Abstract
Tumor necrosis factor (TNF)–related apoptosis-inducing ligand (TRAIL) induces apoptosis in many transformed cells but not in normal cells and, hence, has emerged as a novel anticancer agent. Previously, we showed that although most adult T-cell leukemia/lymphoma (ATLL) cells express the TRAIL death receptor DR4 (TRAIL-R1) or DR5 (TRAIL-R2), they are resistant to TRAIL. Thus, in this study, we tried to find natural products that can overcome TRAIL resistance. Among more than 150 materials screened, a dihydroflavonol that was extracted from Blumea balsamifera (BB-1) exhibited the most striking synergism with TRAIL. Treatment of the TRAIL-resistant ATLL cell line KOB, with a combination of BB-1 and TRAIL, resulted in apparent apoptosis that was not observed on treatment with either agent alone. Furthermore, pretreatment with BB-1 followed by TRAIL further augmented the synergism. BB-1 increased the level of TRAIL-R2 promoter activity and surface protein expression in a p53-independent manner. TRAIL-R2 siRNA inhibited the synergism, indicating that sensitization was caused by the increase of TRAIL-R2 expression. More interestingly, similar effects were observed in other leukemia cell lines by exactly the same mechanisms. These results suggest that combined treatment with BB-1 and TRAIL may be a new strategy for cancer therapy.
Introduction
Tumor necrosis factor (TNF)–related apoptosis-inducing ligand (TRAIL), a member of the TNF superfamily, induces apoptosis in tumor cells of different origins but not in normal cells in vitro or in normal cells of experimental animals, thus providing therapeutic possibilities in human cancer.1-3 The potential importance of the TRAIL pathway for the treatment of cancer is highlighted by the recent introduction of TRAIL receptor (TRAIL-R) agonistic antibodies in human phase 1 trials.4 TRAIL can bind to 2 death receptors (DRs), DR4 (TRAIL-R1) and DR5 (TRAIL-R2), that contain a cytoplasmic functional death domain, and it can bind to 2 antiapoptotic decoys, DcR1 (TRAIL-R3) and DcR2 (TRAIL-R4). TRAIL-R3 lacks a cytoplasmic region, and TRAIL-R4 has a truncated nonfunctional cytoplasmic death domain. Neither receptor, therefore, can mediate death signals; rather, they compete with DRs and are considered to act as decoy receptors (DcRs).5 There are at least 2 fundamental apoptotic pathways, referred to as the extrinsic pathway and the intrinsic pathway. TRAIL can trigger either pathway, depending on the cell type.6 TRAIL-induced apoptosis initiated by the extrinsic pathway involves DR engagement, death-inducing signaling complex (DISC) formation, proteolytic activation of caspase-8, and, consequently, activation of caspase-3.7 Proteolytic caspase-8 further activates Bid, which, in turn, translocates to the mitochondria and activates the intrinsic pathway.8,9 In the intrinsic pathway, death signals lead to changes in mitochondrial outer membrane permeability, subsequently releasing cytochrome c, which forms an apoptosome with Apaf-1 and caspase-9 and activates effector caspases.10 Mechanisms of resistance to TRAIL differ among cancer cells, but DcRs, FLIP, the Bcl-2 family, and the IAP family are all important antiapoptotic effectors.11 Although previous studies have suggested that multiple myeloma cells might be susceptible to TRAIL-induced apoptosis, most hematologic malignancies, especially primary leukemia, are resistant to TRAIL.12,13 Most solid tumor cells are also relatively resistant to TRAIL-induced apoptosis. Nevertheless, numerous studies have shown that TRAIL resistance can be overcome by the combined application of chemotherapeutic drugs and irradiation.14
Natural products have played a highly significant role over the years in the discovery of new drugs. This is particularly evident in the treatment of cancers and infectious diseases in which more than 60% and 75% of drugs, respectively, are of natural origin.15 Therefore, a natural product with strong synergistic activity with TRAIL but minimal toxicity would be a new tool for cancer therapy.
Adult T-cell leukemia/lymphoma (ATLL) is a neoplasm of T-lymphocyte origin etiologically associated with HTLV-I, and it is known to be resistant to standard anticancer therapies.16 In a previous study, we showed that although most ATLL cells express TRAIL-DRs, they are resistant to TRAIL.17 The objective of this study was to not only identify natural products that sensitize cells to TRAIL but to establish an approach for overcoming resistance to TRAIL by using a TRAIL-resistant ATLL cell line.
Here, we show for the first time that a dihydroflavonol from a plant increases sensitivity to TRAIL by enhancing TRAIL-R2 expression not only in ATLL cells but also in other types of leukemia cells.
Materials and methods
Cell preparations
The ATLL cell lines KOB and ST1 were established in our laboratory from patients with ATLL.18 These cell lines are dependent on exogenously added IL-2 and were maintained in RPMI 1640 medium supplemented with 10% FBS and 0.5 U/mL IL-2 (kindly provided by Takeda Chemical Industries, Osaka, Japan). We also used the human T-cell leukemia cell line Jurkat, the monocytic leukemia cell line U-937, the chronic myeloid leukemia cell line K562, the Burkitt lymphoma cell line Ramos, and the acute myeloid leukemia cell line HL-60. These cell lines were maintained in RPMI 1640 medium supplemented with 10% FBS. Peripheral blood mononuclear cells (PBMCs) from healthy donors were purified by density gradient centrifugation using Lympho-prep (Axis-Shield, Oslo, Norway). After approval by the ethics committee at our university hospital, all materials from healthy donors were obtained with informed consent.
Surface expression of TRAIL receptors
The cell-surface expression of TRAIL receptors was examined by flow cytometric analysis (FCM). In brief, cells were incubated for 30 minutes on ice with mouse IgG1 anti–TRAIL-R1, -R2, -R3, or -R4 monoclonal antibodies (Alexis, San Diego, CA). Mouse IgG1 (Dako Japan, Kyoto, Japan) was used as a negative control. Cells were washed 3 times in PBS and then incubated with FITC-conjugated goat anti–mouse IgG1 (Dako Japan) as the secondary antibody. After a wash, 104 cells were analyzed using a FACScalibur flow cytometer and Cellquest software (BD Biosciences Immunocytometry Systems, San Jose, CA).
Screening, cell proliferation, and apoptosis inhibition assay
In the screening assay, KOB cells were incubated with various concentrations of natural products and 0.5 μg/mL recombinant human soluble TRAIL (Biomol Research Laboratories, Plymouth Meeting, PA) in 96-well tissue culture plates for 48 hours. More than 150 natural products from our libraries were used for the screening, and cell proliferation (percentage of control cells) was determined with the Alamar Blue bioassay (Biosource International, Camarillo, CA). For the evaluation of sensitivity to TRAIL, KOB cells were treated with 0.5 μg/mL TRAIL, 12.5 μg/mL BB-1, or both. ST1, Jurkat, U937, K562, HL60, Ramos cells, and PBMCs were treated with 0.2 μg/mL TRAIL, 12.5 μg/mL BB-1, or both. Cell proliferation (percentage of control cells) was determined with the 3-(4,5 dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl-2-(4-sulfophenyl)-2H-tetrazolium (MTS) assay using a Cell Titer 96TMAQueos Cell Proliferation Assay kit (Promega, Madison, WI) in accordance with the manufacturer's instructions. For assaying the inhibition of TRAIL-mediated apoptosis, KOB or Jurkat cells were incubated with or without 2.5 μg/mL recombinant human TRAIL-R2-Fc fusion protein (Alexis) in the presence of graded concentrations of recombinant human soluble TRAIL. All experiments were performed in triplicate.
Detection of apoptotic cells by Annexin V-FITC and propidium iodide staining
Cells were stained simultaneously with FITC-conjugated Annexin V and the nonvital dye PI (Bender Medsystems, Vienna, Austria) to enable the discrimination of intact cells (Annexin V– PI–), early apoptotic cells (Annexin V+ PI–), and late apoptotic or necrotic cells (Annexin V+ PI+). KOB cells were treated with 0.5 μg/mL TRAIL, 12.5 μg/mL BB-1, and a combination of both for 48 hours. Cells harvested at several time points were stained, and 5000 cells per sample were analyzed by FCM.
Western blot analysis, immunoprecipitation, and antibodies
KOB, U937, and K562 cells were treated with TRAIL or BB-1 or a combination of both. Cells were harvested at several time points after treatment, cell lysates (30 μg) were prepared, and Western blot analysis was performed as described previously.17 The cytosolic fraction was prepared using a mitochondrial/cytosol kit in accordance with the manufacturer's instructions (BioVision, Mountain View, CA), and 10 μg of cell lysate was used for Western blotting. In the immunoprecipitation assay, cell lysates (500 μg) were incubated for 1 hour at 4°C with anti–p53 antibody. The immune complex was collected on protein A-Sepharose beads (Sigma, St Louis, MO) and incubated overnight, and the beads were washed 3 times before they were boiled in SDS sample buffer. Immunoprecipitated proteins were resolved and transferred as for the Western blotting. Analysis was performed using antibodies to caspase-8, -9, and -3, Bid, Bax, Bak, Bcl-xL, XIAP, survivin, cytochrome c, phospho-AKT Thr308, phospho-AKT ser473, AKT, phospho-p44/42 MAP kinase (phospho-ERK 1/2), p44/42 MAP kinase (ERK 1/2) and p53 (Cell Signaling Technology, Beverly, MA), FADD (Calbiochem, Darmstadt, Germany), FLIP (Dave-2) (Alexis), Bcl-2 (MBL, Nagoya, Japan), cIAP-1 and cIAP-2 (R&D Systems, Minneapolis, MN), and monoclonal anti–β-actin antibody AC-15 (Sigma).
Measurement of mitochondrial transmembrane potential (ΔΨm)
Variations in mitochondrial transmembrane potential (ΔΨm) during the induction of apoptosis were examined with 3,3′-dihexyloxacarbocyanine iodide (DiOC63 ) (FluoreszenzTechnologie, Grottenhofstr, Austria). Cells were prepared and treated as described for the Western blot analysis. Cells were harvested at several time points, and DiOC63 was added at a final concentration of 40 nM. After 20 minutes of incubation at 37°C, the cells were washed and analyzed using flow cytometry (FCM), and the percentage of cells with low mitochondrial potential was then calculated. For each sample, 104 cells were investigated and all experiments were performed in triplicate.
Protein kinase C activity assay
We quantitated the activity of protein kinase C (PKC) using a PKC Kinase Activity Assay kit (Stressgen Bioreagents, Victoria, BC, Canada) according to the manufacturer's protocol. In short, we used a 96-well plate precoated with the substrate, which was readily phosphorylated by PKC. The protein (2 μg) to be assayed was added and was followed by ATP to initiate the reaction. We used samples without ATP as a negative control in each assay. Next, a phosphospecific substrate antibody that bound specifically to the phosphorylated peptide substrate was added to the wells. Then, a peroxidase-conjugated antibody was added as secondary antibody. The assay was developed with tetramethylbenzidine substrate and was color developed in proportion to PKC phosphotransferase activity. Color intensity was measured in a microplate reader. All experiments were performed in triplicate, and results are given relative (in percentages) to the value for the control cells.
NFκB transcription factor assay
Nuclear extracts from cell lines were obtained using a nuclear/cytosol fractionation kit (BioVision) according to the manufacturer's protocol. Activities of NFκB p50, p65, c-Rel, p52, and RelB were investigated using an NFκB transcription factor assay kit (BD Biosciences) according to the manufacturer's directions. In short, nuclear extract (10 μg/sample) from untreated or treated cells was added to the capture probe, a double-stranded biotinylated oligonucleotide containing the consensus sequence for binding NFκB. After incubation, the sample was transferred to a streptavidin-coated 96-well plate. After washing, the bound NFκB transcription factor subunit was detected with a primary antibody. The plate was incubated with a secondary antibody, a chromogenic substrate was added to the cells, and the absorbance of each sample was read using a microplate reader. All experiments were performed in triplicate, and the results were expressed relative to the value for the positive control—the Raji cell extract contained in the kit.
Inhibition of AKT and ERK activity
The phosphatidylinositol 3-kinase (PI3K) inhibitor LY294002 and the MAPK/MEK inhibitor U0126 (Calbiochem) were used at concentrations of 5 to 10 μM (DMSO stock solution) to inhibit AKT and ERK activity, respectively. Cells were treated with these inhibitors for 24 hours and harvested. Control cultures were treated with an equal volume of DMSO. Cell surface expression of TRAIL-R1 and -R2 was examined by FCM, and the phosphorylation status of AKT and ERK was evaluated by Western blotting as described. All experiments were performed in duplicate.
RNA extraction and reverse transcription PCR
Total RNA was prepared using Isogen (Wako, Osaka, Japan). After contaminating DNA was removed with DNase (Message Clean kit; GenHunter, Nashville, TN), cDNA was constructed from 1 μg total RNA using the Thermoscript RT-PCR System (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Sequences for the relevant primers were as follows: TRAIL-R1, 5′-CTGAGCAACGCAGACTCGCTGTCCAC-3′ and 5′-TCCAAGGACACGGCAGAGCCTGTGCCAT-3′; TRAIL-R2, 5′-GCCTCATGGACAATGAGATAAAGGTGGCT-3′ and 5′-CCAAATCTCAAAGTACGCACAAACGG-3′; and GAPDH, 5′-GGTGAAGGTCGGTGTGAACGGATTT-3′ and 5′-AATGCCAAAGTTGTCATGGATGACC-3′. The reaction was run for 22 to 31 cycles of 94°C for 30 seconds (denaturation), 60°C for 30 seconds (annealing), and 72°C for 30 seconds (extension). Amplification products was subjected to electrophoresis on 2% agarose gel, stained with ethidium bromide and visualized under UV light.
Transfection and luciferase assays
As described previously,19 the digested SacI-NcoI fragment from the TRAIL-R2 promoter region of genomic DNA was subcloned into the SacI-NcoI site of the pGVB2 luciferase assay vector (Toyo Ink, Tokyo, Japan) to produce pDR5/SacI. Then, 2 μg of pDR5/SacI and vacant vector plasmids were transfected into KOB, U937, K562, and HL60 cells. Transfection was performed with a cell line nucleofector kit V and the Nucleofector system (Amaxa Biosystems, Cologne, Germany). The transfection program for KOB, U937, K562, and HL60 cells was T-20, T-16, T-30, and T-30, respectively. Cell preparations were adjusted so that the counts of viable cells were the same in each cell line after 12 hours of transfection. Cells were then incubated with or without 12.5 μg/mL BB-1 for 24 hours. The luciferase activity in 10 μg cell lysate was measured using luciferase assay reagents (Promega) according to the manufacturer's instructions in a TD-20/20 luminometer (Turner Designs, Sunnyvale, CA). The level of activation was obtained by setting the value for the empty vector control as 1.0. Each experiment was carried out in triplicate. Data were analyzed using Student t test, and differences were considered significant from control data when P was below .01.
Small interfering RNAs
The small interfering RNA (siRNA) sequences for targeting TRAIL-R1,20 TRAIL-R2,21 and green fluorescence protein (GFP) 22 were previously described (synthesized by Proligo, Kyoto, Japan). Mock (transfection reagent alone)–transfected cells were used as the negative control. siRNA (20 nM) was introduced into 107 KOB cells in each experiment using the Nucleofector system (Amaxa Biosystems), as described in “Transfection and luciferase assays.” Twenty-four hours after transfection, cells were harvested and the expression of TRAIL receptors was evaluated by RT-PCR, cells were incubated with or without 12.5 μg/mL BB-1 for 24 hours and the expression of TRAIL receptors was evaluated by FCM, or cells were treated with 0.5 μg/mL TRAIL or 12.5 μg/mL BB-1 for 48 hours (or both) and cell proliferation was evaluated by MTS assay. Each siRNA experiment was performed in duplicate.
Data are expressed as the mean plus or minus SD of 3 independent experiments. Data were analyzed using Student t test, and differences were considered significant when P was below .01.
Results
Screening of natural products
We attempted to find natural products that enhance sensitivity to TRAIL by using the TRAIL-resistant ATLL cell line KOB, which faintly expresses TRAIL-R1 and moderately expresses TRAIL-R2 but lacks DcRs (Figure 1A). We found a greater than 20% reduction in cell growth at a high TRAIL concentration of 2 μg/mL (Figure 1B). Jurkat cells, known to be sensitive to TRAIL, showed surface expression of TRAIL receptors similar to that of KOB cells and remarkably reduced cell growth at 0.02 μg/mL TRAIL (Figure 1A-B). In the screening assay, KOB cells were incubated for 48 hours with 0.5 μg/mL TRAIL and various concentrations of natural products and were subjected to an assessment of cell proliferation using the Alamar Blue bioassay. Among the more than 150 natural products in our library (data not shown), 6 showed synergism with TRAIL (Table 1). The most striking effect with TRAIL and minimal damage without TRAIL was observed in an extract from Blumea balsamifera, a family of chrysanthemums from Thailand. We named this extract BB-1. BB-1 is basically a flavonoid, and we eventually identified it as methyl dihydro quercetin (Figure 1C).23,24 We next examined Annexin-V and PI binding by FCM to identify apoptotic cell death. Treatment of KOB cells with 0.5 μg/mL TRAIL, in combination with 12.5 μg/mL BB-1 for 48 hours, resulted in apparent apoptosis that was not observed with TRAIL or BB-1 alone (Figure 1D). The cell proliferation of KOB cells treated with either TRAIL or BB-1 was 98% and 97%, respectively, which decreased to 60% with concurrent treatment (Figure 1E). Of note, sequential treatment, pretreatment with BB-1 and then TRAIL, further augmented the synergism, and the cell proliferation rate decreased to 38%. Importantly, normal PBMCs were not harmed by TRAIL alone, BB-1 alone, or a combination of TRAIL and BB-1 (Figure 1E).
Detection of new natural products that enhance sensitivity to TRAIL
. | . | Concentration of materials, μg/mL . | Cell proliferation, % of control . | . | |
|---|---|---|---|---|---|
| Material . | Plant . | . | TRAIL- . | TRAIL+ . | |
| Voaphylline | Tabernaemontana divaricata | 25 | 71 | 38 | |
| Voafinine | T divaricata | 25 | 79 | 44 | |
| Vincadifformine | Kopsia arborea | 12.5 | 95 | 57 | |
| 5.21-dihydrorhazinilam | K arborea | 3.1 | 67 | 50 | |
| Rhazimal | K arborea | 12.5 | 80 | 51 | |
| Methyldihyroquercetin (BB-1) | Blumera basamifera | 12.5 | 96 | 41 | |
. | . | Concentration of materials, μg/mL . | Cell proliferation, % of control . | . | |
|---|---|---|---|---|---|
| Material . | Plant . | . | TRAIL- . | TRAIL+ . | |
| Voaphylline | Tabernaemontana divaricata | 25 | 71 | 38 | |
| Voafinine | T divaricata | 25 | 79 | 44 | |
| Vincadifformine | Kopsia arborea | 12.5 | 95 | 57 | |
| 5.21-dihydrorhazinilam | K arborea | 3.1 | 67 | 50 | |
| Rhazimal | K arborea | 12.5 | 80 | 51 | |
| Methyldihyroquercetin (BB-1) | Blumera basamifera | 12.5 | 96 | 41 | |
KOB cells (3.5 × 105/mL) were incubated for 48 hours with 0.5 μg/mL TRAIL and various concentrations of natural products; this was followed by assessment of cell proliferation using Alamar Blue bioassay.
Activation of extrinsic and intrinsic pathways in cells treated with BB-1 and TRAIL
In a previous study, we showed the mechanism of resistance to TRAIL in ATLL cells. KOB cells had been demonstrated to be resistant because of insufficient activation of the extrinsic pathway and, more important, because of the blocking of death signaling at the mitochondrial level. Therefore, Western blot analyses of proapoptotic and antiapoptotic factors were performed, presuming that the inhibitory factors in mitochondria were suppressed by combined treatment with BB-1 and TRAIL. Time-dependent cleavage of caspase-3, the effecter caspase, was observed in concurrently treated and sequentially treated KOB cells but was stronger in the latter. In contrast, little or no change was observed in cells treated with a single agent (Figure 2A). Results of analyses of key molecules in the extrinsic pathway are shown in Figure 2B. Decreases in band densities of the full-length caspase-8 and FADD, possibly as a result of the formation of the active DISC, were observed in concurrently and sequentially treated cells. A key regulator of death receptor signaling is the caspase-8 inhibitor c-FLIP, which is often expressed as long (c-FLIPL) and short (c-FLIPS) forms because differential splicing.25 Recent studies have shown that c-FLIPS plays a major role in the sensitivity of cells to TRAIL.26,27 We found faint expression of the full-length (p55) and the truncated (p43) forms of c-FLIPL in control and combined-treatment cells, respectively. Although the strong expression of c-FLIPS suggests its involvement in resistance to TRAIL, the expression was little influenced by BB-1 treatment (Figure 2B). Decreased Bid expression in TRAIL-treated and cotreated cells implies the truncation of full-length Bid and the transference of apoptotic signals from the extrinsic pathway to mitochondria. Apoptotic events in mitochondria were evaluated by measuring the mitochondrial membrane potential (ΔΨm) using FCM. Marked reduction in the ΔΨm that had not occurred in cells treated with TRAIL alone (Figure 2C) was observed in sequentially treated cells in a time-dependent manner. In addition, this process was accompanied by the release of cytochrome c from the mitochondria into the cytosol (Figure 2C). Similarly, caspase-9 was also activated, and the full-length form was decreased after combined treatment (Figure 2D). These results suggest that the inhibitory mechanism of apoptotic signaling at the mitochondrial level was inactivated. In contrast, the levels of Bax, Bak, Bcl-2, Bcl-xL, XIAP, cIAP-1, and cIAP-2 expression showed no change on combined treatment or on BB-1 treatment (Figure 2D). Interestingly, BB-1 decreased survivin expression after 48 hours, which might partly have relieved TRAIL resistance, but it seemed to occur too late to override the mechanisms of resistance (Figure 2D). Our results indicated that combined treatment activated extrinsic and intrinsic pathways, especially in sequential settings.
Screening of new natural products that enhance sensitivity to TRAIL using the TRAIL-resistant ATLL cell line KOB. (A) TRAIL receptor expression on KOB and Jurkat cells. Cells were stained with antibodies to TRAIL receptors or with an isotype-matched control antibody and were analyzed by FCM. Shaded and unshaded peaks correspond to specific and control staining, respectively. RFI was determined as the ratio of mean fluorescence intensity for specific staining to that for control staining and is indicated in each panel. (B) Sensitivity of KOB (▪) and Jurkat (•) cells to TRAIL. KOB cells (5 × 105/mL) and Jurkat cells (106/mL) were cultured for 24 hours in the presence of the indicated concentrations of TRAIL, and cell proliferation was evaluated by MTS assay. Results are expressed relative to the values for the control cells cultured without TRAIL. Data are the mean ± SD of 3 independent experiments. (C) The structure of methyl dihydro quercetin extracted from B balsamifera (BB-1). (D) Evaluation of apoptosis. KOB cells (5 × 105/mL) were incubated for 48 hours with 0.5 μg/mL TRAIL, 12.5 μg/mL BB-1, and a combination of both and apoptotic cells were evaluated by Annexin V binding and propidium iodide (PI) staining. Cells (104) were counted and analyzed by FCM. Percentages of Annexin V+ /PI– cells are shown in each dot-plot graph. (E) Synergism of TRAIL and BB-1. KOB cells (5 × 105/mL) or PBMCs (106/mL) were incubated for 48 hours with 0.5 μg/mL TRAIL (T), 12.5 μg/mL BB-1 (B), BB-1+TRAIL (simultaneous addition, B+T), or BB-1+TRAIL (0.5 μg/mL TRAIL was added after incubation with 12.5 μg/mL BB-1 for 24 hours, B→T). Cell proliferation was assessed by MTS assay, and results are expressed relative to the value for the control cells [C] cultured without inducers. Data are the mean ± SD of 3 independent experiments.
Screening of new natural products that enhance sensitivity to TRAIL using the TRAIL-resistant ATLL cell line KOB. (A) TRAIL receptor expression on KOB and Jurkat cells. Cells were stained with antibodies to TRAIL receptors or with an isotype-matched control antibody and were analyzed by FCM. Shaded and unshaded peaks correspond to specific and control staining, respectively. RFI was determined as the ratio of mean fluorescence intensity for specific staining to that for control staining and is indicated in each panel. (B) Sensitivity of KOB (▪) and Jurkat (•) cells to TRAIL. KOB cells (5 × 105/mL) and Jurkat cells (106/mL) were cultured for 24 hours in the presence of the indicated concentrations of TRAIL, and cell proliferation was evaluated by MTS assay. Results are expressed relative to the values for the control cells cultured without TRAIL. Data are the mean ± SD of 3 independent experiments. (C) The structure of methyl dihydro quercetin extracted from B balsamifera (BB-1). (D) Evaluation of apoptosis. KOB cells (5 × 105/mL) were incubated for 48 hours with 0.5 μg/mL TRAIL, 12.5 μg/mL BB-1, and a combination of both and apoptotic cells were evaluated by Annexin V binding and propidium iodide (PI) staining. Cells (104) were counted and analyzed by FCM. Percentages of Annexin V+ /PI– cells are shown in each dot-plot graph. (E) Synergism of TRAIL and BB-1. KOB cells (5 × 105/mL) or PBMCs (106/mL) were incubated for 48 hours with 0.5 μg/mL TRAIL (T), 12.5 μg/mL BB-1 (B), BB-1+TRAIL (simultaneous addition, B+T), or BB-1+TRAIL (0.5 μg/mL TRAIL was added after incubation with 12.5 μg/mL BB-1 for 24 hours, B→T). Cell proliferation was assessed by MTS assay, and results are expressed relative to the value for the control cells [C] cultured without inducers. Data are the mean ± SD of 3 independent experiments.
Further analysis of modulators of TRAIL sensitivity: PKC, AKT,ERK, and NFκB
Several other factors may modulate the death signal of the TRAIL pathway. It has been reported that the activation of PKC protects against TRAIL-induced apoptosis.28 To determine whether BB-1 modulates PKC, an activity assay was performed, as described in “Materials and methods.” The PKC activity of untreated cells, cells treated with TRAIL or BB-1, and cells treated with a combination of both showed no remarkable changes (Figure 3A). Furthermore, the PKC inhibitor Gö 6976 did not alter cell death after combined treatment (data not shown). These results indicate that BB-1 does not influence PKC activity.
Analysis of extrinsic and intrinsic pathways after treatment with BB-1 and TRAIL. KOB cells (5 × 105/mL) were incubated for 48 hours with 0.5 μg/mL TRAIL [T], 12.5 μg/mL BB-1 [B], BB-1+TRAIL (simultaneous addition [B+T]), or BB-1+TRAIL (0.5 μg/mL TRAIL was added after incubation with 12.5 μg/mL BB-1 for 24 hours [B→T]), or without reagents [C]. Cells were harvested at the indicated time points and analyzed. (A-B,D) Western blot analysis. Using 30 μg cell extract, caspases and proapoptotic and antiapoptotic factors were detected with the antibodies described in “Materials and methods.” (C) Mitochondrial membrane potential (ΔΨm) and cytochrome c translocation. After treatment, cells were evaluated using DiOC6 by FCM, and loss of ΔΨm (%) is indicated in each panel. Using 10 μg cytosolic extract, cytochrome c was detected by Western blotting with the antibodies described in “Materials and methods.”
Analysis of extrinsic and intrinsic pathways after treatment with BB-1 and TRAIL. KOB cells (5 × 105/mL) were incubated for 48 hours with 0.5 μg/mL TRAIL [T], 12.5 μg/mL BB-1 [B], BB-1+TRAIL (simultaneous addition [B+T]), or BB-1+TRAIL (0.5 μg/mL TRAIL was added after incubation with 12.5 μg/mL BB-1 for 24 hours [B→T]), or without reagents [C]. Cells were harvested at the indicated time points and analyzed. (A-B,D) Western blot analysis. Using 30 μg cell extract, caspases and proapoptotic and antiapoptotic factors were detected with the antibodies described in “Materials and methods.” (C) Mitochondrial membrane potential (ΔΨm) and cytochrome c translocation. After treatment, cells were evaluated using DiOC6 by FCM, and loss of ΔΨm (%) is indicated in each panel. Using 10 μg cytosolic extract, cytochrome c was detected by Western blotting with the antibodies described in “Materials and methods.”
AKT and mitogen-activated protein kinase (MAPK p44/42)/extracellular signal–regulated kinase 1/2 (ERK 1/2) have also been found to affect sensitivity to TRAIL.29-31 Western blotting of these molecules was performed in KOB cells treated with BB-1. Constitutive phosphorylation of AKT Thr308 and AKT Ser473 was observed in untreated cells, which decreased on BB-1 treatment within 24 hours (Figure 3B). Constitutive phosphorylation of ERK 1/2 was also observed before treatment, and opposite results—the down-regulation of phospho-ERK1 and the up-regulation of phospho-ERK2—were observed after BB-1 treatment (Figure 3B). It is possible that phospho-AKT and phospho-ERK1 contribute to TRAIL resistance in KOB cells, the effects of which were reduced by BB-1 treatment. Further, it is possible that the activation of ERK2 contributes to the sensitivity to TRAIL.
The constitutive activation of NFκB is thought to be an important factor in antiapoptotic effects on cancer cells, including ATLL,32 and NFκB-targeted therapy has received a great deal of attention.33 Furthermore, a recent report33 suggests that the specific down-regulation of NFκB significantly sensitizes cells to TRAIL. To determine whether BB-1 down-regulates NFκB activities, we performed an assay of transcription factors. Levels of activity of the NFκB p50, p52, c-Rel, and especially p65 subunits were higher in untreated KOB cells than in Raji cells used as positive control (Figure 3C). The level of activity was not altered in p50, was slightly decreased in p65, and was reduced by half in c-Rel and p52, respectively, by BB-1 treatment (Figure 3C). These results suggest that BB-1 influences c-Rel and p52 but not p50 or p65.
Analysis of other factors involved in sensitivity to TRAIL: PKC, AKT, ERK, and NFκB. KOB cells (5 × 105/mL) were incubated with 0.5 μg/mL TRAIL [T], 12.5 μg/mL BB-1 [B], BB-1+TRAIL [B+T], or without reagents [C]. (A) PKC activity. Cells were incubated for 24 hours, and PKC activity was evaluated. ▪, results relative (%) to the value for untreated cells; □, cells treated without ATP. The assay was performed in triplicate, and data are the mean ± SD. (B) Western blot analysis of AKT and ERK 1/2. Cells were harvested at the indicated time points, and Western blotting was performed using 30 μg cell extract. (C) Activities of NFκB transcription factors. Cells were incubated for 24 hours, nuclear extracts were prepared, and activities of p50, p65, c-Rel, and p52 were evaluated. Raji cells were used as a positive control (PC) (□). The fold activation (▪) was obtained by setting the value for the positive control cells as 1.0. The assay was performed in triplicate, and data are the mean ± SD.
Analysis of other factors involved in sensitivity to TRAIL: PKC, AKT, ERK, and NFκB. KOB cells (5 × 105/mL) were incubated with 0.5 μg/mL TRAIL [T], 12.5 μg/mL BB-1 [B], BB-1+TRAIL [B+T], or without reagents [C]. (A) PKC activity. Cells were incubated for 24 hours, and PKC activity was evaluated. ▪, results relative (%) to the value for untreated cells; □, cells treated without ATP. The assay was performed in triplicate, and data are the mean ± SD. (B) Western blot analysis of AKT and ERK 1/2. Cells were harvested at the indicated time points, and Western blotting was performed using 30 μg cell extract. (C) Activities of NFκB transcription factors. Cells were incubated for 24 hours, nuclear extracts were prepared, and activities of p50, p65, c-Rel, and p52 were evaluated. Raji cells were used as a positive control (PC) (□). The fold activation (▪) was obtained by setting the value for the positive control cells as 1.0. The assay was performed in triplicate, and data are the mean ± SD.
Increase of TRAIL-R2 expression by BB-1 contributes to the synergism
It has been recognized that amplification of the intrinsic pathways through engagement of the extrinsic pathways is needed to commit the cell to apoptosis, and the activation of both pathways seems to be vital for TRAIL-induced apoptosis.6 Therefore, we next investigated surface TRAIL receptor expression upstream of the extrinsic pathway because our results indicated that both pathways were activated by combined treatment, especially sequential treatment. As shown in Figure 4A, BB-1 minimally increased TRAIL-R1 expression and almost tripled TRAIL-R2 expression, but it did not affect TRAIL-R3 or TRAIL-R4 expression. To determine whether the increase in TRAIL sensitivity was caused by an increase in TRAIL-R2 expression, we first used TRAIL-R2-Fc chimeric protein, which has a dominant-negative function against TRAIL. Inhibition of TRAIL-induced apoptotic cell death by TRAIL-R2-Fc was clearly documented in Jurkat cells; similarly, synergistic cytotoxicity was abolished in KOB cells (Figure 4B). We next used siRNA of TRAIL-R1 and TRAIL-R2. Transiently transfected TRAIL-R1 or TRAIL-R2 siRNA effectively reduced the quantity of each mRNA (Figure 4C) and the cell surface protein expression (Figure 4D). TRAIL-R2 siRNA profoundly reduced BB-1–induced TRAIL-R2 augmentation (Figure 4D). As a result, siTRAIL-R2 inhibited the synergism of BB-1 and TRAIL, whereas siTRAIL-R1 did not (Figure 4E). These results indicated that sensitization resulted from the increase in TRAIL-R2 expression, and this may explain why extrinsic and intrinsic pathways were activated and why sequential treatment resulted in greater synergism. It has been reported recently that TRAIL-R2 expression is up-regulated through the tumor-suppressor gene p53, and previous reports showed a parallel increase in both p53 and TRAIL-R2 caused by cytotoxic drugs.34,35 However, our immunoprecipitation analysis showed no change in p53 expression on BB-1 treatment in KOB cells (Figure 4F).
Effects of BB-1 on TRAIL-R2 expression, TRAIL sensitivity, and p53 expression. (A) KOB cells (5 × 105/mL) were incubated with or without 12.5 μg/mL BB-1 for 24 hours, and the cells were stained with antibodies to TRAIL receptors or with an isotype-matched control antibody. Cells (104) were counted and analyzed by FCM. Shaded peaks, solid lines, and dotted lines correspond to treated, untreated, and control staining, respectively. RFI was determined as the ratio of mean fluorescence intensity for specific staining to that for control staining. (B) Effects of TRAIL-R2-Fc on TRAIL-induced apoptosis. KOB cells (5 × 105/mL) and Jurkat cells (106/mL) were cultured for 48 hours in the presence of the indicated concentrations of TRAIL (□, ○). KOB cells were incubated with 12.5 μg/mL BB-1 for 24 hours, followed by the indicated concentrations of TRAIL for 24 hours (▪). Inhibition of TRAIL-mediated cell death was examined by adding 2.5 μg/mL TRAIL-R2-Fc fusion protein against each of the above treatments (▪, • with dotted lines). Cell proliferation was assessed by MTS assay, and results are expressed relative to the value for the control cells cultured without TRAIL or BB-1. Data are the mean ± SD from 3 independent experiments. (C) Effect of siRNA determined by RT-PCR. At 24 hours after transfection, cells were harvested and subjected to RT-PCR for the number of cycles indicated. mRNA expression of TRAIL-R1 and -R2 in Mock-, GFP siRNA (siGFP)–, TRAIL-R1 siRNA (siTRAIL-R1)–, and TRAIL-R2 siRNA (siTRAIL-R2)–treated cells is shown with that of GAPDH. (D) Effect of siRNA on DR expression evaluated by FCM. At 24 hours after transfection, cells were incubated with or without 12.5 μg/mL BB-1 for 24 hours and were examined by FCM. Shaded and unshaded areas correspond to specific and control staining, respectively. RFI was determined as the ratio of mean fluorescence intensity for specific staining to that for control staining and is indicated in each panel. (E) Effect of siRNA on cell proliferation assay. At 24 hours after transfection, cells were treated for 48 hours with 0.5 μg/mL TRAIL [T], 12.5 μg/mL BB-1 [B], or BB-1+TRAIL (0.5 μg/mL TRAIL was added after incubation with 12.5 μg/mL BB-1 for 24 hours [B→T]) or without reagents [C]. Cell proliferation was assessed by MTS assay, and results are expressed relative to the value for the control cells. Data are the mean ± SD from 3 independent experiments. *P < .01. (F) p53 expression. KOB cells (5 × 105/mL) were incubated with or without 12.5 μg/mL BB-1 for 24 hours, and cells were harvested at the indicated time points. Immunoprecipitation (IP) was performed as described in “Materials and methods” using p53 monoclonal antibodies.
Effects of BB-1 on TRAIL-R2 expression, TRAIL sensitivity, and p53 expression. (A) KOB cells (5 × 105/mL) were incubated with or without 12.5 μg/mL BB-1 for 24 hours, and the cells were stained with antibodies to TRAIL receptors or with an isotype-matched control antibody. Cells (104) were counted and analyzed by FCM. Shaded peaks, solid lines, and dotted lines correspond to treated, untreated, and control staining, respectively. RFI was determined as the ratio of mean fluorescence intensity for specific staining to that for control staining. (B) Effects of TRAIL-R2-Fc on TRAIL-induced apoptosis. KOB cells (5 × 105/mL) and Jurkat cells (106/mL) were cultured for 48 hours in the presence of the indicated concentrations of TRAIL (□, ○). KOB cells were incubated with 12.5 μg/mL BB-1 for 24 hours, followed by the indicated concentrations of TRAIL for 24 hours (▪). Inhibition of TRAIL-mediated cell death was examined by adding 2.5 μg/mL TRAIL-R2-Fc fusion protein against each of the above treatments (▪, • with dotted lines). Cell proliferation was assessed by MTS assay, and results are expressed relative to the value for the control cells cultured without TRAIL or BB-1. Data are the mean ± SD from 3 independent experiments. (C) Effect of siRNA determined by RT-PCR. At 24 hours after transfection, cells were harvested and subjected to RT-PCR for the number of cycles indicated. mRNA expression of TRAIL-R1 and -R2 in Mock-, GFP siRNA (siGFP)–, TRAIL-R1 siRNA (siTRAIL-R1)–, and TRAIL-R2 siRNA (siTRAIL-R2)–treated cells is shown with that of GAPDH. (D) Effect of siRNA on DR expression evaluated by FCM. At 24 hours after transfection, cells were incubated with or without 12.5 μg/mL BB-1 for 24 hours and were examined by FCM. Shaded and unshaded areas correspond to specific and control staining, respectively. RFI was determined as the ratio of mean fluorescence intensity for specific staining to that for control staining and is indicated in each panel. (E) Effect of siRNA on cell proliferation assay. At 24 hours after transfection, cells were treated for 48 hours with 0.5 μg/mL TRAIL [T], 12.5 μg/mL BB-1 [B], or BB-1+TRAIL (0.5 μg/mL TRAIL was added after incubation with 12.5 μg/mL BB-1 for 24 hours [B→T]) or without reagents [C]. Cell proliferation was assessed by MTS assay, and results are expressed relative to the value for the control cells. Data are the mean ± SD from 3 independent experiments. *P < .01. (F) p53 expression. KOB cells (5 × 105/mL) were incubated with or without 12.5 μg/mL BB-1 for 24 hours, and cells were harvested at the indicated time points. Immunoprecipitation (IP) was performed as described in “Materials and methods” using p53 monoclonal antibodies.
Synergism in other leukemia cell lines
Parallel studies were performed to determine whether synergism occurs in other human hematologic malignant cell types using another ATLL cell line ST1, the monocytic leukemia cell line U-937, the chronic myeloid leukemia cell line K562, the Burkitt lymphoma cell line Ramos, and the acute myeloid leukemia cell line HL-60. Surprisingly, similar synergism was observed in all cell lines except HL60, irrespective of their origins (Figure 5A). Then we investigated the synergistic mechanism, focusing on U937 and K562 cells. Western blot analysis showed the activation of caspase-8, -9, and -3 on combined treatment in accordance with the extent of apoptosis (Figure 5A-B). Similar results were obtained when the loss of mitochondrial membrane permeability and the release of cytochrome c into the cytosol were monitored by FCM and Western blotting, respectively (Figure 5C). Western blot analysis of U937 and K562 cells exposed to BB-1 for specific periods revealed that there are no apparent changes in cFLIP, Bcl-2 family proteins, and IAP family proteins (Figure 5D). In addition, no changes were observed in the level of phospho-AKT Thr308 or phospho-AKT Ser473 in U937 cells (Figure 5E). K562 cells showed a constitutive phosphorylation of ERK 1/2 and were inactivated by BB-1 treatment (Figure 5E).
TRAIL-R2 up-regulation by BB-1 is a common feature of responders
We next investigated the effect of BB-1 on TRAIL receptor expression in U937, K562, and HL60. Similar to KOB cells, BB-1 increased TRAIL-R1 expression (1.2- to 1.5-fold) and TRAIL-R2 expression (3.0-4.0 fold) in U937 and K562 cells. In contrast, this increase was not observed in HL60 cells (Figure 6A). RT-PCR analysis revealed an increase of TRAIL-R2 mRNA in KOB, U937, and K562 but not in HL60 cells, in accordance with the protein expression (Figure 6B and data not shown). Although a weak increase in the expression of TRAIL-R1 after BB-1 treatment was detected at the cell surface, no increase was detected at the mRNA level in any cell lines examined (Figure 6B and data not shown). Thus, BB-1 decreases survivin, ERK1, and phospho-AKT in KOB, decreases ERK 1/2 in K562, and increases TRAIL-R1 in KOB and K562. Consequently, only the up-regulation of TRAIL-R2 was a common feature of cell lines that showed synergism between BB-1 and TRAIL, which was not observed in unresponsive HL60 cells. In addition, BB-1 did not influence TRAIL receptor expression in PBMCs (data not shown). We also investigated the possibility that the decreases in phospho-AKT and phospho-ERK contributed to the increase in DRs. The PI3K inhibitor LY294002 effectively suppressed the expression of phospho-AKT, but it did not influence DRs in KOB and U937 cells (Figure 6C). The MAPK/ERK inhibitor U0126, known to suppress ERK activity, suppressed the expression of phospho-ERK in KOB and K562 cells. Unexpectedly, however, U0126 decreased DR expression in KOB and K562 cells (Figure 6C).
Evaluation of BB-1 in other human leukemia cell lines. ST1, U937, K562, Ramos, and HL60 cells (106 cells/mL) were incubated for 48 hours with 0.2 μg/mL TRAIL [T], 12.5 μg/mL BB-1 [B], BB-1+TRAIL (simultaneous addition [B+T]), or BB-1+TRAIL (0.5 μg/mL TRAIL was added after incubation with 12.5 μg/mL BB-1 for 24 hours [B→T]), or without reagents (C). (A) Sensitivity. Cell proliferation was assessed by MTS assay, and results are expressed relative to the value for the control cells cultured without inducers. Data are the mean ± SD from 3 independent experiments. (B,D-E) Evaluation of the molecules involved in the apoptotic pathway. Cells were harvested at the indicated time points, Western blotting was performed using 30 μg cell extract, and each molecule was detected with the antibodies described in “Materials and methods.” (C) Mitochondrial membrane potential (ΔΨm) and cytochrome c translocation. After treatment, cells were evaluated using DiOC6 by FCM, and loss of ΔΨm (%) is indicated. Data are the mean ± SD of 3 independent experiments. Using 10 μg cytosolic extract, cytochrome c was detected by Western blotting as described in “Materials and methods.”
Evaluation of BB-1 in other human leukemia cell lines. ST1, U937, K562, Ramos, and HL60 cells (106 cells/mL) were incubated for 48 hours with 0.2 μg/mL TRAIL [T], 12.5 μg/mL BB-1 [B], BB-1+TRAIL (simultaneous addition [B+T]), or BB-1+TRAIL (0.5 μg/mL TRAIL was added after incubation with 12.5 μg/mL BB-1 for 24 hours [B→T]), or without reagents (C). (A) Sensitivity. Cell proliferation was assessed by MTS assay, and results are expressed relative to the value for the control cells cultured without inducers. Data are the mean ± SD from 3 independent experiments. (B,D-E) Evaluation of the molecules involved in the apoptotic pathway. Cells were harvested at the indicated time points, Western blotting was performed using 30 μg cell extract, and each molecule was detected with the antibodies described in “Materials and methods.” (C) Mitochondrial membrane potential (ΔΨm) and cytochrome c translocation. After treatment, cells were evaluated using DiOC6 by FCM, and loss of ΔΨm (%) is indicated. Data are the mean ± SD of 3 independent experiments. Using 10 μg cytosolic extract, cytochrome c was detected by Western blotting as described in “Materials and methods.”
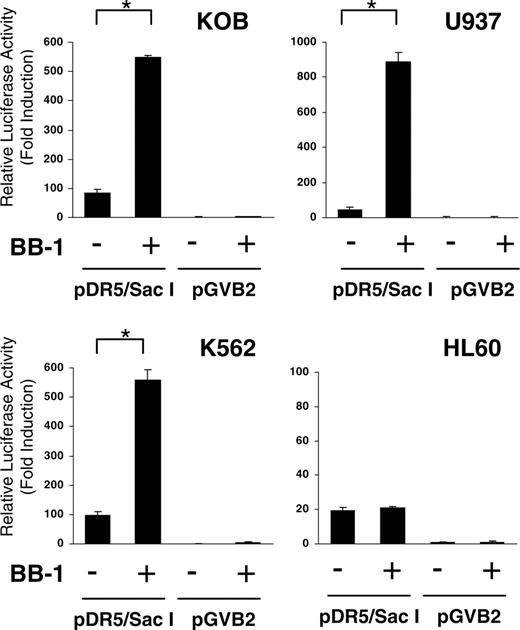
Analysis of TRAIL-R2 promoter activation by luciferase assay
It has been reported that many drugs up-regulate TRAIL-R2 expression after apoptosis. However, it is not clear whether they regulate TRAIL-R2 through the promoter region. We thus investigated the effect of BB-1 on TRAIL-R2 promoter activity. We used the TRAIL-R2 promoter-luciferase reporter plasmid for the transient luciferase assay. As shown in Figure 7, BB-1 increased TRAIL-R2 promoter activity significantly in KOB, U937, and K562 cell—7-, 20-, and 6-fold activation of the reporter gene, respectively—compared with untreated cells. In contrast, HL-60 cells that did not show up-regulation of TRAIL-R2 expression showed almost no gain in promoter activity. These results demonstrated that BB-1 up-regulates TRAIL-R2 expression through activation of its promoter.
Increase of TRAIL-R2 expression by BB-1. (A) Cell surface TRAIL receptors. U937, K562, and HL60 (106 cells/mL) were incubated with or without 12.5 μg/mL BB-1 for 24 hours, and the cells were stained with antibodies to TRAIL receptors or with an isotype-matched antibody. Cells (104) were counted and analyzed by FCM. Shaded peaks, solid lines, and dotted lines correspond to treated, untreated, and control staining, respectively. RFI was determined as the ratio of mean fluorescence intensity for specific staining to that for control staining. (B) TRAIL receptor mRNA. KOB cells (5 × 105/mL), U937 cells (106 cells/mL), and HL60 cells (106 cells/mL) were incubated with or without 12.5 μg/mL BB-1 for 24 hours. Cells were then harvested, cDNAwas constructed from 1 μg total RNA, and RT-PCR (29 cycles) was performed. mRNA expression of TRAIL-R1 and -R2 in untreated and BB-1–treated cells is shown with that of GAPDH. (C) Correlation between receptor expression and activity of AKT and ERK. KOB cells (5 × 105/mL) or cells (106/mL) from the U937 and K562 lines were treated with the indicated concentration of LY294002 or U0126 for 24 hours and were harvested. The phosphorylation status of AKT and ERK was evaluated by Western blotting, and the cell surface expression of TRAIL-R1 and -R2 was examined by FCM as described for panel A.
Increase of TRAIL-R2 expression by BB-1. (A) Cell surface TRAIL receptors. U937, K562, and HL60 (106 cells/mL) were incubated with or without 12.5 μg/mL BB-1 for 24 hours, and the cells were stained with antibodies to TRAIL receptors or with an isotype-matched antibody. Cells (104) were counted and analyzed by FCM. Shaded peaks, solid lines, and dotted lines correspond to treated, untreated, and control staining, respectively. RFI was determined as the ratio of mean fluorescence intensity for specific staining to that for control staining. (B) TRAIL receptor mRNA. KOB cells (5 × 105/mL), U937 cells (106 cells/mL), and HL60 cells (106 cells/mL) were incubated with or without 12.5 μg/mL BB-1 for 24 hours. Cells were then harvested, cDNAwas constructed from 1 μg total RNA, and RT-PCR (29 cycles) was performed. mRNA expression of TRAIL-R1 and -R2 in untreated and BB-1–treated cells is shown with that of GAPDH. (C) Correlation between receptor expression and activity of AKT and ERK. KOB cells (5 × 105/mL) or cells (106/mL) from the U937 and K562 lines were treated with the indicated concentration of LY294002 or U0126 for 24 hours and were harvested. The phosphorylation status of AKT and ERK was evaluated by Western blotting, and the cell surface expression of TRAIL-R1 and -R2 was examined by FCM as described for panel A.
Discussion
Recent studies show that the combined use of conventional cytotoxic drugs and novel molecular targeted agents or irradiation can augment TRAIL-induced apoptosis in tumor cells14 ; thus, efforts to identify agents that activate DRs or block antiapoptotic effectors may improve therapeutic design. Until recently, however, little was known about the synergy among natural products and apoptosis-inducing ligands such as TNF-α, Fas, and TRAIL. This is the first study to screen a number of natural products for new agents that enhance sensitivity to TRAIL. Here we found BB-1, which is classified a natural flavonoid quercetin, as a novel TRAIL enhancer in human leukemia cells.
Flavonoids are a large chemical class in the plant kingdom comprising more than 5000 types. Quercetin, found in vegetables such as onions and apples and a common component of most human diets, is one of the most studied flavonoids because of its potent antiproliferative effect and ability to induce apoptosis in cancer cells.36,37 Genistein, an isoflavone, apigenin, a flavone, flavopiridol (FP), a semisynthetic compound, and quercetin, a flavonol, receive the greatest attention today as potent anticancer agents.38,39 Similarly, phytochemicals such as curcumin, resveratrol, indole-3-carbinol, and triterpenoids have received attention recently as anticancer agents, and it is reported that they augment TRAIL-mediated apoptosis in cancer cells.40-42 Recent reports investigating mechanisms of synergy between flavonoids and apoptosis-inducing ligands suggest that PKC, casein kinase II,43 XIAP, and NFκB play important roles, though some of them produced contrary results. In our study, PKC activity showed no apparent correlation with the synergistic effects of BB-1 and TRAIL; however, one recent report44 showed that the activation of PKC inhibited TRAIL-induced apoptosis, and another showed that quercetin sensitized cells to Fas-mediated apoptosis by activating PKC.45 Rosato et al46 reported that FP sensitized U937 cells to TRAIL through the down-regulation of XIAP, but we did not observe this in our experiment. Recent reports suggest that the inhibition of NFκB activity by FP or apigenin resulted in a synergistic effect on TNF-α–induced apoptosis in a human lung cell carcinoma cell line and a prostate cancer cell line, respectively.47,48 The physiologic role of NFκB in cell survival and apoptosis has led to conflicting views,49,50 and a recent report showed that the dual function of NFκB, as an inhibitor or an activator of apoptosis, depends on the relative level of p65 or c-Rel subunit, respectively.51 This report and a previous report suggested that the overexpression of p65 inhibits TRAIL-induced apoptosis, whereas the overexpression of c-Rel can enhance sensitivity to TRAIL by inducing the expression of TRAIL DRs.50 Our results suggest that the overexpression of p65 contributes to TRAIL resistance in KOB cells. However, KOB cells treated with BB-1 still showed higher (4 times) levels of p65 activity than positive control cells; thus, a moderate down-regulation of p65 by BB-1 treatment may not help to relieve TRAIL resistance. When we considered c-Rel as a regulator of DRs, we obtained paradoxic results. BB-1 up-regulated c-Rel in U937 cells but down-regulated c-Rel in KOB and K562 cells (Figure 3C and data not shown).
Analysis of TRAIL-R2 promoter activity by luciferase assay. The TRAIL-R2 promoter region plasmid pDR5/SacI and empty vector pGVB2 were transfected into KOB, U937, K562, and HL60 cells. Cells were then incubated with or without 12.5 μg/mL BB-1 for 24 hours. Luciferase activity in 10 μg cell lysate was measured using luciferase assay reagents and a luminometer. Fold activation was obtained by setting the value for the empty vector control as 1.0. Each experiment was carried out in triplicate, and data are the mean ± SD. *P < .01.
Analysis of TRAIL-R2 promoter activity by luciferase assay. The TRAIL-R2 promoter region plasmid pDR5/SacI and empty vector pGVB2 were transfected into KOB, U937, K562, and HL60 cells. Cells were then incubated with or without 12.5 μg/mL BB-1 for 24 hours. Luciferase activity in 10 μg cell lysate was measured using luciferase assay reagents and a luminometer. Fold activation was obtained by setting the value for the empty vector control as 1.0. Each experiment was carried out in triplicate, and data are the mean ± SD. *P < .01.
Previous reports suggest that flavonoids have the potential to act as AKT inhibitors.52 The results with KOB cells in our study coincide with this idea, but the results in U937 cells argue against it. Interestingly, our results indicated that the MAPK/ERK inhibitor decreased DR expression, though it has not been reported that this inhibitor is the DR regulator.
Although no report indicates the direct involvement of flavonoids in DR expression, it has been reported that certain anticancer drugs up-regulate DRs and thereby enhance TRAIL-induced apoptosis. In some studies, this synergistic action was attributed to the inhibition of the antiapoptotic factors or the induction of the proapoptotic p53 pathway or to a combination of both. However, in many studies, the molecular basis of these synergistic actions was poorly explained.14,34 Chemotherapeutic agents are known to induce the expression of TRAIL-R2 in various cancer cells,53 including a breast cancer cell line,54 glioblastoma cells,55 prostate cancer cells,56 renal cell carcinoma cells,57 colon cancer cells,58 and acute leukemia cells.59 In addition, the synthetic retinoid CD437,60 interferon-α,61 interferon-γ, dexamethasone,62 sulindac sulfide,63,64 thapsigargin,65 proteasome inhibitors,66,67 and 2-methoxyestradiol68 are also reported to be potent inducers of TRAIL-R2. Importantly, the p53 tumor-suppressor gene has been reported to regulate TRAIL-R2 gene expression.69 Apoptosis in response to most DNA-damaging agents usually requires the function of p53, which engages primarily the intrinsic pathway. In most human cancers, however, p53 is inactivated, and conventional treatments eventually result in resistance to therapy. Therefore, further analysis from the perspective of p53 status is required in TRAIL-R2–based cancer therapies. Although one attractive feature of TRAIL is its ability to kill cancers with mutations in the p53 gene, the combination of TRAIL with chemotherapeutic agents has been found to be particularly effective in killing cancers with wild-type p53.35 Thus far, TRAIL-R2–based cancer therapies can be summarized as follows. First, in tumors that retain some responsiveness to conventional therapy, up-regulation of TRAIL-R2 might lead to synergistic apoptosis and might reduce the possibility of tumor cells becoming resistant to treatment. Second, in tumors that have lost p53 function, targeting of TRAIL-R2 might, therefore, help circumvent resistance to conventional therapies. Third, efforts to identify agents that up-regulate TRAIL-R2 independently of p53 may be useful, especially for killing tumor cells that are resistant to conventional therapies. In this regard, recent reports70-72 showed the possibility of a new p53-independent TRAIL-R2 enhancer. As described, various potent inducers of TRAIL-R2 have been identified, but few investigators have performed promoter analysis.70,73 It is worth mentioning that the TRAIL-R2 promoter plasmid we used does not contain an intronic p53-binding site. Furthermore, KOB cells harbor an inactivated p53 gene with a point mutation (our unpublished data, April 2003). Consequently, we found a new agent, BB-1, among more than 150 natural products. BB-1 up-regulates TRAIL-R2 through a p53-independent mechanism and sensitizes TRAIL-resistant cells. Its broad action on various leukemia cell lines may open the way for a new therapeutic strategy in cancer.
Prepublished online as Blood First Edition Paper, October 4, 2005; DOI 10.1182/blood-2005-05-1982.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

![Figure 1. Screening of new natural products that enhance sensitivity to TRAIL using the TRAIL-resistant ATLL cell line KOB. (A) TRAIL receptor expression on KOB and Jurkat cells. Cells were stained with antibodies to TRAIL receptors or with an isotype-matched control antibody and were analyzed by FCM. Shaded and unshaded peaks correspond to specific and control staining, respectively. RFI was determined as the ratio of mean fluorescence intensity for specific staining to that for control staining and is indicated in each panel. (B) Sensitivity of KOB (▪) and Jurkat (•) cells to TRAIL. KOB cells (5 × 105/mL) and Jurkat cells (106/mL) were cultured for 24 hours in the presence of the indicated concentrations of TRAIL, and cell proliferation was evaluated by MTS assay. Results are expressed relative to the values for the control cells cultured without TRAIL. Data are the mean ± SD of 3 independent experiments. (C) The structure of methyl dihydro quercetin extracted from B balsamifera (BB-1). (D) Evaluation of apoptosis. KOB cells (5 × 105/mL) were incubated for 48 hours with 0.5 μg/mL TRAIL, 12.5 μg/mL BB-1, and a combination of both and apoptotic cells were evaluated by Annexin V binding and propidium iodide (PI) staining. Cells (104) were counted and analyzed by FCM. Percentages of Annexin V+ /PI– cells are shown in each dot-plot graph. (E) Synergism of TRAIL and BB-1. KOB cells (5 × 105/mL) or PBMCs (106/mL) were incubated for 48 hours with 0.5 μg/mL TRAIL (T), 12.5 μg/mL BB-1 (B), BB-1+TRAIL (simultaneous addition, B+T), or BB-1+TRAIL (0.5 μg/mL TRAIL was added after incubation with 12.5 μg/mL BB-1 for 24 hours, B→T). Cell proliferation was assessed by MTS assay, and results are expressed relative to the value for the control cells [C] cultured without inducers. Data are the mean ± SD of 3 independent experiments.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/107/2/10.1182_blood-2005-05-1982/4/m_zh80020689690001.jpeg?Expires=1769094734&Signature=rlNUnQkEuTw39eM9ozWEFX7bTMT2iyox3m2v0xAl-gIZWYBdRdikfl-6p0U-P1Hvm~G4byYwoahGlVj8Txu1Pkqq7oBdCjaWatTYN1EFB2AWkQGNn~VnUjgTOo6hTwcJ6nhizD5RB2KzzUMGbPeE3jfwO1WXPHOPKHTCmqJtLTQX2yYZu04F5dP50Be6z4bc54N7KzJXvyfnrHYcGTEwQ3vtMApEPIZse-8QwyzFOQkmg~np5jvhqq5dlo9ykjTDSeQgGpiCTCty5rvhRV4fCnOkuU-R07WGnAt2BHBugjIpnbQgOoBela9ItdTIhwvuOltbijZHIfAPsgFUCrh4XQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 2. Analysis of extrinsic and intrinsic pathways after treatment with BB-1 and TRAIL. KOB cells (5 × 105/mL) were incubated for 48 hours with 0.5 μg/mL TRAIL [T], 12.5 μg/mL BB-1 [B], BB-1+TRAIL (simultaneous addition [B+T]), or BB-1+TRAIL (0.5 μg/mL TRAIL was added after incubation with 12.5 μg/mL BB-1 for 24 hours [B→T]), or without reagents [C]. Cells were harvested at the indicated time points and analyzed. (A-B,D) Western blot analysis. Using 30 μg cell extract, caspases and proapoptotic and antiapoptotic factors were detected with the antibodies described in “Materials and methods.” (C) Mitochondrial membrane potential (ΔΨm) and cytochrome c translocation. After treatment, cells were evaluated using DiOC6 by FCM, and loss of ΔΨm (%) is indicated in each panel. Using 10 μg cytosolic extract, cytochrome c was detected by Western blotting with the antibodies described in “Materials and methods.”](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/107/2/10.1182_blood-2005-05-1982/4/m_zh80020689690002.jpeg?Expires=1769094734&Signature=dCkI5RkBlIP5spVNVilanVy7cZl-IJIwX~T-KPCnewfO1xN~yb5~eOu6rlubNjh2eZlJmkOYwgEFEGuxycDgchQawbGwr~C6KhWz0HrPDUh2X6YTcv6Wo-sNG47KqzNZQwkquufCcAyvK-FAHbfs4L1tS3f5PL5VbrXEKCCSiNBKPOzvaJYNB7NmLr0El~WF9H5cSB3ziv6pYAo~3trqUnP8TPQ70ElN62roZo2ux63z6vEDO1AEJ2hxkwmwt3ufXQ85XG4Z2iQYA6jxINehTnUMBnpehcPl7eujFPV0cDq40CnZLT99ar~U~WTHq0L~QCqsN7yab~LUyOkJBHfAEQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 3. Analysis of other factors involved in sensitivity to TRAIL: PKC, AKT, ERK, and NFκB. KOB cells (5 × 105/mL) were incubated with 0.5 μg/mL TRAIL [T], 12.5 μg/mL BB-1 [B], BB-1+TRAIL [B+T], or without reagents [C]. (A) PKC activity. Cells were incubated for 24 hours, and PKC activity was evaluated. ▪, results relative (%) to the value for untreated cells; □, cells treated without ATP. The assay was performed in triplicate, and data are the mean ± SD. (B) Western blot analysis of AKT and ERK 1/2. Cells were harvested at the indicated time points, and Western blotting was performed using 30 μg cell extract. (C) Activities of NFκB transcription factors. Cells were incubated for 24 hours, nuclear extracts were prepared, and activities of p50, p65, c-Rel, and p52 were evaluated. Raji cells were used as a positive control (PC) (□). The fold activation (▪) was obtained by setting the value for the positive control cells as 1.0. The assay was performed in triplicate, and data are the mean ± SD.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/107/2/10.1182_blood-2005-05-1982/4/m_zh80020689690003.jpeg?Expires=1769094734&Signature=kADdPfHIu~h7RB~maa3GuQO~Gl9axmZlMMHgTIB~PO9FiLgo5LvBPF4mTL01RCXOOCfT-6T2J-fZAKmn0Gi0bgOv4tKJN~qWD9~upxCm7fbjRjtt96QvsBq5am3q0l2SJF6YgsBOk6-Nc-Sx9M-MxQPiHNszDuuqbt--U2HyR7lBdEdQsqtV8n9~0JGKa1RjjDQewtLwJkGKykS-vIdVXUL3EhmFPF0EP3ik9F7ZpUrJlp96uO78r0FKTGJt4~4KKDR6e3NJ2g8O7ZeMl3~mbVHN7-MtcZSle8hWfOTCGDcnUkxn3KDvm70JaRkX-b330O82rX-zpT-TujrWRdrmVA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 4. Effects of BB-1 on TRAIL-R2 expression, TRAIL sensitivity, and p53 expression. (A) KOB cells (5 × 105/mL) were incubated with or without 12.5 μg/mL BB-1 for 24 hours, and the cells were stained with antibodies to TRAIL receptors or with an isotype-matched control antibody. Cells (104) were counted and analyzed by FCM. Shaded peaks, solid lines, and dotted lines correspond to treated, untreated, and control staining, respectively. RFI was determined as the ratio of mean fluorescence intensity for specific staining to that for control staining. (B) Effects of TRAIL-R2-Fc on TRAIL-induced apoptosis. KOB cells (5 × 105/mL) and Jurkat cells (106/mL) were cultured for 48 hours in the presence of the indicated concentrations of TRAIL (□, ○). KOB cells were incubated with 12.5 μg/mL BB-1 for 24 hours, followed by the indicated concentrations of TRAIL for 24 hours (▪). Inhibition of TRAIL-mediated cell death was examined by adding 2.5 μg/mL TRAIL-R2-Fc fusion protein against each of the above treatments (▪, • with dotted lines). Cell proliferation was assessed by MTS assay, and results are expressed relative to the value for the control cells cultured without TRAIL or BB-1. Data are the mean ± SD from 3 independent experiments. (C) Effect of siRNA determined by RT-PCR. At 24 hours after transfection, cells were harvested and subjected to RT-PCR for the number of cycles indicated. mRNA expression of TRAIL-R1 and -R2 in Mock-, GFP siRNA (siGFP)–, TRAIL-R1 siRNA (siTRAIL-R1)–, and TRAIL-R2 siRNA (siTRAIL-R2)–treated cells is shown with that of GAPDH. (D) Effect of siRNA on DR expression evaluated by FCM. At 24 hours after transfection, cells were incubated with or without 12.5 μg/mL BB-1 for 24 hours and were examined by FCM. Shaded and unshaded areas correspond to specific and control staining, respectively. RFI was determined as the ratio of mean fluorescence intensity for specific staining to that for control staining and is indicated in each panel. (E) Effect of siRNA on cell proliferation assay. At 24 hours after transfection, cells were treated for 48 hours with 0.5 μg/mL TRAIL [T], 12.5 μg/mL BB-1 [B], or BB-1+TRAIL (0.5 μg/mL TRAIL was added after incubation with 12.5 μg/mL BB-1 for 24 hours [B→T]) or without reagents [C]. Cell proliferation was assessed by MTS assay, and results are expressed relative to the value for the control cells. Data are the mean ± SD from 3 independent experiments. *P < .01. (F) p53 expression. KOB cells (5 × 105/mL) were incubated with or without 12.5 μg/mL BB-1 for 24 hours, and cells were harvested at the indicated time points. Immunoprecipitation (IP) was performed as described in “Materials and methods” using p53 monoclonal antibodies.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/107/2/10.1182_blood-2005-05-1982/4/m_zh80020689690004.jpeg?Expires=1769094734&Signature=jhxnXWe9s6pwtFNdG6FY60GitYUhoKaxmRHIIxDk45bYZfS14rb2TtNf9wVn0SjiTovSAWehhZsqOeJZcCN57iGNPXq9TiSpziuZaKKFNFWBnO7~OyZFR4YI6Q1hAzWwoCKn3I7qd2SNHJdFbtpQme53mYGeH9bY0~Rv3dP8eDX6uf9zzkpMusJlAloctTs2ENAW80ljanCHyBtGRysfaLu4v7Dlbu-q87yCd-RH7dSma9QPVtwAYNZxY2jAQHJ7o05k3B8C-HAtoU-723aDfnM8tV9fQ6S7wjfaRDKUWNw0q~LwueSNMeKe51Xu~awDixSFlkHAPQY~Wy~CJtmdlQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 5. Evaluation of BB-1 in other human leukemia cell lines. ST1, U937, K562, Ramos, and HL60 cells (106 cells/mL) were incubated for 48 hours with 0.2 μg/mL TRAIL [T], 12.5 μg/mL BB-1 [B], BB-1+TRAIL (simultaneous addition [B+T]), or BB-1+TRAIL (0.5 μg/mL TRAIL was added after incubation with 12.5 μg/mL BB-1 for 24 hours [B→T]), or without reagents (C). (A) Sensitivity. Cell proliferation was assessed by MTS assay, and results are expressed relative to the value for the control cells cultured without inducers. Data are the mean ± SD from 3 independent experiments. (B,D-E) Evaluation of the molecules involved in the apoptotic pathway. Cells were harvested at the indicated time points, Western blotting was performed using 30 μg cell extract, and each molecule was detected with the antibodies described in “Materials and methods.” (C) Mitochondrial membrane potential (ΔΨm) and cytochrome c translocation. After treatment, cells were evaluated using DiOC6 by FCM, and loss of ΔΨm (%) is indicated. Data are the mean ± SD of 3 independent experiments. Using 10 μg cytosolic extract, cytochrome c was detected by Western blotting as described in “Materials and methods.”](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/107/2/10.1182_blood-2005-05-1982/4/m_zh80020689690005.jpeg?Expires=1769094734&Signature=xFsPEJJvlPT07tFueyvEK~ueZY~utl72iFT2xK3a9K2Zhjbi02GsxE0Ge8E7T60nnqFvwUHAw8hz~Uujp3Aq4tZOb9JCvWygGk3vUaLkHV2hP9fcIzUZl0K5vG9bCiinn4X2sbuxy9sadC9K-bldUDiS~U2eKRLLAG5k8L0FSohmfzmkuoIVPv0zZVJV2K5DSqOk2HW9owMin~cz4-uNMcyRxxqBprE6VaVUr34KEQ9k5lJ2T3Y7-kdh5wE8aL4058olgsv-4J~hB1Ea9WU46IdST1b-9H9sHUox6l~rwMOia7t0sU~OX13uf8cZxaRcJ5bq1IxRulXLeJ~i0qyX2g__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal