulp, resulting in loss of the boundaries (marginal zones) that separate white pulp from red pulp (Figure 4B). Extramedullary myelopoiesis was apparent in the spleen of BUBR1+/– mice. Notably, the spleen of most BUBR1+/– mice contained an increased number of apparently mature megakaryocytes (Figure 4B). The increase in the number of megakaryocytes in BUBR1+/– mice revealed by light microscopy was confirmed by flow cytometric analysis of the DNA content of splenic cells stained with propidium iodide (Figure 4C). Whereas most splenic cells of wild-type mice were either 2N or 4N, those of BUBR1+/– mice exhibited a marked increase in the proportion of cells with a DNA content greater than 4N. These large cells were megakaryocytic as they were all positive for AChE (Figure 4D), a unique marker for murine megakaryocytes.
Splenomegaly and extramedullary megakaryopoiesis. (A) Mean (± standard deviation) mass of the spleen freshly dissected from adult BUBR1+/– and wild-type mice. The mean body mass for these animals is also shown. (B) The spleen from BUBR1+/– and wild-type mice were fixed, embedded in paraffin, sectioned, stained with H&E, and examined by light microscopy at low (scale bar = 0.2 μm) and high magnification (scale bar = 5 nm) (upper and lower panels, respectively). (C) Spleen cells from BUBR1+/– and wild-type mice stained with propidium iodide (PI) were subjected to analysis of DNA content by flow cytometry. (D) Sections of freshly frozen BUBR1+/– spleen were stained in situ for AChE+ cells. (E) Spleen and peripheral blood cells from BUBR1+/– and wild-type mice were subjected to flow cytometric analysis of CD41 and Ter119 expression, with an isotype-matched IgG used as a negative control. Data in all panels are representative of 3 independent experiments. These observations were confirmed in multiple pairs of BUBR1+/– and wild-type littermates.
Splenomegaly and extramedullary megakaryopoiesis. (A) Mean (± standard deviation) mass of the spleen freshly dissected from adult BUBR1+/– and wild-type mice. The mean body mass for these animals is also shown. (B) The spleen from BUBR1+/– and wild-type mice were fixed, embedded in paraffin, sectioned, stained with H&E, and examined by light microscopy at low (scale bar = 0.2 μm) and high magnification (scale bar = 5 nm) (upper and lower panels, respectively). (C) Spleen cells from BUBR1+/– and wild-type mice stained with propidium iodide (PI) were subjected to analysis of DNA content by flow cytometry. (D) Sections of freshly frozen BUBR1+/– spleen were stained in situ for AChE+ cells. (E) Spleen and peripheral blood cells from BUBR1+/– and wild-type mice were subjected to flow cytometric analysis of CD41 and Ter119 expression, with an isotype-matched IgG used as a negative control. Data in all panels are representative of 3 independent experiments. These observations were confirmed in multiple pairs of BUBR1+/– and wild-type littermates.
To determine whether the increase in the ploidy of splenic cells in BUBR1+/– mice was accompanied by megakaryocytic differentiation, we examined the expression of surface markers specific for megakaryocytic as well as for the erythroid lineage by flow cytometry. The spleen of BUBR1+/– mice contained an increased population of cells highly positive for CD41 (Figure 4E), a megakaryocytic surface marker.29 This increase in the proportion of megakaryocytes in the spleen was associated with an apparent decrease in the percentage of CD41+ cells in peripheral blood of BUBR1+/– mice. Moreover, BUBR1+/+ spleen contained a population of cells highly positive for Ter119, and this cell population was essentially absent from the spleen of BUBR1+/– mice (Figure 4E).
Our results thus suggested that BUBR1 haploinsufficiency was associated with increased splenic megakaryocytes. To investigate whether this increase was associated with a modulation in megakaryocytic progenitor cells in bone marrow, we examined CFU-Mk's as well as of the CFU-Es. Compared with wild-type animals, CFU-Mk's were significantly increased in BUBR1+/– mice (Figure 5A) whereas CFU-Es were decreased in BUBR1-deficient mice (Figure 5B). The platelet counts in peripheral blood of BUBR1+/– mice were 1.77 ± 0.87 × 106/μL (mean ± standard deviation, n = 20), which were not significantly elevated compared with those (1.46 ± 0.67 × 106/μL, mean ± standard deviation, n = 18) of wild-type animals. Consistent with the reduced CFU-Es in bone marrow cells, peripheral blood red cell (RBC) counts were significantly reduced (Table 2) (below the normal range, bold and italicized numbers) in half of BUBR1+/– mice although a couple of wild-type mice (12%) also exhibited peripheral RBC counts below the normal range. In addition, the average white blood cell counts were 5.1 ± 2.5 × 103/μL (mean ± standard deviation, n = 9) and 6.9 ± 2.4 × 103/μL (mean ± standard deviation, n = 10) for wild-type and BUBR1+/– mice, respectively.
Effect of modulation of BUBR1 expression on hematopoiesis. Modulation of BUBR1 expression affected hematopoiesis in vitro. (A) Bone marrow cells from BUBR1+/– and wild-type mice were assayed for CFU-Mk's (P < .001, n = 12) as described in “Materials and methods.” (B) Bone marrow cells from BUBR1+/– and wild-type mice were assayed for CFU-Es (P < .0005, n = 8). Error bars represent means ± standard deviation.
Effect of modulation of BUBR1 expression on hematopoiesis. Modulation of BUBR1 expression affected hematopoiesis in vitro. (A) Bone marrow cells from BUBR1+/– and wild-type mice were assayed for CFU-Mk's (P < .001, n = 12) as described in “Materials and methods.” (B) Bone marrow cells from BUBR1+/– and wild-type mice were assayed for CFU-Es (P < .0005, n = 8). Error bars represent means ± standard deviation.
Peripheral blood cell counts
. | RBC count, × 106/μL . | . | Platelet count, × 103/μL . | . | ||
|---|---|---|---|---|---|---|
| Animal no. . | BUBR1+/+ . | BUBR1+/- . | BUBR1+/+ . | BUBR1+/- . | ||
| 1 | 9.2 | 6.1* | 850 | 1029 | ||
| 2 | 11.0 | 13.3 | 332* | 734 | ||
| 3 | 11.7 | 7.2 | 593 | 789 | ||
| 4 | 9.4 | 10.9 | 749 | 608 | ||
| 5 | 9.7 | 9.3 | 1910 | 1512 | ||
| 6 | 8.1 | 6.4* | 1100 | 2814 | ||
| 7 | 9.6 | 6.3* | 2368 | 3412 | ||
| 8 | 9.2 | 5.2* | 2245 | 2512 | ||
| 9 | 9.7 | 9.8 | 2467 | 2716 | ||
| 10 | 9.7 | 3.4* | 921 | 598 | ||
| 11 | 5.5* | 9.3 | 780 | 2400 | ||
| 12 | 9.0 | 7.7 | 1200 | 1700 | ||
| 13 | 8.6 | 9.7 | 1610 | 800 | ||
| 14 | 8.6 | 3.3* | 2100 | 1760 | ||
| 15 | 5.5* | 4.0* | 1460 | 1940 | ||
| 16 | 9.9 | 4.6* | 2075 | 2810 | ||
| 17 | ND | 2.1* | 1937 | 2250 | ||
| 18 | ND | 9.6 | 1150 | 1804 | ||
| 19 | ND | ND | ND | 590* | ||
| 20 | ND | ND | ND | 2000 | ||
| Mean | 9.0 | 7.1 | 1463 | 1771 | ||
| ± SD | 1.6 | 3.0 | 665 | 870 | ||
. | RBC count, × 106/μL . | . | Platelet count, × 103/μL . | . | ||
|---|---|---|---|---|---|---|
| Animal no. . | BUBR1+/+ . | BUBR1+/- . | BUBR1+/+ . | BUBR1+/- . | ||
| 1 | 9.2 | 6.1* | 850 | 1029 | ||
| 2 | 11.0 | 13.3 | 332* | 734 | ||
| 3 | 11.7 | 7.2 | 593 | 789 | ||
| 4 | 9.4 | 10.9 | 749 | 608 | ||
| 5 | 9.7 | 9.3 | 1910 | 1512 | ||
| 6 | 8.1 | 6.4* | 1100 | 2814 | ||
| 7 | 9.6 | 6.3* | 2368 | 3412 | ||
| 8 | 9.2 | 5.2* | 2245 | 2512 | ||
| 9 | 9.7 | 9.8 | 2467 | 2716 | ||
| 10 | 9.7 | 3.4* | 921 | 598 | ||
| 11 | 5.5* | 9.3 | 780 | 2400 | ||
| 12 | 9.0 | 7.7 | 1200 | 1700 | ||
| 13 | 8.6 | 9.7 | 1610 | 800 | ||
| 14 | 8.6 | 3.3* | 2100 | 1760 | ||
| 15 | 5.5* | 4.0* | 1460 | 1940 | ||
| 16 | 9.9 | 4.6* | 2075 | 2810 | ||
| 17 | ND | 2.1* | 1937 | 2250 | ||
| 18 | ND | 9.6 | 1150 | 1804 | ||
| 19 | ND | ND | ND | 590* | ||
| 20 | ND | ND | ND | 2000 | ||
| Mean | 9.0 | 7.1 | 1463 | 1771 | ||
| ± SD | 1.6 | 3.0 | 665 | 870 | ||
Normal range for RBC count, 6.4-9.4 × 106/μL. Normal range for platelet count, 592-2972 × 103/μL.
ND indicates not done.
Counts below the normal range.
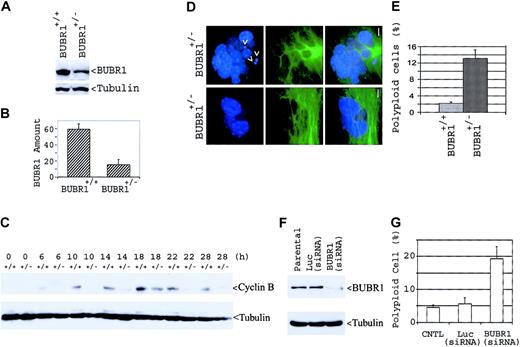
To confirm that the phenotypic characteristics of BUBR1+/– mice were due to a reduction in BUBR1 expression, we examined the abundance of BUBR1 in MEFs derived from E14.5 embryos produced from intercrosses of BUBR1+/– mice. The genotype of each MEF line was determined by nested PCR. Immunoblot analysis revealed that the amount of BUBR1 in BUBR1+/– MEFs was smaller than that in wild-type MEFs (Figure 6A). Interestingly, the average level of BUBR1 in BUBR1+/– MEFs was about 25% (rather than the expected 50%) of that in wild-type MEFs (Figure 6B). Consistent with the notion that BUBR1+/– cells are defective in spindle checkpoint control, the abundance of cyclin B in asynchronous cells was markedly lower in BUBR1+/– MEFs than in wild-type cells after nocodazole treatment (Figure 6C); cyclin B expression failed to accumulate in BUBR1+/– cells to the high level apparent in wild-type MEFs following spindle checkpoint activation, strongly suggesting a slippage of mitotic arrest in the mutant cells, leading to abnormal mitotic exit.
Compromised spindle checkpoint function in BUBR1+/– MEFs. (A) Wild-type and BUBR1+/– MEF cells were lysed, and an equal amount of cell lysates was blotted for BUBR1 and for α-tubulin. (B) Independent BUBR1 signals from paired MEFs were quantified via densitometry, and average amounts of BUBR1 signals after normalization by α-tubulin signals were shown. (C) BUBR1+/– and wild-type MEFs were incubated with nocodazole (0.2 μg/mL) for the indicated times, after which cell lysates were subjected to immunoblot analysis with antibodies to cyclin B or to α-tubulin. (D) BUBR1+/– MEFs stained with antibody to α-tubulin (green) and with DAPI (blue) were examined by fluorescent microscopy. Two representative polyploid cells were shown. Arrows denote micronuclei. Bar = 5 μm. (E) Polyploid cells in both BUBR1+/– and wild-type MEFs were scored from 500 cells, and the percentage of cells with polyploidy is shown. (F) MEF cells were transfected with siRNA targeting human BUBR1 or luciferase gene mRNA for 3 days, after which equal amounts of cell lysates, along with untransfected parental lysates, were subjected to immunoblot analysis with antibodies to BUBR1 or to α-tubulin. (G) Wild-type MEFs were transfected with siRNA targeting BUBR1 or luciferase (Luc) mRNA for 6 days, after which the transfected cells, along with parental MEFs, were fixed and stained with antibody to α-tubulin and with DAPI. Polyploid cells were scored from 500 MEFs. Error bars represent means ± standard deviation.
Compromised spindle checkpoint function in BUBR1+/– MEFs. (A) Wild-type and BUBR1+/– MEF cells were lysed, and an equal amount of cell lysates was blotted for BUBR1 and for α-tubulin. (B) Independent BUBR1 signals from paired MEFs were quantified via densitometry, and average amounts of BUBR1 signals after normalization by α-tubulin signals were shown. (C) BUBR1+/– and wild-type MEFs were incubated with nocodazole (0.2 μg/mL) for the indicated times, after which cell lysates were subjected to immunoblot analysis with antibodies to cyclin B or to α-tubulin. (D) BUBR1+/– MEFs stained with antibody to α-tubulin (green) and with DAPI (blue) were examined by fluorescent microscopy. Two representative polyploid cells were shown. Arrows denote micronuclei. Bar = 5 μm. (E) Polyploid cells in both BUBR1+/– and wild-type MEFs were scored from 500 cells, and the percentage of cells with polyploidy is shown. (F) MEF cells were transfected with siRNA targeting human BUBR1 or luciferase gene mRNA for 3 days, after which equal amounts of cell lysates, along with untransfected parental lysates, were subjected to immunoblot analysis with antibodies to BUBR1 or to α-tubulin. (G) Wild-type MEFs were transfected with siRNA targeting BUBR1 or luciferase (Luc) mRNA for 6 days, after which the transfected cells, along with parental MEFs, were fixed and stained with antibody to α-tubulin and with DAPI. Polyploid cells were scored from 500 MEFs. Error bars represent means ± standard deviation.
In addition, BUBR1+/– MEFs contained a large number of polyploid cells (Figure 6D-E); frequently, these polyploid cells also contained micronuclei (Figure 6D, arrows). To confirm that the nuclear abnormalities such as polyploidy formation were a consequence of BUBR1 deficiency, wild-type MEF cells were transfected with siRNA targeting either BUBR1 mRNA or luciferase mRNA. Immunoblot analysis showed that BUBR1 was efficiently down-regulated via RNA interference (Figure 6F). Moreover, a significant fraction (almost 20%) of the cells transfected with BUBR1-specific siRNA, but not with the control siRNA, was polyploid (Figure 6G), indicating that BUBR1 down-regulation leads to polyploidy formation.
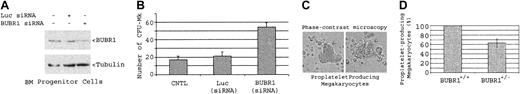
We further carried out experiments in which enriched bone marrow progenitor cells were targeted for BUBR1 silencing via RNAi. At 2 days after transfection with siRNAs, bone marrow progenitor cells were analyzed for down-regulation of BUBR1 via Western blotting and assayed for CFU-Mk's in a semisolid culture. BUBR1 levels in bone marrow progenitor cells were significantly down-regulated after 2 days' transfection of siRNA targeting BUBR1 but not luciferase (Figure 7A). There was a dramatic increase in the number of CFU-Mk's when bone marrow progenitor cells were transfected with siRNAs targeting BUBR1 but not luciferase (Figure 7B), indicating that BUBR1 down-regulation plays an essential role in polyploidy formation as well as in murine megakaryopoiesis. It was noted that the RNAi-mediated down-regulation of BUBR1 was not as pronounced in bone marrow cells (Figure 7A) as with MEF cells (Figure 6E), which was partly due to a shorter time (2 days) of transfection. As the RNAi effect can be achieved by the presence of a few double-stranded RNAs in the cell, it is reasonable to speculate that BUBR1 down-regulation had continued during CFU-Mk assays.
Effect of down-regulation of BUBR1 via RNAi. Down-regulation of BUBR1 in vitro via RNAi stimulates CFU-Mk's although BUBR1 haploinsufficiency causes an inhibition in platelet production. (A) Enriched bone marrow progenitor cells from wild-type mice were transfected with or without siRNAs targeting BUBR1 or luciferase for 2 days. Equal amounts of cell lysates were blotted for BUBR1 or α-tubulin. (B) Bone marrow progenitor cells transfected with siRNAs targeting BUBR1 or luciferase for 2 days were split for CFU-Mk assays as described in “Materials and methods.” (C) Bone marrow cells from both wild-type or BUBR1+/– mice were used for purification of murine megakaryocytes and proplatelet assays as described in “Materials and methods.” Two typical proplatelet-producing megakaryocytes are shown. Original magnification, × 200. (D) The percentage of proplatelet-producing megakaryocytes in bone marrow cell–derived cultures. The proplatelet-producing megakaryocytes in wild-type cell cultures were arbitrarily set at 100%. The data are summarized from 3 independent experiments. Error bars represent means ± standard deviation.
Effect of down-regulation of BUBR1 via RNAi. Down-regulation of BUBR1 in vitro via RNAi stimulates CFU-Mk's although BUBR1 haploinsufficiency causes an inhibition in platelet production. (A) Enriched bone marrow progenitor cells from wild-type mice were transfected with or without siRNAs targeting BUBR1 or luciferase for 2 days. Equal amounts of cell lysates were blotted for BUBR1 or α-tubulin. (B) Bone marrow progenitor cells transfected with siRNAs targeting BUBR1 or luciferase for 2 days were split for CFU-Mk assays as described in “Materials and methods.” (C) Bone marrow cells from both wild-type or BUBR1+/– mice were used for purification of murine megakaryocytes and proplatelet assays as described in “Materials and methods.” Two typical proplatelet-producing megakaryocytes are shown. Original magnification, × 200. (D) The percentage of proplatelet-producing megakaryocytes in bone marrow cell–derived cultures. The proplatelet-producing megakaryocytes in wild-type cell cultures were arbitrarily set at 100%. The data are summarized from 3 independent experiments. Error bars represent means ± standard deviation.
As enhanced megakaryopoiesis in BUBR1+/– mice did not result in significantly elevated platelet counts in peripheral blood, we investigated whether a defect existed in platelet formation by directly comparing proplatelet-producing megakaryocytes in wild-type and BUBR1+/– mice. Bone marrow progenitor cells were cultured in vitro to observe proplatelet-producing megakaryocytes as described,27,28 and typical proplatelet cells (Figure 7C) were counted. As expected, the proplatelet-producing megakaryocytes were significantly reduced (about 60%) in bone marrow cells isolated from BUBR1+/– mice compared with those from wild-type mice (Figure 7D), suggesting a defect of BUBR1+/– megakaryocytes in making platelets. This may also contribute to the observed increase in splenic megakaryocytes in BUBR1+/– mice.
Discussion
Our data demonstrate that BUBR1 is an essential gene, its absence resulting in death during early embryonic development. This conclusion is supported by the observations that (1) no BUBR1–/– animals were detected among more than 300 live births of BUBR1+/– intercrosses; (2) BUBR1–/– blastocysts did not proliferate in vitro beyond E6.5; (3) BUBR1–/– embryos were detected in the expected Mendelian ratio before implantation (E3.5), whereas about a quarter of embryos at E6.5 had either developed abnormally or were resorbed and these latter embryos exhibited extensive apoptosis at E8.5; and (4) the spindle checkpoint was compromised in BUBR1+/– MEFs, resulting in a marked decrease in the abundance of cyclin B and slippage of mitotic arrest. Given the importance of BUBR1 in spindle checkpoint control,5,31 the early embryonic death of BUBR1–/– mice is most likely due to severe chromosomal instability. Our observations are thus consistent with the previous demonstrations that deficiency of either MAD2 or BUB3,23,32 both of which play an important role in spindle checkpoint regulation in mammals, results in embryonic death.
The importance of BUBR1 for normal mammalian development is also reflected by our observation that haploinsufficiency of this gene results in splenomegaly as well as extramedullary megakaryopoiesis in the spleen. The increase in splenic megakaryocytes appeared to be associated with an increase in bone marrow megakaryocytic progenitor cells. Several lines of evidence suggest that the enhanced splenic megakaryopoiesis in BUBR1+/– mice is a direct result of a reduced abundance of BUBR1 and consequent spindle checkpoint failure: (1) The development of polyploidy was much more pronounced in BUBR1+/– MEFs than in wild-type cells (Figure 6D-E). (2) Silencing BUBR1 in wild-type MEFs via RNAi caused a significant increase in polyploidy cells (Figure 6F-G). (3) More importantly, down-regulation of BUBR1 in wild-type bone marrow progenitor cells via RNAi resulted in a great increase in the number of CFU-Mk's (Figure 7B).
A weakened spindle checkpoint would lead to frequent chromosomal missegregation, a condition that daughter cells would not tolerate. It is conceivable that, in the presence of chromosomal missegregation, daughter cells would rather fuse to form bionucleated or multinucleated cells than yield individual cells with wrongly partitioned genetic contents. It is also conceivable that cells of various developmental origins would be equally susceptible to the development of polyploidy when the spindle checkpoint is compromised. Developmentally, most cell types are, however, programmed not to allow polyploidy formation in vivo, and these cells would conceivably be eliminated through apoptosis. Consistent with this notion, inhibition of spindle checkpoint function by expression of dominant negative BUB1 leads to enhanced apoptosis.12 The observation that BUBR1 deficiency primarily results in an increased number of bone marrow megakaryocytic progenitors and splenic megakaryocytes also indirectly supports this notion because in mammals, megakaryocytes are the major cell type with a polyploid DNA content under physiologic conditions. In fact, the proteasome inhibitor PS-341 specifically inhibits platelet production in humans,33 underscoring the important role of ubiquitin-dependent proteolysis in megakaryocyte development.
The enhanced splenic megakaryopoiesis caused by BUBR1 haploinsufficiency was associated with a greatly reduced population of cells positive for the red cell marker Ter119. Consistently, about half of BUBR1+/– mice are anemic, having low red cell counts in peripheral blood (Table 2). This observation can be explained by the possibility that enhanced differentiation of one lineage of blood cells depletes or greatly reduces the availability of bipotential progenitor cells for differentiation into the other lineage. The fact that bone marrow cells from BUBR1+/– mice contain significantly fewer erythroid progenitors than those of wild-type ones is consistent with the “progenitor-steal” effect, as megkaryocytes and erythroid cells are believed to share a common bipotential progenitor.34 The enhanced splenic megakaryopoiesis in BUBR1+/– mice did not correlate with a significant increase in the proportion of CD41+ cells and platelet counts in peripheral blood. In this scenario, the increase in the number of splenic megakaryocytes would be due to a prolonged retention and perhaps subsequent destruction of defective megakaryocytes (eg, cells with nonphysiologic ploidy levels). An alternative explanation is that there exists a defect in the terminal differentiation, resulting in a failure in producing functional platelets. This notion is supported by our study that bone marrow cells from BUBR1+/– mice contained fewer proplatelet-producing megakaryocytes than those from wild-type mice (Figure 7D) even though they had significantly more megakaryocytic progenitor cells (Figure 5A).
Prepublished online as Blood First Edition Paper, October 23, 2003; DOI 10.1182/blood-2003-06-2158.
Supported in part by a grant from the National Institutes of Health (RO1-CA90658).
Q.W. and T.L. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Dr Jianhua Zhang for assistance in blastocyst culture; Dr Frank Traganos for helpful discussions; Dr Ming Xu's laboratory for various helps in genotyping; Dr Robert Benezra and Dr Jasminder Weinstein for MAD and Cdc20 antibodies, respectively; and Dr Jan Kunicki for assistance in flow cytometric analysis. We are also grateful to Dr Stuart Newman for allowing us to use his facility for image documentation; to Dr Chung-Xiong Wang and Dr Robert Russell for discussions about mouse histology; and to Dr Tao Cheng's laboratory for assistance in analysis of murine spleen, bone marrow, and blood cells.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal