Abstract
Activating mutations in the FLT3 tyrosine kinase (TK) occur in approximately 35% of patients with acute myeloid leukemia (AML). Therefore, targeting mutated FLT3 is an attractive therapeutic strategy, and early clinical trials testing FLT3 TK inhibitors (TKI) showed measurable clinical responses. Most of these responses were transient; however, in a subset of patients blast recurrence was preceded by an interval of prolonged remission. The etiology of clinical resistance to FLT3-TKI in AML is unclear but is of major significance for the development of future therapeutic strategies. We searched for mechanisms of resistance in 6 patients with AML who had relapses upon PKC412 treatment. In an index AML patient, an algorithm of analyses was applied using clinical material. In vivo and in vitro investigation of primary blasts at relapse revealed persistent TK phosphorylation of FLT3 despite sufficient PKC412 serum levels. Through additional molecular analyses, we identified a single amino acid substitution at position 676 (N676K) within the FLT3 kinase domain as the sole cause of resistance to PKC412 in this patient. Reconstitution experiments expressing the N676K mutant in 32D cells demonstrated that FLT3-ITD-N676K was sufficient to confer an intermediate level of resistance to PKC412 in vitro. These studies point out that a genetically complex malignancy such as AML may retain dependence on a single oncogenic signal.
Introduction
The FLT3 receptor tyrosine kinase (RTK) is expressed in 70% to 90% of patients with acute myeloid leukemia (AML). Activating FLT3 gene mutations are present in the leukemic blasts of approximately 35% of patients with AML. Two distinct classes of FLT3 mutations have been recognized: 20% to 27% are internal tandem duplications, located in the juxtamembrane (JM) region,1-3 and 7% are point mutations in the tyrosine kinase domain at position D835 of FLT3.4,5 Recently, additional activating mutations at positions 836, 840, 841, and 842 have been described.6-9 FLT3 gene mutations lead to autophosphorylation and enhanced receptor signaling, promoting ligand-independent cell proliferation and inhibition of apoptosis of the malignant clone. However, others and we10,11 have recently shown that FLT3-ITD and FLT3-TKD mutations display significant differences in their signaling properties that could account for observed differences in their transforming capacity.
Treatment with FLT3 tyrosine kinase inhibitors (TKIs) is a novel and promising tool in the therapy for AML. Recently, several FLT3 kinase inhibitors have been developed and tested in early clinical trials.12-14 The kinase inhibitor PKC412 is a derivative of the alkaloid staurosporine15 and an inhibitor of several kinases, including PKC-α, -β, and -γ, VEGF-R2, c-kit, and PDGF-α and -β. Moreover, it is described as a potent inhibitor of mutant FLT3 receptors and has an IC50 lower than 10 nM.16 Preliminary results investigating FLT3-TKI PKC412 in patients with relapsed/refractory AML revealed that 23 of 35 (66%) patients with mutated FLT3 and 23 of 57 (40%) patients with wild-type FLT3 showed a 50% or greater blast response in peripheral blood.17 However, a common theme in all studies reporting clinical results using FLT3-TKI is the observation that despite remarkable efficacy in reducing the leukemic clone in a subset of patients with AML, remission in patients who have had single-agent therapy tends to be short and secondary resistance develops rapidly.12-14,17,18 Therefore, current strategies are to use FLT3-TKI in combination with chemotherapy, with FLT3 siRNA approaches, or with HSP-90 inhibitors.19-21
Little is known about the mechanisms of clinical resistance with single-agent therapy using FLT3-TKI. Thus, the goal of the present report was to investigate molecular mechanisms of clinical resistance in AML patients treated within a clinical phase 2 study using PKC412.
Patients, materials, and methods
Mutation analysis
Blood samples, bone marrow samples, or both were obtained from 6 AML patients enrolled in a clinical phase 2 study (PKC412A-2104 trial) investigating the efficacy and toxicity of PKC412.12 Informed consent was obtained from all patients. The clinical trial was conducted in accordance with the Declaration of Helsinki and with institutional review board approval. RNA from peripheral blood (PB) or bone marrow (BM) was extracted using the RNeasy Mini-Kit (Qiagen, Hilden, Germany), and cDNA was generated with the SuperScript First-Strand Synthesis System for reverse transcription–polymerase chain reaction (RT-PCR) according to the manufacturer's protocol (Invitrogen, Groningen, The Netherlands). Screening of the N676 region corresponding to amino acids 647-684 (complete exon 16) was performed on genomic DNA using the following primers: N676 forward, ccagtcttgaacttctgacctc; N676 reverse, tgttgttgtttttaagcaggtg.
The FLT3 coding region of patient UPN1 was amplified by PCR and sequenced using the following primers (covering the following amino acid [AA] codons): FLT3mRNA forward, tgccgctgtcgttgtttt; FLT3mRNA reverse, agaaggccttggatgcaga (located in the 5′ and 3′ nontranslated regions); FLT3mRNA forward 2 (FLT3mf2), caaactcctcagaccacattg (covering AA 250-257); FLT3mr2, catgatatctcgagccaatcc (covering AA 830-837); FLT3mf3, aggagttgtttccatggtc (covering AA 123-129); FLT3mr3, gtgtaattggtagcttcactctg (covering AA145-152); FLT3mf4, acggatacagcatatccaag (covering AA 399-405); FLT3mf5, gcactcatgtcagaactcaag (covering AA 656-663).
DNA constructs and vectors
A human FLT3-ITD construct, subcloned into the pAL expression vector under control of the 5' long terminal repeat (LTR) of the Moloney murine sarcoma virus (MoMSV) and the plasmid pMAM/BSD were used.22 The N676K point mutation was introduced into this FLT3-ITD construct by Medigenomix (Martinsried, Germany). Vector constructs were confirmed by nucleotide sequencing.
Transfection of 32D cells
Parental 32D cells were maintained in RPMI 1640 with 10% FCS and 10% WEHI-conditioned medium as a source of IL-3. Ten micrograms plasmid DNA and 1 μg pMAM/BSD were cotransfected into 32D cells by electroporation. Cells were selected with 15 μg/mL blasticidine (Invitrogen) in IL-3–supplemented culture. Polyclonal cell lines were used for experiments after a period of IL-3 starvation of at least 7 days. FLT3-receptor surface expression was confirmed by fluorescence-activated cell sorter (FACS) analysis.
MTS assay
In the presence of varying concentrations of PKC412A (kindly provided by T. Meyer, Novartis, Basel, Switzerland), 2 × 103 32D cells expressing FLT3-ITD and FLT3-ITD-N676K were seeded in triplicate in 96-well plates and grown in RPMI 1640 medium. Relative numbers of viable cells were determined after 48, 72, and 96 hours using the Aqueous One Solution (Promega, Madison, WI).
Cell cycle
Cell cycle analysis was performed as described previously.23
Isolation of primary AML blasts and cell culture
Bone marrow (BM) and peripheral blood (PB) samples with heparin as the anticoagulant were obtained from 6 patients with AML after informed consent at different time points during study treatment and at relapse; in patients with peripheral pancytopenia, only BM samples were taken. Mononuclear cells (MNCs) were isolated immediately by means of Ficoll-Hypaque (Seromed, Berlin, Germany) density gradient centrifugation. For immunoblotting, freshly isolated MNCs were lysed either directly or after incubation in RPMI 1640 supplemented with 20 mM HEPES (pH 7.3), 50 mM β-mercaptoethanol, and 2 mM L-glutamine containing varying amounts of tyrosine kinase inhibitors (PKC412 or AST487). For cell cycle analysis, MNCs were maintained in RPMI 1640 medium supplemented as described with the addition of 10% FCS.
Ex vivo bioassay
The human leukemia cell line MV4-11 harboring an FLT3-ITD mutation was incubated with 1 mL serum spiked with or without 100 nM PKC412 at 37°C for 30 minutes. Cells were washed, lysed, and analyzed for FLT3-tyrosine phosphorylation by immunoblotting.
Protein extract preparation and Western blotting
Cells (2 × 106) were incubated in the presence of different concentrations of PKC412A and flat ligand (FL) (R&D Systems, Minneapolis, MN), different concentrations of the novel tyrosine kinase inhibitor AST487 (kindly provided by J. Roesel, Novartis, Basel, Switzerland), and in combination for 30 minutes at 37°C. Preparation of cellular lysates was performed as described previously.24 Protein lysates were subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and blotted onto nitrocellulose membrane (Amersham, Freiburg, Germany), as previously described.23 The following antibodies were used: anti–phospho-Flt3, anti–phospho-STAT5 (Cell Signaling Technology, Frankfurt, Germany), anti–Flt3, anti–STAT5, and anti–β-actin (Santa Cruz, Heidelberg, Germany). Densitometric analysis was performed using the program Gel-Pro Analyzer (Media Cybernetics/Meyer Instruments, Houston, TX).
Colony assays
Transfected 32D cells were pelleted, washed extensively in PBS, and seeded in 6-well culture plates at a fixed density (3000 cells/mL) in complete RPMI 1640/methylcellulose (Methocult; Stem Cell Technologies, Vancouver, Canada) medium without growth factors containing 10% FCS and incubated for 7 days at 37°C. Colony counts were determined at 3 different time points using the Gel-Pro Analyzer (Media Cybernetics) software. Images were taken using the chemiluminescence software CSX-1400M (Cybertech, Berlin, Germany).
Quantitative real-time PCR (TaqMan)
Total RNA was isolated using Trizol reagent (Invitrogen, Karlsruhe, Germany) according to the manufacturer's recommendations. One microgram total RNA was used for reverse transcription. The cDNA was diluted to 200 μL with ddH2O, and 2.5 μL was used for each PCR reaction. Quantification of mRNA levels was carried out using a real-time fluorescence detection method with an ABI 4700 analyzer (ABI Prism, Foster City, CA), as previously described.22 Relative gene expression levels were calculated using standard curves generated by serial dilutions of cDNA from 32D cells, with expression of GAPDH serving as the reference. All samples were independently analyzed at least twice for each gene.
Measurement of plasma levels of PKC412 and its metabolites
Pharmacokinetic analysis of PKC412 and of its metabolites was performed as previously described.12 Additionally, at various times during PKC412 therapy, plasma levels of PKC412 and its metabolites were measured by extraction from plasma using diethylether in the presence of an internal standard. An aliquot of the ether phase was dehumidified, the residue was dissolved in the mobile phase, and native fluorescence was measured after isocratic high-performance liquid chromatography (HPLC) separation. The sensitivity threshold of this assay was 10 ng/mL, and the intra-assay and interassay variances were 6%.
Results
Clinical results
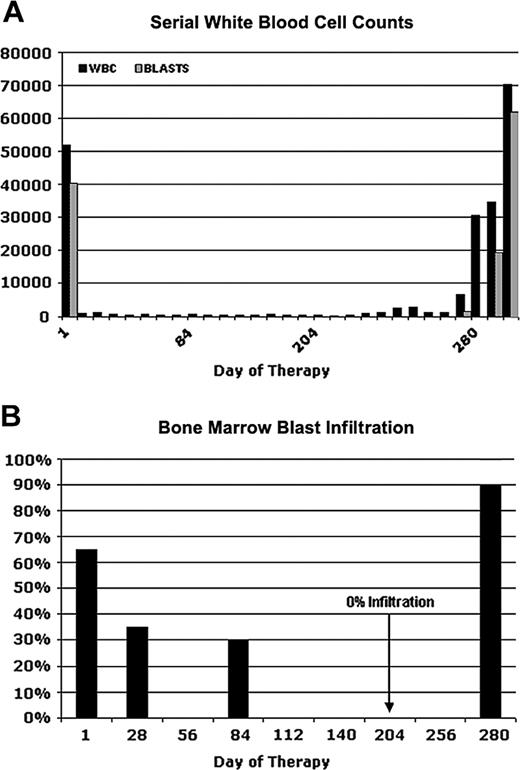
Figure 1 depicts serial blood counts and percentages of bone marrow blast infiltration in a 60-year-old male patient (UPN1) who was enrolled in an open label, multicenter phase 2 study with the tyrosine kinase inhibitor PKC412.12,17,18 An initial diagnosis of AML had been made 3 months before the start of the study medication. At that time, cytogenetic analysis revealed a normal karyotype. The patient received induction chemotherapy with mitoxantrone, cytarabine, and etoposide but had persistent leukemic blasts in PB and BM at day 14 after the start of chemotherapy. Therefore, reinduction therapy with amsacrine and high-dose cytarabine was started. However, with the reconstitution of hematopoiesis, he again had 30% leukemic blasts in BM. Because FLT3 screening revealed an internal tandem duplication, he was enrolled in the PKC412-2104 trial and started therapy with 100 mg PKC412 twice a day.
After 1 week of PKC412 treatment, the patient had a complete peripheral blast response (Figure 1A) but still had leukopenia and neutropenia. In addition, he still had thrombocytopenia and frequently required platelet transfusion. BM aspirates showed a gradual reduction of blast infiltration with complete elimination of blasts on day 204 (Figure 1B). On day 280 of PKC412 treatment, the patient again had bone marrow infiltration of more than 90% blasts with accompanying peripheral leukocytosis as high as 70 000/μL (Figure 1A). Further dose escalation of PKC412 did not produce any benefit. The patient was taken off study on day 296 of PKC412 treatment.
Clinical response and relapse in a patient (UPN1) receiving PKC412. Serial white blood cell counts (A) and percentages of bone marrow blast infiltration (B) at several time points are depicted. On day 280, the patient had a fulminant relapse while on PKC412 treatment. Bone marrow aspiration was performed on days 1, 28, 84, 204, and 280.
Clinical response and relapse in a patient (UPN1) receiving PKC412. Serial white blood cell counts (A) and percentages of bone marrow blast infiltration (B) at several time points are depicted. On day 280, the patient had a fulminant relapse while on PKC412 treatment. Bone marrow aspiration was performed on days 1, 28, 84, 204, and 280.
Identification of FLT3-N676K mutation in primary AML blasts and investigation of its functional role
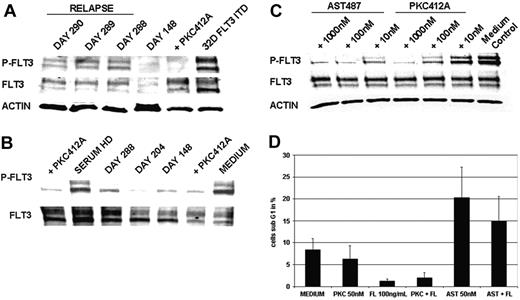
As a first approach to elucidate molecular mechanisms of clinical resistance, we investigated FLT3-kinase activity in vivo at the time of relapse by Western blot analysis of cellular lysates from primary AML blasts of patient UPN1. This analysis revealed persistent tyrosine phosphorylation of FLT3 in 3 consecutive PB samples at the time of relapse (Figure 2A). A PB sample drawn during hematologic remission on day 148 showed a faint FLT3-specific band corresponding to almost complete clearance of leukemic blasts from PB (Figure 2A). Thus, this analysis revealed that, at the time of relapse, FLT3 tyrosine kinase activity was not suppressed.
Trough levels of PKC412 and of its active metabolite, CGP62221, were obtained during therapy, as indicated in Table 1. This analysis showed that serum concentrations of PKC412 and of its active metabolite, CGP62221, decreased after day 7, similar to what has been reported in an earlier trial investigating PKC412 in AML patients.12 However, at relapse (day 288), the actual level of the sum of PKC412 and CGP62221 was similar (44.6 nM) to trough levels obtained earlier (day 148 and day 176), when the patient was in continued remission. Assuming that CGP62221 has an IC50 for FLT3-ITD receptors similar to that of PKC412, this level suggests adequate FLT3-ITD inhibition at the time of relapse (day 288).
Trough drug levels of PKC412 and of its active metabolite GCP62221 during therapy with PKC412 in patient UPN1
Day of treatment . | PKC412 trough level, ng/mL . | PKC412 trough level, nM* . | CGP62221 trough level, ng/mL . | CGP62221 trough level, nM* . | Sum of trough levels of PKC412 and CGP62221* . |
|---|---|---|---|---|---|
| 0 | 0 | 0 | 0 | 0 | 0 |
| 1 | 8950 | 157.0 | 313 | 5.6 | 162.6 |
| 7 | 8660 | 151.9 | 4950 | 89.0 | 240.9 |
| 28 | 1070 | 18.8 | 1360 | 24.5 | 43.3 |
| 56 | 804 | 14.1 | 1190 | 21.4 | 35.5 |
| 84 | 1230 | 21.6 | 1240 | 22.3 | 43.9 |
| 119 | 815 | 14.3 | 1130 | 20.3 | 34.6 |
| 148 | 1129 | 19.8 | 1171 | 21.1 | 40.9 |
| 176 | 1547 | 27.1 | 1090 | 19.6 | 46.7 |
| 288 | 707 | 12.4 | 1790 | 32.2 | 44.6 |
Day of treatment . | PKC412 trough level, ng/mL . | PKC412 trough level, nM* . | CGP62221 trough level, ng/mL . | CGP62221 trough level, nM* . | Sum of trough levels of PKC412 and CGP62221* . |
|---|---|---|---|---|---|
| 0 | 0 | 0 | 0 | 0 | 0 |
| 1 | 8950 | 157.0 | 313 | 5.6 | 162.6 |
| 7 | 8660 | 151.9 | 4950 | 89.0 | 240.9 |
| 28 | 1070 | 18.8 | 1360 | 24.5 | 43.3 |
| 56 | 804 | 14.1 | 1190 | 21.4 | 35.5 |
| 84 | 1230 | 21.6 | 1240 | 22.3 | 43.9 |
| 119 | 815 | 14.3 | 1130 | 20.3 | 34.6 |
| 148 | 1129 | 19.8 | 1171 | 21.1 | 40.9 |
| 176 | 1547 | 27.1 | 1090 | 19.6 | 46.7 |
| 288 | 707 | 12.4 | 1790 | 32.2 | 44.6 |
Corrected for 99% protein binding12
As a screening test for potential mechanisms of clinical resistance that would prevent FLT3-TKI from reaching the target, we performed an ex vivo bioassay that determined the bioactivity of serum from patients treated with PKC412. Incubation of FLT3-ITD–positive MV4-11 cells with the serum of patient UPN1 resulted in dephosphorylation of FLT3 at all time points tested (Figure 2B). In particular, this analysis showed that serum obtained at the time of relapse (day 288) still provided adequate levels of PKC412 and of active metabolites to cause more than 95% inhibition of tyrosine phosphorylated FLT3 levels. This result also ruled out potential inhibitory activity mediated by serum proteins as α-acid glycoprotein (α-AGP).25,26
To further investigate the degree of resistance to PKC412 in comparison with a second-generation TKI, such as AST487, we performed ex vivo treatment of primary AML blasts obtained from patient UPN1. AST487 is an inhibitor of several class 3 tyrosine kinases, including FLT3 and c-KIT.27 Treatment with PKC412 did not cause significant dephosphorylation of FLT3 at pharmacologic levels as high as 10 nM. However, this resistance could be overcome using higher doses of PKC412 at 100 nM and 1000 nM (Figure 2C). In contrast, treatment with the TKI AST487 clearly inhibited tyrosine phosphorylation of FLT3 at a dose of 10 nM and exhibited complete suppression at a dose of 100 nM.
As functional analysis, we monitored the induction of apoptosis in primary AML blasts on in vitro treatment with PKC412 (50 nM) and AST487 (50 nM), respectively. Effects of PKC412 did not differ from medium control, whereas there was an apparent increase in the proportion of apoptotic cells on treatment with AST487 (Figure 2D). FL stimulation resulted in partial rescue of the apoptotic effect, probably because of the activation of FLT3-WT receptors by FL stimulation given that the patient's blasts were positive for FLT3-ITD and FLT3-WT receptors by PCR analysis (data not shown). Together, these results indicate that primary AML blasts isolated at the time of relapse did not exhibit major defects in apoptosis pathways.
Analysis of sensitivity to PKC412 in primary AML blasts obtained from patient UPN1. (A) In vivo tyrosine phosphorylation status of FLT3. MNCs were isolated from patient UPN1 at the time of remission (day 148) and at the time of recurrence of PB blasts (days 288, 289, 290). Whole cell lysates were prepared and were analyzed by Western blotting. Constitutive tyrosine phosphorylation of FLT3 was detected at the time points of relapse, but not in remission, on day 148.As a control, 32D cells harboring FLT3-ITD treated with or without PKC412 (100 nM, 30 minutes at 37°C) were used. To control equal loading, the blot was stripped and reprobed using an antiactin antibody. (B) Ex vivo bioassay for FLT3 tyrosine kinase inhibition. This immunoblot was derived from MV4;11 cells harboring the FLT3-ITD mutation exposed to serum from patient UPN1 and to serum from a healthy donor (HD) spiked with and without PKC412 (100 nM, 30 minutes at 37°C). Serum was obtained from patient UPN1 at various time points while he was on therapy with PKC412. Whole cell lysates were analyzed by Western blotting using FLT3-specific antiphosphotyrosine antibodies. Inhibition of the constitutive tyrosine phosphorylation was detectable using patient serum derived from all time points and healthy donor serum spiked with PKC412. Densitometric analysis revealed more than 95% inhibition of tyrosine-phosphorylated FLT3 levels at day 288. (C) FLT3 tyrosine phosphorylation in primary AML blasts treated with and without PKC412 and AST487. MNCs isolated from peripheral blood were incubated in RPMI 1640 with 10% FCS and were treated with increasing doses of PKC412 and the novel FLT3-TKI AST487. Whole cell lysates were analyzed by Western blotting using FLT3-specific antiphosphotyrosine antibodies. (D) Analysis of induction of apoptosis in primaryAML blasts using PKC412 andAST487. MNCs obtained from patient UPN1 at the time relapse were maintained in RPMI 1640 without any growth factor supplementation and were incubated with or without PKC412 (50 nM), FL (100 ng/mL), AST487 (50 nM), or in combination for 90 hours, as indicated. After this, the percentage of sub-G1 cells corresponding to apoptotic cells was determined in triplicate by flow cytometry. Error bars correspond to standard deviation.
Analysis of sensitivity to PKC412 in primary AML blasts obtained from patient UPN1. (A) In vivo tyrosine phosphorylation status of FLT3. MNCs were isolated from patient UPN1 at the time of remission (day 148) and at the time of recurrence of PB blasts (days 288, 289, 290). Whole cell lysates were prepared and were analyzed by Western blotting. Constitutive tyrosine phosphorylation of FLT3 was detected at the time points of relapse, but not in remission, on day 148.As a control, 32D cells harboring FLT3-ITD treated with or without PKC412 (100 nM, 30 minutes at 37°C) were used. To control equal loading, the blot was stripped and reprobed using an antiactin antibody. (B) Ex vivo bioassay for FLT3 tyrosine kinase inhibition. This immunoblot was derived from MV4;11 cells harboring the FLT3-ITD mutation exposed to serum from patient UPN1 and to serum from a healthy donor (HD) spiked with and without PKC412 (100 nM, 30 minutes at 37°C). Serum was obtained from patient UPN1 at various time points while he was on therapy with PKC412. Whole cell lysates were analyzed by Western blotting using FLT3-specific antiphosphotyrosine antibodies. Inhibition of the constitutive tyrosine phosphorylation was detectable using patient serum derived from all time points and healthy donor serum spiked with PKC412. Densitometric analysis revealed more than 95% inhibition of tyrosine-phosphorylated FLT3 levels at day 288. (C) FLT3 tyrosine phosphorylation in primary AML blasts treated with and without PKC412 and AST487. MNCs isolated from peripheral blood were incubated in RPMI 1640 with 10% FCS and were treated with increasing doses of PKC412 and the novel FLT3-TKI AST487. Whole cell lysates were analyzed by Western blotting using FLT3-specific antiphosphotyrosine antibodies. (D) Analysis of induction of apoptosis in primaryAML blasts using PKC412 andAST487. MNCs obtained from patient UPN1 at the time relapse were maintained in RPMI 1640 without any growth factor supplementation and were incubated with or without PKC412 (50 nM), FL (100 ng/mL), AST487 (50 nM), or in combination for 90 hours, as indicated. After this, the percentage of sub-G1 cells corresponding to apoptotic cells was determined in triplicate by flow cytometry. Error bars correspond to standard deviation.
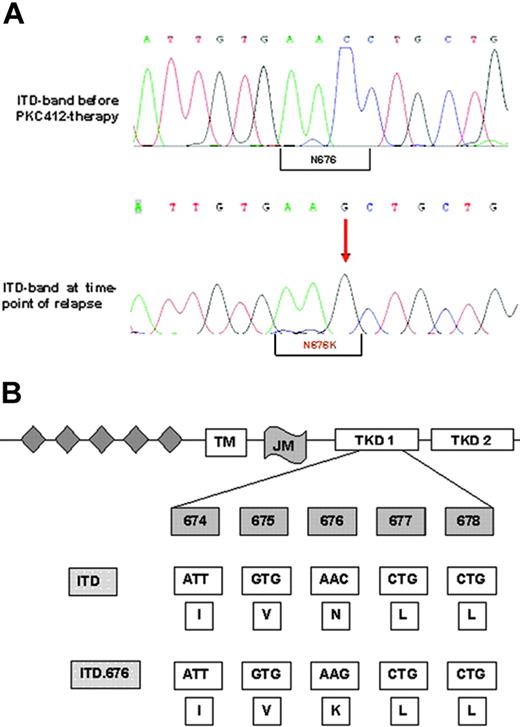
These assays ruled out common resistance mechanisms such as drug metabolism, drug efflux, inhibition by serum proteins, and FLT3 amplification at a protein level. Therefore, we performed FLT3 cDNA sequencing. This analysis revealed a single novel point mutation in the N-lobe of the kinase domain that resulted in the substitution of asparagine for lysine (N676K; Figure 3A-B). This mutation was located within the FLT3 allele harboring the ITD mutation (data not shown). As a confirmatory experiment, we amplified the FLT3 region using cDNA from AML blasts, subcloned the PCR product into the pAL vector, and transformed competent Escherichia coli cells with these constructs. Again, sequence analysis of cDNA clones revealed that the N676K mutation was located within the FLT3-ITD allele and not within the FLT3 wild-type sequence (data not shown). Importantly, the amino acid exchange was detectable in the FLT3-ITD sequence of patient material obtained at the time of relapse only but not in clinical material obtained before PKC412 therapy (Figure 3A).
Expression of the FLT3-ITD-N676K mutant in 32D cells results in persistent FLT3-TK phosphorylation on PKC412A treatment
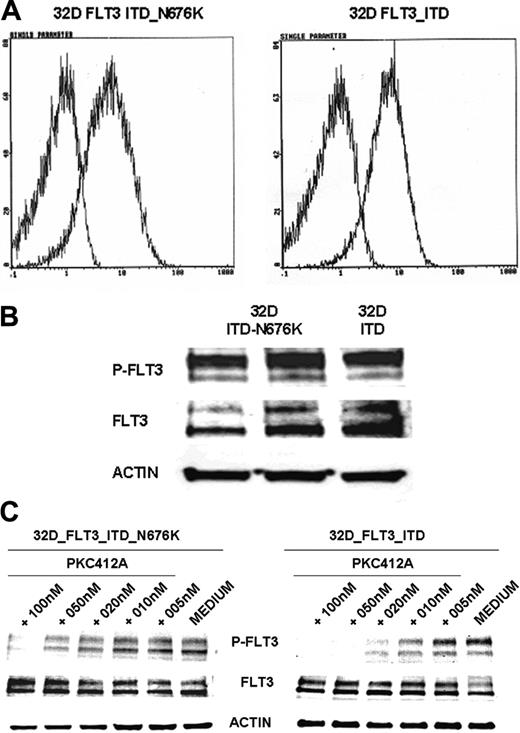
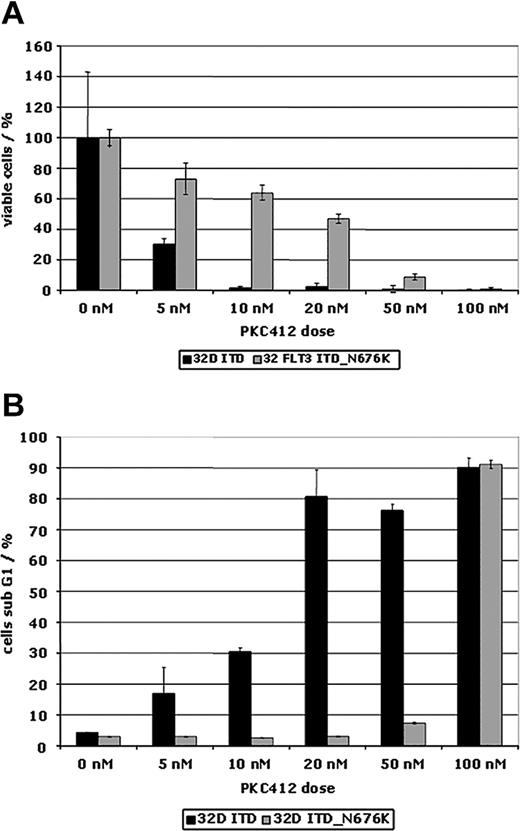
To further characterize the functional role of the N676K mutation, we generated FLT3-ITD– and FLT3-ITD-N676K–expressing 32D cells. Comparable membrane and total protein expression of FLT3-ITD and FLT3-ITD-N676K receptors was confirmed by FACS analysis and Western blot analysis (Figure 4A-B), respectively. On the basis of antiphosphotyrosine immunoblot analysis, the kinase activities of FLT3-ITD and of the FLT3-ITD-N676K mutant appear similar. However, there was a significant difference in sensitivity to PKC412. Although FLT3-ITD kinase activity was completely inhibited by PKC412 at a dose of 20 nM, the FLT3-ITD-N676K mutant retained relatively high levels—up to a concentration of 50 nM—of phosphotyrosine (Figure 4C).
Nucleotide and protein sequences of the FLT3-ITD-N676K mutation. (A-B) Genomic sequence shows the exchange of cytosine to guanine leading to an amino acid exchange at position 676 from asparagine to lysine. The N676K mutation was detectable in the larger FLT3-ITD band only, and not in the smaller wild-type allele, as confirmed by gel extraction and direct sequencing and by sequencing of FLT3-cDNA clones (data not shown).
Nucleotide and protein sequences of the FLT3-ITD-N676K mutation. (A-B) Genomic sequence shows the exchange of cytosine to guanine leading to an amino acid exchange at position 676 from asparagine to lysine. The N676K mutation was detectable in the larger FLT3-ITD band only, and not in the smaller wild-type allele, as confirmed by gel extraction and direct sequencing and by sequencing of FLT3-cDNA clones (data not shown).
Altered sensitivity of FLT3-ITD-N676K expressing 32D cells toward treatment with PKC412
There was also a differential effect of PKC412 on proliferation of FLT3-ITD– and FLT3-ITD-N676K–expressing 32D cells. Using an MTS-based proliferation assay, in FLT3-ITD-N676K cells a 10-fold higher concentration of PKC412 was needed for complete inhibition of cellular proliferation than was needed in FLT3-ITD cells (Figure 5A). This was confirmed by MTS-based proliferation assays looking at various time points (data not shown). The FLT3-ITD-N676K mutant cells were also consistently resistant to induction of apoptosis up to a PKC412 concentration of 50 nM, whereas FLT3-ITD cells already exhibited a significant increase in the ratio of apoptotic cells on incubation with 5 nM PKC412 (Figure 5B). This result corresponded exactly with results obtained using primary AML blasts harboring the N676K mutant (Figure 2D). The relative resistance to TKI-induced apoptosis in FLT3-ITD-N676K–expressing 32D cells could be overcome by increasing the dose to 100 nM PKC412A (Figure 5B).
Biologic phenotypes and gene expression profile of 32D-FLT3-ITD cells and 32D-FLT3-ITD-N676K cells do not show apparent differences
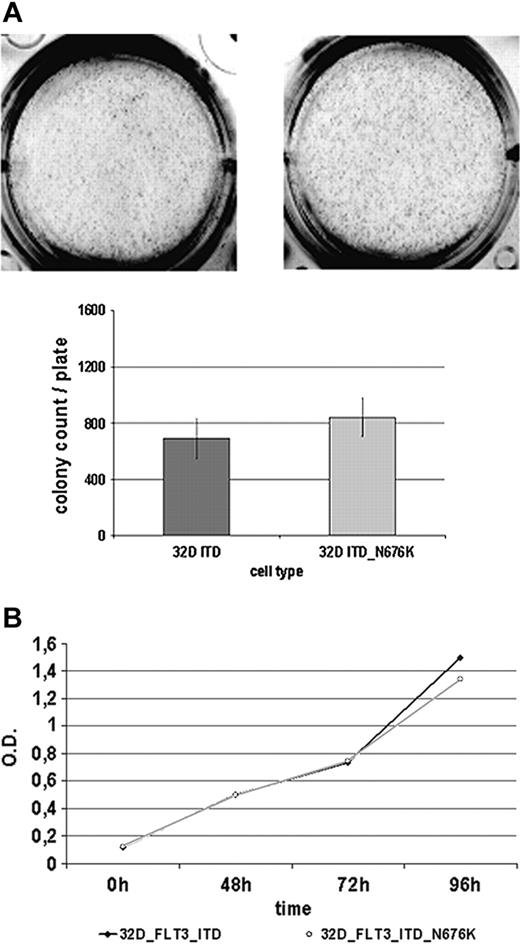
Given that it has been suggested that certain mutants of oncogenic tyrosine kinases create a more potent oncogene—such as Tyr-253 mutants of Bcr-Abl selected on imatinib therapy of CML28 —we sought to determine the basic biologic features of 32D-FLT3-ITD-N676K cells in comparison with 32D-FLT3-ITD cells. As a first approach, their colony-forming potential was investigated but revealed no significant differences (Figure 6A). Both cell lines were able to form a comparable number of colonies of similar sizes within 7 days of incubation. Further, there were no apparent differences in cellular proliferation using an MTS-based assay. Until 96 hours of incubation, no difference between the 2 cell lines was detectable (Figure 6B). Additionally, by quantitative PCR, we analyzed a set of 8 genes (PIM1, PIM2, SOCS1, SOCS2, SOCS3, CIS, RGS2, GADD45) that were shown to be differentially regulated in cells expressing FLT3-ITD rather than FLT3-wt.22 In this analysis, only SOCS3 expression was found to be significantly different in 32D FLT3-ITD-N676K compared with 32D FLT3-ITD cells (data not shown). Additional studies investigating the functional role of this observation are under way.
Analysis of potential mechanisms of resistance in 5 more AML patients at the time of relapse to PKC412
Because these studies demonstrated that the FLT3-ITD-N676K mutation was the sole cause of clinical resistance in patient UPN1, we screened for this mutation in 5 more patients with AML who harbored ITD length mutations or D835 point mutations and who initially responded to PKC412 treatment (50% or greater decrease in leukemic blasts in PB, BM, or both) and then had a sudden relapse after weeks to months. In this cohort, sequence analysis of the 676 region of FLT3 corresponding to amino acids 647-684 (complete exon 16) for a mutation at this position was found to be negative.
FLT3 receptor membrane expression, tyrosine phosphorylation, and inhibition of receptor tyrosine phosphorylation by PKC412 in 32D cells transfected with FLT3-ITD and FLT3-ITD-N676K. Murine 32D cells expressing FLT3-ITD and FLT3-ITD-N676K were generated as described in “Materials and methods.” (A) Membrane expression of FLT3 was determined by flow cytometry using a specific anti-FLT3 antibody. FLT3 receptors were expressed at comparable levels in 32D-ITD and 32D-ITD-N676K cells (B). FLT3 tyrosine phosphorylation was analyzed by Western blotting using phosphotyrosine-specific FLT3 antibodies. FLT3 tyrosine phosphorylation levels in 32D-FLT3-ITD and 32D-FLT3-ITD-N676K cells did not reveal apparent differences. (C) Inhibition of FLT3 tyrosine phosphorylation by PKC412 was analyzed in 32D cells expressing FLT3-ITD or FLT3-ITD-676 mutant receptors. Cells were treated with or without PKC412 at various concentrations for 30 minutes, and FLT3 tyrosine phosphorylation was analyzed by immunoblotting using phosphotyrosine-specific FLT3 antibodies. To control equal loading, the blot was stripped and reprobed using anti-FLT3 and antiactin antibodies, respectively. On treatment with 10 nM PKC412, densitometric analysis revealed more than 80% inhibition of tyrosine-phosphorylated FLT3 levels in 32D-FLT3-ITD cells, whereas in 32D-FLT3-ITD-N676K cells inhibition was 19%.
FLT3 receptor membrane expression, tyrosine phosphorylation, and inhibition of receptor tyrosine phosphorylation by PKC412 in 32D cells transfected with FLT3-ITD and FLT3-ITD-N676K. Murine 32D cells expressing FLT3-ITD and FLT3-ITD-N676K were generated as described in “Materials and methods.” (A) Membrane expression of FLT3 was determined by flow cytometry using a specific anti-FLT3 antibody. FLT3 receptors were expressed at comparable levels in 32D-ITD and 32D-ITD-N676K cells (B). FLT3 tyrosine phosphorylation was analyzed by Western blotting using phosphotyrosine-specific FLT3 antibodies. FLT3 tyrosine phosphorylation levels in 32D-FLT3-ITD and 32D-FLT3-ITD-N676K cells did not reveal apparent differences. (C) Inhibition of FLT3 tyrosine phosphorylation by PKC412 was analyzed in 32D cells expressing FLT3-ITD or FLT3-ITD-676 mutant receptors. Cells were treated with or without PKC412 at various concentrations for 30 minutes, and FLT3 tyrosine phosphorylation was analyzed by immunoblotting using phosphotyrosine-specific FLT3 antibodies. To control equal loading, the blot was stripped and reprobed using anti-FLT3 and antiactin antibodies, respectively. On treatment with 10 nM PKC412, densitometric analysis revealed more than 80% inhibition of tyrosine-phosphorylated FLT3 levels in 32D-FLT3-ITD cells, whereas in 32D-FLT3-ITD-N676K cells inhibition was 19%.
PKC412 drug levels at the time of relapse were measured in another 3 patients included in this cohort: PKC412 levels corrected for 99% protein binding were 17.56 nM (UPN2), 30.08 nM (UPN3), and 34.17 nM (UPN4), respectively. Corresponding levels for free concentrations of the active metabolite CGP62221 were 19.37 nM, 43.97 nM, and 25.43 nM, respectively. Thus, these actual drug levels strongly suggest that patients were able to achieve levels sufficient to suppress normal FLT3-ITD–mutated disease at the time of relapse. This conclusion is supported by results of an additional ex vivo bioassay performed in patient UPN4 showing greater than 95% inhibition of tyrosine-phosphorylated FLT3 levels on incubation with the patient's serum obtained at relapse (data not shown).
Discussion
For AML patients younger than 55 years of age, the outcome of therapy has improved from a 5-year overall survival rate of 11% in the 1970s to 37% in the 1990s.29 This was achieved by a treatment regimen consisting of cytarabine and daunorubicin for induction followed by more intensive postremission therapy. However, in older patients with AML, the 5-year survival rate using this regimen is less than 15%, and progress in the past 3 decades has been modest.30 In addition, in older AML patients, current chemotherapy regimens are associated with significant morbidity and mortality. Clearly, there is an urgent medical need for more effective and less toxic therapies in this patient population.
PKC412 dose-response curves of 32D cells expressing FLT3-ITD and FLT3-ITD-N676K. Cells were treated with and without PKC412 at increasing doses for 48 hours. (A) Proliferation using an MTS-based assay was measured at 0 and at 48 hours of incubation, and the percentage of proliferating cells relative to the control (no inhibitor) was plotted. (B) 32D cells expressing FLT3-ITD and FLT3-ITD-N676K were treated with and without PKC412A for 48 hours before assessing apoptosis using cell cycle analysis. Depicted is the proportion of cells in sub-G1.
PKC412 dose-response curves of 32D cells expressing FLT3-ITD and FLT3-ITD-N676K. Cells were treated with and without PKC412 at increasing doses for 48 hours. (A) Proliferation using an MTS-based assay was measured at 0 and at 48 hours of incubation, and the percentage of proliferating cells relative to the control (no inhibitor) was plotted. (B) 32D cells expressing FLT3-ITD and FLT3-ITD-N676K were treated with and without PKC412A for 48 hours before assessing apoptosis using cell cycle analysis. Depicted is the proportion of cells in sub-G1.
32D FLT3-ITD and 32D FLT3-ITD-N676K cells show a similar biologic phenotype. (A) Analysis of colony formation. 32D cells expressing FLT3-ITD and FLT3-ITD-N676K were seeded in methylcellulose medium and incubated for 7 days at 37°C. Thereafter, numbers of colonies were counted and plotted. (B) Proliferation was measured at various time points from 0 to 96 hours using an MTS-based assay.
32D FLT3-ITD and 32D FLT3-ITD-N676K cells show a similar biologic phenotype. (A) Analysis of colony formation. 32D cells expressing FLT3-ITD and FLT3-ITD-N676K were seeded in methylcellulose medium and incubated for 7 days at 37°C. Thereafter, numbers of colonies were counted and plotted. (B) Proliferation was measured at various time points from 0 to 96 hours using an MTS-based assay.
In addition, the success of targeting specific oncogenes in leukemia, such as PML-RARα, by all-trans retinoic acid and BCR-ABL by imatinib has initiated an intensive search for alternative and more specific therapies to treat AML.
At the heart of this search lie genetic defects that are critically involved in pathogenesis of AML. However, oncogenesis in AML is multifactorial and complicated by a multitude of genetic defects. Nevertheless, recent studies revealed that the class 3 receptor tyrosine kinase FLT3 is mutated in approximately 35% of AML patients. Mutations are located in the juxtamembrane domain and in the kinase domain of FLT3. Recently, novel-activating FLT3 mutations have been described and suggest that the rate of FLT3 mutations in AML may be underestimated.8,9 Thus, FLT3 is the most common single gene affected by mutations in AML; therefore, inhibition of mutated FLT3 kinase activity by a pharmacologic agent is an attractive therapeutic strategy.
We have recently reported on the clinical results of a phase 2 clinical trial using the small molecule tyrosine kinase inhibitor (TKI) PKC412.12,17 Overt complete or partial hematologic remissions were infrequent, but biologic activity, defined as a 50% or greater decrease in PB or BM blasts, was observed in 23 (66%) of 35 patients with mutated FLT3 and in 23 (40%) of 57 patients with wild-type FLT3. Most of these responses were transient; however, in a subset of patients, blast recurrence was preceded by an interval of prolonged response. The etiology of primary and secondary clinical resistance to FLT3-TKI in AML is poorly understood, but detailed information on this topic is clearly needed for the rational development of future therapeutic strategies using these compounds.
We performed a search for potential mechanisms of resistance in an index patient (UPN1) who experienced complete clearance of leukemic blasts for a prolonged period of time. The algorithm of assays applied in this study was feasible and effective, and we believe it represents a rational handle for further investigation of primary and secondary mechanisms of resistance in AML.
In patient UPN1, the absolute sensitivity to PKC412 is decreased because levels of 100 nM are needed to overcome the relative resistance in this patient. The actual level of the sum of PKC412 and CGP62221 measured at relapse (44.6 nM) is still expected to adequately inhibit FLT3-ITD receptors because the IC50 of PKC412 is 5 nM to 10 nM and the IC50 of CGP62221 is assumed to be similar. This is supported by an ex vivo bioassay (Figure 2B) demonstrating that serum obtained at the time of relapse still provides adequate levels of PKC412 and enough active metabolites (such as GCP62221) to cause more than 95% inhibition of tyrosine-phosphorylated FLT3 levels.
Thus, our analyses ruled out mechanisms preventing PKC412 from reaching its target and identified a single amino acid substitution in the kinase domain of FLT3 (N676K) as the sole cause of clinical resistance in this patient. We did not identify any other AML patient harboring this mutation at relapse to PKC412. However, the cohort of patients analyzed in this study was limited, suggesting that the identified mechanism of resistance, which is attributed to a point mutation within the ATP binding site of FLT3, may not be infrequent. Thus, further studies looking at a larger cohort of patients and at additional amino acid positions are warranted.
Recently, results of an in vitro screen designed to identify mutations in the ATP binding pocket of FLT3 that confer resistance to TKI have been reported.31 Interestingly, mutations identified included position N676, which confers resistance to PKC412. FLT3 proteins mutated at N676 revealed relative resistance to PKC412 and remained sensitive to higher concentrations, with IC50 of 235 nM for the N676D mutant and 162 nM for the N676S mutant. Devised from a structural model of the FLT3 kinase domain in complex with PKC412, it was concluded that the mutation of N676 might destabilize the conformation of the hinge segment, which makes H-bonds with the lactam ring of PKC412.31 It is conceivable that the degree of destabilization of this hinge segment conformation critically depends on the amino acid residue introduced at position 676. In principle, this may explain the differences in IC50 observed for N676D, for N676S, and for the N676K mutation.
Our results confirm that the N676 position of FLT3 is critical for sensitivity to PKC412 and point out that an in vitro screen for resistance using random mutagenesis is a powerful tool to predict relevant mutations involved in the development of clinical resistance.
It has been proposed that certain mutations of oncogenic kinases conferring resistance to TKI are associated with a more oncogenic phenotype.28 However, for the N676K mutant of FLT3-ITD, our preliminary analysis investigating proliferation, colony formation, and induction of FLT3-ITD–dependent genes does not indicate a major difference in phenotype compared with FLT3-ITD.
In conclusion, through biochemical and molecular analysis of clinical material, we identified a single amino acid substitution at the 676 asparagine residue of the tyrosine kinase domain of FLT3 as the sole cause of clinical resistance to the TKI PKC412 in 1 of 6 patients investigated. PKC412 drug levels obtained at the time of relapse in 4 patients showed that these patients were able to achieve levels sufficient to suppress normal FLT3-ITD–mutated disease. This conclusion is supported by results of additional ex vivo bioassays demonstrating more than 95% inhibition of tyrosine-phosphorylated FLT3 levels on incubation with the patients' serum obtained at relapse. Together, these points, combined with the rest of the data outlined, demonstrate that inadequate drug levels most likely cannot explain resistance to PKC412 in patients included in this cohort.
Reconstitution experiments revealed that N676K mutation of FLT3-ITD was sufficient to confer an intermediate level of resistance in vitro. The fact that resistance is mediated by FLT3 mutation strongly suggests that the mechanism of action of PKC412 is indeed inhibition of FLT3 kinase activity rather than inhibition of other kinases targeted by PKC412. These studies also point out that a genetically complex malignancy such as AML may retain dependence on a single oncogenic signal. It is tempting to speculate that in these patients a strategy using FLT3-TKI, combined with therapeutic entities such as chemotherapy, antibody therapy, inhibition of HSP90, or inhibition of histone deacetylases, may significantly improve clinical efficacy.
Prepublished online as Blood First Edition Paper, September 8, 2005; DOI 10.1182/blood-2005-06-2469.
Supported in part by a grant from the Deutsche Krebshilfe Foundation (T.F.).
Three of the authors (J.R., Y.W., P.S.C.) are employed by a company or competitor of a company (Novartis Pharma Inc) whose product is discussed in this article.
F.H. performed Western blotting experiments and apoptosis assays, participated in the generation of transfected cells and the design of the experiments, and participated in writing the manuscript; F.K.S. performed proliferation assays and contributed to the generation of transfected cell lines expressing the N676K mutant; F.B. contributed to mutation screening, design of the experiments, sequence analysis, and writing of the manuscript; S.K. participated in the generation of transfected cell lines and the generation of colony assays; D.B.L. contributed to Western blotting experiments and transfection of 32D cells; M.H.T. performed the PKC412 dose level analysis in patient material; J.R. provided pharmacologic reagents; E.F., G.E., G.J.S., S.N., F.G., and R.M.S. contributed to the design of the phase 2 clinical study and were involved in patient care; C.B. and H.S. performed the quantitative gene expression analysis; Y.W. performed analysis of dose levels of PKC412 in patient material; T.K. contributed to the generation of transfected cells; P.S.C. coordinated the clinical phase 2 trial and provided pharmacokinetic data; C.H. contributed to the clinical phase 2 study; and T.F. designed and supervised the experimental work and data analysis and wrote the manuscript.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal