Overexpression of Hoxb4 in bone marrow cells promotes expansion of hematopoietic stem cell (HSC) populations in vivo and in vitro, indicating that this homeoprotein can activate the genetic program that determines self-renewal. However, this function cannot be solely attributed to Hoxb4 because Hoxb4-/- mice are viable and have an apparently normal HSC number. Quantitative polymerase chain reaction analysis showed that Hoxb4-/- c-Kit+ fetal liver cells expressed moderately higher levels of several Hoxb cluster genes than control cells, raising the possibility that normal HSC activity in Hoxb4-/- mice is due to a compensatory up-regulation of other Hoxb genes. In this study, we investigated the competitive repopulation potential of HSCs lacking Hoxb4 alone, or in conjunction with 8 other Hoxb genes. Our results show that Hoxb4-/- and Hoxb1-b9-/- fetal liver cells retain full competitive repopulation potential and the ability to regenerate all myeloid and lymphoid lineages. Quantitative Hox gene expression profiling in purified c-Kit+Hoxb1-b9-/- fetal liver cells revealed an interaction between the Hoxa, b, and c clusters with variation in expression levels of Hoxa4,-a11, and -c4.Together, these studies show a complex network of genetic interactions between several Hox genes in primitive hematopoietic cells and demonstrate that HSCs lacking up to 30% of the active Hox genes remain fully competent. (Blood. 2006;108:116-122)
Introduction
Expression of the clustered homeobox (Hox) genes is positional and temporally orchestrated during embryonic development. This stringent regulation provides the basis for their function as determinants of cell fate. Several fundamental studies over the past years have highlighted the importance of homeodomain-containing proteins in the regulation of hematopoiesis.1-4 Hoxb4 is normally expressed in human and mouse hematopoietic progenitor cells in bone marrow (BM)5 and fetal liver (FL),6 and engineered overexpression of this gene has been shown by several groups to be sufficient to induce expansion of hematopoietic stem cells (HSCs) both in vitro and in vivo.7-10 Importantly, Hoxb4-transduced HSCs regenerated the HSC pools of recipients up to, but not above, normal levels and have never been associated with leukemic transformation.7,8
Establishment of definitive hematopoiesis is likely characterized by pronounced HSC self-renewal activity,11 thus providing HSC populations sufficient for steady-state hematopoiesis throughout adult life. Of interest, Hoxb4 is expressed in expanding HSCs.12 Engineered overexpression of Hoxb4 in embryonic stem (ES) cells enhances their hematopoietic potential.13,14 Together, these findings support a physiologic role for Hoxb4 in the regulation of HSC self-renewal.
Although overexpression of Hoxb4 induces noticeable expansions of mouse HSC populations, Hoxb4-/- mice are healthy and fertile,15,16 suggesting that this gene is dispensable for establishment of definitive hematopoiesis. Hoxb4-/- mice also exhibit no gross abnormalities during steady-state hematopoiesis and appear to have normal to subnormal HSC compartments.16
Studies using compound Hox knock-out (KO) mice have revealed the functional redundancy between several paralogs or orthologs.17-19 One obvious hypothesis is that compensatory mechanisms intrinsic to the homeotic network explain the absence of overt functional defects in HSCs lacking Hoxb4.
To explore this possibility, we examined the competitive repopulation ability and differentiation potential of Hoxb4-/- HSCs, as well as HSCs derived from Hoxb1-b9 mutant mice. Our results show that not only Hoxb4, but all Hoxb cluster genes normally expressed in c-Kit+ E14.5 FL cells are dispensable for hematopoiesis. Hoxb4-/- and Hoxb1-b9-/- HSCs are as competitive as wild-type cells in long-term repopulation of lethally irradiated hosts and are fully competent in reconstitution of myeloid and lymphoid lineages, suggesting that Hoxb gene expression is not essential for HSC functions. Expression analysis of the complete “Hoxome” in these mutant cells showed important changes in expression levels of genes from the Hoxa and Hoxc clusters, reflecting the existence of a complex cross-regulation network within the “Hoxome”20,21 and suggesting potential roles for other Hox genes in the regulation of HSC self-renewal.
Materials and methods
Animals
Mutant mice for Hoxb4 and Hoxb1-b9 were generated by Ramirez-Solis et al.15,22 Engineering of the Hoxb4 mutants was achieved by standard targeting procedure, and Hoxb1-b9 mutants were produced by introducing a series of loxP sites in ES cells followed by Cre-induced recombination.
Hoxb4 and Hoxb1-b9 mutant mice were backcrossed at least 5 times in the C57Bl/6J strain and analyzed for the presence of the mutation by Southern blotting on genomic tail DNA digested with BamHI (Hoxb4) or EcoRV (Hoxb1-b9). Membranes were hybridized with either a probe for Hoxb4 (Hoxb4-/-) or for the region of Hoxb1 still present in the Hoxb1-b9 mutant (Hoxb1-b9-/-).23 The generation of homozygous Hoxb4 and Hoxb1-b9 mutant E14.5 embryos was obtained by breeding homozygous and heterozygous mice, respectively. Females with vaginal plugs the next morning were considered at day 0.5 of pregnancy (E0.5). FLs of E14.5 embryos were dissected, passed through a 70-μm cell strainer (Falcon, BD Bioscience, Mississauga, ON, Canada) and individually frozen in FCS with 10% DMSO. gDNA isolated from each embryo was genotyped by Southern blotting as described for the tail gDNA.
Competitive repopulation assay
Mutant FL cells (containing the locus Ly5.2) were thawed and mixed with competitor wild-type FL (Hoxb4) or BM (Hoxb1-b9) cells derived from Pep3b mice (Ly5.1 for Pep3b and Ly5.2 for C57Bl/6J). A total of 5 × 105 cells (4 × 105 mutant and 1 × 105 wild-type cells) were transplanted intravenously per mouse via the tail veins of congenic recipients (Pep3b) irradiated (800 cGy) using a cesium source. Competition inoculates of each mutant FL were transplanted into 4 recipients. For each genotype, 4 FLs were tested for their competitive repopulation properties. Mutant E14.5 FL and wild-type FL or BM cells were distinguished by fluorescence-activated cell-sorting (FACS) analysis using antibodies specific to the leukocytic surface antigens Ly5.1 and Ly5.2 (BD Biosciences Pharmingen, San Diego, CA), respectively.
In vitro clonogenic progenitor assays
For myeloid clonogenic progenitor assays, cells were plated in 35-mm dishes in semisolid medium, containing 1% methylcellulose in α-medium supplemented with 10% FCS, 5.7% bovine serum albumin, 10-5 β-mercaptoethanol (β-ME), 5 U/mL erythropoietin (Epo), 10 ng/mL IL-3, 10 ng/mL IL-6, 50 ng/mL steel factor, 2 mM glutamine, and 200 mg/mL transferrin. FL cells of mutant and control embryos were plated at concentrations of 0.5 × 105 cells/mL. BM and spleen from Hoxb4-/- mice were plated at 3 × 104 and 106 cells/mL, respectively. Colonies were scored on day 12 to 14 of incubation and identified according to standard criteria. Pre-B cell clonogenic progenitor assays were performed only with Hoxb4-/- and control cells. For this assay, 3 × 105 BM cells or 106 spleen cells were plated in 1% methylcellulose in α-medium supplemented with 30% FCS, 10-4 β-ME, 2 mM glutamine, and 0.2 ng/mL IL-7. Colonies were scored on day 8.
FACS analysis and sorting
Myeloid and lymphoid populations in BM, spleen, and thymus of 3- to 4-month-old healthy Hoxb4-/- mice were determined using the following antibodies: CD4-FITC, CD8-PE, B220-PE, CD43-FITC, Mac1-bio, IgMbio (BD Bioscience Pharmingen), IgD-FITC (Southern Biotechnology Associates, Birmingham, AL), and Gr1-FITC labeled, which was kindly provided by P. Lansdorp (BC Cancer Research Center, Terry Fox, Vancouver, BC, Canada). Biotinylated antibodies were detected with PE- or APC-conjugated (BD Bioscience Pharmingen) streptavidin. Fluorescence was analyzed using the FACS Calibur (BD Bioscience, San Jose, CA) or the LSR II (BD Bioscience), using DIVA software. FACS data were analyzed with WinMDI or FCS Express (De Novo Software) software. For Hox gene expression analysis, fresh FL cells of E14.5 embryos were incubated with anti-c-Kit antibody conjugated to APC (BD Bioscience Pharmingen) and sorted with a MoFlo (Cytomation, Fort Collins, CO) using Summit software.
Quantitative RT-PCR
Total RNA was isolated by TRIzol, DNase-I treated, and cDNA was prepared (MMLV-reverse transcriptase, random primers) according to the manufacturer's instructions (Invitrogen, Paisley, United Kingdom). Quantitative polymerase chain reaction (QRT-PCR) was carried out using TaqMan probe-based chemistry (Applera, Foster City, CA). Oligonucleotides for all 39 murine Hox genes were designed from nucleotide sequences deposited in murine genome databases (GenBank, http://www.psc.edu/general/software/packages/genbank/genbank.html; RefSeq, www.ncbi.nlm.nih.gov/RefSeq/; and EMBL, www.ebi.ac.uk/embl/using Primer Express [Applera]). Reactions, analysis, and validation of the Hox amplicons were carried out as previously described.24
Results
Hoxb4 gene expression in E14.5 FL cell fractions enriched for HSCs
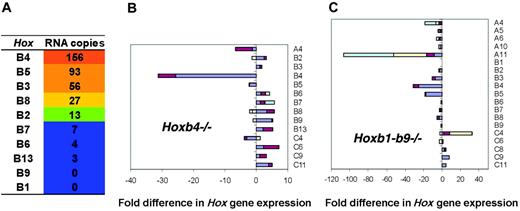
The expression of all Hoxb cluster genes was analyzed in “stem-cellenriched” c-Kit+ fraction of E14.5 mouse FL cells using QRT-PCR. Five of 10 Hoxb genes were expressed, from highest to lowest levels: Hoxb4, Hoxb5, Hoxb3, Hoxb8, and Hoxb2 (Figure 1A). Interestingly, Hoxb4 was expressed at the highest level (156 copies per 50 ng total RNA), followed by its direct neighbors Hoxb5 and Hoxb3 (93 and 56 copies, respectively). Hoxb8 and Hoxb2 were expressed at much lower levels (27 and 13 copies, respectively), whereas expression of the remaining Hoxb genes was barely detectable (copy number < 10).
Complete Hox expression profiling of Hoxb4-/- c-Kit+ FL cells
To evaluate whether the absence of Hoxb4 expression was compensated for with an increase in expression levels of other Hoxb genes, expression profiles of all 39 murine Hox genes in the c-Kit+ fraction of Hoxb4-/- FL cells were obtained (Figure 1B). Lack of Hoxb4 expression validated the mutant model (Figure 1B). All Hox genes centromeric to Hoxb5 showed a 2.3- to 3.6-fold increase in expression compared with wild-type cells (Figure 1B and Figure S1, which is available on the Blood website; see the Supplemental Figures link at the top of the online article). Interestingly, the expression of all Hoxa cluster genes was altered less than 2-fold compared with wild type. In the C cluster, the only noticeable change was increase in expression levels of Hoxc6 (3.4-fold) and Hoxc11 (4.5-fold) compared with controls. Hoxd genes were expressed at very low levels, if at all, in wild-type and Hoxb4-/- cells.
Perhaps most surprisingly, the expression levels of the Hoxb4 paralogs, that is, Hoxa4, Hoxc4, and Hoxd4, located within their respective clusters at a position identical to Hoxb4 within the Hoxb cluster, were either unaltered (Hoxa4 and Hoxd4), or decreased below control values as determined for Hoxc4 (2.2-fold). To assess the potential for partial cumulative compensation within the Hoxb cluster, the hematopoietic system of mutant mice harboring a large deletion encompassing most Hoxb cluster genes (Hoxb1-Hoxb9) was evaluated with a specific emphasis on the stem cell compartment.
Hox expression profiling in purified c-Kit+Hoxb1 to Hoxb9-/- FL cells
The most notable finding from the QRT-PCR analysis of Hox gene expression in Hoxb1-b9-/- cells was a dramatic decrease in expression of Hoxa11 (23-fold) compared to wild-type cells (Figure 1C). This result suggests that expression of Hoxa11 is regulated by one or more Hoxb genes or by the transactivating regulatory elements located in the region spanning the Hoxb1 to Hoxb9 loci.20,21,25 In addition to the decrease in Hoxa11 expression, all Hoxa genes, except Hoxa13, were expressed at levels lower than those determined for the wild-type cells, ranging from 1.7- to 5.8-fold differences in copy numbers. In contrast, 3 genes from the Hoxc cluster showed a substantial increase in expression: Hoxc4 (6.9-fold), Hoxc9 (4.5-fold), and Hoxc11 (4.2-fold). Similar to Hoxb4-/- cells, the basal levels of the Hoxd cluster expression were negligible. Overall, deletion of 9 Hoxb genes resulted in moderate to noticeable alterations in the expression of several distinct Hox genes located in other clusters. The lack of a simple compensatory event underscores the complexity of the “Hoxome” and suggests that multiple cross-talk events are involved in the integrated output of the Hox gene expression network.
Evaluation of Hox gene expression in wild-type and mutant c-Kit+ FL cells. Expression of Hox genes in c-Kit+ FL cells derived from 6 to 8 E14.5 FLs from littermates (wt) or from mutant embryos of different litters was evaluated by QRT-PCR. Analysis was carried out in triplicate on cDNA from total RNA, and data processing was done as described by Thompson et al.24 (A) Expression of Hoxb genes in wild-type FL cells in copy numbers, ordered according to the highest levels of RNA copies per 0.05 μg cDNA. Values below 10 copies were considered as not expressed. Fold difference in expression of the complete set of Hox genes in c-Kit+ cells of Hoxb4-/- (B) and Hoxb1-b9-/- (C) FLs. Two sets of independent data were generated for wild-type FL cells. Hox gene expression in KO FL cells was compared to both data sets. For genes that showed more that a 3-fold change in expression, a second independent experiment was performed. The bar graph includes the fold difference of both values for mutant cells (when available) compared to both independent data sets of wild-type cells. Amplitudes of changes in expression determined by comparison of various data sets are indicated by different colors. Note that no expression was detected for Hoxb4 and Hoxb1 to Hoxb9 in the Hoxb4 and Hoxb1-b9 mutants. Of all 39 clustered Hox genes, no appreciable values were obtained for Hoxc12,-c13,-d1,-d8,-d10,-d11,-d12, and -d13 in the Hoxb4 mutant and for Hoxc12 and -d1 in Hoxb1-Hoxb9-/- cells.
Evaluation of Hox gene expression in wild-type and mutant c-Kit+ FL cells. Expression of Hox genes in c-Kit+ FL cells derived from 6 to 8 E14.5 FLs from littermates (wt) or from mutant embryos of different litters was evaluated by QRT-PCR. Analysis was carried out in triplicate on cDNA from total RNA, and data processing was done as described by Thompson et al.24 (A) Expression of Hoxb genes in wild-type FL cells in copy numbers, ordered according to the highest levels of RNA copies per 0.05 μg cDNA. Values below 10 copies were considered as not expressed. Fold difference in expression of the complete set of Hox genes in c-Kit+ cells of Hoxb4-/- (B) and Hoxb1-b9-/- (C) FLs. Two sets of independent data were generated for wild-type FL cells. Hox gene expression in KO FL cells was compared to both data sets. For genes that showed more that a 3-fold change in expression, a second independent experiment was performed. The bar graph includes the fold difference of both values for mutant cells (when available) compared to both independent data sets of wild-type cells. Amplitudes of changes in expression determined by comparison of various data sets are indicated by different colors. Note that no expression was detected for Hoxb4 and Hoxb1 to Hoxb9 in the Hoxb4 and Hoxb1-b9 mutants. Of all 39 clustered Hox genes, no appreciable values were obtained for Hoxc12,-c13,-d1,-d8,-d10,-d11,-d12, and -d13 in the Hoxb4 mutant and for Hoxc12 and -d1 in Hoxb1-Hoxb9-/- cells.
Hoxb4 mutant mice have no gross hematopoietic anomalies
Subsequently, the hematopoietic compartments of Hoxb4-/- mice were analyzed for the presence of any detectable anomalies. Homozygous Hoxb4-/- mice were healthy and fertile. Cellularity in BM, spleen, and thymus was comparable to that of control littermates (Table 1). Subpopulations of lymphoid and myeloid lineages were evaluated by flow cytometry. There was no over- or under-representation of mature B-cell, T-cell, or myeloid populations (Table 2), indicating that the absence of Hoxb4 neither affected the output of mature end cells nor the commitment of multipotent cells to lymphoid and myeloid lineages. Furthermore, FACS analysis revealed no differences between the sizes of stem-cell-enriched fractions (Sca1+c-Kit+ lineage-negative cells) in Hoxb4-/- mice and wild-type controls (not shown). To determine whether mature cells were derived from quantitatively comparable progenitor pools, myeloid and B-cell progenitor cell numbers were evaluated by clonogenic progenitor assays. Comparable numbers of colony-forming cells were obtained for BM and spleen of Hoxb4-/- mice and wild-type controls (Table 1). Taken together, these and previously published observations16 suggested Hoxb4 is dispensable for the establishment and maintenance of steady-state hematopoiesis.
Hematopoietic parameters of Hoxb4 mutants and control littermates
. | Cellularity . | . | . | B-CFC . | . | M-CFC . | . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
. | BM, × 106 . | Spleen, × 106 . | Thymus, × 106 . | BM, × 106 . | Spleen, × 106 . | BM, × 106 . | Spleen, × 106 . | ||||
| Hoxb4−/− | 29 ± 2.4 | 141 ± 74 | 97 ± 27 | 6.6 ± 2.4 | 0.4 ± 0.1 | 54 ± 2.4 | 3.8 ± 2.2 | ||||
| Control | 25 ± 5.3 | 152 ± 18 | 86 ± 13 | 4.4 ± 0.8 | 0.4 ± 0.3 | 41 ± 14 | 2.9 ± 0.4 | ||||
. | Cellularity . | . | . | B-CFC . | . | M-CFC . | . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
. | BM, × 106 . | Spleen, × 106 . | Thymus, × 106 . | BM, × 106 . | Spleen, × 106 . | BM, × 106 . | Spleen, × 106 . | ||||
| Hoxb4−/− | 29 ± 2.4 | 141 ± 74 | 97 ± 27 | 6.6 ± 2.4 | 0.4 ± 0.1 | 54 ± 2.4 | 3.8 ± 2.2 | ||||
| Control | 25 ± 5.3 | 152 ± 18 | 86 ± 13 | 4.4 ± 0.8 | 0.4 ± 0.3 | 41 ± 14 | 2.9 ± 0.4 | ||||
n = 3 mice for each group. Total cell numbers and numbers of B-cell and myeloid progenitors (B-cell and myeloid colony-forming cells, B-CFCs and M-CFCs, respectively) in BM, spleen, and thymus of Hoxb4−/− and control littermates.
Lymphoid and myeloid population in hematopoietic organs of Hoxb4 mutants and control littermates
. | Hoxb4−/−, % . | . | . | Control, % . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|---|
. | BM . | Spleen . | Thymus . | BM . | Spleen . | Thymus . | ||||
| B cells | ||||||||||
| B220+/CD43 | 24 ± 3.7 | 36 ± 13 | — | 30 ± 1.8 | 55 ± 4.8 | — | ||||
| B220+/CD43+ | 3.0 ± 0.2 | 8.4 ± 1.6 | — | 4.1 ± 0.8 | 6.9 ± 0.7 | — | ||||
| B220+/lgM+ | 8.0 ± 3.3 | 49 ± 13 | — | 15 ± 1.5 | 56 ± 3.7 | — | ||||
| B220+/lgM− | 17 ± 1.0 | 2.7 ± 19 | — | 17 ± 0.9 | 6.1 ± 2.2 | — | ||||
| T cells | ||||||||||
| CD4 | — | 26 ± 5.1 | 6.9 ± 1.3 | — | 20 ± 1.1 | 6.6 ± 0.5 | ||||
| CD8 | — | 17 ± 3.5 | 7.2 ± 0.8 | — | 12 ± 0.9 | 4.4 ± 0.4 | ||||
| CD4/CD8 | — | 0 | 84 ± 2.2 | — | < 0.01 | 87 ± 0.9 | ||||
| Myeloid cells | ||||||||||
| Mac1 | 17 ± 1.3 | 5.6 ± 1.6 | — | 18.6 ± 1.1 | 5.4 ± 0.8 | — | ||||
| Mac1/Gr1 | 11 ± 1.8 | 0.8 ± 0.5 | — | 9.2 ± 1.2 | 0.4 ± 0.1 | — | ||||
. | Hoxb4−/−, % . | . | . | Control, % . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|---|
. | BM . | Spleen . | Thymus . | BM . | Spleen . | Thymus . | ||||
| B cells | ||||||||||
| B220+/CD43 | 24 ± 3.7 | 36 ± 13 | — | 30 ± 1.8 | 55 ± 4.8 | — | ||||
| B220+/CD43+ | 3.0 ± 0.2 | 8.4 ± 1.6 | — | 4.1 ± 0.8 | 6.9 ± 0.7 | — | ||||
| B220+/lgM+ | 8.0 ± 3.3 | 49 ± 13 | — | 15 ± 1.5 | 56 ± 3.7 | — | ||||
| B220+/lgM− | 17 ± 1.0 | 2.7 ± 19 | — | 17 ± 0.9 | 6.1 ± 2.2 | — | ||||
| T cells | ||||||||||
| CD4 | — | 26 ± 5.1 | 6.9 ± 1.3 | — | 20 ± 1.1 | 6.6 ± 0.5 | ||||
| CD8 | — | 17 ± 3.5 | 7.2 ± 0.8 | — | 12 ± 0.9 | 4.4 ± 0.4 | ||||
| CD4/CD8 | — | 0 | 84 ± 2.2 | — | < 0.01 | 87 ± 0.9 | ||||
| Myeloid cells | ||||||||||
| Mac1 | 17 ± 1.3 | 5.6 ± 1.6 | — | 18.6 ± 1.1 | 5.4 ± 0.8 | — | ||||
| Mac1/Gr1 | 11 ± 1.8 | 0.8 ± 0.5 | — | 9.2 ± 1.2 | 0.4 ± 0.1 | — | ||||
Percentages of lymphoid and myeloid cell populations in BM, spleen, and thymus of mutant and control animals using FACS analysis. Comparison between mutant and control cell populations and total cell numbers in Tables 1 and 2 was evaluated using a 2-tailed Student t test; P > .05 for all groups. n = 3 mice per group.
— indicates not done.
Cellularity of Hoxb4 mutant and Hoxb1-b9 mutant FLs is reduced
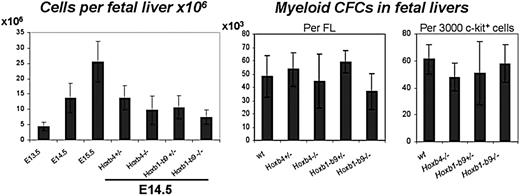
Homozygous Hoxb1-b9-/- mice are not viable23 and die around birth, most likely due to inadequate chest skeleton development, brain hemorrhage, edema, and heart problems. At E14.5, Hoxb1-b9-/- embryos already exhibit these defects but are viable and present in normal mendelian ratios. They typically show an open thoracic cavity, allowing direct visualization of their heart, and edema.23 Hematopoietic parameters of Hoxb1-b9-/- and Hoxb4-/- mutants were analyzed using E14.5 FL cells. FLs of Hoxb4 homozygous and Hoxb1-9 heterozygous and homozygous mutants were smaller and had significantly reduced cell numbers compared with control embryos (Figure 2, left panel). Importantly, the proportion of these mutant cells that stained positive for annexin V was identical to wild-type cells, indicating that apoptosis is not responsible for this reduction in FL cellularity (data not shown). Evaluation of absolute progenitor numbers per wild-type versus mutant FLs revealed comparable values (Figure 2, center panel). Moreover, the proliferation (ie, colony size) and differentiation potential of these mutant progenitors were also similar to wild-type controls (Figure 2, right panel, for myeloid progenitors; data not shown for erythroid progenitors). Together these results reveal the absence of a cell autonomous defect in our mutant progenitors and point to extrahematopoietic anomalies for the observed reduction in FL cellularity.
Hoxb-mutant cells are competitive in reconstitution of irradiated hosts
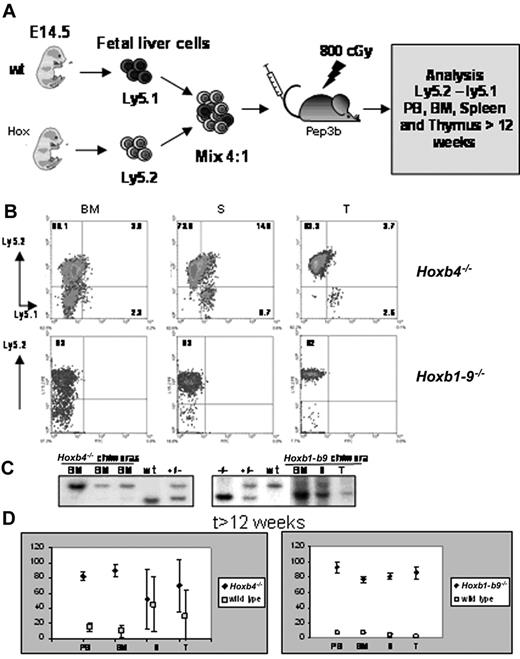
The existence of Hoxb4 mutant mice and survival to full-term of Hoxb1-b9 mutants implies that Hoxb genes are not critical for specification of HSCs or for the onset of definitive hematopoiesis. We therefore raised the question whether maintenance of definitive hematopoiesis is affected by the absence of either Hoxb4 alone or most Hoxb cluster genes. Because the Hoxb1-b9 mutants are not viable, we developed a competitive repopulation strategy that exploits the ability of FL cells to reconstitute hematopoietic systems of lethally irradiated mice26 (Figure 3A). Mutant (Ly5.2+) cells were mixed with wild-type (Ly5.1+) cells in a 4:1 ratio and transplanted into irradiated congenic (Ly5.1+) recipients. At 6 to 7 weeks after transplantation, the Hoxb4-/-- and Hoxb1-b9-/--derived Ly5.2+ cells represented approximately 75% (72% ± 5%) of peripheral blood mononucleated cells (data not shown). Contribution of mutant cells to reconstitution of recipients thus reflected the composition of the transplant, suggesting that Hoxb gene expression is not essential for the competitive reconstitution of the hematopoietic systems.
Hematopoietic characterization of Hox mutants. FL cellularity (left panel) from 13.5-, 14.5-, and 15.5-day-old wild-type embryos and from homozygous and heterozygous Hoxb4 and Hoxb1-Hoxb9 14.5-day-old embryos (n = 6, 22, 8, 19, 20, 17, and 10 from left to right). P values are .01, .03, and < .001 for FL cellularity of Hoxb4-/-, Hoxb1-b9+/-, and Hoxb1-b9-/- mice, respectively. Myeloid progenitors per fetal liver (center panel, n = 3, 4, 3, 3, and 3) and per 3000 sorted c-kit+ cells (right panel, n = 3) of E14.5 mutant and control FLs. Values are expressed as the mean ± SD from 3 samples per test group (exact number shown in Tables 1 and 2). Statistical analysis was performed using a 2-tailed Student t test.
Hematopoietic characterization of Hox mutants. FL cellularity (left panel) from 13.5-, 14.5-, and 15.5-day-old wild-type embryos and from homozygous and heterozygous Hoxb4 and Hoxb1-Hoxb9 14.5-day-old embryos (n = 6, 22, 8, 19, 20, 17, and 10 from left to right). P values are .01, .03, and < .001 for FL cellularity of Hoxb4-/-, Hoxb1-b9+/-, and Hoxb1-b9-/- mice, respectively. Myeloid progenitors per fetal liver (center panel, n = 3, 4, 3, 3, and 3) and per 3000 sorted c-kit+ cells (right panel, n = 3) of E14.5 mutant and control FLs. Values are expressed as the mean ± SD from 3 samples per test group (exact number shown in Tables 1 and 2). Statistical analysis was performed using a 2-tailed Student t test.
The competitive nature of mutant HSCs was evaluated again at 12 weeks after transplantation, at which time the peripheral cell output is considered to reflect the activity of the transplanted HSCs. At 12 weeks after transplantation, Hoxb4-/- (Ly5.2+) cells represented on average more than 80% of the peripheral blood, BM, spleen, and thymus-derived cell populations, indicating that Hoxb4-/- stem cells contributed to reconstitution of myeloablated mice in a competitive manner (Figure 3B-C). The contribution of Hoxb4-/- cells in spleen was, on average, only 59% ± 37% (Figure 3D), which was above the contribution determined for the wild-type cells (38% ± 35%) and could reflect slower turnover of residual lymphoid cells of the host that still could be present in the spleen.
Of interest, analysis of Hoxb1-b9-/- recipients suggested that both the hematopoietic progenitors as well as stem-cell compartments were adequate. At 7 weeks after transplantation, 70% to 80% of the peripheral blood leukocytes (PBLs) were derived from the mutant (Ly5.2+) cells. At 24 weeks after transplantation, mutant cells contributed to 80% to 90% reconstitution of BM, spleen, and thymius of recipients (Figure 3B-C). Thus, although the transplantation inocula in this group comprised 75% mutant (Ly5.2+) and only 25% wild-type (Ly5.1+) cells, Hoxb1-b9-/- cells contributed to between 80% and 90% reconstitution of BM, spleen, and thymus, indicating that Hoxb1-b9-/- cells out-competed wild-type cells in long-term reconstitution of recipients. The BM origin of competing cells likely explains their lower competitive repopulating activity than the mutant cells, which were isolated from FLs.27
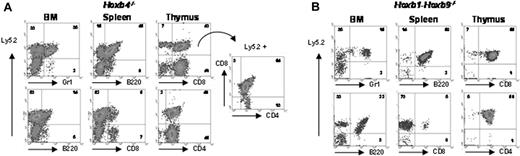
To determine whether mutant Hoxb4 and Hoxb1-b9 cells also retained their full differentiation ability, BM, spleen, and thymic cells were stained for surface markers B220 (B-cell lineage), CD4 and CD8 (T-cell lineage), and Mac1 (myeloid lineage), in combination with the Ly5.2 marker. Double-positive cells for Ly5.2 and each of these markers demonstrated the full potential of the mutant Hoxb4-/- and Hoxb1-b9-/- stem cells to differentiate into both lymphoid and myeloid cells (Figure 4). Together, these data identify cross-regulation of Hox gene expression in primitive hematopoietic cells and demonstrate the nonessential role for the Hoxb cluster locus in HSC activity.
Competitive reconstitution assays. (A) Experimental outline of the competition assay between control and mutant FL cells to reconstitute hematopoietic organs of an adult myeloablated recipient on intravenous transplantation. Mutant and wildtype FL cells differ in their Ly5 locus and can be distinguished by FACS analysis using antibodies directed against Ly5.1 (wild-type cells and residual cells from Pep3b recipient) and Ly5.2 (mutant cells). (B) FACS profiles from hematopoietic chimeras for Hoxb4 and Hoxb1-b9 mutants assaying the contribution of mutant (Ly5.2) and wild-type (Ly5.1) cells. (C) Southern blots of BM, spleen (S), or thymus (T) derived from recipients of a mixed Hoxb4-/- or Hoxb1-9 and wild-type FL transplants. (D) Percentages of mutant and wild-type hematopoietic cells in BM, S, T, and peripheral blood (PB) of mice given transplants with mixed mutant and wild-type FL or BM cells. Each value represents the mean percentage of mutant or wild-type populations measured in 16 different mice. As described in “Materials and methods,” each mutant FL tested for competitive repopulation with wild-type cells was transplanted into 4 recipients and therefore these 16 mice represent 4 mutant FLs. Because not all hematopoietic cells in BM present the leukocyte marker CD45 (Ly5), the sum of Ly5.1 and Ly5.2 cells was assumed to be 100% and the percentages for these populations was recalculated. A color version of this figure is available online as Supplemental Figure S2.
Competitive reconstitution assays. (A) Experimental outline of the competition assay between control and mutant FL cells to reconstitute hematopoietic organs of an adult myeloablated recipient on intravenous transplantation. Mutant and wildtype FL cells differ in their Ly5 locus and can be distinguished by FACS analysis using antibodies directed against Ly5.1 (wild-type cells and residual cells from Pep3b recipient) and Ly5.2 (mutant cells). (B) FACS profiles from hematopoietic chimeras for Hoxb4 and Hoxb1-b9 mutants assaying the contribution of mutant (Ly5.2) and wild-type (Ly5.1) cells. (C) Southern blots of BM, spleen (S), or thymus (T) derived from recipients of a mixed Hoxb4-/- or Hoxb1-9 and wild-type FL transplants. (D) Percentages of mutant and wild-type hematopoietic cells in BM, S, T, and peripheral blood (PB) of mice given transplants with mixed mutant and wild-type FL or BM cells. Each value represents the mean percentage of mutant or wild-type populations measured in 16 different mice. As described in “Materials and methods,” each mutant FL tested for competitive repopulation with wild-type cells was transplanted into 4 recipients and therefore these 16 mice represent 4 mutant FLs. Because not all hematopoietic cells in BM present the leukocyte marker CD45 (Ly5), the sum of Ly5.1 and Ly5.2 cells was assumed to be 100% and the percentages for these populations was recalculated. A color version of this figure is available online as Supplemental Figure S2.
Lineage reconstitution of mutant cells in competition with wild-type cells. FACS profiles of BM, spleen, and thymus from hematopoietic chimeric mice, indicating whether mutant cells (Ly5.2) differentiate into myeloid (Gr1), B-cell (B220+), or T-cell populations (CD4 and CD8) by simultaneous staining with anti-Ly5.2 antibody. Ly5.2+Hoxb4-/- cells of the thymus were gated and analyzed on distribution of CD4 and CD8 single- and double-positive populations (not available for Hoxb1-b9 mutant). (A) Hoxb4-/- chimeras and (B) Hoxb1-b9 chimeras. A color version of this figure is available online as Supplemental Figure S3.
Lineage reconstitution of mutant cells in competition with wild-type cells. FACS profiles of BM, spleen, and thymus from hematopoietic chimeric mice, indicating whether mutant cells (Ly5.2) differentiate into myeloid (Gr1), B-cell (B220+), or T-cell populations (CD4 and CD8) by simultaneous staining with anti-Ly5.2 antibody. Ly5.2+Hoxb4-/- cells of the thymus were gated and analyzed on distribution of CD4 and CD8 single- and double-positive populations (not available for Hoxb1-b9 mutant). (A) Hoxb4-/- chimeras and (B) Hoxb1-b9 chimeras. A color version of this figure is available online as Supplemental Figure S3.
Discussion
Although overexpression of Hoxb4 promotes HSC self-renewal, the function of the endogenous gene appears dispensable for HSC activity, thus raising an intriguing possibility of redundancy of HOX proteins in modulation of HSC behavior. In this study, we confirm the nonessential role of Hoxb4 in hematopoiesis and report that the absence of Hoxb4 expression results in alterations in expression levels of several neighboring Hoxb genes in c-Kit+ FL cells. Our results also demonstrate that HSCs retain their full competitive reconstitution and differentiation potential in the absence of the majority of the Hoxb gene cluster (b1-b9) and show that this deficiency correlates with suppression of Hoxa4 and Hoxa11, and increase in Hoxc4 expression levels, suggesting an overlapping and redundant function for HOX proteins during stress hematopoiesis.
The noticeable change in expression levels of most Hoxb genes in our Hoxb4 mutant hematopoietic cells merits attention. It may result from the influence of the residual pgk promoter in the locus or, alternatively, from direct or indirect cross-regulation. The normal HSC activity in Hoxb1-b9 mutant mice indicates that all 9 Hoxb genes are dispensable for the competitive reconstitution ability of HSCs. This suggests that the apparent normal abilities of Hoxb4-/- HSCs do not result from the compensatory activities of other Hoxb genes.
Complete Hox gene expression profiling was applied to c-Kit+ FL cells deficient for either Hoxb4 alone or the genetic region encompassing Hoxb1 to Hoxb9. Two important observations have been made: (1) expression of most Hoxa genes, and Hoxa11 in particular, was decreased in Hoxb1-b9-/- cells compared with wild-type cells, and (2) the expression levels of Hoxa genes in all cohorts are at least one log higher than those determined for genes of the B and C cluster, respectively. From these data we conclude that there is no direct compensation for the loss of Hoxb4 in regulation of HSC self-renewal by other Hox genes. Moreover, decreased expression of Hoxa11 and other Hoxa genes is indicative of either direct cross talk between Hoxb genes and Hoxa genes, or the presence of regulatory sequences within the Hoxb cluster that act on particular Hoxa genes. Because Hoxb-deficient HSCs are able to replenish completely the hematopoietic compartment of irradiated recipients, and expression levels of Hoxa genes are higher than those determined for Hoxb4, it is possible that the physiologic determinant of HSC self-renewal might be one or more Hoxa genes. The relative absence of Hoxa11 expression in Hoxb mutant cells rules out this gene as the potential candidate.
A possible candidate is the Hoxb4 paralog gene Hoxa4. The property of at least some paralog genes to execute identical biologic functions was demonstrated by Greer et al28 by engineering mice in which the coding region of Hoxa3 was swapped with that of Hoxd3. From the same group of paralogs, Hoxc4 was shown to induce a clonogenic expansion of human early progenitors.29 Based on this, it will be interesting to determine the effect of other Hox paralog 4 group gene ablation on hematopoiesis.
Furthermore, overexpression studies from our laboratory demonstrated that Hoxa9 also induces self-renewal within the HSC pool prior to development of myeloid leukemia,30 suggesting that Hoxa9 could contribute to regulation of HSC self-renewal under physiologic conditions.
In contrast to our data, Brun et al16 reported mild reductions in cellularity of the hematopoietic compartment in Hoxb4-deficient mice resulting from marginally decreased numbers of HSCs, with reduced proliferation capacity. This phenotype was more pronounced in a Hoxb3/Hoxb4-deficient model from the same group.31 Such disparity might be a result of the targeting strategy used, that is, their mice were engineered using the Cre-loxP system, which removed the complete gene and brought Hoxb3 and Hoxb5 physically closer, whereas Hoxb4-/- mice used in this study were targeted in the first exon with a selection cassette driven by the pgk promoter. The concept that targeting procedures can affect the phenotype is supported by differences in skeleton formation in the Hoxb4-deficient models.15,16 Moreover, the expression of Hoxb2, -b3, and -b5 genes in Cre/loxP Hoxb4-deficient mice is significantly reduced, whereas the expression of these genes does not change in our mutant mice. Although the different targeting procedure might influence the expression of the neighboring Hox genes, this can also be a result of the difference in the sorted population (c-Kit+ versus Ter119 depleted).
Hoxa11 is the only gene that showed a dramatic change in expression in Hoxb1-b9-/-. The levels decreased 23-fold from more than 1000 copies to an average of 50 copies of mRNA. There is one report describing 2 families suffering from amegakaryocytic thrombocytopenia with absent radii associated with a mutation in the HOXA11 gene.32 Interestingly, the hematopoietic Hoxb1-b9-/- chimeras had normal megakaryocytes numbers in their marrow (data not shown). The donor origin of these cells in our recipient animals was not tested. In contrast to an earlier report showing that Hoxb6 mutant mice have increased elements derived from the erythroid lineage, our results with the Hoxb1-b9 mutant fail to show an effect on this lineage.33 However due to cross-regulation between different Hox genes, a direct comparative analysis remains potentially flawed without the complete assessment of their expression in other Hox mutant mice.
Collectively our studies suggest that HSC proliferation and differentiation are insensitive to important changes in Hoxb gene expression and point to the existence of a complex cross-regulatory network of Hox gene expression in primitive hematopoietic cells. Thus, the question whether Hox genes are involved in physiologic HSC activity remains unsolved and similar strategies perhaps with a focus on Hoxa cluster genes are needed for a final assessment.
Prepublished online as Blood First Edition Paper, December 8, 2005; DOI 10.1182/blood-2005-06-2245.
Supported by National Institutes of Health grants RO165430 (G.S.) and DK48642 (H.J.L.). A.T. is supported by a fellowship from the American Cancer Society for Beginning Investigators (ACSBI) under the administration of the Union Internationale Contre le Cancer (UICC). G.S. is a recipient of a Canada Research Chair in molecular genetics of stem cells and a scholar of the Leukemia Lymphoma Society of America.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We want to thank in particular Melanie Frechette for the excellent animal care. Special thanks are owed to Nathalie Tessier, Eric Massicotte, and Martine Dupuis from the flow cytometry service of the IRCM in Montreal. Sylvie Provost and Claude Belisle are thanked for their assistance with Q-PCR.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal