Abstract
The involvement of platelets in the pathogenesis of atherosclerosis has recently gained much attention. Platelet factor 4 (PF4), a platelet-specific chemokine released on platelet activation, has been localized to atherosclerotic lesions, including macrophages and endothelium. In this report, we demonstrate that E-selectin, an adhesion molecule involved in atherogenesis, is up-regulated in human umbilical vein endothelial cells exposed to PF4. Induction of E-selectin RNA is time and dose dependent. Surface expression of E-selectin, as measured by flow cytometry, is also increased by PF4. PF4 induces E-selectin expression by activation of transcriptional activity. Activation of nuclear factor-κB is critical for PF4-induced E-selectin expression, as demonstrated by promoter activation studies and electrophoretic mobility shift assays. Further, we have identified the low-density lipoprotein receptor-related protein as the cell surface receptor mediating this effect. These results demonstrate that PF4 is able to increase expression of E-selectin by endothelial cells and represents another potential mechanism by which platelets may participate in atherosclerotic lesion progression.
Introduction
Recruitment of leukocytes into arterial subendothelium is an important pathogenic event in atherogenesis.1 Leukocyte recruitment involves increased expression of adhesion molecules and their corresponding receptors on the surface of arterial wall cells.2 The increased expression of adhesion molecules and their receptors is mediated by the release of various inflammatory mediators, including chemokines.2
Selectins are a family of adhesion receptors that function in leukocyte capture, rolling, and firm adhesion, particularly under the increased shear stress of arteries (for a review, see Ley3 ). Knockout of P-selectin and E-selectin decreases atherosclerosis in murine models.4,5 In addition, E-selectin is expressed in the endothelium of human atherosclerotic lesions,6-9 particularly when lesions are associated with subendothelial leukocytic infiltrates.6 In view of the inflammatory model of atherosclerosis, E-selectin is thought to be an important player in lesion formation.
Lipoprotein receptor-related protein (LRP) is a 420-kDa10 member of the low-density lipoprotein (LDL) receptor superfamily.11,12 It is well established that LRP functions as a multiligand endocytosis receptor, involved in the endocytosis of a wide range of molecules, including tissue plasminogen activator (tPA), urokinase plasminogen activator (uPA), and plasminogen activator inhibitor type 1 (PAI-1).12,13 Soluble ligands endocytosed through LRP are rapidly taken up by lysosomes, where they are released and degraded, whereas the unoccupied receptor is recycled to the plasma membrane.14 Recently, emerging studies demonstrated that the binding of certain ligands to LRP initiates intracellular signal transduction15-19 mediating contractility of blood vessels15,17,18 as well as permeability of the blood-brain barrier.19 Although it is appreciated that LRP contributes to atherosclerosis as a scavenger receptor, the importance of LRP as a signaling receptor in atherosclerosis or inflammation is largely unexplored.
Platelets are now thought to be involved in atherogenesis.20-23 It has been known for many years that activated platelets release mediators that promote smooth muscle cell proliferation24-28 and are involved in the accumulation of lipoproteins in vascular cells and macrophages.25,29-31 Activated platelets augment the recruitment of monocytes to the vessel wall by inducing endothelial cells to synthesize monocyte chemoattractant protein 1 (MCP-1) and intercellular adhesion molecule 1 (ICAM-1),32 and they promote monocyte adherence to the vascular at sites of high shear stress.25 Activation of platelets causes sustained translation and release of interleukin 1β (IL-1β), which activates endothelial cells, increasing their adhesiveness toward neutrophils.33 Platelet adhesion has been found to precede leukocyte recruitment to the vessel wall in apolipoprotein E (apoE) knockout mice,20 and such adherent platelets may deliver platelet factor 4 (PF4) to vascular cells.22,34 Taken together, these studies suggest that the contribution of platelets in the atherogenic process may begin at an earlier stage than previously appreciated.
Toward this end, our laboratory has been studying the effects of PF4, a platelet-specific chemokine, in atherosclerosis.34-36 We have previously shown that PF4 is localized to atherosclerotic lesions, including the macrophages of fatty streaks, and that the presence of PF4 in carotid atherosclerotic lesions correlates with symptomatic coronary artery disease.34 We have also shown that PF4 decreases the degradation of LDL36 and increases the uptake and esterification of oxidized LDL by macrophages.35 These studies suggest that PF4 represents a proatherogenic platelet molecule.
We hypothesized that PF4, which binds avidly to endothelial cells via cell surface glycosaminoglycans, may be involved in promoting leukocyte adhesion by increasing expression of adhesion molecules on the cell surface. In the current study, we demonstrate that PF4 increases E-selectin RNA and cell surface protein in human umbilical vein endothelial cells (HUVECs) in a time- and dose-dependent manner. Further, we demonstrate that PF4 induces the expression of the E-selectin via activation of nuclear factor-κB (NF-κB). In addition, we have identified LRP as the cell surface receptor that mediates this effect. These studies demonstrate a novel mechanism by which PF4 may promote atherosclerosis as well as other inflammatory vascular diseases.
Materials and methods
Reagents
Recombinant PF4 was purified from bacteria as previously described.36 Purified PF4 was demonstrated to be free of endotoxin by testing in the University of Pennsylvania Cell Center. HUVECs and umbilical artery smooth muscle cells were obtained from Cambrex (Walkersville, MD). The murine embryonic fibroblasts (MEF-1, MEF-2) were obtained from the American Type Culture Collection (ATCC; Manassas, VA). Antihuman E-selectin monoclonal antibody and fluorescein isothiocyanate (FITC)–conjugated antihuman CD31 monoclonal antibody (to identify endothelial cells) were from BD Biosciences (San Diego, CA). Anti–NF-κB P50, P65 polyclonal antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA). Anti–P-IKKα, anti-IκBα, and anti–P-IκBα antibodies were from Cell Signaling Technology (Beverly, MA). The monoclonal anti-LRP (catalogue no. 3402) antibody was from American Diagnostica (Greenwich, CT). Polymyxin B was obtained from Sigma (St Louis, MO).
Plasmids
Control plasmid pSV-β-galactosidase control vector, pGL3B, containing the promoterless luciferase gene, and pGL3-SV40, containing the SV-40 promoter luciferase gene, were obtained from Promega Biotechnology (Madison, WI). Plasmid pGL3-840(ELAM-Luc), kindly provided by Dr Sonia C. Flores (University of Colorado, Denver, CO),37 contains sequences 840 bp upstream of the transcriptional start site of the human E-selectin gene driving firefly luciferase transcription. Deletion mutants were constructed by polymerase chain reaction (PCR) using the reverse primer 5′-GCGAAGCTTCTGCTGCCTCTG-3′ paired with the forward primer 5′-GCAGCTAGCATAACTGCCGTG-3′ for pGL3-540(ELAM-Luc), 5′-GCAGCTAGCGAGAGACACTAC-3′ for pGL3-310(ELAM-Luc), 5′-GCAGCTAGCGTGGATATTCC-3′ for pGL3-130(ELAM-Luc), or 5′-A GAGCTAGCGCAATCCCTCC-3′ for pGL3-40(ELAM-Luc) using plasmid pGL3-840(ELAM-Luc) as the template. Restriction sites included in the primers (HindIII for reverse and NheI for forward) are underlined. Figure 4 presents a schematic representation of promoters.
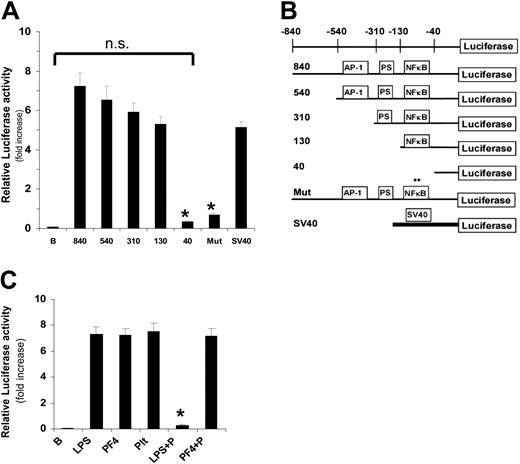
Activation of the E-selectin promoter by PF4. Plasmids containing either the full-length (-840) E-selectin promoter or variants (B) driving expression of luciferase were transfected into HUVECs. (A) HUVECs were incubated in the absence or presence of PF4 (20 μg/mL) for 4 hours prior to measurement of luciferase activity. Data are expressed as fold increase in the presence of PF4 compared to buffer alone and are the mean ± SEM of 3 independent experiments performed in triplicate; n.s. indicates lack of statistical significance by 2-tailed t test (P > .05); asterisk, statistical significance compared to the full-length (840) promoter (P < .001); B, promoterless luciferase activity; Luc, luciferase coding sequence; E-selectin and SV40 indicate the promoters present. (B) Schematic diagram depicting promoter construct used in these experiments. The 5′ promoter region of the E-selectin gene is depicted (top). Consensus binding sites for AP-1 and NF-κB are shown as well as a palindromic site (PS) thought to be important for E-selectin expression. (C) Polymyxin B inhibition of LPS (100 ng/mL), but not PF4 (20 μg/mL), demonstrates that PF4 is not contaminated with LPS. P indicates polymyxin B treatment. Ability of platelet releasate to activate the E-selectin promoter is also shown (Plt). Data are mean ± SEM of 3 independent experiments performed in triplicate.
Activation of the E-selectin promoter by PF4. Plasmids containing either the full-length (-840) E-selectin promoter or variants (B) driving expression of luciferase were transfected into HUVECs. (A) HUVECs were incubated in the absence or presence of PF4 (20 μg/mL) for 4 hours prior to measurement of luciferase activity. Data are expressed as fold increase in the presence of PF4 compared to buffer alone and are the mean ± SEM of 3 independent experiments performed in triplicate; n.s. indicates lack of statistical significance by 2-tailed t test (P > .05); asterisk, statistical significance compared to the full-length (840) promoter (P < .001); B, promoterless luciferase activity; Luc, luciferase coding sequence; E-selectin and SV40 indicate the promoters present. (B) Schematic diagram depicting promoter construct used in these experiments. The 5′ promoter region of the E-selectin gene is depicted (top). Consensus binding sites for AP-1 and NF-κB are shown as well as a palindromic site (PS) thought to be important for E-selectin expression. (C) Polymyxin B inhibition of LPS (100 ng/mL), but not PF4 (20 μg/mL), demonstrates that PF4 is not contaminated with LPS. P indicates polymyxin B treatment. Ability of platelet releasate to activate the E-selectin promoter is also shown (Plt). Data are mean ± SEM of 3 independent experiments performed in triplicate.
Generation of pGL3-840-NF-κBmut(ELAM-Luc) containing a 3-bp mutation in the NF-κB consensus sequence was accomplished using the QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) and using plasmid pGL3-840(ELAM-Luc) as the template and the oligonucleotides NF-κBm1 (5′-GGATGCCATTGCTCATTTCCTCTTTACTGGATGTGG-3′) and NF-κBm2 (5′-CCACATCCAGTAAAGAGGAAATGAGCAATGGCATCC-3′). Mutations introduced are underlined. All constructs were confirmed by DNA sequencing to ensure fidelity.
Probes for ribonuclease protection assay
A 332-bp fragment of the E-selectin coding sequence (from 1130-1462) was cloned into pCRII-TOPO (Invitrogen Life Technologies, Carlsbad, CA) by reverse transcription-PCR (RT-PCR) from HUVEC RNA using 5′-CTTCATGTTGCAGGGACCAG-3′ and 5′-CCAATAGGGGAATGAGCACA-3′ as primers to form pTOPO-ELAM. Similarly, pTOPO-GAPDH was created by cloning a 147-bp fragment of glyceraldehyde phosphate dehydrogenase (GAPDH; from 135-281) using 5′-GGCTGCTTTTAACTCTGGTA-3′ and 5′-ATGACAAGCTTCCCGTTCTC-3′ as primers.
Antisense RNA probes were synthesized by transcription of the linearized plasmids (pTOPO-ELAM or pTOPO-GAPDH) using the MAXI-script in vitro transcription kit (Ambion, Austin, TX) following the manufacturer's instructions. The reaction included biotin-cytidine triphosphate (CTP) at a 6:4 ratio with CTP for incorporation of the biotin tag. Following transcription, the DNA template was digested with DNAse, and the probes were purified using NucAway Spin Columns (Ambion).
Ribonuclease protection assay
For all experiments, HUVECs were grown to 80% confluence (EGM-2 Bulletkit [Cambrex]; EGM-2 medium supplemented with 2% fetal bovine serum [FBS], human epidermal growth factor, hydrocortisone, vascular endothelial growth factor [VEGF], human fibroblast growth factor-β, R3-insulin-like growth factor 1 [IGF-1], heparin, ascorbic acid, and geneticin/amphotericin), and tested at P5 or P6. Cells were exposed to serum-free growth media with or without PF4. The ribonuclease protection assay (RPA) was performed using 4 μg total RNA (isolated using the Micro-to Midi Total RNA Purification System; Invitrogen Life Technologies) using the SuperSignal RPA III chemiluminescent detection kit (Pierce Biotechnology, Rockford, IL) following the manufacturer's standard protocol. After transfer to a nylon membrane, protected bands were detected with streptavidin-horseradish peroxidase (HRP) and luminal by exposure to x-ray film. Bands were quantitated using the Kodak 1D Image Analysis system. E-selectin RNA levels were normalized to GAPDH expression in identical samples.
Flow cytometry
Flow cytometry was performed using a FACSCalibur cytometer (BD Biosciences). HUVECs were stained with phycoerythrin (PE)–conjugated antihuman E-selectin and FITC-conjugated antihuman CD31 monoclonal antibodies prior to cytometric analysis. Background was measured using FITC- and PE-conjugated preimmune isotype antibodies. Data were analyzed using CellQuest software. E-selectin expression was calculated on cells expressing CD31.
Measurement of E-selectin promoter activation
HUVECs were seeded at a density of 5 × 106 cells/well on 12-well plates and grown to 70% to 80% confluence. Cells were washed with sterile phosphate-buffered saline (PBS) and cotransfected with 1 μg LUC containing plasmid (pGL3-840(ELAM-Luc) or variant) and 0.5 μg pSV-β-galactosidase (internal transfection control) using Lipofectamine 2000 (Invitrogen Life Technologies). Following transfection, cells were grown for 18 hours. For some experiments, cell were incubated for 1 hour in medium containing polymyxin B (20 μg/mL) or anti-LRP (10 μg/mL) monoclonal antibody. Next, cells were treated with or without PF4 (20 μg/mL) for 3 hours. The cells were harvested and lysed and centrifuged at 2000g for 5 minutes at room temperature. Then, 10 μL cell lysate was combined with 50 μL luciferase assay buffer and luciferase activity was measured using the luciferase assay system (Promega, Madison, WI) and Minilumat LB 9506 luminometer (EG & G Berthold, Bad Wildbad, Germany). For β-galactosidase activity assay, the luminescent β-galactosidase detection kit II was used (Clontech User Manual PT210601, Clontech, Palo Alto, CA).
Electrophoretic mobility shift assay
HUVECs were grown and treated as described (see “Measurement of E-selectin promoter activation”) after which they were harvested with versene and washed by PBS. Nuclei were isolated using the NE-PER nuclear extraction reagents (Pierce Biotechnology) following the manufacturer's instructions. For gel shift studies, a 25-bp oligonucleotide containing the human E-selectin NF-κB site was synthesized (5′-GCCATTGGGGATTTCCTCTTTACTG-3′ and complement, κB site underlined). Mutant human E-selectin NF-κB probe containing the sequence 5′-GCCATTGCTCATTTCCTCTTTACTG-3′ and complement also were synthesized (mutant base pair underlined). The oligonucleotides were labeled using the biotin 3′ end DNA labeling kit (Pierce Biotechnology) and annealed before use in gel shift assays. Nuclear extract (10 μg) was incubated with the biotinylated DNA probe at room temperature for 30 minutes in 1 × binding buffer (Pierce Biotechnology). Complexes were then separated on a 6% acrylamide nondenaturing gel and transferred to a nylon membrane for detection using the light-shift electrophoretic mobility shift assay (EMSA) kit (Pierce Biotechnology). In some studies, unlabeled DNA or antibodies to specific DNA-binding protein were incubated for 20 minutes prior to addition of biotinylated DNA.
Western blot assay
HUVECs were incubated with PF4 as described (see “Measurement of E-selectin promoter activation”). The cells were washed with 1 × PBS, and the cytoplasmic proteins were extracted with NE-PER nuclear extraction reagents (Pierce Biotechnology) following the manufacturer's protocol. Then 12 μg protein was separated on a 4% to 12% NuPAGE Bis-Tris gel (Invitrogen), transferred to a nylon membrane. Primary antibodies (anti–PIKKα; anti–P-IκBα; anti-IκBα) were used at 1:1000 dilution. Goat anti–rabbit HRP antibodies (1:10 000) were used for detection on combination with the enhanced chemiluminescence (ECL) plus Western blotting detection system (Amersham Biosciences, Uppsala, Sweden).
Statistics
Comparisons were made using a nonpaired t test as implemented in Graphpad Prism (V 4.0). Statistical significance was defined as P = .05.
Results
PF4 increases E-selectin RNA in HUVECs
As a first step in determining the involvement of PF4 in leukocyte recruitment to the vessel wall, total RNA from HUVECs exposed to PF4 (5 μg/mL) was subjected to transcriptional profiling using the Affymetrix U133A human gene chip. These experiments demonstrated that E-selectin RNA levels increased (>10-fold) on PF4 exposure, peaking at about 3 hours (data not shown). Neither L-selectin nor P-selectin was detected in these experiments, even after exposure to PF4.
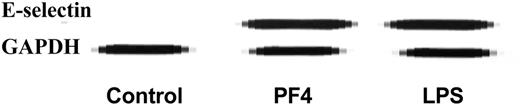
To confirm this observation, HUVECs were exposed to medium alone, PF4 (20 μg/mL), or lipopolysaccharide (LPS; 100 ng/mL). Total RNA was isolated and examined by the RPA. As seen in Figure 1, treatment of HUVECs with PF4 resulted in a comparable increase in E-selectin RNA as compared to LPS. As expected, no E-selectin was detected in human vascular smooth muscle cells (HVSMCs) in the absence or presence of PF4 (not shown).
PF4 increases E-selectin RNA in HUVECs. RPA of E-selectin RNA in HUVECs exposed to media alone (control), PF4 (20 μg/mL), or LPS (100 ng/mL). GAPDH was also measured as an internal control. Data are representative of 3 experiments performed.
PF4 increases E-selectin RNA in HUVECs. RPA of E-selectin RNA in HUVECs exposed to media alone (control), PF4 (20 μg/mL), or LPS (100 ng/mL). GAPDH was also measured as an internal control. Data are representative of 3 experiments performed.
To determine the concentration dependence of this effect, HUVECs were exposed to various concentrations of PF4 for 3 hours and E-selectin RNA expression was again measured by the RPA. As shown in Figure 2A-B, E-selectin RNA levels increased with exposure to as little as 2.5 μg/mL PF4, reaching maximal levels at 20 μg/mL. As expected, similar experiments using smooth muscle cells failed to demonstrate the presence of E-selectin RNA (not shown). Neutrophil-activating protein 2 (NAP-2), a homologous platelet-specific chemokine, had no effect on E-selectin RNA levels under identical conditions (not shown).
Dose response and time course of increased E-selectin RNA expression by PF4. (A) RPA of E-selectin RNA in HUVECs exposed to increasing concentrations of PF4. GAPDH was also measured as an internal control. (B) Relative expression of E-selectin RNA from the experiment in panel A as compared to GAPDH. Data were quantitated as described in “Materials and methods.” (C) RPA demonstrating time course of E-selectin expression after exposure to PF4. (D) Quantitation of data in panel C. Data shown are representative of 3 independent experiments.
Dose response and time course of increased E-selectin RNA expression by PF4. (A) RPA of E-selectin RNA in HUVECs exposed to increasing concentrations of PF4. GAPDH was also measured as an internal control. (B) Relative expression of E-selectin RNA from the experiment in panel A as compared to GAPDH. Data were quantitated as described in “Materials and methods.” (C) RPA demonstrating time course of E-selectin expression after exposure to PF4. (D) Quantitation of data in panel C. Data shown are representative of 3 independent experiments.
It is known that induction of E-selectin expression by cytokines during inflammation is transient, with maximal RNA levels seen at 2 to 4 hours, and a return to baseline by 24 hours.38 To determine the time course of E-selectin RNA induction by PF4, HUVECs were exposed to PF4 (20 μg/mL) for various times and E-selectin RNA levels determined by RPA. As seen in Figure 2C-D, PF4 increased E-selectin RNA expression in a time-dependent manner, with maximal stimulation at 3 hours, and returning to baseline by 8 hours. These data demonstrate that PF4 induction of E-selectin RNA expression occurs with a similar time course as with inflammatory cytokines such as IL-1 and tumor necrosis factor α (TNF-α).38
PF4 increases cell surface E-selectin in HUVECs
Increased RNA levels do not always result in increased protein expression. We therefore examined the ability of PF4 to increase surface E-selectin levels on HUVECs by flow cytometry. As shown in Figure 3, HUVECs exposed to PF4 (20 μg/mL) increased cell surface E-selectin (CD 62E) in a time-dependent manner. This increase was due to increased numbers of cells expressing E-selectin (increasing number of positive cells) as well as increased expression levels on individual cells (increasing brightness of the positive cells). The time course of increased protein expression closely follows the increase in RNA levels seen (Figure 2).
PF4 increases E-selectin surface expression on HUVECs. (A) Contour plots of flow cytometric analysis of HUVECs before (0 hours) and after (2 and 6 hours) PF4 exposure. Data are gated on CD31+ cells as described in “Materials and methods.” Data shown are representative of 3 or more independent experiments. (B) Quantitation of E-selectin expression. GMFI indicates the geometric mean of the fluorescence intensity. Data are presented as the mean ± SEM of 3 to 4 independent experiments.
PF4 increases E-selectin surface expression on HUVECs. (A) Contour plots of flow cytometric analysis of HUVECs before (0 hours) and after (2 and 6 hours) PF4 exposure. Data are gated on CD31+ cells as described in “Materials and methods.” Data shown are representative of 3 or more independent experiments. (B) Quantitation of E-selectin expression. GMFI indicates the geometric mean of the fluorescence intensity. Data are presented as the mean ± SEM of 3 to 4 independent experiments.
Activation of the E-selectin promoter by PF4
To examine the mechanism by which PF4 increases E-selectin levels, we asked if PF4 is able to activate the E-selectin promoter. To do this, we transfected HUVECs with DNA corresponding to the 840 bp upstream of E-selectin37 driving the expression of luciferase (-840; Figure 4), and determined luciferase activity as a measure of promoter activity. Incubation of HUVECs with PF4 increased E-selectin promoter activity 7-fold (Figure 4A), demonstrating that PF4 can activate this promoter.
As a first step in determining the transcriptional mechanism of this activation, we constructed deletion mutants of the promoter region and examined their activity using a luciferase expression assay. Three subdomains within this promoter have been identified as potentially important for cytokine induction of E-selectin expression.39-41 These subdomains are binding sites for the transcription factors activator protein 1 (AP-1) and NF-κB, as well as a palindromic site within this region. A schematic of these constructs is shown in Figure 4B. Sequential deletion of the 5′ end of the promoter up to -130 bp had no significant effect on PF4 activation (Figure 4A). However, removal of the sequence between -130 and -40 completely eliminated the ability of PF4 to activate this promoter. This suggested that the PF4-sensitive cis-acting element in the promoter was the binding site for NF-κB.
Because endotoxin (LPS) is a potent activator of NK-κB, we next examined the possibility that LPS contamination of our purified PF4 was responsible for our observations. All PF4 and NAP-2 preparations were demonstrated to be free of endotoxin by testing in the University of Pennsylvania Cell Center. However, to more formally exclude LPS activity, E-selectin expression was induced in the presence of polymyxin B (Figure 4C). In these experiments, polymyxin B completely inhibited LPS induction of E-selectin, whereas it had no effect on the activity of PF4. In addition, boiling of PF4 prior to incubation with cells completely destroyed the ability of PF4 to induce NF-κB activity (data not shown). Finally, platelet releasate induced E-selectin as well as purified PF4 (Figure 4C). Taken together, these data exclude LPS contamination of PF4.
PF4 activation of NF-κB
To gain direct support for the importance of the NF-κB–binding site in PF4 activation of E-selectin expression, we created a construct (Mut in Figure 4B) that consisted of the full E-selectin promoter with 3 nucleotides mutated in the NF-κB–binding site. As demonstrated by a lack of luciferase activity, PF4 was unable to activate this mutant promoter (Figure 4A). These data provide direct evidence that the NF-κB–binding site is required for PF4 activation of the E-selectin promoter.
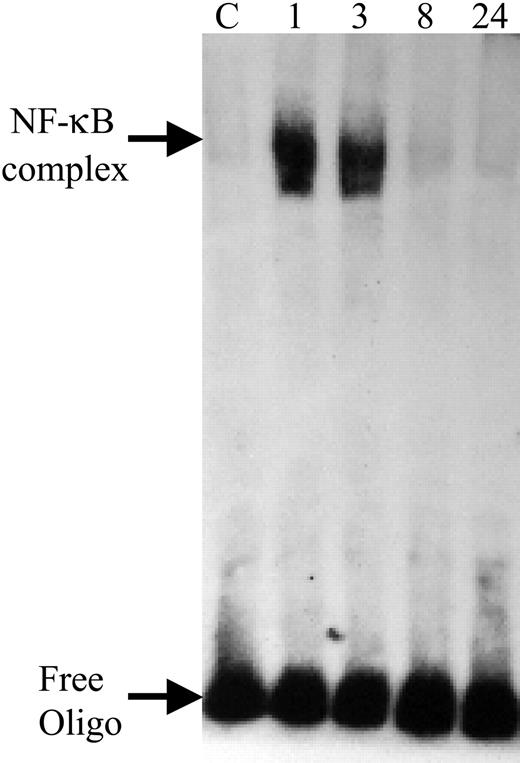
We next examined the ability of PF4 to activate and promote the binding of NF-κB to DNA. To do this, we treated HUVECs with PF4 as described, isolated nuclear extracts from these cells, and performed EMSAs. Figure 5 demonstrates that PF4 treatment of HUVECs results in binding of NF-κB to its DNA-binding site, as manifested by a shift in the migration of biotin-labeled DNA containing this sequence. The binding of NF-κB occurs with a time course similar to the induction of E-selectin RNA (Figure 2).
Time course of PF4 activation of NF-κB. EMSA demonstrating the binding of nuclear proteins to an oligonucleotide containing the NF-κB–binding site. HUVECs were incubated in the absence (C) or presence of PF4 (20 μg/mL) for 1, 3, 8, or 24 hours prior to assay. NF-κB–complex denotes shifted band corresponding to the binding of NF-κB to the oligonucleotide; free oligo denotes unbound oligonucleotide. Data are representative of at least 3 independent experiments.
Time course of PF4 activation of NF-κB. EMSA demonstrating the binding of nuclear proteins to an oligonucleotide containing the NF-κB–binding site. HUVECs were incubated in the absence (C) or presence of PF4 (20 μg/mL) for 1, 3, 8, or 24 hours prior to assay. NF-κB–complex denotes shifted band corresponding to the binding of NF-κB to the oligonucleotide; free oligo denotes unbound oligonucleotide. Data are representative of at least 3 independent experiments.
To investigate the specificity of this gel shift, a variety of additional EMSA experiments were performed (Figure 6). Observation of the gel shift was dependent on the presence of nuclear extract (lane 1). LPS, a known activator of endothelial cells and NF-κB, activated NF-κB in our assay (lane 2). The presence of the NF-κB/DNA complex (shifted band) required the presence of the wild-type NF-κB–binding site, as demonstrated by lack of direct binding to or competition by the mutant oligonucleotide (lanes 5 and 6, respectively), whereas excess unlabeled wild-type oligonucleotide was able to effectively compete with biotinylated oligonucleotide for NF-κB binding. These data demonstrate that the NF-κB–binding site is required for the shift induced by PF4.
Specificity of the NF-κB–binding site. EMSAs were performed using nuclear protein from HUVECs and biotin-labeled probes (WT [wild-type] oligo, biotin or Mut oligo, biotin) as described in Figure 5. Symbols above each lane (+/-) denote the presence or absence of various reagents. For competition experiments (lanes 4 and 6), a 50-fold excess of unlabeled oligo (WT oligo or Mut oligo) was used. Data are representative of at least 3 independent experiments.
Specificity of the NF-κB–binding site. EMSAs were performed using nuclear protein from HUVECs and biotin-labeled probes (WT [wild-type] oligo, biotin or Mut oligo, biotin) as described in Figure 5. Symbols above each lane (+/-) denote the presence or absence of various reagents. For competition experiments (lanes 4 and 6), a 50-fold excess of unlabeled oligo (WT oligo or Mut oligo) was used. Data are representative of at least 3 independent experiments.
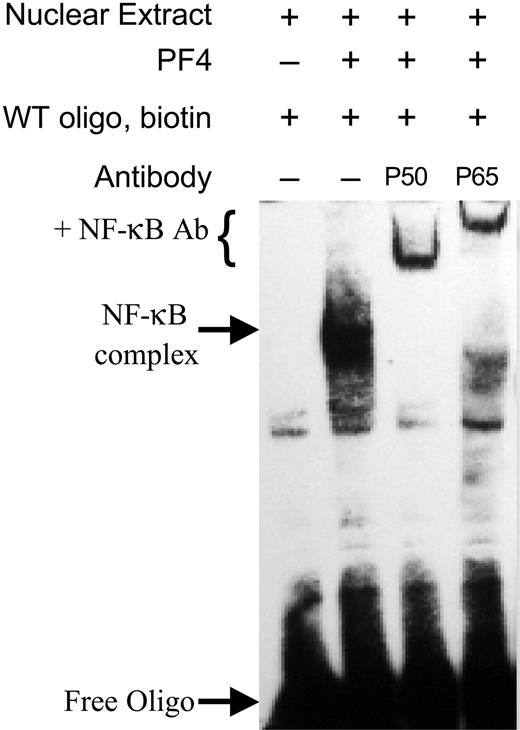
To demonstrate that the protein binding to the NF-κB–binding site is NF-κB, we performed experiments where antibodies to NF-κB were incubated with the NF-κB/DNA complex prior to gel electrophoresis (super shift assays). Such experiments performed with antibodies to either the P50 or P65 subunit of NF-κB resulted in the expected super shift of the NF-κB/DNA complex (Figure 7). No such shift was seen in the presence of nonspecific control antibodies (not shown). These data confirm the identity of the nuclear-binding component at NF-κB.
Specificity of NF-κB binding. EMSAs were performed using nuclear protein from HUVECs as described in Figure 5. Prior to electrophoresis, complexes were incubated with antibodies to either the P50 or P65 subunit of NF-κB. Binding of these antibodies results in a super shift (denoted by +NF-κB Ab) due to the increased mass of the antibody. Data are representative of at least 3 independent experiments.
Specificity of NF-κB binding. EMSAs were performed using nuclear protein from HUVECs as described in Figure 5. Prior to electrophoresis, complexes were incubated with antibodies to either the P50 or P65 subunit of NF-κB. Binding of these antibodies results in a super shift (denoted by +NF-κB Ab) due to the increased mass of the antibody. Data are representative of at least 3 independent experiments.
To investigate the pathway of NF-κB activation by PF4, we examined the phosphorylation of IκBα and IKKα, 2 important regulatory proteins in the NF-κB activation pathway.42 Phosphorylation of these proteins is associated with receptor-mediated activation of NK-κB. As can be seen in Figure 8, phosphorylation of IκBα is observed with a time course consistent with NF-κB activation (Figure 8). As expected, the increase in phosphorylated IκBα is accompanied by a concomitant decrease in native IκBα. The subsequent decrease in phosphorylated IκBα is likely due to proteosomal degradation, which is typically observed for this protein.42 Interestingly, levels of IκBα did not return to normal until after 6 hours (Figure 8). This is somewhat delayed compared to that reported by others.43 Further studies will be required to determine the significance and mechanism of this phenomenon and to determine if it is agonist or cell specific.
Phosphorylation of IκBα and IKKα. Western blot analysis of cytoplasmic proteins extracted from control (con) and PF4-exposed HUVECs. HUVECs were incubated with PF4 for the indicated times. Data are representative of 3 independent experiments.
Phosphorylation of IκBα and IKKα. Western blot analysis of cytoplasmic proteins extracted from control (con) and PF4-exposed HUVECs. HUVECs were incubated with PF4 for the indicated times. Data are representative of 3 independent experiments.
We also observed that IKKα, part of the IKK complex responsible for phosphorylation of IκBα, is phosphorylated with a similar time course. Taken together, these data further support the notion that PF4 induction of E-selectin is via NF-κB activation.
LRP mediates PF4 induction of E-selectin
Finally, we wished to identify the cellular receptor responsible for PF4 induction of E-selectin expression in HUVECs. We have previously demonstrated that PF4 binds to the LDL receptor.36 Although this receptor is not known to mediate intracellular signaling, LRP is.15-19 Further, although the presence of LRP on endothelial cells is not a universal finding, LRP on endothelial cells has been reported.44,45
To examine whether or not E-selectin induction by PF4 is mediated through LRP in HUVECs we studied the effects of PF4 on the LRP-deficient murine embryonic fibroblasts (due to targeted gene disruption; MEF-2) in comparison to the wild-type fibroblasts (MEF-1),46,47 using the luciferase reporter as described in Figure 4. Whereas PF4 activated the E-selectin promoter in the wild-type MEF-1 cells to the levels seen in HUVECs, it has no effect on MEF-2 cells (Figure 9A), suggesting that LRP is essential for this function of PF-4.
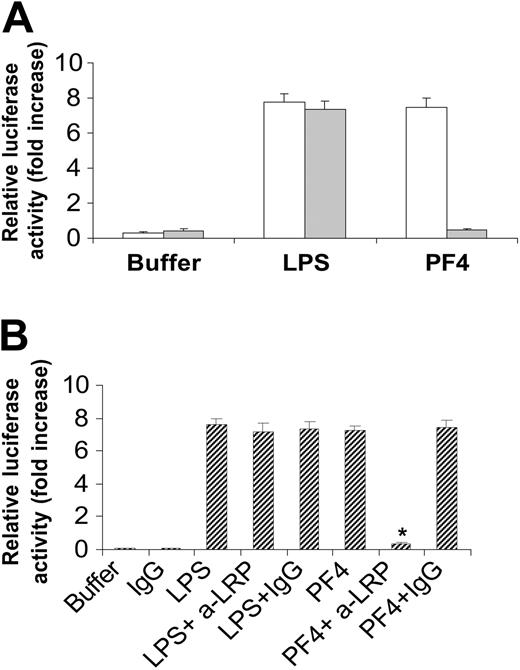
LRP is required for PF4 induction of E-selectin in HUVECs. (A) MEF-1 (□) or MEF-2 (▦) cells were incubated in the presence of either PF4 (20 μg/mL) or LPS (100 ng/mL) for 4 hours prior to measurement of luciferase activity as described in Figure 4. Similar experiments were also performed in HUVECs (B). Buffer indicates no PF4 control; α-LRP, anti-LRP antibody; IgG, isotype control. The asterisk indicates statistical significance compared to PF4 alone (P < 0.001). Data are the mean ± SEM of 3 independent experiments performed in triplicate.
LRP is required for PF4 induction of E-selectin in HUVECs. (A) MEF-1 (□) or MEF-2 (▦) cells were incubated in the presence of either PF4 (20 μg/mL) or LPS (100 ng/mL) for 4 hours prior to measurement of luciferase activity as described in Figure 4. Similar experiments were also performed in HUVECs (B). Buffer indicates no PF4 control; α-LRP, anti-LRP antibody; IgG, isotype control. The asterisk indicates statistical significance compared to PF4 alone (P < 0.001). Data are the mean ± SEM of 3 independent experiments performed in triplicate.
To demonstrate that LRP mediates PF4 activation of the E-selectin promoter in HUVECs, we examined the effect of anti-LRP antibodies on PF4-treated HUVECs. Anti-LRP, but not the isotype control immunoglobulin G (IgG), added at a concentration of 10 μg/mL abolished (> 90% inhibition) the ability of PF4 to activate the E-selectin promoter (Figure 9B). Furthermore, anti-LRP antibodies did not affect the LPS-mediated activation. Although we cannot rule out cross-reactivity of this antibody to other LDL receptor family members, such as apoER2, taken together with the lack of response by MEF-2 cells, these results strongly suggest that LRP is the major cellular receptor that directly or indirectly mediates this PF4 signal transduction activity. This activity differs from the typical LPS pathway and supports the exclusion of endotoxin contamination in our assays.
Discussion
The importance of leukocyte recruitment in the pathogenesis of atherosclerosis is viewed as a seminal event.1 Adhesion molecules that promote leukocyte-endothelial interactions, including E-selectin, are known to be expressed by the endothelium of atherosclerotic lesions in humans.6-8 Further, endothelial selectins (P and E) are important for the development of atherosclerotic lesions in LDL receptor knockout mice.4 Stimuli that promote selectin expression, and therefore leukocyte recruitment, are expected to accentuate atherosclerotic disease.
In the current study, we have demonstrated for the first time that a platelet-specific chemokine, PF4, increases the expression of an important adhesion molecule, E-selectin, by endothelium. The mechanism for this increased expression involves both binding to LRP on the cell surface and activation of NF-κB, resulting in activation of the E-selectin promoter. This conclusion comes from the observations that: (1) PF4 increases E-selectin RNA and cell surface protein expression in HUVECs, (2) PF4 activates the E-selectin promoter, (3) this activation is dependent on the NF-κB consensus binding site, (4) activated NF-κB is present in the nuclei of PF4-treated HUVECs as demonstrated by specific DNA binding, and (5) PF4 does not increase E-selectin expression in cells lacking LRP or that have been treated with anti-LPR antibodies. Finally, we have also demonstrated that E-selectin surface protein expression is increased by endothelial cells exposed to PF4.
Our data are consistent with the well-established facts that the 5′ flanking region of the E-selectin gene is critical for E-selectin expression39-41 and that NF-κB activation is central to E-selectin gene activation.39,41,48,49 It is interesting to note that a recent report suggests that activation of NF-κB is sufficient for promotion of E-selectin expression,48 although earlier work suggest that other factors also modulate expression.39,41 Therefore, activation of NF-κB by PF4 is a highly significant finding.
Platelets are now thought to play important roles in the pathogenesis of atherosclerotic lesions. The absence of P-selectin (which is important for platelet-endothelial interactions) protects against development of atherosclerotic lesions in both LDL receptor50 and apoE5,51 knockout mice, especially early in lesion development.50 In apoE knockout mice, P-selectin expressed by both platelets and endothelium is important for atherosclerotic lesion formation, although endothelial P-selectin seems to be of relatively greater importance,23 possibly due to its involvement in platelet rolling along the endothelium.52
More recently it has been demonstrated that, in apoE-deficient mice, platelet adhesion to atherosclerosis prone regions of the carotid artery precedes both leukocyte adhesion and visible lesion formation.20 Blockade of platelet adhesion in this model, using either α-glycoprotein (GP) IIb-IIIa or α-GPIbα, decreased platelet adhesion and leukocyte recruitment to these regions and decreased lesion size. It has also been shown that activated platelets, as well as platelet-leukocyte aggregates, interact with atherosclerotic lesions in apoE-deficient mice.22
PF4 is stored in platelet α-granules and released on platelet activation. The serum concentrations of PF4 exceeds 8 μg/mL during platelet activation.53 Because the concentration of PF4 within α-granules is estimated to be 1500 μg/mL,54 even higher concentrations of PF4 may be present at the site of platelet activation. It is therefore likely that PF4 levels at sites of platelet activation would be sufficient to increase E-selectin expression by endothelial cells under pathophysiologic conditions such as vascular inflammation or atherosclerotic vascular disease or both. Importantly, it has been shown (in both apoE-deficient mice and in isolated cell systems) that platelet-derived chemokines (RANTES and PF4) are delivered to endothelial cells and macrophages by activated platelets in a P-selectin–dependent manner.22,55 Further, we have shown that PF4 is present in the endothelium of human atherosclerotic lesions,34 demonstrating that PF4 is present at the appropriate location for E-selectin induction.
Data presented in our study are consistent with recent studies suggesting that LRP is not only an endocytosis receptor but also mediates signal transduction.15-19 Furthermore, although the presence of LRP on endothelial cells is not a uniform finding,44,45 our data show that LRP is the cellular receptor on endothelial cells that (directly or indirectly) mediate this effect of PF4 and suggest LRP for the first time as proinflammatory receptor. The fact that anti-LRP antibodies totally inhibited PF4-mediated signal transduction in endothelial cells and that cells that do not express LRP were resistant to PF4 but not to LPS, strongly suggest that LRP directly mediates PF4 signaling on endothelial cells. Although we cannot exclude the possibility that LRP mediates these effects of PF4 indirectly by forming a triple complex with another signal transducing receptor as it has been suggested for tPA,19 the fact that PF4 has no enzymatic activity suggests this scenario is unlikely. In any case, in future studies we will explore whether PF4/LRP directly initiates intracellular signaling or PF4/LRP signaling is initiated on endocytosis.
In summary, we have identified a new signal transduction function of PF4, activation of NF-κB, resulting in up-regulation of E-selectin on the surface of endothelium. We have also identified the cellular receptor that mediates this function.
We propose PF4 as a link between platelets, endothelium, and leukocytes in inflammatory disease like atherosclerosis. PF4 released from activated platelets in the vasculature may interact with LRP on endothelial cells in atherosclerotic prone regions of the vasculature or over early atherosclerotic lesions. Once bound, PF4 may increase the expression of E-selectin, promoting leukocyte adhesion and lesion expansion. Other potentially proatherogenic properties of PF4, including alteration of LDL metabolism35,36 and monocyte recruitment,56 may also promote atherosclerotic lesion formation. Future studies in murine models are required to determine the in vivo consequences of PF4 activation of NF-κB in atherosclerosis and inflammatory vascular disorders.
Prepublished online as Blood First Edition Paper, December 9, 2004; DOI 10.1182/blood-2004-07-2617.
Supported by National Institutes of Health grant K08 HL04245 (B.S.S.) and HL068631 (K.B.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.



![Figure 6. Specificity of the NF-κB–binding site. EMSAs were performed using nuclear protein from HUVECs and biotin-labeled probes (WT [wild-type] oligo, biotin or Mut oligo, biotin) as described in Figure 5. Symbols above each lane (+/-) denote the presence or absence of various reagents. For competition experiments (lanes 4 and 6), a 50-fold excess of unlabeled oligo (WT oligo or Mut oligo) was used. Data are representative of at least 3 independent experiments.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/105/9/10.1182_blood-2004-07-2617/6/m_zh80090577870006.jpeg?Expires=1769111174&Signature=SWVFJkWb6zIyVzZM03j0MWHT9asgicqLZMsKxL1qMvwDaEsqeEy~cV1tVLXDjbPWSTCSF0gcDGrBHREAxN3b2xSfJQgJ9OkkDqx4vjg47whfQU166i9sHcSLUAgwv4JsF0hzNPIC7sOVhu0TfFQyqG4IpJS3CmAlNnr7TXFOub0dI-cx80wvocEcdOlTU4YuiY-7CzuUvGUInGz97hqZZGRWrWQMmB4lEKA-PX4EwYWqIrv7g4S2DyqzTwnQNEEf47lKxoQM8gYAljROfhqxijquVLXY5i5UO9yR2O9avmGmJOm9A5Vbh9JNucEMxVKSawLAGWwOkqXjt6PDqQpEDw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal