Abstract
C/EBPα is an essential transcription factor required for myeloid differentiation. While C/EBPα can act as a cell fate switch to promote granulocyte differentiation in bipotential granulocyte-macrophage progenitors (GMPs), its role in regulating cell fate decisions in more primitive progenitors is not known. We found increased numbers of erythroid progenitors and erythroid cells in C/EBPα–/– fetal liver (FL). Also, enforced expression of C/EBPα in hematopoietic stem cells resulted in a loss of erythroid progenitors and an increase in myeloid cells by inhibition of erythroid development and inducing myeloid differentiation. Conditional expression of C/EBPα in murine erythroleukemia (MEL) cells induced myeloid-specific genes, while inhibiting erythroid-specific gene expression including erythropoietin receptor (EpoR), which suggests a novel mechanism to determine hematopoietic cell fate. Thus, C/EBPα functions in hematopoietic cell fate decisions by the dual actions of inhibiting erythroid and inducing myeloid gene expression in multipotential progenitors.
Introduction
Hematopoiesis is the developmental process whereby hematopoietic stem cells (HSCs) undergo proliferation to self-renew and/or differentiate into more committed progenitor cells that generate mature blood cells. These progenitors have been purified and include common lymphoid progenitors (CLPs) that differentiate into lymphoid cells, and common myeloid progenitors (CMPs), which differentiate into more committed progenitors including the granulocyte-macrophage progenitor (GMP) and megakaryocyte-erythrocyte progenitors (MEPs), which give rise to nonlymphoid cells.1,2
Several lines of evidence indicate that cell fate determination during hematopoietic development is dependent on the level of transcription factor expression. For example, the levels of E2A are critical for B- or T-cell fate determination,3,4 and the levels of PU.1 determine B-cell or myeloid development.5,6 In addition, it has been proposed that HSCs express low levels of transcriptional programs for many cell lineages, and that cell fate is determined by up-regulation of transcription factors that promote the differentiation of specific cell lineages and the concomitant down-regulation of transcriptional regulators required for the development of other cell lineages.7 For example, CCAAT enhancer binding protein α (C/EBPα) and PU.1 are expressed in HSCs and are up-regulated in GMPs during granulocyte and macrophage development; however, they are down-regulated in MEPs.8,9 In comparison, GATA-1 and FOG-1, which are required for erythroid differentiation, are up-regulated in MEPs but not GMPs or CLPs.8-10 Furthermore, inappropriate expression of transcription factors can reverse cell fate. For instance, enforced expression of GATA-1 in CLPs or GMPs promotes erythroid differentiation,11 and enforced C/EBPα expression in B-cell progenitors leads to their reprogramming into macrophages.11,12
C/EBPα is a basic leucine zipper transcription factor, which can positively or negatively affect gene transcription depending on the specific promoter or enhancer element that it binds.13,14 In this regard, C/EBPα regulates the expression of a number of genes involved in cell growth and differentiation, and has pivotal roles in lineage commitment in the hematopoietic system.15-18 For example, overexpression of C/EBPα in hematopoietic progenitor cell (HPC) lines induces granulocyte differentiation. In addition, C/EBPα also promotes neutrophil maturation at the expense of macrophage differentiation in U937 cells and in PU.1–/– cell lines, indicating that C/EBPα may play a role in cell fate decisions in GMPs.6,19,20 C/EBPα is also a potent inhibitor of cell proliferation, and is thought to play a critical role in regulating the balance between proliferation, growth arrest, and differentiation in GMPs.21-23 However, C/EBPα mRNA is also expressed in HSCs and CMPs, and C/EBPα is required for CMPs to differentiate into GMPs, suggesting that C/EBPα might regulate the proliferation and differentiation of progenitors more primitive than GMPs.1,8,24,25 Furthermore, the expression of oncogenic BCR-ABL protein in normal hematopoietic progenitors decreases C/EBPα expression and promotes erythropoiesis, suggesting that C/EBPα may affect CMP differentiation.26,27 Based on these observations, we proposed that C/EBPα regulates cell fate decisions in progenitors more primitive than GMPs, where it may influence the commitment to myeloid or erythroid differentiation.
In the present report, we demonstrate that C/EBPα inhibits the differentiation and development of erythroid cells while promoting myeloid development. Moreover, C/EBPα induces myeloid differentiation of erythroid-restricted progenitor cells. Altogether, our data show C/EBPα affects hematopoietic cell fate decisions in 2 ways: by inducing myeloid differentiation and inhibiting erythroid differentiation in progenitors more primitive than GMPs.
Materials and methods
Mice
The C/EBPα+/– mice (mixed background of 129sv and C57Bl/6) were backcrossed onto a pure C57Bl/6 background (Ly5.1).28 Mice and embryos were genotyped by polymerase chain reaction (PCR) of genomic DNA. Four- to 8-week-old C57Bl/6-Ly 5.2 mice were obtained for transplantation experiments from the animal production area at the National Cancer Institute (NCI)–Frederick (Frederick, MD). Animal care was provided in accordance with the procedures outline in the “Guide for Care and Use of Laboratory Animals.”29
Soft agar colony assays
To measure erythroid progenitors, FL from C/EBPα–/–, C/EBPα+/–, and C/EBPα+/+ mice or fluorescence-activated cell sorter (FACS)–sorted cells were plated in 35-mm Lux Petri dishes (Nalge Nunc, Rochester, NY) at the indicated cell densities in Iscoves modified Dulbecco medium (IMDM). Dishes were incubated in a fully humidified atmosphere at 37°C, 5% CO2 and then scored for colony formation after 7 to 10 days.
Retroviral transduction, FACS analysis, and transplantation
Recombinant mouse stem cell virus (MSCV) vectors expressing C/EBPα and green fluorescent protein (GFP) were generated as previously described.30 pMSCV-GFP and pMSCV-C/EBPα-GFP were transfected into Phoenix packaging cells (a gift from Dr Gary Nolan, Stanford University) with pCL-eco plasmids by FuGENE 6 (Roche Applied Science, Indianapolis, IN) to produce infectious ecotropic retrovirus. Medium was changed 24 hours after transfecting Phoenix cells, and viral supernatants were collected 48 hours after transfection. GFP+ cells were cultured in vitro, or GFP+ FL (4 × 105) and BM (2 × 105) cells were transplanted to 8-week-old recipient mice (C57BL/6-Ly5.2) exposed to 11 Gy (1100 rad) from 137Cs source. Three transplantation cohorts were set up for GFP+ FL and BM cells. Repopulation was determined 4 months after transplantation by analyzing GFP+ peripheral blood or bone marrow cells (BMCs) using a dual laser FACSCAN (BD Biosciences, San Jose, CA). BD CELLQuest was used for FACS analysis. The mean plus or minus standard deviation (SD) was obtained from all mice within a group, and then statistical differences between the groups were determined by the Student t test. All lineage-specific antibodies and Tri-color streptavidin were purchased from BD Pharmingen (San Diego, CA). Gr-1 and TER119 were PE-conjugated antibodies, while Mac-1 and CD71 were biotin-conjugated antibodies. Images of cell cytospin preparation were visualized using a Leica DMLB microscope (Leica Microsystems, Bannockburn, IL) and a 100 ×/1.25-0.60 numeric aperture oil objective and a 10 ×/20 eyepiece. Images were captured using DVC C-view Epix Imaging software, version 2.1 (Epix, Buffalo Grove, IL).
Cell lines
The MEL cell line was maintained in complete Dulbecco modified Eagle medium (cDMEM) without phenol red with 10% FCS, P/S/G. MEL cells that express conditionally active C/EBPα were established by infecting MEL cells with retroviral vectors that express C/EBPα/hER or the control construct lacking C/EBPα/hER coding regions. C/EBPα/hER-infected MEL cells were cloned by limiting dilution in the presence of 2.0 μg/mL puromycin. The expression of C/EBPα was monitored by Western blot using anti-C/EBPα antibody (SC-61; Santa Cruz Biotechnology, Santa Cruz, CA). MEL cells were seeded at 4 × 105/mL in cDMEM containing 3 mM hexamethylene bisacetamide (HMBA) and incubated at 37°C. Cells were stained for hemoglobin expression with benzidine at acid pH. The SCF-dependent EML cell line was maintained in IMDM supplemented with 20% horse serum, 8% conditioned medium from BHK/MKL cells containing mSCF, and P/S/G. EML cells were cultured with mSCF (100 ng/mL) and hEpo (40 U/mL) 24 hours prior to transduction with retrovirus containing pMSCV-C/EBPα-GFP or pMSCV-GFP.
Western blot analysis
Cell lysates were resolved on gradient sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) gels (Invitrogen, Frederick, MD) and transferred to Immobilon-P membrane (Millipore, Billerica, Spain). The membrane was blocked at room temperature in 5% milk/Tris-buffered saline-Tween (TBS-T) solution, and then probed with anti–rabbit C/EBPα antibody, anti–rat GATA-1 antibody (sc-265), anti–rabbit EpoR antibody (sc-697), anti–mouse α-actin antibody (sc-8432), anti–rabbit Id1 (sc-488), or anti–rabbit PU.1 antibody (sc-352; Santa Cruz Biotechnology). All primary antibodies were diluted to 1:200. Bound antibody was detected using a chemiluminescence system (Amersham Biosciences UK Limited, Buckinghamshire, United Kingdom).
Reverse-transcription (RT)–PCR
Total RNA was purified using RNeasy (Qiagen, Valencia, CA) and converted to cDNA with Moloney murine leukemia virus reverse transcriptase (Applied Biosystems, Foster City, CA). The PCR was conducted using ampli-Taq DNA polymerase (PerkinElmer, Shelton, CT) at 4 different sets with different cycle numbers to confirm that the reaction was in the exponential phase. PCR amplification cycles consisted of 30 seconds at 94°C, 30 seconds at 58°C, and 1 minute at 72°C as indicated. The primers used are as follows: Id1 5′-GATCATGAAGGTCGCCAGTG-3′, 5′-TCCATCTGGTCCCTCAGTGC-3′; PU.1 5′-TGGAAGGGTTTTCCCTCACC-3′, 5′-TGCTGTCCTTCATGTCGCCG-3′; SCL 5′-TATGAGATGGAGATTTCTGATG-3′, 5′-GCTCCTCTGTGTAACTGTCC-3′; E2A 5′-CATCCATGTCCTGCGAAGCCA-3′, 5′-TTCTTGTCCTCTTCGGCGTC-3′; GATA-1 5′-CATTGGCCCCTTGTGAGGCCAGAGA-3′,5′-ACCTGATGGAGCTTGAAATAGAGGC-3′; EpoR 5′-ACGAAACAGGGGCGCTGGAG-3′, 5′-ACACGTCCACTTCATATCGG-3′; β-globin 5′-ATGGTGCACCTGACTGATGCTG-3′, 5′-GGTTTAGTGGTACTTGTGAGCC-3′; β-actin 5′-GTGGGCCGCTCTAGGCACCAA-3′, 5′-CTCTTTGATGTCACGCACGATTTC-3′. PCR products were resolved in a 1% agarose gel containing ethidium bromide. The intensity of the bands was quantified by using the National Institutes of Health Image program (ImageJ, V1.32; National Institutes of Health, Bethesda, MD).
Northern blot analysis
Total RNA was purified using RNeasy (Qiagen) as outlined by the manufacturer. Separation of RNA samples (10 μg) by electrophoresis was performed on 1% agarose, 5.2% formaldehyde (37% solution), 1 × MOPS gels. The running buffer was 1 × MOPS, 5% formaldehyde. RNA was transferred to nylon membrane (MSI) and hybridized at 68°C in QuickHyb hybridization solution (Stratagene, La Jolla, CA). Murine EpoR and β-globin cDNA probes were labeled with the random priming Prime-It II kit (Stratagene) following the manufacturer's instructions.
Construction of luciferase reporter plasmids and reporter assay
The EpoR 5′-flanking sequence (bp –1694 to –1) was amplified by polymerase chain reaction using mouse genomic DNA as a template. This fragment was ligated into pCR 2.1 vector (Invitrogen) and subsequently inserted into the SacI and XhoI sites of the luciferase reporter plasmid pGL3-Basic (Promega, Madison, WI), yielding the reporter construct mEpoR-Luc. The 2xC/EBP-Luc reporter contains 2 canonical C/EBP binding sites. For transfection assays, L cells (ATCC CCL-1: murine fibroblasts) were plated at a density of 8 × 104 cells/well in 6-well plates containing complete DMEM medium.
Results
Erythroid development is increased in C/EBPα–/– fetal liver
To evaluate if C/EBPα regulates cell fate decisions in multipotential progenitors, we examined erythroid development in C/EBPα–/– mice, where absence of C/EBPα expression in CMPs might affect erythroid development. While the red cell numbers are normal in PB of C/EBPα–/– newborn embryos,17 a detailed analysis of erythroid progenitors has not been reported in these mice. Therefore, we examined hematopoietic progenitor cells in the fetal liver of C/EBPα–/– embryos at E15.5, since C/EBPα–/– mice die shortly after birth due to defects in glucose metabolism. First, differential cell counts of Wright-Giemsa–stained cytocentrifuge preparations from C/EBPα–/– FL showed an increase in the number of erythroblasts compared with C/EBPα+/+ FL (394.0 ± 1.7/400 cells in C/EBPα–/– FL cells versus 315.0 ± 8.0/400 cells in C/EBPα+/– FL cells versus 274.0 ± 13.3 in C/EBPα+/+ FL cells, P < .05; Figure 1A). In agreement with previous data, there was a concomitant decrease in neutrophils in C/EBPα–/– FL.17 To determine if the increase in erythroid cell number correlated with increased erythroid progenitor activity, we plated FL cells in erythroid colony assays. There was a 2-fold increase in the number of erythroid burst-forming unit (BFU-E) colonies in C/EBPα–/– FL cells compared with C/EBPα+/– and C/EBPα+/+ FL cells (207.4 ± 12.8 vs 98.7 ± 10.4 and 120.4 ± 23.7, respectively; P < .05; Figure 1B). Thus, C/EBPα–/– FL showed increased numbers of erythroid progenitors and erythroid cells compared with C/EBPα+/+ and C/EBPα+/– FL.
Increased erythropoiesis is an intrinsic property of C/EBPα–/– FL cells
Increased erythroid development in C/EBPα–/– mice could be due to the deletion of C/EBPα in hematopoietic progenitor cells or a defect in the cells that constitute the hematopoietic microenvironment of fetal liver. To distinguish between these possibilities, we transplanted C/EBPα–/– FL cells into irradiated recipient mice and evaluated erythroid development. To track donor reconstitution, FL cells were transduced with retroviral vectors that express GFP (pMSCV-GFP). Equivalent numbers of GFP+ C/EBPα–/– and C/EBPα+/+ FL cells were cotransplanted with normal mouse BMCs into lethally irradiated mice. Four months after transplantation, BMCs were analyzed for donor (GFP+)–derived erythroid (TER119 and CD71) and myeloid (Mac-1 and Gr-1) cell reconstitution by flow cytometry and colony assay (Figure 1C-D). There was a significant increase in percentage of TER119+/CD71+ erythroid cells in BMCs of mice that received a transplant of C/EBPα–/– FL cells compared with mice that received a transplant of C/EBPα+/+ FL cells (63.5% ± 4.6% versus 44.2% ± 2.6%; P < .05). In addition, mice that received a transplant of C/EBPα–/– FL cells showed the expected defect in Mac-1+ myeloid cell differentiation compared with control mice that received a transplant of C/EBPα+/+ FL cells (28.1% ± 3.5% versus 58.5% ± 4.2%; P < .05; Figure 1D). To evaluate if there was an increase in erythroid progenitors in vivo, we sorted GFP+ BMCs from mice that received a transplant of C/EBPα–/– or C/EBPα+/+ FL cells and plated them in erythroid and myeloid progenitor cell assays. There was a 2-fold increase in the number of BFU-E colonies in mice that received a transplant of C/EBPα–/– FL compared with control mice that underwent transplantation (15.7 ± 1.3 versus 7.3 ± 0.8; P < .05; Figure 1E). In comparison, there was a defect in the development of CFU-GM and CFU-M progenitors in mice that received a transplant of C/EBPα–/– FL cells as previously reported.24 Thus, increased erythroid development in C/EBPα–/– mice was most likely intrinsic to the progenitor cells and not dependent on the microenvironment.
Increased erythroid development in C/EBPα–/– mice. (A) Cytocentrifuge preparations of C/EBPα–/– FL cells were stained with Wright-Giemsa, and differential cell counts were performed on 400 cells from 3 individual mice. The data are reported as the mean number of cells ± standard error (SE). (B) FL cells (2.5 × 104) from C/EBPα+/+, C/EBPα +/–, and C/EBPα–/– embryos at E15.5 were cultured in complete IMDM (cIMDM) containing 1.1% methylcellulose, 25% FCS, 100 U/mL penicillin, 100 μg/mL streptomycin, and 2 mM l-glutamine (P/S/G), and cultures were supplemented with murine (m) IL-3 (30 ng/mL), stem cell factor (mSCF, 100 ng/mL), and human erythropoietin (hEpo, 5 U/mL). The mean number of BFU-E colonies ± SE of triplicate plates is shown. (C-D) C/EBPα–/– and C/EBPα+/+ FL cells were transduced with MSCV-GFP retroviral vectors to track donor erythroid reconstitution. Fetal liver cells were resuspended at 2 × 105/mL in cIMDM and cultured with 8 μg/mL polybrene, 100 ng/mL mSCF, 100 ng/mL thrombopoietin (hTpo), 100 ng/mL hFlt-3L, and 50 ng/mL IL-6. A total of 3 retrovirus infections were performed, after which the cells were cultured with the same cytokine mixture. GFP-positive (GFP+) cells were isolated 2 days after the last infection by FACS sorting (FACSAria [BD Biosciences] or MoFlo [Cytomation, Fort Collins, CO]). BM cells were harvested 4 months after transplantation and then analyzed for the expression of the erythroid markers (TER119 and CD71) and myeloid cell markers (Gr-1 and Mac-1). (D) Horizontal bars indicate median values. (E) GFP+ BM cells were purified by FACS and cultured in methylcellulose (7.5 × 104/plate) or soft agar (1 × 105/plate) to determine the number of erythroid or myeloid progenitors. For myeloid progenitors, cells were plated in cIMDM containing 10% FCS, P/S/G, and 0.35% sea plaque agarose in the presence or absence of 50 ng/mL hG-CSF, 100 ng/mL M-CSF, 100 ng/mL mSCF, 20 ng/mL mGM-CSF, 30 ng/mL mIL-3. The mean number of BFU-E and CFU-c colonies ± SE of triplicate plates is shown.
Increased erythroid development in C/EBPα–/– mice. (A) Cytocentrifuge preparations of C/EBPα–/– FL cells were stained with Wright-Giemsa, and differential cell counts were performed on 400 cells from 3 individual mice. The data are reported as the mean number of cells ± standard error (SE). (B) FL cells (2.5 × 104) from C/EBPα+/+, C/EBPα +/–, and C/EBPα–/– embryos at E15.5 were cultured in complete IMDM (cIMDM) containing 1.1% methylcellulose, 25% FCS, 100 U/mL penicillin, 100 μg/mL streptomycin, and 2 mM l-glutamine (P/S/G), and cultures were supplemented with murine (m) IL-3 (30 ng/mL), stem cell factor (mSCF, 100 ng/mL), and human erythropoietin (hEpo, 5 U/mL). The mean number of BFU-E colonies ± SE of triplicate plates is shown. (C-D) C/EBPα–/– and C/EBPα+/+ FL cells were transduced with MSCV-GFP retroviral vectors to track donor erythroid reconstitution. Fetal liver cells were resuspended at 2 × 105/mL in cIMDM and cultured with 8 μg/mL polybrene, 100 ng/mL mSCF, 100 ng/mL thrombopoietin (hTpo), 100 ng/mL hFlt-3L, and 50 ng/mL IL-6. A total of 3 retrovirus infections were performed, after which the cells were cultured with the same cytokine mixture. GFP-positive (GFP+) cells were isolated 2 days after the last infection by FACS sorting (FACSAria [BD Biosciences] or MoFlo [Cytomation, Fort Collins, CO]). BM cells were harvested 4 months after transplantation and then analyzed for the expression of the erythroid markers (TER119 and CD71) and myeloid cell markers (Gr-1 and Mac-1). (D) Horizontal bars indicate median values. (E) GFP+ BM cells were purified by FACS and cultured in methylcellulose (7.5 × 104/plate) or soft agar (1 × 105/plate) to determine the number of erythroid or myeloid progenitors. For myeloid progenitors, cells were plated in cIMDM containing 10% FCS, P/S/G, and 0.35% sea plaque agarose in the presence or absence of 50 ng/mL hG-CSF, 100 ng/mL M-CSF, 100 ng/mL mSCF, 20 ng/mL mGM-CSF, 30 ng/mL mIL-3. The mean number of BFU-E and CFU-c colonies ± SE of triplicate plates is shown.
C/EBPα inhibits erythroid development in vivo
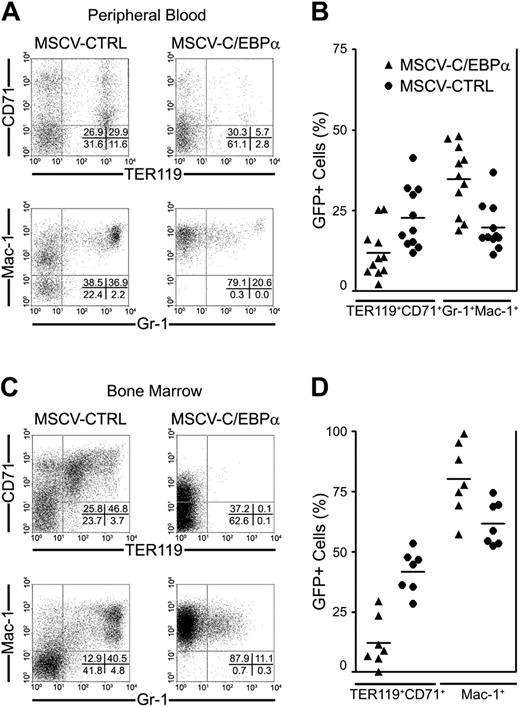
The increase in erythroid development and absence of myeloid cells in C/EBPα–/– mice suggested that C/EBPα might regulate erythroid and myeloid development in CMPs. To examine this hypothesis, we transduced BMCs from 5-FU–treated mice with retroviral vectors that express C/EBPα (pMSCV-C/EBPα-GFP) or pMSCV-GFP, and transplanted these cells into lethally irradiated mice. Four months after transplantation, PB and BMCs were analyzed for donor-derived (GFP+) erythroid (TER119 and CD71) and myeloid (Mac-1 and Gr-1) cell reconstitution using fluorochrome-labeled antibodies. Overexpression of C/EBPα in bone marrow cells resulted in profound inhibition of erythroid development in PB (11.9% ± 2.3% versus 22.7% ± 2.9%; P < .05; Figure 2B) and BM (12.2% ± 3.9% versus 41.8% ± 3.3%; P < .05; Figure 2D) 4 months after transplantation, while it induced the expansion of Mac-1+ myeloid cells in the bone marrow of recipient mice (80.3% ± 5.6% versus 61.7% ± 3.4%; P < .05; Figure 2D), as well as granulocytes that coexpress Mac-1 and Gr-1 in peripheral blood of recipient mice (34.74% ± 3.2% versus 19.7% ± 2.2%; P < .05; Figure 2B). These data demonstrate that overexpression of C/EBPα in hematopoietic progenitor cells inhibited erythroid development while promoting myeloid development in vivo.
C/EBPα promotes myeloid development versus erythroid development in vivo. Femoral BMCs were prepared from 8-week-old inbred female C57Bl/6 mice that had been treated with 5-fluorouracil (5-FU) 4 days earlier to enrich for hematopoietic stem cells, and then infected with retrovirus containing MSCV-GFP or MSCV-C/EBPα-GFP. GFP+ BM cells (1 × 106) were transplanted into lethally irradiated mice with host bone marrow cells (2 × 105) to promote radioprotection. Four months after transplantation, the presence of donor-derived (GFP+) hematopoietic cells was determined by flow cytometry. Granulocytes and macrophages (Gr-1 and Mac-1) and erythroid cells (CD71 and TER119) in peripheral blood are presented in panels A-B, while myeloid (Mac-1) and erythroid (CD71 and TER119) cells in bone marrow are presented in panels C-D. These flow cytometry data were collected from 3 different transplantation experiments. (B, D) Horizontal bars indicate median values.
C/EBPα promotes myeloid development versus erythroid development in vivo. Femoral BMCs were prepared from 8-week-old inbred female C57Bl/6 mice that had been treated with 5-fluorouracil (5-FU) 4 days earlier to enrich for hematopoietic stem cells, and then infected with retrovirus containing MSCV-GFP or MSCV-C/EBPα-GFP. GFP+ BM cells (1 × 106) were transplanted into lethally irradiated mice with host bone marrow cells (2 × 105) to promote radioprotection. Four months after transplantation, the presence of donor-derived (GFP+) hematopoietic cells was determined by flow cytometry. Granulocytes and macrophages (Gr-1 and Mac-1) and erythroid cells (CD71 and TER119) in peripheral blood are presented in panels A-B, while myeloid (Mac-1) and erythroid (CD71 and TER119) cells in bone marrow are presented in panels C-D. These flow cytometry data were collected from 3 different transplantation experiments. (B, D) Horizontal bars indicate median values.
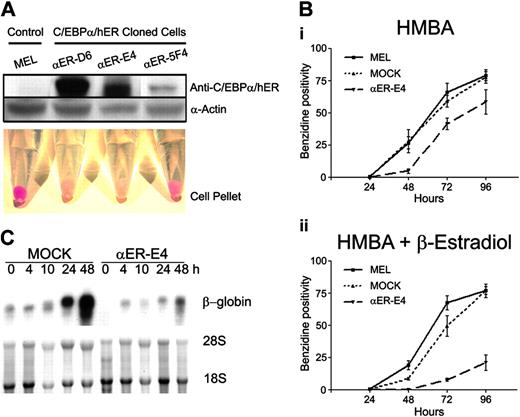
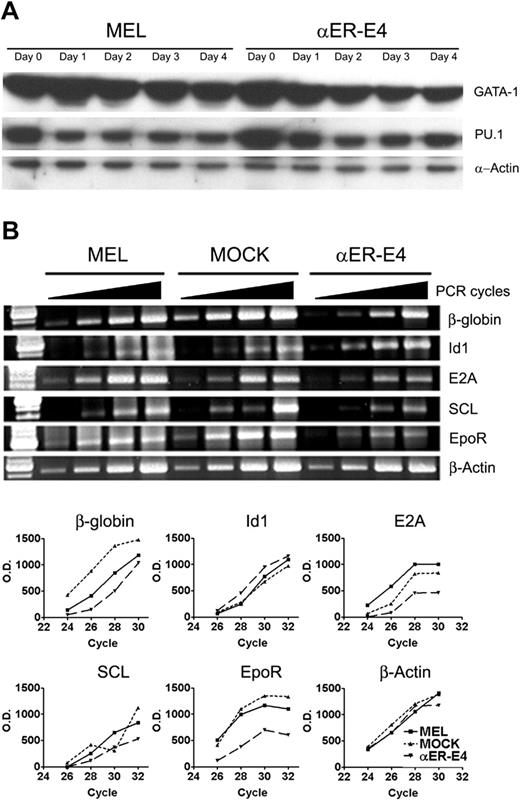
C/EBPα inhibits erythroid differentiation of MEL cells. (A) MOCK and C/EBPα/hER-transduced MEL clones were incubated in the presence of HMBA (3 mM) and β-estradiol (1 μM) for 96 hours. Red cell pellets indicate terminal differentiation of MEL cells. C/EBPα/hER and α-actin protein expression of each clone was confirmed by Western blot analysis as described in “Materials and methods.” (B) Hemoglobinization of MEL, MOCK, and αER-E4 cells was evaluated at the indicated times after treatment with (i) HMBA or (ii) HMBA + β-estradiol by benzidine staining. The mean benzidine positivity of each cell group ± SE of triplicate experiments is shown. (C) β-Globin mRNA levels were measured in HMBA-treated MOCK and αER-E4 cells by Northern blot analysis as described in “Materials and methods” using 447-bp fragment of mouse β-globin cDNA. Control lanes show 18S and 28S ribosomal RNA.
C/EBPα inhibits erythroid differentiation of MEL cells. (A) MOCK and C/EBPα/hER-transduced MEL clones were incubated in the presence of HMBA (3 mM) and β-estradiol (1 μM) for 96 hours. Red cell pellets indicate terminal differentiation of MEL cells. C/EBPα/hER and α-actin protein expression of each clone was confirmed by Western blot analysis as described in “Materials and methods.” (B) Hemoglobinization of MEL, MOCK, and αER-E4 cells was evaluated at the indicated times after treatment with (i) HMBA or (ii) HMBA + β-estradiol by benzidine staining. The mean benzidine positivity of each cell group ± SE of triplicate experiments is shown. (C) β-Globin mRNA levels were measured in HMBA-treated MOCK and αER-E4 cells by Northern blot analysis as described in “Materials and methods” using 447-bp fragment of mouse β-globin cDNA. Control lanes show 18S and 28S ribosomal RNA.
C/EBPα inhibits erythroid differentiation of MEL cells
The results of studies examining overexpression of C/EBPα and deletion of C/EBPα expression in hematopoietic cells indicate that C/EBPα might inhibit erythroid development. To test this possibility, we examined if C/EBPα could inhibit the differentiation of MEL cells, which are transformed erythroid precursors blocked at the proerythroblast stage of differentiation.31 Since C/EBPα has been reported to be growth inhibitory,21-23 we established inducible MEL cell lines (αER-D6, αER-E4, and αER-5F4) that conditionally express C/EBPα. MEL cells were transduced with a retroviral vector that expresses a fusion protein of C/EBPα and human estrogen receptor alpha ligand binding domain (C/EBPα/hER), which can be activated by treating cells with β-estradiol or 4-hydroxy tamoxifen (Figure 3A). Treatment of MEL cells with HMBA induces terminal erythroid differentiation, including the accumulation of hemoglobin and other erythrocyte-specific proteins.32,33 Treatment of parent MEL cells, mock vector–transduced control MEL cell clones (MOCK), and the established MEL clones with HMBA for 4 days induced erythroid differentiation, as indicated by the direct visualization of red cell pellets (hemoglobin-positive cells; Figure 3A) and by increased benzidine staining (Figure 3B). By comparison, HMBA-induced erythroid differentiation of αER-5F4, αER-D6, and αER-D4 clone cells was inhibited by treatment with β-estradiol, which activates the C/EBPα fusion protein, as demonstrated by the absence of hemoglobin-positive cells in the cell pellets (Figure 3A). There was a correlation between the level of C/EBPα fusion protein expression and inhibition of erythroid differentiation. Specifically, HMBA-treated αER-5F4 clone cells, which expressed the lowest levels of C/EBPα/hER by Western blot analysis, showed 47% benzidine-positive cells (Figure 3A), while αER-E4 clone cells with the highest level of C/EBPα/hER expression showed 17% benzidine-positive cells. These findings suggest that inhibition of erythroid differentiation is dependent on the level of C/EBPα expression. When the cells were analyzed for the presence of benzidine-positive cells, we observed that MEL and MOCK cells readily differentiated into hemoglobin-positive cells (76%) within 96 hours (Figure 3B). By comparison, the αER-E4 clone showed delayed kinetics of hemoglobin expression and only 17% of the cells were benzidine-positive after 96 hours (Figure 3B). In agreement with this observation, we found that C/EBPα/hER protein also inhibited the induction of β-globin mRNA in HMBA-treated MEL cells (Figure 3C). Finally, C/EBPα also significantly inhibited the proliferation of MEL cell clones compared with parental MEL and MOCK cells (data not shown). Taken together, our data show that C/EBPα/hER inhibits erythroid differentiation of MEL cells, suggesting that C/EBPα is a negative regulator of erythroid differentiation. Furthermore, the erythroid differentiation inhibition was inversely correlated with the expression levels of C/EBPα.
C/EBPα induces lineage switching of MEL cells, EML cells, and normal MEPs to the myeloid lineage
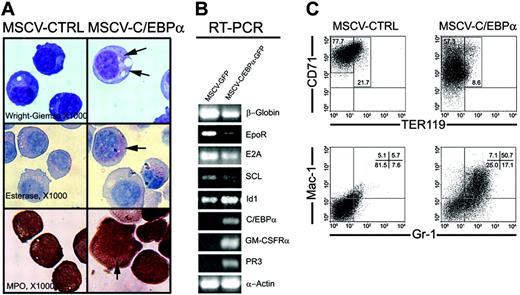
Since it was previously shown that C/EBPα reprograms B-cell precursors to macrophages,12 we determined if C/EBPα might act as a molecular switch to inhibit erythroid differentiation and promote myeloid development in committed erythroid progenitor cells that were transduced with pMSCV-GFP and pMSCV-C/EBPα-GFP. First, we examined if C/EBPα could promote myeloid development in erythroid-committed MEL cells. We performed flow cytometry analysis with erythroid-specific cell surface marker proteins, TER119 and CD71, and found that TER119 was not expressed on MEL cells before or after overexpression of C/EBPα, while CD71 expression was decreased by C/EBPα (data not shown). Cytocentrifuge preparations of MEL cells, transduced with pMSCV-GFP or pMSCV-C/EBPα-GFP vectors, showed an increase in cell size and the presence of myeloid granules, while control MEL cells maintained an erythroblast morphology, indicating that C/EBPα induced myeloid differentiation of MEL cells (Figure 4A). C/EBPα also induced the expression of myeloid-specific genes in MEL cells, including the neutrophil granule protein PR3 and GM-CSFRα, while β-globin, EpoR, E2A, and SCL were down-regulated (Figure 4B). These data indicate that C/EBPα initiated a myeloid differentiation program in MEL cells.
EML cells are representative of primitive multipotential progenitors that can be induced to differentiate into myeloid, lymphoid, and erythroid lineage cells.34 We have previously shown that C/EBPα is induced in EML cells during myeloid differentiation by treatment with all-trans retinoic acid (ATRA) and hematopoietic growth factor (HGF).35 Therefore, to evaluate if C/EBPα inhibits erythroid development while promoting myeloid development in multipotential EML cells, EML cells were transduced with MSCV-GFP and MSCV-C/EBPα-GFP retroviral vectors. Overexpression of C/EBPα in EML cells promoted a more rapid and complete differentiation of granulocytes compared with EML cells treated with HGF plus ATRA. Specifically, we observed numerous segmented neutrophils after 5 days of C/EBPα overexpression (12.4% ± 2.1%) compared with few or no segmented neutrophils in EML cells (0.75% ± 0.5%) treated with ATRA and HGF over the same time period (data not shown). Furthermore, overexpression of C/EBPα significantly inhibited mSCF and hEpo-induced erythroid differentiation of EML cells. Specifically, 22% of control EML cells expressed the erythroid-specific cell surface antigen TER119 after 3 days in culture, while EML cells transduced with MSCV-C/EBPα-GFP expressed only 8.6% TER119-positive cells (Figure 4C). In addition, the level of CD71 expression was decreased and 50.7% of MSCV-C/EBPα-GFP–transduced cells expressed Mac-1 and Gr-1. These data indicate that C/EBPα acts as a switch to promote myeloid differentiation of multipotential EML cells, while inhibiting erythroid differentiation.
C/EBPα switches lineage potential of MEL and EML cells toward myeloid development. (A) MEL cells were transduced by retrovirus containing MSCV-C/EBPα-GFP or MSCV-GFP as described in “Materials and methods.” After sorting GFP+ cells, MEL cells were cultured in vitro for an additional 3 days, after which cell morphology was determined by Wright-Giemsa staining, and cell lineage by naphthol AS-D chloroacetate esterase and MPO staining (× 1000). Arrows indicate stained myeloid granules. (B) RT-PCR was performed using total RNA harvested from the cultured MEL cells 3 days after transduction with retrovirus containing MSCV-C/EBPα-GFP or MSCV-GFP. (C) The SCF-dependent EML cell line (gift of Dr Schickwann Tsai) was maintained in IMDM supplemented with 20% horse serum, 8% conditioned medium from BHK/MKL cells containing mSCF, and P/S/G. EML cells were cultured with mSCF (100 ng/mL) and hEpo (40 U/mL) 24 hours prior to transduction with retrovirus containing pMSCV-C/EBPα-GFP or pMSCV-GFP. EML cells were transduced with the retroviral vectors that express MSCV-C/EBPα-GFP or MSCV-GFP and then cultured for 3 days with mSCF (100 ng/mL) and hEpo (40 U/mL) to promote erythroid development. The expression of (GFP+) granulocytes and monocytes (Gr-1 and Mac-1) and erythroid cells (CD71 and TER119) was determined by flow cytometry.
C/EBPα switches lineage potential of MEL and EML cells toward myeloid development. (A) MEL cells were transduced by retrovirus containing MSCV-C/EBPα-GFP or MSCV-GFP as described in “Materials and methods.” After sorting GFP+ cells, MEL cells were cultured in vitro for an additional 3 days, after which cell morphology was determined by Wright-Giemsa staining, and cell lineage by naphthol AS-D chloroacetate esterase and MPO staining (× 1000). Arrows indicate stained myeloid granules. (B) RT-PCR was performed using total RNA harvested from the cultured MEL cells 3 days after transduction with retrovirus containing MSCV-C/EBPα-GFP or MSCV-GFP. (C) The SCF-dependent EML cell line (gift of Dr Schickwann Tsai) was maintained in IMDM supplemented with 20% horse serum, 8% conditioned medium from BHK/MKL cells containing mSCF, and P/S/G. EML cells were cultured with mSCF (100 ng/mL) and hEpo (40 U/mL) 24 hours prior to transduction with retrovirus containing pMSCV-C/EBPα-GFP or pMSCV-GFP. EML cells were transduced with the retroviral vectors that express MSCV-C/EBPα-GFP or MSCV-GFP and then cultured for 3 days with mSCF (100 ng/mL) and hEpo (40 U/mL) to promote erythroid development. The expression of (GFP+) granulocytes and monocytes (Gr-1 and Mac-1) and erythroid cells (CD71 and TER119) was determined by flow cytometry.
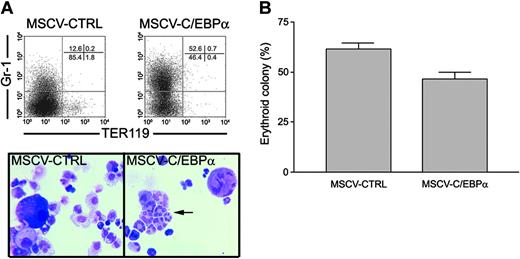
Finally, to evaluate if C/EBPα could induce lineage switching of normal erythroid progenitors to myeloid cells, we purified MEPs and transduced these cells with pMSCV-GFP– or pMSCV-C/EBP-GFP–expressing retroviruses, and then sorted GFP+ MEPs by flow cytometry. Retroviral vectors that express C/EBPα/hER fusion proteins were not used for MEPs, as these vectors would require further selection in puromycin. The differentiation of MEPs was analyzed by cell morphology, flow cytometry, and growth in soft agar colony assays. We found that induction of C/EBPα in MEPs increased myeloid lineage development as evidenced by induction of Gr-1 expression and increased numbers of segmented neutrophils, and the loss of TER119 expression and erythroid colony formation. Specifically, C/EBPα induced more than half of the MEPs (52.6%) to express the granulocyte-specific marker Gr-1 in liquid culture, while 12% of the MEPs transduced with pMSCV-GFP expressed Gr-1 (Figure 5A). In addition, C/EBPα decreased the TER119-positive cells in MEPs to 0.4%. These results were confirmed by the morphologic examination of GFP+ MEPs, where 79.3% of the cells were myeloid and 20.5% were erythroid in MSCV-C/EBPα-GFP–transduced MEPs in comparison with 77.3% erythroid for MSCV-GFP–transduced MEPs (Figure 5A). Overexpression of C/EBPα decreased erythroid colony formation of MEPs by 25% (61.4% versus 46.6%), while the percentage of myeloid colonies was increased (Figure 5B). The results were presented as percentage of erythroid colonies versus total colony number due to the growth inhibitory function of C/EBPα in MEPs. In addition, the FACS-purified MEPs were more than 60% erythroid progenitors by colony formation assay. Therefore, it is possible that C/EBPα induced myeloid differentiation of the 40% contaminating myeloid progenitors. However, we found that 77% of the cells in liquid culture were myeloid cells after C/EBPα transduction, suggesting that some of the erythroid progenitors had undergone myeloid differentiation. Furthermore, these results agreed with previous studies showing that C/EBPα overexpression in human CD71+ cells decreased erythroid colony number.36 Finally, transduction of CMPs with retroviral vectors that express C/EBPα promoted myeloid development and inhibited erythroid differentiation (data not shown). Taken together, these data suggest that C/EBPα can switch the lineage differentiation potential of committed erythroid progenitors to myeloid lineage.
C/EBPα induces myeloid differentiation of primary MEPs. (A) Lineage-negative (Linneg) cell population was stained with the following cocktail of directly conjugated monoclonal antibodies: PE anti-cKit, FITC anti-CD34, APC anti–Sca-1, PE-Cy7 anti-FcRII/III (2.4G2), and PE-Cy5 anti–IL-7Rα. These cells were then sorted by high-speed sorter (MoFlo; Cytomation) for the IL-7Rα–/c-Kit+/Sca-1–/CD34–/FcRII/IIIlo cell population identified as MEPs. Normal MEPs were transduced with retrovirus containing pMSCV-C/EBPα-GFP or pMSCV-GFP. GFP+ MEPs were isolated and cultured for 3 days in cIMDM containing mSCF (100 ng/mL), hEpo (10 U/mL), and hTpo (100 ng/mL). Lineage differentiation was analyzed by flow cytometry using Gr-1 and TER119 antibodies and differential cell counts of Wright-Giemsa–stained cytocentrifuge preparations. Arrow indicates segmented neutrophils. (B) GFP+ MEPs (5 × 104/plate) were also cultured in 1.1% methyl-cellulose containing mSCF (100 ng/mL), hEpo (5 U/mL) to detect erythroid progenitors as described in “Materials and methods.” The mean number of erythroid colonies ± SE of triplicate plates is shown.
C/EBPα induces myeloid differentiation of primary MEPs. (A) Lineage-negative (Linneg) cell population was stained with the following cocktail of directly conjugated monoclonal antibodies: PE anti-cKit, FITC anti-CD34, APC anti–Sca-1, PE-Cy7 anti-FcRII/III (2.4G2), and PE-Cy5 anti–IL-7Rα. These cells were then sorted by high-speed sorter (MoFlo; Cytomation) for the IL-7Rα–/c-Kit+/Sca-1–/CD34–/FcRII/IIIlo cell population identified as MEPs. Normal MEPs were transduced with retrovirus containing pMSCV-C/EBPα-GFP or pMSCV-GFP. GFP+ MEPs were isolated and cultured for 3 days in cIMDM containing mSCF (100 ng/mL), hEpo (10 U/mL), and hTpo (100 ng/mL). Lineage differentiation was analyzed by flow cytometry using Gr-1 and TER119 antibodies and differential cell counts of Wright-Giemsa–stained cytocentrifuge preparations. Arrow indicates segmented neutrophils. (B) GFP+ MEPs (5 × 104/plate) were also cultured in 1.1% methyl-cellulose containing mSCF (100 ng/mL), hEpo (5 U/mL) to detect erythroid progenitors as described in “Materials and methods.” The mean number of erythroid colonies ± SE of triplicate plates is shown.
C/EBPα inhibits erythropoietin receptor expression in differentiating MEL cells and fetal liver cells
To determine the molecular mechanisms by which C/EBPα regulates erythroid differentiation, we examined the expression of transcription factors that promote erythroid differentiation, including GATA-1, E2A and SCL,37-43 and those that inhibit erythroid development, including PU.132,33 and Id1.44,45 In agreement with previous results,32,33 the expression of PU.1 and GATA-1 protein was decreased during HMBA-induced erythroid differentiation of MEL cells and αER-E4 clone cells that were blocked for erythroid differentiation (Figure 6A). Activation of C/EBPα in αER-E4 cells did not affect PU.1 and GATA-1 protein expression during HMBA-induced erythroid differentiation (Figure 6A), indicating that the C/EBPα-mediated block in erythroid differentiation is likely due to mechanisms other than reduced GATA-1 or increased PU.1 expression. In comparison, C/EBPα down-regulated the expression of E2A and SCL mRNA in MEL cells as determined by semiquantitative RT-PCR, while Id1, which inhibits erythroid differentiation in MEL cells, was up-regulated by C/EBPα during erythroid differentiation (Figure 6B).
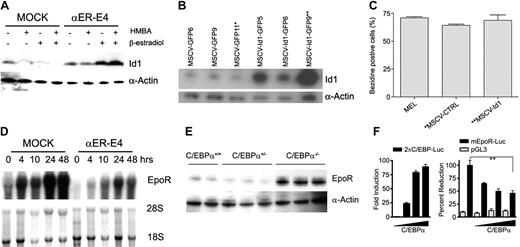
To confirm that Id1 protein expression was induced by C/EBPα in MEL cells, we evaluated Id1 protein expression by Western blot analysis. Id1 protein expression was induced in αER cells treated with β-estradiol in the presence or absence of HMBA, while Id1 expression was not induced in MEL or MOCK cells (Figure 7A). To evaluate if Id1 might act downstream of C/EBPα to inhibit erythroid differentiation, we infected MEL cells with retroviral vectors that express GFP (pMSCV-GFP) or Id1 (pMSCV-Id1-GFP), and compared the response of control and Id1-expressing single-cell clones to undergo HMBA-induced erythroid differentiation (Figure 7B). MEL cells that overexpress Id1 (MSCV-Id1-GFP9 clone) or MOCK cells (MSCV-GFP11 clone) were induced to undergo erythroid differentiation to the same extent by HMBA (71.0% versus 68.7% benzidine-positive cells; Figure 7C). These data indicate that even though Id1 expression was induced by C/EBPα, Id1 overexpression is not sufficient to suppress erythropoiesis, suggesting there may be other mechanisms by which C/EBPα blocks erythroid development and differentiation.
Regulation of gene expression in MEL cells by C/EBPα. (A) MEL cells and αER-E4 clone cells were treated with 3 mM HMBA and 1.0 μM β-estradiol. Total cell lysates were prepared from treated cells at the indicated times and 30 μg was analyzed for GATA-1, PU.1, and α-actin expression by Western blot analysis as described in “Materials and methods.” (B) Total RNA was also obtained from MEL cell clones 10 hours after treatment with HMBA and β-estradiol. Expression of the indicated genes was evaluated by RT-PCR using the indicated cycle numbers (24, 26, 28, and 30 cycles for β-globin, or 26, 28, 30, and 32 cycles for all other genes). The line graph represents densitometry scan of each band on an agarose gel.
Regulation of gene expression in MEL cells by C/EBPα. (A) MEL cells and αER-E4 clone cells were treated with 3 mM HMBA and 1.0 μM β-estradiol. Total cell lysates were prepared from treated cells at the indicated times and 30 μg was analyzed for GATA-1, PU.1, and α-actin expression by Western blot analysis as described in “Materials and methods.” (B) Total RNA was also obtained from MEL cell clones 10 hours after treatment with HMBA and β-estradiol. Expression of the indicated genes was evaluated by RT-PCR using the indicated cycle numbers (24, 26, 28, and 30 cycles for β-globin, or 26, 28, 30, and 32 cycles for all other genes). The line graph represents densitometry scan of each band on an agarose gel.
Erythropoietin signaling is required for the proliferation and survival of erythroid cells, and absence of EpoR expression results in impaired erythroid development.46,47 In addition, C/EBPα increases the expression of cytokine receptors while inducing myeloid lineage differentiation.15-18,24 Based on these findings, we hypothesized that C/EBPα might regulate EpoR expression. To confirm the RT-PCR results (Figure 6B), we evaluated EpoR expression by Northern blot analysis and found that C/EBPα significantly inhibited HMBA-induced EpoR mRNA expression in MEL cells (Figure 7D). These results suggested that EpoR expression might be altered in C/EBPα–/– mice. Therefore, we determined EpoR protein levels by Western blot analysis in C/EBPα–/– FL lysates. C/EBPα–/– FL lysates showed a 10-fold increase in EpoR expression compared with C/EBPα+/– and C/EBPα+/+ lysates (Figure 7E). It is unlikely that the increased levels of EpoR expression in C/EBPα–/– FL are the result of increased numbers of erythroid cells (Figure 1A), as these cells were increased by only 1.5-fold compared with C/EBPα+/+ FL. Thus, the data indicate that C/EBPα suppresses EpoR expression in vitro and in vivo.
To confirm that C/EBPα decreases EpoR gene transcription, increasing amounts of murine C/EBPα expression plasmid were coexpressed with an EpoR promoter-reporter construct (mEpoR-Luc), the pGL3 basic control vector, the positive control vector (2x C/EBP-Luc), and mEpoR-Luc in L cells. We observed inhibitory effect of C/EBPα on EpoR promoter activity by C/EBPα, while C/EBPα activated the positive control reporter, 2xC/EBP-Luc (Figure 7F). Thus, C/EBPα may inhibit erythropoiesis through direct inhibition of EpoR gene transcription.
Regulation of EpoR and Id1 expression by C/EBPα during erythroid differentiation. (A) MEL cells, MOCK cells, and αER-E4 cells were cultured in 3 mM HMBA and/or 1.0 μM β-estradiol as indicated. Id1 protein expression was determined 24 hours after treatment by Western blot analysis as described in “Materials and methods.” (B) MEL cells were transduced with control (pMSCV-GFP) or Id1-expressing (pMSCV-Id1-GFP) retroviral vectors. Stable cell lines were cloned and Id1 expression was determined by Western blot analysis. Asterisks indicate the cell lines used for induction of erythroid differentiation. (C) Cloned MEL cells were seeded at 4 × 105/mL in cDMEM containing 3 mM hexamethylene bisacetamide (HMBA) and incubated at 37°C. Erythroid differentiation of Id1-expressing cell lines was determined by benzidine staining 4 days after HMBA treatment (3 mM). (D) Total RNA was obtained from MOCK and αER-E4 cell clones at the indicated times after treatment with HMBA (3 mM) and β-estradiol (1 μM). EpoR mRNA expression in MOCK and αER-E4 cells was analyzed by Northern blot analysis before and after treatment with HMBA and β-estradiol. The EpoR probe was an XhoI 1450-bp fragment of the murine EpoR cDNA (gift of Dr Sandy Ruscetti, NCI-Fredrick. (E) Total cellular extracts were prepared from C/EBPα–/–, C/EBPα+/–, C/EBPα+/+ FL cells, and EpoR expression was determined by Western blot analysis of 30 μg protein. (F) L cells were transfected with the pGL3, 2xC/EBPα-Luc, mEpoR-Luc, alone or with increasing amounts (20, 50, 100 ng) of C/EBPα expression vector by liposomal method (FuGENE 6). Cell extracts were assayed after 48 hours of incubation with complete DMEM media. The histograms represent means ± SE of the luciferase activity from triplicates normalized for transfection efficiency using renilla luciferase. **P < .05 t test.
Regulation of EpoR and Id1 expression by C/EBPα during erythroid differentiation. (A) MEL cells, MOCK cells, and αER-E4 cells were cultured in 3 mM HMBA and/or 1.0 μM β-estradiol as indicated. Id1 protein expression was determined 24 hours after treatment by Western blot analysis as described in “Materials and methods.” (B) MEL cells were transduced with control (pMSCV-GFP) or Id1-expressing (pMSCV-Id1-GFP) retroviral vectors. Stable cell lines were cloned and Id1 expression was determined by Western blot analysis. Asterisks indicate the cell lines used for induction of erythroid differentiation. (C) Cloned MEL cells were seeded at 4 × 105/mL in cDMEM containing 3 mM hexamethylene bisacetamide (HMBA) and incubated at 37°C. Erythroid differentiation of Id1-expressing cell lines was determined by benzidine staining 4 days after HMBA treatment (3 mM). (D) Total RNA was obtained from MOCK and αER-E4 cell clones at the indicated times after treatment with HMBA (3 mM) and β-estradiol (1 μM). EpoR mRNA expression in MOCK and αER-E4 cells was analyzed by Northern blot analysis before and after treatment with HMBA and β-estradiol. The EpoR probe was an XhoI 1450-bp fragment of the murine EpoR cDNA (gift of Dr Sandy Ruscetti, NCI-Fredrick. (E) Total cellular extracts were prepared from C/EBPα–/–, C/EBPα+/–, C/EBPα+/+ FL cells, and EpoR expression was determined by Western blot analysis of 30 μg protein. (F) L cells were transfected with the pGL3, 2xC/EBPα-Luc, mEpoR-Luc, alone or with increasing amounts (20, 50, 100 ng) of C/EBPα expression vector by liposomal method (FuGENE 6). Cell extracts were assayed after 48 hours of incubation with complete DMEM media. The histograms represent means ± SE of the luciferase activity from triplicates normalized for transfection efficiency using renilla luciferase. **P < .05 t test.
Discussion
This study provides evidence for the first time that C/EBPα can function to promote myeloid (GMP) versus erythroid (MEP) development in primitive (CMP) progenitors. Furthermore, our results indicate that C/EBPα functions to determine cell fate in primitive progenitors by positively regulating myeloid-specific gene expression and negatively regulating erythroid-specific gene expression. Specifically, mice lacking C/EBPα had increased numbers of erythroid progenitors and erythroid cells. Aberrant erythroid development was most likely intrinsic to the hematopoietic progenitors and not due to a defect in the microenvironment (Figure 1). Conversely, transplantation of hematopoietic progenitors overexpressing C/EBPα showed increased myeloid and decreased erythroid lineage development in long-term reconstitution assays (Figure 2). In addition, C/EBPα induced myeloid differentiation of MEL and EML cell lines (Figure 4) and primary MEPs (Figure 5), suggesting that C/EBPα acts as a cell fate switch to promote myeloid versus erythroid development in CMPs. Similarly, increased levels of C/EBPα in HSCs might promote CMP versus CLP differentiation. In this regard, overexpression of C/EBPα in CLPs promoted macrophage differentiation and inhibited B-cell development.12 We propose that HGF or other extrinsic and intrinsic signals that increase or decrease the expression of C/EBPα in CMPs will determine myeloid versus erythroid development.
The zinc finger transcription factor GATA-1 is required for erythroid development. PU.1 is a known antagonist of GATA-1 function and can block erythroid differentiation, including the differentiation of MEL cells.9,10 Thus, it has been proposed that expression levels of GATA-1 and PU.1 can promote erythroid or myeloid cell fate in CMPs. We found that C/EBPα inhibits erythroid differentiation without affecting the expression of PU.1 or GATA-1 in MEL cells (Figure 6A), suggesting that C/EBPα inhibits erythroid differentiation by alternative mechanisms. In this regard, C/EBPα inhibited the transcription of erythroid-specific genes transactivated by GATA-1, including β-globin and EpoR, suggesting a potential interaction between GATA-1 and C/EBPα. However, it remains to be determined if C/EBPα represses GATA-1 activity in MEL cells by preventing the binding of GATA-1 to its target DNA sequences, or if C/EBPα alters the interaction of GATA-1 with coregulators such as E2A, SCL, LMO2, and Ldb1.39 In this regard, high levels of C/EBPα in hematopoietic progenitor cells might inhibit GATA-1 activity and activate myeloid-specific genes in collaboration with PU.1 to promote myeloid differentiation and inhibit erythroid differentiation.
Id1 acts as a negative regulator of E2A by inhibiting E2A/SCL heterodimer formation required to bind E-box sequences in erythroid promoters.39,42 C/EBPα induced Id1 expression in MEL cells, suggesting that C/EBPα might inhibit erythroid differentiation through Id1 function. However, we found that MEL cells overexpressing Id1 can be induced to undergo erythroid differentiation by HMBA (Figure 7B-C). While our results differ from previous reports,44,45 they are in agreement with a recent study demonstrating that overexpression of murine Id1 in human hematopoietic progenitor cells does not inhibit normal erythroid differentiation in clonogenic assays.36 Furthermore, we found that mice that received a transplant of hematopoietic progenitors overexpressing Id1 did not exhibit defects in erythroid development (data not shown). Thus, C/EBPα-induced Id1 expression alone does not explain inhibition of erythroid differentiation by C/EBPα. We found that C/EBPα inhibits the induction of EpoR expression during erythroid differentiation of MEL cells and that C/EBPα–/– FL cells express increased levels of EpoR, suggesting that C/EBPα negatively modulates erythroid development by down-regulating EpoR expression (Figure 7D-F). C/EBPα is required to activate myeloid-specific genes, including growth factor receptor genes such as M-CSFR, G-CSFR, and GM-CSFR.15-18 Therefore, C/EBPα promotes myeloid development and inhibits erythroid development in hematopoietic progenitors at least partly by regulating HGF receptor expression, including induction of G-CSFR, GM-CSFR, and M-CSFR and suppression of EpoR. In MEL cells, C/EBPα also decreased the mRNA expression of E2A and SCL, which are important transcription factors for erythropoiesis (Figure 6B). These data are consistent with previous reports that C/EBPα can function as a positive or negative regulator of transcriptional activity depending on the individual promoter or enhancer element.13,14 In this regard, we showed here that expression of C/EBPα inhibits an EpoR promoter reporter construct in vitro, suggesting that C/EBPα may regulate EpoR expression. Since there is a potential C/EBPα binding site (–1129 to –1120) in a previously reported repressor segment of the mouse EpoR promoter,48 C/EBPα may directly bind to the EpoR promoter to inhibit gene expression. While it is not known if C/EBPα directly or indirectly regulates EpoR promoter activity, it is also possible that C/EBPα exerts its activation or inhibition function by formation of different coregulator complexes in myeloid or erythroid-specific gene promoter.14,49
We have found that C/EBPα–/– FL HPCs show increased cell proliferation and a block in the ability of CMPs to differentiate to GMPs.24 Mutations in at least one allele of the C/EBPα gene, which result in loss of normal C/EBPα function, are present in 10% of acute myelogenous leukemia (AML) cases, suggesting a role for C/EBPα in the development of these leukemias.50-52 However, HPCs from C/EBPα–/– mice give rise to myelodysplastic syndrome (MDS), but not AML, when transplanted in vivo,24 suggesting that additional mutations are required for the development of AML. We show here that HPCs from C/EBPα–/– FL have an enhanced ability to differentiate into erythroid cells, which would prevent the expansion of undifferentiated progenitor populations and development of AML. Thus, additional mutations resulting in a defect in erythroid differentiation, for example, would be required for the development of AML. Furthermore, our observation that C/EBPα can regulate the balance between myeloid and erythroid cell determination in multipotential progenitors expands the therapeutic relevance of C/EBPα. For example, in polycythemia vera (PV), which results from increased erythropoiesis due to clonal expansion of multipotent progenitors, overexpression of C/EBPα may decrease erythroid cell production by inhibiting erythroid development and proliferation. Progression of chronic myelogenous leukemia (CML) from chronic phase to blast phase crisis is associated with down-regulation of C/EBPα by the BCR-ABL fusion protein.27 Thus, drugs that increase C/EBPα expression levels could be used not only to treat AML,53 but also to treat chronic myeloproliferative diseases such as PV or CML, thereby, expanding the utility of C/EBPα as a potential therapeutic target.
Prepublished online as Blood First Edition Paper, February 9, 2006; DOI 10.1182/blood-2005-06-2216.
Supported in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract no. NO1-CO-12400.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We gratefully acknowledge the technical support of Steve Stull, Barbara Shankle, Kathleen Noer, Roberta Matthai, and Mehrnoosh Abshari. We also thank Drs Joost Oppenheim, Sandra Ruscetti, and Kimberly Klarmann for critically reviewing this paper. We also appreciate editorial assistance of the NCI Fellows Editorial Board.
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US government.

![Figure 1. Increased erythroid development in C/EBPα–/– mice. (A) Cytocentrifuge preparations of C/EBPα–/– FL cells were stained with Wright-Giemsa, and differential cell counts were performed on 400 cells from 3 individual mice. The data are reported as the mean number of cells ± standard error (SE). (B) FL cells (2.5 × 104) from C/EBPα+/+, C/EBPα +/–, and C/EBPα–/– embryos at E15.5 were cultured in complete IMDM (cIMDM) containing 1.1% methylcellulose, 25% FCS, 100 U/mL penicillin, 100 μg/mL streptomycin, and 2 mM l-glutamine (P/S/G), and cultures were supplemented with murine (m) IL-3 (30 ng/mL), stem cell factor (mSCF, 100 ng/mL), and human erythropoietin (hEpo, 5 U/mL). The mean number of BFU-E colonies ± SE of triplicate plates is shown. (C-D) C/EBPα–/– and C/EBPα+/+ FL cells were transduced with MSCV-GFP retroviral vectors to track donor erythroid reconstitution. Fetal liver cells were resuspended at 2 × 105/mL in cIMDM and cultured with 8 μg/mL polybrene, 100 ng/mL mSCF, 100 ng/mL thrombopoietin (hTpo), 100 ng/mL hFlt-3L, and 50 ng/mL IL-6. A total of 3 retrovirus infections were performed, after which the cells were cultured with the same cytokine mixture. GFP-positive (GFP+) cells were isolated 2 days after the last infection by FACS sorting (FACSAria [BD Biosciences] or MoFlo [Cytomation, Fort Collins, CO]). BM cells were harvested 4 months after transplantation and then analyzed for the expression of the erythroid markers (TER119 and CD71) and myeloid cell markers (Gr-1 and Mac-1). (D) Horizontal bars indicate median values. (E) GFP+ BM cells were purified by FACS and cultured in methylcellulose (7.5 × 104/plate) or soft agar (1 × 105/plate) to determine the number of erythroid or myeloid progenitors. For myeloid progenitors, cells were plated in cIMDM containing 10% FCS, P/S/G, and 0.35% sea plaque agarose in the presence or absence of 50 ng/mL hG-CSF, 100 ng/mL M-CSF, 100 ng/mL mSCF, 20 ng/mL mGM-CSF, 30 ng/mL mIL-3. The mean number of BFU-E and CFU-c colonies ± SE of triplicate plates is shown.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/107/11/10.1182_blood-2005-06-2216/4/m_zh80110696180001.jpeg?Expires=1767717488&Signature=Hb4gpsGMUae7wSk9HP32Ra2bdAkiHTWHHmRunaDphxQRPblFozVGKy0hpNT3RgaSsTofLaRrc47nJ-R-PRT0aj6sRLVoXEEpMMIOHGD25e5ijdUHPKVSC9osCgfQjJ6eHEVzTbt0YkYLj9RVtmzKUytzX9ft4dU-y1hVOjtirdVTxdxiENhPiPrrfbbihACV-1zqhcMg6s3vcIpF7lMBscTq7VN86UC7uHlHrHvH-nep3VdKaiQYnxBU7q3tMou7Y8-ZQE-SKAvJmTw4faPmVC77I2qrXDKRkgT2JKbsMXfv4P-g487nqPSQ1K4CIWRZLg4t8lQ1jlOm60pMJDpWbw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal