The cellular reservoir for Kaposi sarcoma-associated herpesvirus (KSHV) infection in the hematopoietic compartment and mechanisms governing latent infection and reactivation remain undefined. To determine susceptibility of human CD34+ hematopoietic progenitor cells (HPCs) to infection with KSHV, purified HPCs were exposed to KSHV, and cells were differentiated in vitro and in vivo. Clonogenic colony-forming activity was significantly suppressed in KSHV-infected CD34+ cells, and viral DNA was predominantly localized to granulocyte-macrophage colonies differentiated in vitro. rKSHV.219 is a recombinant KSHV construct that expresses green fluorescent protein from a cellular promoter active during latency and red fluorescent protein from a viral lytic promoter. Infection of CD34+ HPCs with rKSHV.219 showed similar patterns of infection, persistence, and hematopoietic suppression in vitro in comparison with KSHV. rKSHV.219 infection was detected in human CD14+ and CD19+ cells recovered from NOD/SCID mouse bone marrow and spleen following reconstitution with rKSHV.219-infected CD34+ HPCs. These results suggest that rKSHV.219 establishes persistent infection in NOD/SCID mice and that virus may be disseminated following differentiation of infected HPCs into the B-cell and monocyte lineages. CD34+ HPCs may be a reservoir for KSHV infection and may provide a continuous source of virally infected cells in vivo. (Blood. 2006;108:141-151)
Introduction
Kaposi sarcoma-associated herpesvirus (KSHV), also known as human herpesvirus 8 (HHV-8), is the first human virus identified belonging to the γ2-herpesvirus family.1-3 KSHV infection has been linked to the pathogenesis of all 4 forms of Kaposi sarcoma (KS).1,4-6 In addition to KS, KSHV infection has been associated with the pathogenesis of body cavity-based B-cell lymphoma (BCBL), also called primary effusion lymphoma (PEL), and a subset of multicentric Castleman disease.4-6 Little is known of the pathobiology of KSHV infections. Previous investigations have demonstrated KSHV infection in circulating monocyte/macrophages and B lymphocytes of KS patients, suggesting that these cells may participate in the dissemination of viral infection in vivo.7-9 KSHV can establish limited infections in a variety of cells in vitro, including human B cells, endothelial cells, epithelial cells, fibroblast cells, mesenchymal stem cells, dendritic cells, and macrophages.4-6,10-16 In vivo, KSHV infection has been localized to human B cells, macrophages, keratinocytes, endothelial cells, and epithelial cells.4-6
CD34+ hematopoietic progenitor stem cells (HPCs) are a heterogeneous cell population that includes pluripotent stem cells and cells in the early stages of lineage commitment. HPCs continuously migrate to and from the bone marrow (BM) in normal adult animals.17 CD34+ HPCs can differentiate to produce all hematopoietic cell lineages found in the circulation.18 CD34+ HPCs have been shown to be susceptible to infection with a number of viruses, including HIV-1,19,20 hepatitis C virus,21 JC virus,22 HCMV (HHV-5),23-28 HHV-6,29,30 HHV-7,31 and HTLV-1.32 Suppression of hematopoiesis has been documented to occur following infection of CD34+ HPCs with HCMV/HHV-5,23,26,33-36 HHV-6,29,30 HIV-1,37,38 and measles virus39 either by direct infection of CD34+ HPCs or by indirect mechanisms such as disruption of cytokine expression or stromal cells. Notably, KSHV previously has been detected in CD34+ cells from peripheral blood lymphocyte (PBL) samples of KS patients.40
Little is known about the characteristics of KSHV persistence in vivo with respect to cell types that hold latent virus and function as reservoirs of viral infection. Here we demonstrate that KSHV can infect CD34+ HPCs, resulting in the concomitant expression of lytic and latent genes and in the suppression of hematopoiesis in vitro. KSHV infection is maintained during differentiation of CD34+ HPCs in vitro, using clonogenic colony-formation assays (CFA), and in vivo following inoculation of NOD/SCID mice. These results suggest that KSHV infection of CD34+ HPCs may result in the suppression of hematopoiesis and that CD34+ cells may function as viral reservoirs that play a role in KSHV viral persistence and dissemination in vivo.
Materials and methods
Cell lines
All cells were maintained in Iscove modified Dulbecco media (IMDM) (Gibco BRL, Grand Island, NY) supplemented with 10% heat inactivated fetal bovine serum (FBS) (Gibco BRL), penicillin (100 U/mL), streptomycin (100 μg/mL), and L-glutamine (292 μg/mL) (Gibco BRL), in a humidified incubator at 37°C, 5% CO2.
KSHV preparation
KSHV was prepared as previously described.41 Briefly, BCBL-1 cells (5 × 105 cells/mL) were cultivated in complete IMDM (10% FBS + penicillin/streptomycin/glutamine) with 12-O-tetradecanoylphorbor-13-acetate (TPA, 20 ng/mL) (Sigma Aldrich, St Louis, MO) for 12 to 20 hours. Cells were pelleted (302 g, 5 minutes) and resuspended in complete IMDM without TPA and allowed to grow for 4 to 5 days at 37°C, 5% CO2. BCBL-1 cells were pelleted and discarded. The supernatant was filtered using a 0.45-μm filter to remove remaining cells. Filtered supernatant was then centrifuged at 53 000 g for 2 hours with a SW-27 rotor to pellet virus. Virus was resuspended in 1 mL serum-free IMDM. An aliquot of virus stock was collected, and viral DNA was purified and analyzed by real time polymerase chain reaction (PCR) using KS330233 (ORF 26, minor capsid protein) primers to determine viral genomic copy number.
rKSHV.219 preparation
Preparation of recombinant virus was performed as previously described.42 Briefly, a fragment of the KSHV genome containing an enhanced green fluorescent protein (GFP) gene expressed by the elongation factor 1-α promoter, a Ds red fluorescent protein (RFP) expressed by the polyadenylated nuclear (PAN) RNA promoter, and puromycin resistance gene was electroporated into JSC-1 cells.43 Recombinant JSC-1 cells were selected and expanded by cultivating in puromycin (0.5 μg/mL). rKSHV.219 production in recombinant JSC-1 cells was induced by TPA (15 ng/mL), and harvested virus was used to infect Vero cells. Individual GFP-positive Vero cells were isolated and expanded in the presence of puromycin. Vero cultures containing latent rKSHV.219 were infected with BacK50,42 a recombinant baculovirus expressing KSHV RTA to produce rKSHV.219 viral stocks.
Isolation of CD34+ HPCs
Human CD34+ cells were prepared as previously described.32 CD34+ cells were purified by passage of the cell suspension through a MidiMACS magnetic column (Miltenyi Biotec, Auburn, CA). The purity of isolated CD34+ cells was analyzed with a phycoerythrin (PE)-conjugated monoclonal mouse antibody (mAb) against CD34+ (Becton Dickinson, Mountain View, CA) and found to be at least 92% CD34 positive.
Infection of CD34+ HPCs with KSHV and rKSHV.219
Purified CD34+ cells (5 × 106 ) were infected with KSHV (25-100 viral genome equivalents per cell) in a final volume of 1.0 mL serum-free AIM-V medium (Gibco BRL) containing hexadimethrine bromide (polybrene, 1 μg/mL) (Sigma Aldrich). KSHV and CD34+ HPCs were co-incubated and centrifuged at 839 g, 4°C for 4 hours. Supernatant was discarded after centrifugation, and pelleted CD34+ HPCs were washed 3 times with phosphate-buffered saline (PBS). Washed CD34+ HPCs were plated into 2 mL MethoCult H4433 (StemCell Technologies, Vancouver, British Columbia, Canada) (3000 cells per 35 mm × 10-mm dish). CD34+ HPCs infected with UV-irradiated KSHV (10 μW/cm2 for 1 hour, 1000 mJ energy equivalent) or mock-infected with serum-free AIM-V medium were plated in MethoCult H4433 in a similar fashion. Purified CD34+ cells (5 × 106 ) were infected with rKSHV.219 (660-775 viral genome equivalents per cell)42 in a final volume of 1.0 mL serum-free AIM-V medium (Gibco BRL) containing polybrene (1 μg/mL) (Sigma Aldrich). rKSHV.219 and CD34+ HPCs were co-incubated and centrifuged at 839 g, 4°C for 4 hours. Supernatant was discarded after centrifugation, and pelleted CD34+ HPCs were washed 3 times with PBS. CD34+ HPCs infected with UV-irradiated rKSHV.219 (10 μW/cm2 for 1 hour, 1000 mJ energy equivalent), heat-inactivated rKSHV.219 (65°C for 1 hour), or mock-infected with serum-free AIM-V medium also were plated in MethoCult H4433.
Real-time PCR genomic copy standard preparation and cycle conditions
To prepare KSHV genomic DNA standards, the KS330233 fragment PCR-amplified from BCBL-1 DNA was cloned into the pCR2.1 plasmid using a TA cloning kit (Invitrogen, Carlsbad, CA) to create pCR2.1-KS330233. For the preparation of human β-globin DNA standards, a 585-bp fragment (huBGLOB585) amplified by primers 5′-GAAGAGCCAAGGACAGGTAC-3′ (GH20)44 and 5′-GCAAAGGTGCCCTTGAGGT-3′45 was cloned into pCR2.1 to create pCR2.1-huBGLOB585. pCR2.1-KS330233 and pCR2.1-huBGLOB585 were linearized with BamHI and serially diluted into a salmon sperm DNA carrier (2 μg/μL) to provide copy number standards. Real-time PCR assay used KSHV KS330233 primers (within ORF 26 minor capsid protein) (5′-AGCCGAAAGGATTCCACCAT-3′, 5′-TCCGTGTTGTCTACGTCCAG-3′),1 human β-globin primers 5′-TGAGCCTTCACCTTAGGGTTGCCCA-3′ and 5′-GCCCTGGGCAGGTTGGTATCAAGGT-3′, which amplify a 240-bp region within huB-GLOB585, and the QuantiTech SYBR Green PCR kit (Qiagen, Valencia, CA). Cycle conditions on an iCycler (Bio-Rad Laboratories, Hercules, CA) were set as follows: 15 minutes at 95°C; 55 cycles of 30 seconds at 94°C, 30 seconds at 58°C, 1 minute at 72°C, 30 seconds of detection at 80.5°C; 55°C to 95°C melting curve. Data were collected, and KSHV and human β-globin DNA copy number per sample was calculated using iCycler iQ Real-Time Detection System Software (Bio-Rad Laboratories). KSHV DNA copies were normalized to 0.5 × human β-globin DNA copies, representing 1 human cell.
Clonogenic colony analysis
Clonogenic colony assays were performed as previously described.46 Briefly, CD34+ HPCs were infected with KSHV or rKSHV.219, washed 3 times with PBS, cultured in 2 mL MethoCult H4433 medium (StemCell Technologies), and incubated at 37°C in a humidified atmosphere with 5% CO2. Clonogenic granulocyte-macrophage (CFU-GM), erythroid burst (BFU-E), and high proliferative pluripotent (HPP) colonies were identified by morphology at 10 to 15 days after plating and counted under an inverted fluorescent microscope (Leica DMIL).47 Colonies were randomly isolated by aspiration, DNA purified, and subjected to real-time PCR analysis for KSHV and human β-globin sequences.
Real-time PCR analysis of DNA from cultured CD34+ HPCs
CD34+ HPCs infected with KSHV or rKSHV.219 were cultured in IMDM supplemented with 10% FBS, penicillin, streptomycin, and L-glutamine. Twenty-five microliter aliquots were collected from cultures daily. Collected samples were centrifuged, washed once with PBS, and subjected to phenol-chloroform extraction to isolate DNA. Isolated DNA was subjected to real-time PCR analysis for KSHV and human β-globin sequences. CD34+ HPCs infected with UV-irradiated KSHV, UV-irradiated rKSHV.219, and heat-inactivated rKSHV.219 also were cultured in IMDM, and extracted DNA was analyzed by PCR in an identical manner.
SCID mice
NOD.CB17-Prkdcscid/J (NOD/SCID) mice were kept under specific pathogen-free conditions at the SCID Mouse Core Facility at SUNY Upstate Medical University, Syracuse, NY. NOD/SCID mice were housed in micro-isolator cages and all food, water, and bedding were autoclaved prior to use. NOD/SCID breeder mice originally were purchased from the Jackson Laboratory (Bar Harbor, ME). All experiments were approved by the Institutional Animal Care and Use Committee of SUNY Upstate Medical University. NOD/SCID mice were anesthetized with isoflurane (Minrad, Buffalo, NY) and/or methoxyflurane (Lancaster Synthesis, Pelham, NH) prior to all manipulations, as previously described.48
Inoculation of rKSHV.219-infected CD34+ HPCs into NOD/SCID mice
rKSHV.219-infected CD34+ HPCs (0.1 to 2 × 107 cells/mouse) were intravenously inoculated into NOD/SCID mice by tail vein injection. Some of the NOD/SCID mice were sublethally irradiated (300 rads) 2 days prior to inoculation to promote CD34+ HPC engraftment. Briefly, mice were anesthetized with methoxyflurane (Lancaster Synthesis), temporarily restrained, and heated with a heat lamp to induce vasodilation. Mice received 200 μL of sample per tail vein injection. Mice were monitored, and moribund animals were euthanized with isoflurane (Minrad) and cervical dislocation. After 6 to 19 weeks, mice were killed, and tissue samples were collected from spleen, bone marrow, and peripheral blood for analysis by real-time PCR, reverse transcriptase-polymerase chain reaction (RT-PCR), flow cytometry, indirect fluorescence antibody assay (IFA), and fluorescence microscopy.
Fluorescence microscopy
GFP and RFP expression from clonogenic colonies and NOD/SCID mouse samples were detected using a Nikon Eclipse TE300 fluorescence microscope equipped with a SPOT digital camera (Diagnostic Instruments, Sterling Heights, MI). Images were captured using SPOT software (Diagnostic Instruments), and stored images were analyzed with Adobe Photoshop (Adobe Systems, San Jose, CA) software and adjusted using the autolevel and brightness functions.
Flow cytometry
CD34+ HPCs infected with KSHV or rKSHV.219 were cultured in IMDM supplemented with 10% FBS, penicillin, streptomycin, L-glutamine, and StemSpan CC100 cytokine cocktail (StemCell Technologies). Fifty-microliter aliquots were collected from cultures daily. Collected samples were centrifuged, washed once with PBS, and treated with 1:1 acetone/methanol for 20 minutes at -20°C. KSHV protein products were detected by labeling with 10 μg/mL of rat monoclonal antibody (mAb) to ORF-73 (LANA-1), rabbit polyclonal antibody to ORF K2 (vIL-6), and mouse mAbs to ORF 59 (PF-8) and ORF K8.1A (all from ABI, Columbia, MD) and 20 μg/mL of the following secondary antibodies: 7-amino-4-methylcoumarin-3-acetic acid (AMCA)-conjugated rabbit anti-rat immunoglobulin G (IgG) (Vector Laboratories, Burlingame, CA), Cy5-conjugated goat anti-rabbit IgG, and Cy5-conjugated goat anti-mouse IgG (Jackson ImmunoResearch, West Grove, PA). Samples recovered from murine spleen and bone marrow were incubated with allophycocyanin (APC)-conjugated mAb directed against human CD19, and PE-Cy7-conjugated mAb against human CD14 (all from BD Biosciences Pharmingen, San Diego, CA) according to manufacturer's specifications. Dead cells were excluded by staining with 7-amino-actinomycin D (7-AAD) (Calbiochem-Novabiochem, La Jolla, CA) staining (1 μL/mL). Cells were washed twice with 3 mL PBS and then resuspended in 1 mL PBS for analysis. Samples were run on a FACStarplus flow cytometer (Becton Dickinson). Data analysis was performed using WinMDI 2.8 software.
Indirect fluorescence antibody assays
IFAs were performed on KSHV-infected CD34+ HPCs and ex vivo-cultured cells recovered from rKSHV.219-infected NOD/SCID-hu mice. Cells recovered from NOD/SCID-hu mice were fixed onto siliconized glass slides by Cytospin (252 g, 5 minutes), and then slides with cells were immersed in 1:1 acetone/methanol for 20 minutes at -20°C. Viral protein products were detected by single-label IFA with a rat mAb to KSHV ORF-73 (LANA-1) (10 μg/mL) (ABI) and AMCA-labeled rabbit anti-rat IgG secondary antibody (20 μg/mL) (Vector Laboratories). Stained slides were viewed and photographed using a Nikon Eclipse TE300 fluorescence microscope with SPOT digital camera and software (Diagnostic Instruments). Images were analyzed with Adobe Photoshop (Adobe Systems) software and adjusted using the autolevel function. CD34+ HPCs infected with KSHV in vitro were fixed onto glass slides and treated with 1:1 acetone/methanol as described earlier in this paragraph and stained for IFA. KSHV protein products were detected by labeling with 10 μg/mL of rat mAb to ORF-73 (LANA-1), rabbit polyclonal antibody to ORF K2 (vIL-6), and mouse mAbs to ORF 59 (PF-8) and ORF K8.1A (all from ABI), and 20 μg/mL of the following secondary antibodies: tetramethyl rhodamine isothiocyanate (TRITC)-conjugated goat anti-rat immunoglobulin (IgG), fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit IgG, and FITC-conjugated goat anti-mouse IgG (all from Jackson ImmunoResearch). Cells were washed with PBS and stained with 5 μM TO-PRO-3 iodide nuclear stain (Molecular Probes, Eugene, OR). Stained cells were mounted using Vectashield mounting medium (Vector Laboratories) and stored at 4°C in darkness prior to analysis.
Confocal microscopy
Confocal micrographs were taken through the 100 × oil immersion PA objective of a Nikon Eclipse E600 microscope equipped with a MRC1024ES/60-WL red/green/far-red laser illumination system (Bio-Rad Laboratories). The 3 wavelengths were excited sequentially. Images were gathered and merged using LaserSharp2000 v5.2 software (Bio-Rad Laboratories). Image resolution was 1024 × 1024, and a 4-pass Kalman filter was applied to minimize digital noise.
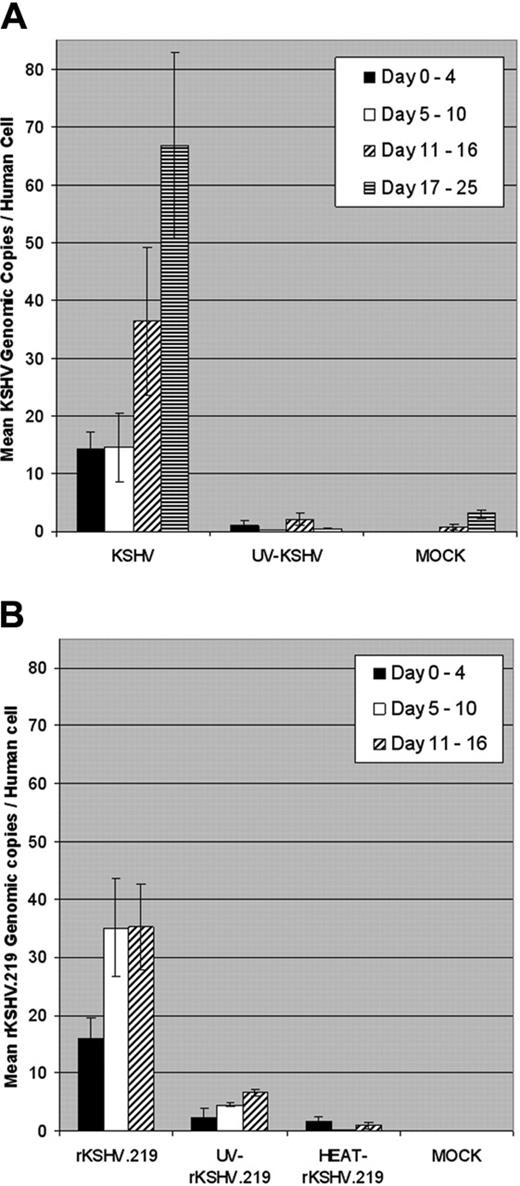
Quantitation of KSHV and rKSHV.219 infection of CD34+cells cultured in vitro. (A) CD34+ HPCs were infected with wild-type KSHV or UV-irradiated KSHV (UV, 10 μW/cm2 for 1 hour) (100 viral genome equivalents per cell) or were mock-infected. Infected CD34+ HPCs (5 × 106 cells) were cultivated in IMDM supplemented with 10% FBS at 37°C, 5% CO2 for up to 25 days. Cell aliquots were collected daily, and DNA was extracted and analyzed by RQ-PCR using KSHV KS330233 (ORF 26 minor capsid protein, 233 bp) and human β-globin (240 bp) primers. KSHV genomic copy number was normalized to 0.5 × β-globin copy number, representing one human cell. Bars represent mean KSHV genomic copies per human cell at indicated periods after infection, and error bars represent standard error of the mean (SEM). (B) CD34+ HPCs were infected with rKSHV.219, UV-irradiated rKSHV.219 (UV, 10 μW/cm2 for 1 hour), heat-inactivated rKSHV.219 (HEAT, 65°C for 1 hour) (600 viral genome equivalents per cell), or mock-infected. Infected CD34+ HPCs were cultivated, cell aliquots collected, and DNA was extracted and analyzed by RQ-PCR as in panel A. Infected CD34+ HPCs were cultivated for up to 16 days. Bars represent mean rKSHV.219 genomic copies per human cell at indicated time periods after infection, and error bars represent SEM.
Quantitation of KSHV and rKSHV.219 infection of CD34+cells cultured in vitro. (A) CD34+ HPCs were infected with wild-type KSHV or UV-irradiated KSHV (UV, 10 μW/cm2 for 1 hour) (100 viral genome equivalents per cell) or were mock-infected. Infected CD34+ HPCs (5 × 106 cells) were cultivated in IMDM supplemented with 10% FBS at 37°C, 5% CO2 for up to 25 days. Cell aliquots were collected daily, and DNA was extracted and analyzed by RQ-PCR using KSHV KS330233 (ORF 26 minor capsid protein, 233 bp) and human β-globin (240 bp) primers. KSHV genomic copy number was normalized to 0.5 × β-globin copy number, representing one human cell. Bars represent mean KSHV genomic copies per human cell at indicated periods after infection, and error bars represent standard error of the mean (SEM). (B) CD34+ HPCs were infected with rKSHV.219, UV-irradiated rKSHV.219 (UV, 10 μW/cm2 for 1 hour), heat-inactivated rKSHV.219 (HEAT, 65°C for 1 hour) (600 viral genome equivalents per cell), or mock-infected. Infected CD34+ HPCs were cultivated, cell aliquots collected, and DNA was extracted and analyzed by RQ-PCR as in panel A. Infected CD34+ HPCs were cultivated for up to 16 days. Bars represent mean rKSHV.219 genomic copies per human cell at indicated time periods after infection, and error bars represent SEM.
Real-time RT-PCR analysis of RNA extracted from rKSHV.219-infected murine samples
RNA was isolated from rKSHV.219-infected NOD/SCID-hu mouse spleen and bone marrow samples using TRIzol reagent (Invitrogen). Isolated RNA was subjected to real-time RT-PCR analysis for the presence of KSHV LANA-1 (ORF 73) and Rta (ORF 50) transcripts. Real-time RT-PCR assay used LANA-1 primers (5′-GAAGTGGATTACCCTGTTGTTAGC-3′,49 5′-TTGGATCTCGTCTTCCATCC-3′50 ), Rta primers (5′-CACAAAAATGGCGCAAGATGA-3′,51 5′-TGGTAGAGTTGGGCCTTCAGTT-3′51 ), and the QuantiTech SYBR Green RT-PCR kit (Qiagen). Cycle conditions on an iCycler (Bio-Rad Laboratories) were set as follows: 30 minutes at 50°C, 15 minutes at 95°C, 55 cycles of 15 seconds at 95°C, 15 seconds at 64°C, 30 seconds at 72°C, 15 seconds of detection at 80.5°C; 55°C to 95°C melting curve. Data were collected and analyzed using iCycler iQ Real-Time Detection System Software (Bio-Rad Laboratories).
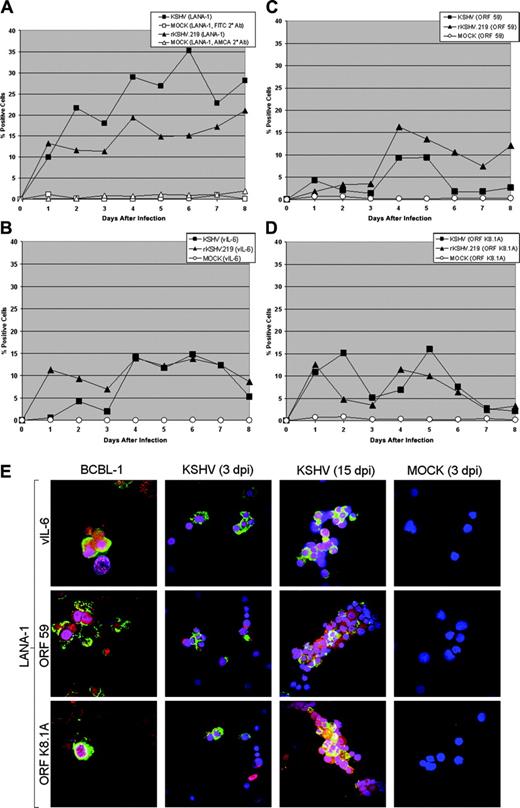
Latent and lytic viral gene expression in KSHV- and rKSHV.219-infected CD34+HPCs. (A) CD34+ HPCs infected with KSHV or rKSHV.219 and cells were permeabilized and incubated with primary rat monoclonal antibody (mAb) directed against KSHV ORF 73 (LANA-1) followed by incubation with either an FITC-labeled goat anti-rat IgG secondary antibody (KSHV-infected cells) or AMCA-labeled rabbit anti-rat IgG secondary antibody (rKSHV.219-infected cells). Samples were analyzed by flow cytometry for the presence of LANA-1, and cells were gated according to patterns demonstrated by staining with secondary antibody alone. (B-D) CD34+ HPCs infected with KSHV and rKSHV.219 were incubated with primary rabbit polyclonal antibodies directed against (B) ORF K2 (vIL-6), primary mouse mAbs directed against (C) ORF 59 (PF-8) and (D) ORF K8.1A of KSHV, followed by incubation with Cy5-conjugated goat anti-rabbit or goat anti-mouse IgG secondary antibody. Samples were analyzed by flow cytometry for the presence of vIL-6, ORF 59, and ORF K8.1A, and gated as described in panel A. (E) CD34+ HPCs infected with KSHV were fixed onto siliconized glass slides, permeabilized, and incubated with primary rat mAb directed against KSHV LANA-1, primary rabbit polyclonal antibodies directed against KSHV vIL-6, and primary mouse mAbs directed against KSHV ORF59 and ORF K8.1A. Cells were washed with PBS, followed by incubation with TRITC-conjugated goat anti-rat immunoglobulin (IgG), FITC-conjugated goat anti-rabbit IgG, or FITC-conjugated goat anti-mouse IgG. Cells were washed and counterstained with TO-PRO-3 iodide nuclear stain. KSHV LANA-1 staining is represented by red. KSHV vIL-6, ORF 59, and ORF K8.1A staining are represented by green. TO-PRO-3 nuclear staining is represented by blue. KSHV-infected CD34+ HPCs were analyzed at 3 and 15 days after infection (dpi) and are from the cultures presented in panels A-D. TPA-treated BCBL-1 cells and mock-infected CD34+ HPCs were similarly analyzed. Cells were visualized through a 100 ×1.40 NA oil-immersion Plan-Apochromat objective lens (Nikon, Tokyo, Japan); total magnification was × 1000.
Latent and lytic viral gene expression in KSHV- and rKSHV.219-infected CD34+HPCs. (A) CD34+ HPCs infected with KSHV or rKSHV.219 and cells were permeabilized and incubated with primary rat monoclonal antibody (mAb) directed against KSHV ORF 73 (LANA-1) followed by incubation with either an FITC-labeled goat anti-rat IgG secondary antibody (KSHV-infected cells) or AMCA-labeled rabbit anti-rat IgG secondary antibody (rKSHV.219-infected cells). Samples were analyzed by flow cytometry for the presence of LANA-1, and cells were gated according to patterns demonstrated by staining with secondary antibody alone. (B-D) CD34+ HPCs infected with KSHV and rKSHV.219 were incubated with primary rabbit polyclonal antibodies directed against (B) ORF K2 (vIL-6), primary mouse mAbs directed against (C) ORF 59 (PF-8) and (D) ORF K8.1A of KSHV, followed by incubation with Cy5-conjugated goat anti-rabbit or goat anti-mouse IgG secondary antibody. Samples were analyzed by flow cytometry for the presence of vIL-6, ORF 59, and ORF K8.1A, and gated as described in panel A. (E) CD34+ HPCs infected with KSHV were fixed onto siliconized glass slides, permeabilized, and incubated with primary rat mAb directed against KSHV LANA-1, primary rabbit polyclonal antibodies directed against KSHV vIL-6, and primary mouse mAbs directed against KSHV ORF59 and ORF K8.1A. Cells were washed with PBS, followed by incubation with TRITC-conjugated goat anti-rat immunoglobulin (IgG), FITC-conjugated goat anti-rabbit IgG, or FITC-conjugated goat anti-mouse IgG. Cells were washed and counterstained with TO-PRO-3 iodide nuclear stain. KSHV LANA-1 staining is represented by red. KSHV vIL-6, ORF 59, and ORF K8.1A staining are represented by green. TO-PRO-3 nuclear staining is represented by blue. KSHV-infected CD34+ HPCs were analyzed at 3 and 15 days after infection (dpi) and are from the cultures presented in panels A-D. TPA-treated BCBL-1 cells and mock-infected CD34+ HPCs were similarly analyzed. Cells were visualized through a 100 ×1.40 NA oil-immersion Plan-Apochromat objective lens (Nikon, Tokyo, Japan); total magnification was × 1000.
Statistical analysis
All statistical analyses were performed using single factor analysis of variance (ANOVA). A P value of less than .05 was considered significant.
Results
De novo infection of CD34+ hematopoietic progenitor cells by KSHV
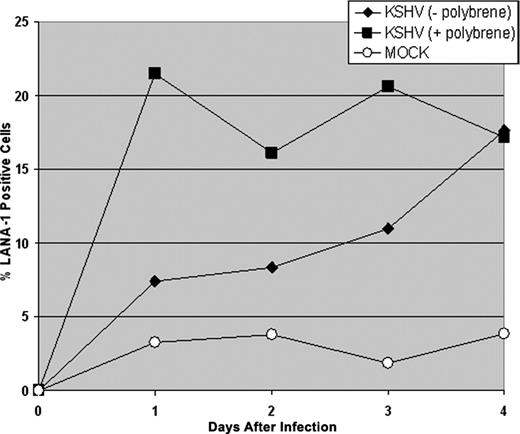
CD34+ HPCs are susceptible to infection by a number of viruses, including HIV-1,19,20 hepatitis C virus,21 JC virus,22 and lymphotropic herpesviruses such as HCMV,23-28 HHV-6,29,30 HHV-7,31 and HTLV-1.32 A report previously has shown that KSHV is detected in circulating CD34+ cells in Kaposi sarcoma patients and that KSHV is able to infect fetal mesenchymal stem cells in vitro.15,40 To investigate the susceptibility of CD34+ HPCs to KSHV infection, purified human CD34+ HPCs were exposed to KSHV in vitro. Infected CD34+ HPCs were cultured in vitro for up to 25 days, and DNA was isolated from aliquots of cells collected daily from cultures to determine the relative levels of KSHV infection and replication. Real-time quantitative PCR (RQ-PCR) analysis of DNA extracted from KSHV-infected CD34+ HPCs displayed consistently elevated levels of KSHV genomic copies in comparison to HPC cultures exposed to UV-irradiated KSHV or mock-infected HPCs (Figure 1A). At 17 to 25 days after infection, KSHV-infected CD34+ HPCs exhibited a 141-fold increase in KSHV genomic copies per human cell over UV-irradiated controls. KSHV-infected CD34+ HPCs also expressed KSHV latent antigen LANA-1 and lytic antigens vIL-6, ORF 59, and ORF K8.1A when analyzed by flow cytometry (Figure 2A-D) and by IFA and confocal microscopy (Figure 2E), suggesting concomitant latent and lytic viral gene expression. These results are similar to a recent report demonstrating concurrent lytic and latent KSHV gene expression initially following infection of primary endothelial and fibroblast cells.52,53 Polybrene enhances receptor-independent infection54 and can artificially expand the cellular host range of KSHV infection to include cells normally resistant to KSHV infection.55 To confirm that KSHV could naturally target and infect CD34+ HPCs, CD34+ cells were infected with KSHV in the absence of polybrene and analyzed for LANA-1 expression. Similar levels of LANA-1 were detected in CD34+ HPCs infected with KSHV in the absence of polybrene at 4 days after infection, suggesting that CD34+ cells are capable of sustaining infection by KSHV (Figure 3).
KSHV infection of CD34+HPCs in the absence of polybrene. CD34+ HPCs were infected with KSHV in the presence or absence of polybrene. CD34+ HPCs infected with KSHV were permeabilized and incubated with primary rat monoclonal antibody (mAb) directed against KSHV LANA-1 followed by incubation with AMCA-labeled rabbit anti-rat IgG secondary antibody. Samples were analyzed by flow cytometry for the presence of LANA-1, and cells were gated according to patterns demonstrated by staining with secondary antibody alone.
KSHV infection of CD34+HPCs in the absence of polybrene. CD34+ HPCs were infected with KSHV in the presence or absence of polybrene. CD34+ HPCs infected with KSHV were permeabilized and incubated with primary rat monoclonal antibody (mAb) directed against KSHV LANA-1 followed by incubation with AMCA-labeled rabbit anti-rat IgG secondary antibody. Samples were analyzed by flow cytometry for the presence of LANA-1, and cells were gated according to patterns demonstrated by staining with secondary antibody alone.
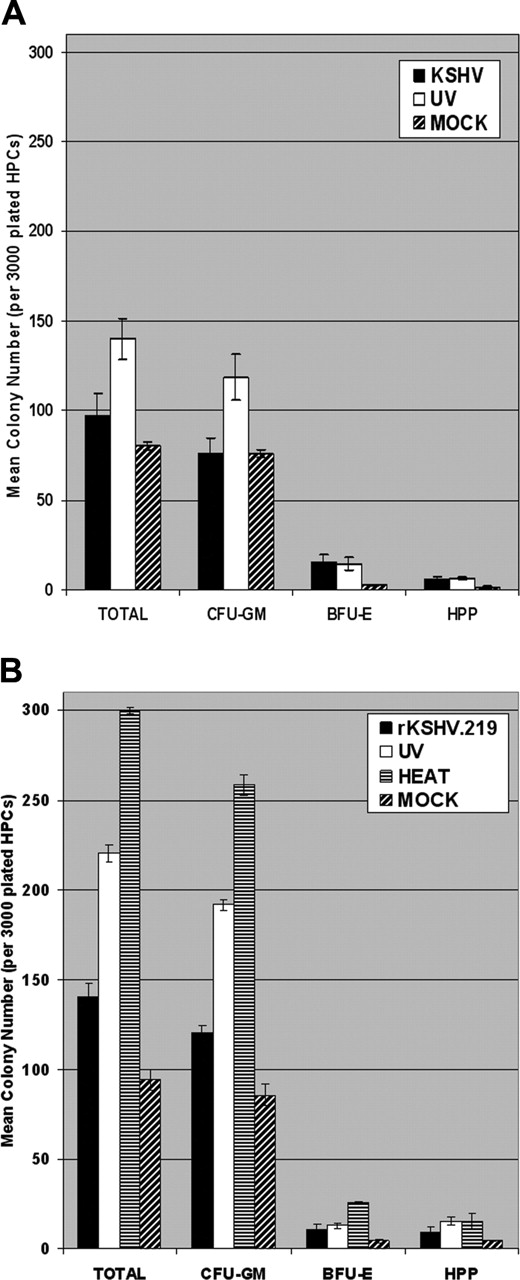
KSHV and rKSHV.219 infection suppresses clonogenic colony-forming potential of CD34+HPCs in vitro. (A) CD34+ HPCs were infected with KSHV or with UV-irradiated KSHV (10 μW/cm2 for 1 hour). Cells were washed 3 times with PBS and plated in 2 mL MethoCult H4433 medium (3000 cells per 35mm × 10mm plate) in triplicate at 4 hours after infection. Colonies were visually scored and enumerated at 12 to 15 days after plating. Experiments were repeated 3 times using purified CD34+ HPCs from different donor tissues. The mean CFU-GM, BFU-E, HPP, and total colony numbers are presented. Statistically significant differences in the mean total colony numbers between KSHV and UV-KSHV (P = .021) groups were determined by single-tail ANOVA analysis. (B) CD34+ HPCs were infected with rKSHV.219, UV-irradiated rKSHV.219 (10 μW/cm2 for 1 hour), or heat-inactivated rKSHV.219 (65°C for 1 hour). Infected HPCs were washed and plated in MethoCult H4433 as described in panel A. Colonies were visually scored at 12 to 15 days after plating and used to calculate mean CFU-GM, BFU-E, HPP, and total colony numbers. Statistically significant differences in the mean total colony numbers between the rKSHV.219, UV-rKSHV.219, and HEAT-rKSHV.219 (P = 2.31 × 10-6) groups were determined by single-tail ANOVA analysis. Experiments were repeated twice. Error bars represent standard error of the mean (SEM).
KSHV and rKSHV.219 infection suppresses clonogenic colony-forming potential of CD34+HPCs in vitro. (A) CD34+ HPCs were infected with KSHV or with UV-irradiated KSHV (10 μW/cm2 for 1 hour). Cells were washed 3 times with PBS and plated in 2 mL MethoCult H4433 medium (3000 cells per 35mm × 10mm plate) in triplicate at 4 hours after infection. Colonies were visually scored and enumerated at 12 to 15 days after plating. Experiments were repeated 3 times using purified CD34+ HPCs from different donor tissues. The mean CFU-GM, BFU-E, HPP, and total colony numbers are presented. Statistically significant differences in the mean total colony numbers between KSHV and UV-KSHV (P = .021) groups were determined by single-tail ANOVA analysis. (B) CD34+ HPCs were infected with rKSHV.219, UV-irradiated rKSHV.219 (10 μW/cm2 for 1 hour), or heat-inactivated rKSHV.219 (65°C for 1 hour). Infected HPCs were washed and plated in MethoCult H4433 as described in panel A. Colonies were visually scored at 12 to 15 days after plating and used to calculate mean CFU-GM, BFU-E, HPP, and total colony numbers. Statistically significant differences in the mean total colony numbers between the rKSHV.219, UV-rKSHV.219, and HEAT-rKSHV.219 (P = 2.31 × 10-6) groups were determined by single-tail ANOVA analysis. Experiments were repeated twice. Error bars represent standard error of the mean (SEM).
KSHV-infected CD34+ HPCs were plated in MethoCult H4433 semisolid medium to allow for the development of clonogenic hematopoietic colonies, including granulocyte/monocyte precursors (CFU-GM), erythroid precursors (BFU-E), and progenitor cells with high proliferative potential (HPP). KSHV-infected CD34+ HPCs displayed a marked suppression of hematopoiesis, as demonstrated by reduction in clonogenic CFAs in comparison with CD34+ cells infected with UV-irradiated KSHV (Figure 4A). KSHV infection of CD34+ HPCs resulted in a 31% reduction in clonogenic colony formation in comparison to UV-irradiated controls (Figure 4A). Interestingly, CFA was significantly elevated in UV-irradiated or heat-inactivated CD34+ cell cultures in comparison to mock-infected cultures, implying the presence of secreted viral cytokines or growth factors in virion-inactivated supernatant virus preps. Clonogenic hematopoietic colonies arising from KSHV-infected CD34+ HPCs were visually scored after 12 to 15 days. KSHV sequences were detected in 17% of CFU-GM and in 4% of BFU-E colonies by RQ-PCR analysis, but virus was not detected in HPP (Table 1). KSHV-infected CFU-GM and BFU-E demonstrated approximately 17 and 3.3 KSHV genomic copies per human cell, respectively, when analyzed by RQ-PCR (Table 1). These results demonstrate that KSHV can persistently infect human CD34+ HPCs, and viral infection results in suppression of hematopoiesis in vitro. Furthermore, KSHV infection is predominantly detected in CFU-GMs that differentiate from KSHV-infected CD34+ cells.
Detection of KSHV genomic DNA in clonogenic colonies derived from infected CD34+ HPCs
. | KSHV+ colonies/total colonies analyzed . | . | . | ||
|---|---|---|---|---|---|
. | CFU-GM . | BFU-E . | HPP . | ||
| KSHV | 17% (15 of 90)* | 4% (1 of 23)† | 0% (0 of 20) | ||
| UV-KSHV | 1% (1 of 67)‡ | 0% (0 of 12) | 0% (0 of 17) | ||
| Mock | 0% (0 of 25) | 0% (0 of 7) | 0% (0 of 8) | ||
. | KSHV+ colonies/total colonies analyzed . | . | . | ||
|---|---|---|---|---|---|
. | CFU-GM . | BFU-E . | HPP . | ||
| KSHV | 17% (15 of 90)* | 4% (1 of 23)† | 0% (0 of 20) | ||
| UV-KSHV | 1% (1 of 67)‡ | 0% (0 of 12) | 0% (0 of 17) | ||
| Mock | 0% (0 of 25) | 0% (0 of 7) | 0% (0 of 8) | ||
CD34+ HPCs were exposed to KSHV or UV-irradiated KSHV (10 μW/cm2 for 1 hour). Cells were washed 3 times with PBS and plated into MethoCult H4433. Colonies were visually scored, randomly isolated, and DNA was analyzed by real-time PCR analysis at 12 to 15 days after plating. Experiments were performed 3 times. Colonies with less than 1.0 calculated viral copy per cell were scored as negative for KSHV infection. Colonies demonstrating less than 2 human β-globin copies were excluded from analysis.
17 ± 5.6 mean KSHV genomic copies per human cell in positive CFU-GM colonies.
3.3 KSHV genomic copies per human cell in positive BFU-E colony.
2.7 KSHV genomic copies per human cell in positive CFU-GM colony.
rKSHV.219 infection of CD34+ hematopoietic progenitor cells in vitro
rKSHV.219 is a recombinant KSHV that expresses GFP and RFP from the latent elongation factor-1α promoter and lytic PAN promoter, respectively.42 To compare the characteristics of KSHV and rKSHV.219 infection, CD34+ HPCs were incubated with rKSHV.219 and cultured in vitro. Similar to KSHV, rKSHV.219-infected CD34+ HPCs displayed an elevation in viral genomic copies per human cell when compared to mock, UV-irradiated, or heat-treated control cultures (Figure 1B). rKSHV.219-infected HPCs also demonstrated detectable GFP expression at 15 days after infection when analyzed by flow cytometry in contrast to heattreated and mock-infected cultures, which did not show GFP expression (data not shown). Furthermore, rKSHV.219-infected CD34+ HPCs also showed concurrent expression of LANA-1, vIL-6, ORF 59, and ORF K8.1A when analyzed by flow cytometry (Figure 2A-D), similar to patterns seen in KSHV-infected CD34+ HPCs. Infection of CD34+ HPCs with rKSHV.219 resulted in a 36% and 53% reduction in total CFA, in comparison with CFA displayed by CD34+ HPCs infected by UV-irradiated or heat-inactivated virus, respectively (Figure 4B). RQ-PCR analysis demonstrated that rKSHV.219 infection was predominantly localized in CFU-GMs, similar to what was displayed with wild-type KSHV infection of CD34+ HPCs (Table 2). A limited pattern of rKSHV.219 infection also was detected in HPP, which represent progenitor cells with pluripotent potential. When analyzed by RQ-PCR, rKSHV.219-infected CFU-GMs displayed 5.3 rKSHV.219 genomic copies per human cell, and infected HPP showed 1.7 viral copies per human cell (Table 2). rKSHV.219 sequences were not detected in BFU-E colonies. Clonogenic colonies derived from rKSHV.219-infected CD34+ cells demonstrated expression of GFP and RFP at 28 days after plating when analyzed by fluorescence microscopy (data not shown). Although rKSHV.219 virus genomic copy load was significantly lower in CFU-GMs compared to KSHV infection, these data demonstrate that rKSHV.219 shows an infection and persistence pattern in CD34+ HPCs that is similar to wild-type KSHV.
Detection of rKSHV.219 DNA in clonogenic colonies derived from infected CD34+ HPCs
. | rKSHV.219+ colonies/total colonies analyzed . | . | . | ||
|---|---|---|---|---|---|
. | CFU-GM . | BFU-E . | HPP . | ||
| rKSHV.219 | 21% (12 of 56)* | 0% (0 of 9) | 40% (4 of 10)† | ||
| UV-rKSHV.219 | 0% (0 of 33) | 0% (0 of 10) | 0% (0 of 7) | ||
| Heat-treated rKSHV.219 | 0% (0 of 35) | 0% (0 of 7) | 0% (0 of 7) | ||
| Mock | 0% (0 of 36) | 0% (0 of 3) | 0% (0 of 2) | ||
. | rKSHV.219+ colonies/total colonies analyzed . | . | . | ||
|---|---|---|---|---|---|
. | CFU-GM . | BFU-E . | HPP . | ||
| rKSHV.219 | 21% (12 of 56)* | 0% (0 of 9) | 40% (4 of 10)† | ||
| UV-rKSHV.219 | 0% (0 of 33) | 0% (0 of 10) | 0% (0 of 7) | ||
| Heat-treated rKSHV.219 | 0% (0 of 35) | 0% (0 of 7) | 0% (0 of 7) | ||
| Mock | 0% (0 of 36) | 0% (0 of 3) | 0% (0 of 2) | ||
CD34+ HPCs were exposed to rKSHV.219, UV-irradiated rKSHV.219 (10 μW/cm2 for 1 hour), or heat-inactivated rKSHV.219 (65°C for 1 hour), and plated into MethoCult H4433. Colonies were visually scored, randomly isolated, and DNA was analyzed by real-time PCR analysis at 12 to 15 days after plating. Experiments were performed twice. Colonies with less than 1.0 calculated viral copy per cell were scored as negative for KSHV infection. Colonies demonstrating less than 2 human β-globin copies were excluded from analysis.
5.3 ± 1.2 mean rKSHV.219 genomic copies per human cell in positive CFU-GM colonies.
1.7 ± 0.4 mean rKSHV.219 genomic copies per human cell in positive HPP colonies.
Differentiation of rKSHV.219-infected CD34+ HPCs in vivo
Human hematopoiesis can be reconstituted in NOD/SCID mice following intravenous inoculation of human CD34+ HPCs. Murine bone marrow has been shown to support the development of human monocytes, B lymphocytes, T lymphocytes, erythrocytes, and megakaryocytes,56,57 and significant B lymphopoiesis is detected in the spleen.56 To evaluate the ability of rKSHV.219-infected CD34+ HPCs to reconstitute hematopoiesis in vivo, NOD/SCID mice were injected intravenously with rKSHV.219-infected CD34+ HPCs. Cell samples were recovered from spleen, bone marrow, and peripheral blood 3 to 29 weeks after inoculation, and extracted DNA was analyzed by RQ-PCR for KSHV genomic sequences. rKSHV.219-infected cells were detected by real-time PCR in the spleen, bone marrow, and peripheral blood of NOD/SCID-hu mice at 3 to 29 weeks after reconstitution (Table 3). GFP expression was detected in cells recovered from the spleen and bone marrow of 10 mice inoculated with rKSHV.219-infected CD34+ HPCs, when analyzed by flow cytometry. Subpopulations of human CD14+/GFP+ cells were detected in the bone marrow and spleen samples of mice inoculated with rKSHV.219-infected CD34+ HPCs. Human CD19+/GFP+ cells, representing rKSHV.219-infected B cells, were identified in the spleen and bone marrow samples recovered from 9 mice inoculated with rKSHV.219-infected HPCs. All rKSHV.219-infected NOD/SCID-hu mice also showed detectable expression of LANA-1 (ORF 73) and Rta (ORF 50), when RNA from spleen and bone marrow was analyzed by real-time RT-PCR (Table 4). The bone marrow cells from mouse D6 also displayed LANA-1 expression when analyzed by IFA analysis (Figure 5). Three mice infected with rKSHV.219 developed pleural effusions at 14 to 23 weeks after inoculation (mouse D17, D19, and D21). The majority of cells recovered from the pleural cavity of these mice were phenotyped as murine B or pro-B lymphocytes (mu-CD45+/mu-CD19+), although rKSHV.219-infected human monocyte (hu-CD14+/GFP+) cells were present at detectable levels (∼2% to 4% of cells) in each effusion sample. None of the control mice inoculated with heat-treated rKSHV.219- or mock-infected CD34+ HPCs developed this condition. Notably, the murine CD19+ cells from each of these pleural effusions were immortalized for growth in vitro. Intraperitoneal inoculation of unpurified cells from pleural effusions into naive NOD/SCID mice resulted in rapid development of neoplasms 2 to 4 weeks after inoculation (data not shown). These data suggest that rKSHV.219 infection may be actively disseminated during differentiation of infected CD34+ HPCs into human monocytes and B cells in the NOD/SCID-hu mouse bone marrow and spleen, respectively. Importantly, these results demonstrate that rKSHV.219 can establish persistent infection in NOD/SCID-hu mice following intravenous inoculation of infected CD34+ HPCs.
rKSHV.219 infection in NOD/SCID-hu mice
. | . | . | KSHV PCRd . | . | . | CD14+/GFP+e, % . | . | CD19+/GFP+f, % . | . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Groupa, mouse ID . | No. CD34+ cellsb, × 107 . | Weeksc . | Spleen . | Bone marrow . | Peripheral blood . | Spleen . | Bone marrow . | Spleen . | Bone marrow . | ||||
| MOCK | |||||||||||||
| D8g | 0.1 | 8 | − | − | − | 0 | 0 | 0 | 0 | ||||
| D9g | 0.1 | 20 | − | − | − | 0 | 0 | 0 | 0 | ||||
| D10g | 0.5 | 8 | − | − | − | 0 | 0 | 0 | 0 | ||||
| D11gh | 0.5 | 7 | nd | nd | − | nd | nd | nd | nd | ||||
| D51g | 0.5 | 17 | − | − | − | 0 | 0 | 0 | 0 | ||||
| D52g | 0.5 | 3 | − | − | nd | 0 | 0 | 0 | 0 | ||||
| D55g | 0.5 | 17 | − | − | − | 0 | 0 | 0 | 0 | ||||
| D57g | 0.5 | 4 | − | − | nd | 0 | 0 | 0 | 0 | ||||
| D59g | 0.5 | 5 | − | − | nd | 0 | 0 | 0 | 0 | ||||
| D13g | 1.0 | 12 | − | − | − | 0 | 0 | 0 | 0 | ||||
| D1 | 2.0 | 8 | − | − | nd | 0 | 0 | 0 | 0 | ||||
| D2 | 2.0 | 10 | − | − | nd | 0 | 0 | 0 | 0 | ||||
| HEAT | |||||||||||||
| D3 | 2.0 | 8 | − | − | nd | 0 | 0 | 0 | 0 | ||||
| D4 | 2.0 | 10 | − | − | nd | 0 | 0 | 0 | 0 | ||||
| rKSHV.219 | |||||||||||||
| D14gi | 0.1 | 3 | +++ | +++ | nd | nd | nd | nd | nd | ||||
| D15g | 0.1 | 8 | + | + | + | 0 | 0 | 0 | 0 | ||||
| D16g | 0.1 | 29 | + | + | + | 3.6 | 0.6 | 2.5 | 0.6 | ||||
| D17gj | 0.1 | 19 | ++ | +++ | +++ | 0.5 | 0.9 | 0.3 | 0.9 | ||||
| D18gk | 0.5 | 8 | +++ | ++ | + | 3.8 | 0.8 | 0 | 0 | ||||
| D19gj | 0.5 | 14 | ++ | ++ | + | 1.4 | 0.3 | 0 | 0 | ||||
| D20g | 0.5 | 14 | − | − | − | 0 | 0 | 0 | 0 | ||||
| D21gj | 0.5 | 23 | + | + | + | 2.7 | 2.7 | 2.3 | 2.7 | ||||
| D67g | 0.5 | 17 | + | + | + | 8.9 | 3.4 | 2.8 | 1.2 | ||||
| D73g | 0.5 | 17 | + | + | + | 9.7 | 1.7 | 2.7 | 0.5 | ||||
| D22g | 1.0 | 8 | + | ++ | + | 0 | 0 | 0 | 0 | ||||
| D23g | 1.0 | 14 | + | + | ++ | 0 | 0 | 0 | 0 | ||||
| D25g | 1.0 | 13 | − | − | − | 0 | 0 | 0 | 0 | ||||
| D5 | 2.0 | 8 | ++ | ++ | nd | 2.7 | 4.3 | 4.5 | 6.4 | ||||
| D6 | 2.0 | 10 | ++ | +++ | nd | 4.5 | 2.6 | 0.5 | 2.0 | ||||
| D7 | 2.0 | 10 | +++ | ++ | nd | 5.9 | 0 | 0.4 | 0 | ||||
. | . | . | KSHV PCRd . | . | . | CD14+/GFP+e, % . | . | CD19+/GFP+f, % . | . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Groupa, mouse ID . | No. CD34+ cellsb, × 107 . | Weeksc . | Spleen . | Bone marrow . | Peripheral blood . | Spleen . | Bone marrow . | Spleen . | Bone marrow . | ||||
| MOCK | |||||||||||||
| D8g | 0.1 | 8 | − | − | − | 0 | 0 | 0 | 0 | ||||
| D9g | 0.1 | 20 | − | − | − | 0 | 0 | 0 | 0 | ||||
| D10g | 0.5 | 8 | − | − | − | 0 | 0 | 0 | 0 | ||||
| D11gh | 0.5 | 7 | nd | nd | − | nd | nd | nd | nd | ||||
| D51g | 0.5 | 17 | − | − | − | 0 | 0 | 0 | 0 | ||||
| D52g | 0.5 | 3 | − | − | nd | 0 | 0 | 0 | 0 | ||||
| D55g | 0.5 | 17 | − | − | − | 0 | 0 | 0 | 0 | ||||
| D57g | 0.5 | 4 | − | − | nd | 0 | 0 | 0 | 0 | ||||
| D59g | 0.5 | 5 | − | − | nd | 0 | 0 | 0 | 0 | ||||
| D13g | 1.0 | 12 | − | − | − | 0 | 0 | 0 | 0 | ||||
| D1 | 2.0 | 8 | − | − | nd | 0 | 0 | 0 | 0 | ||||
| D2 | 2.0 | 10 | − | − | nd | 0 | 0 | 0 | 0 | ||||
| HEAT | |||||||||||||
| D3 | 2.0 | 8 | − | − | nd | 0 | 0 | 0 | 0 | ||||
| D4 | 2.0 | 10 | − | − | nd | 0 | 0 | 0 | 0 | ||||
| rKSHV.219 | |||||||||||||
| D14gi | 0.1 | 3 | +++ | +++ | nd | nd | nd | nd | nd | ||||
| D15g | 0.1 | 8 | + | + | + | 0 | 0 | 0 | 0 | ||||
| D16g | 0.1 | 29 | + | + | + | 3.6 | 0.6 | 2.5 | 0.6 | ||||
| D17gj | 0.1 | 19 | ++ | +++ | +++ | 0.5 | 0.9 | 0.3 | 0.9 | ||||
| D18gk | 0.5 | 8 | +++ | ++ | + | 3.8 | 0.8 | 0 | 0 | ||||
| D19gj | 0.5 | 14 | ++ | ++ | + | 1.4 | 0.3 | 0 | 0 | ||||
| D20g | 0.5 | 14 | − | − | − | 0 | 0 | 0 | 0 | ||||
| D21gj | 0.5 | 23 | + | + | + | 2.7 | 2.7 | 2.3 | 2.7 | ||||
| D67g | 0.5 | 17 | + | + | + | 8.9 | 3.4 | 2.8 | 1.2 | ||||
| D73g | 0.5 | 17 | + | + | + | 9.7 | 1.7 | 2.7 | 0.5 | ||||
| D22g | 1.0 | 8 | + | ++ | + | 0 | 0 | 0 | 0 | ||||
| D23g | 1.0 | 14 | + | + | ++ | 0 | 0 | 0 | 0 | ||||
| D25g | 1.0 | 13 | − | − | − | 0 | 0 | 0 | 0 | ||||
| D5 | 2.0 | 8 | ++ | ++ | nd | 2.7 | 4.3 | 4.5 | 6.4 | ||||
| D6 | 2.0 | 10 | ++ | +++ | nd | 4.5 | 2.6 | 0.5 | 2.0 | ||||
| D7 | 2.0 | 10 | +++ | ++ | nd | 5.9 | 0 | 0.4 | 0 | ||||
Real-time PCR and flow cytometric analysis of spleen, bone marrow, and peripheral blood samples recovered from NOD/SCID mice intravenously inoculated with CD34+ HPCs infected with rKSHV.219.
nd indicates not done.
CD34+ HPCs were infected with rKSHV.219, heat-treated rKSHV.219 (HEAT), or mock-infected (MOCK).
Number of CD34+ HPCs inoculated intravenously via tail vein injection.
Weeks after inoculation at which mouse was killed.
Real-time PCR analysis of DNA extracted from indicated tissues using primers specific for KSHV (KS330233). KSHV PCR signal was standardized to the number of human cells detected as determined by human β-globin gene signal. Calculated KSHV genome equivalents per human cell: −, (0); +, (<0.1); ++, (0.1-1.0); +++, (>1.0).
Flow cytometric analysis of cells recovered from indicated tissues using antibodies directed against human CD14. Numbers indicate percentage of gated cells that co-express human CD14 and GFP.
Flow cytometric analysis of cells recovered from indicated tissues using antibodies directed against human CD19. Numbers indicate percentage of gated cells that co-express human CD19 and GFP.
Mouse sublethally irradiated (300 rad) 2 days prior to tail vein inoculation.
Peripheral blood sample collected prior to death. Tissues not available due to decomposition.
Sufficient spleen and bone marrow samples for PCR analysis only.
Pleural effusion. Fluid (400-1000 μl) was recovered from pleural cavity of mouse and contained transformed murine B cells.
Ocular inoculation of rKSHV.219-infected CD34+ HPCs.
KSHV latent and lytic gene expression in rKSHV.219-infected NOD/SCID-hu mice
. | LANA-1 (ORF 73)† . | . | . | . | RTA (ORF 50)‡ . | . | . | . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Group*mouse ID . | Spleen . | . | Bone marrow . | . | Spleen . | . | Bone marrow . | . | ||||||
| . | Exp 1 . | Exp 2 . | Exp 1 . | Exp 2 . | Exp 1 . | Exp 2 . | Exp 1 . | Exp 2 . | ||||||
| MOCK | ||||||||||||||
| D1 | − | − | − | − | − | − | − | − | ||||||
| D2 | − | − | − | − | − | − | − | − | ||||||
| D8 | − | − | − | − | − | − | − | − | ||||||
| D9 | − | − | − | − | − | − | − | − | ||||||
| D10 | − | − | − | − | − | − | − | − | ||||||
| D11§ | nd | nd | nd | nd | nd | nd | nd | nd | ||||||
| D13 | − | − | − | − | − | − | − | − | ||||||
| D51 | − | − | − | − | − | − | − | − | ||||||
| D52 | − | − | − | − | − | − | − | − | ||||||
| D55 | − | − | − | − | − | − | − | − | ||||||
| D57 | − | − | − | − | − | − | − | − | ||||||
| D59 | − | − | − | − | − | − | − | − | ||||||
| HEAT | ||||||||||||||
| D3 | − | − | − | − | − | − | − | − | ||||||
| D4 | − | − | − | − | − | − | − | − | ||||||
| rKSHV.219 | ||||||||||||||
| D5 | + | − | + | − | − | + | − | + | ||||||
| D6 | − | + | − | + | − | + | − | + | ||||||
| D7 | − | + | − | + | − | + | − | + | ||||||
| D14 | − | + | − | + | − | + | − | + | ||||||
| D15 | − | + | − | + | − | + | − | + | ||||||
| D16 | + | − | − | + | + | − | − | + | ||||||
| D17 | + | − | + | − | + | − | + | + | ||||||
| D18 | − | + | + | + | − | + | + | + | ||||||
| D19 | + | + | + | + | + | + | + | + | ||||||
| D20 | − | − | − | − | − | − | − | − | ||||||
| D21 | + | + | + | − | + | + | + | + | ||||||
| D22 | − | + | − | + | − | + | − | + | ||||||
| D23 | − | + | + | + | + | − | + | − | ||||||
| D25 | − | − | − | − | − | − | − | − | ||||||
| D67 | + | − | − | + | + | + | + | − | ||||||
| D73 | + | − | − | + | − | + | + | − | ||||||
. | LANA-1 (ORF 73)† . | . | . | . | RTA (ORF 50)‡ . | . | . | . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Group*mouse ID . | Spleen . | . | Bone marrow . | . | Spleen . | . | Bone marrow . | . | ||||||
| . | Exp 1 . | Exp 2 . | Exp 1 . | Exp 2 . | Exp 1 . | Exp 2 . | Exp 1 . | Exp 2 . | ||||||
| MOCK | ||||||||||||||
| D1 | − | − | − | − | − | − | − | − | ||||||
| D2 | − | − | − | − | − | − | − | − | ||||||
| D8 | − | − | − | − | − | − | − | − | ||||||
| D9 | − | − | − | − | − | − | − | − | ||||||
| D10 | − | − | − | − | − | − | − | − | ||||||
| D11§ | nd | nd | nd | nd | nd | nd | nd | nd | ||||||
| D13 | − | − | − | − | − | − | − | − | ||||||
| D51 | − | − | − | − | − | − | − | − | ||||||
| D52 | − | − | − | − | − | − | − | − | ||||||
| D55 | − | − | − | − | − | − | − | − | ||||||
| D57 | − | − | − | − | − | − | − | − | ||||||
| D59 | − | − | − | − | − | − | − | − | ||||||
| HEAT | ||||||||||||||
| D3 | − | − | − | − | − | − | − | − | ||||||
| D4 | − | − | − | − | − | − | − | − | ||||||
| rKSHV.219 | ||||||||||||||
| D5 | + | − | + | − | − | + | − | + | ||||||
| D6 | − | + | − | + | − | + | − | + | ||||||
| D7 | − | + | − | + | − | + | − | + | ||||||
| D14 | − | + | − | + | − | + | − | + | ||||||
| D15 | − | + | − | + | − | + | − | + | ||||||
| D16 | + | − | − | + | + | − | − | + | ||||||
| D17 | + | − | + | − | + | − | + | + | ||||||
| D18 | − | + | + | + | − | + | + | + | ||||||
| D19 | + | + | + | + | + | + | + | + | ||||||
| D20 | − | − | − | − | − | − | − | − | ||||||
| D21 | + | + | + | − | + | + | + | + | ||||||
| D22 | − | + | − | + | − | + | − | + | ||||||
| D23 | − | + | + | + | + | − | + | − | ||||||
| D25 | − | − | − | − | − | − | − | − | ||||||
| D67 | + | − | − | + | + | + | + | − | ||||||
| D73 | + | − | − | + | − | + | + | − | ||||||
Real-time RT-PCR analysis of spleen and bone marrow samples recovered from NOD/SCID mice intravenously inoculated with rKSHV.219-infected CD34+ HPCs. + indicates transcript detected; −, transcript not detected; nd, not done; and exp, experiment.
CD34+ HPCs were infected with rKSHV.219, heat-treated rKSHV.219 (HEAT), or mock-infected (MOCK).
Real-time RT-PCR analysis of RNA extracted from indicated tissues using primers specific for KSHV LANA-1 (ORF 73).
Real-time RT-PCR analysis of RNA extracted from indicated tissues using primers specific for KSHV Rta (ORF 50).
Spleen and bone marrow samples not available for analysis.
Discussion
The characterization of cell types that harbor KSHV in vivo is crucial for establishing the pathogenesis of KSHV-associated disease. Our results show that KSHV and rKSHV.219 are capable of infecting CD34+ HPCs and that viral infection persists during differentiation of HPCs in vitro and in vivo. Previous studies have found evidence of KSHV in a number of cell types in tissue culture, including human B cells, endothelial cells, epithelial cells, fibroblast cells, and mesenchymal stem cells.4-6,11-15 CD34+ HPCs, including circulating CD34+ cells from KS patients, previously have been reported to harbor KSHV infection.40,58 Clinical investigators have demonstrated the occurrence of bone marrow failure and peripheral blood cytopenia as a result of primary infection and reactivation of KSHV.59,60 These reports suggest that CD34+ HPCs may be target cells for the establishment of KSHV infection in vivo. Our laboratory has previously shown that HTLV-1 infects CD34+ cells and that proviral sequences are maintained during differentiation.32 HCMV recently has been shown to infect human CD34+ cells and to establish a latent infection in primitive subsets of HPCs (ie, CD34+/CD38- cells).23,61 We speculate that CD34+ HPCs may serve as a reservoir for latent KSHV infection and that these cells may provide a renewable source of newly infected cells, disseminating virus into the B-cell and monocyte/macrophage lineages in vivo.
Previous studies have reported suppression of hematopoiesis following infection with HCMV,23,26,33-36 HHV-6,29,30 HIV-1,19,37,38 and measles virus.39 A recent report indicated that KSHV infection of bone marrow mononuclear cells has a suppressive effect on in vitro colony formation,62 similar to our observations. Bone marrow aplasia, pancytopenia, and marrow failure with plasmacytosis associated with KSHV infection and reactivation have been reported following autologous stem cell transplantation60 and renal transplantation.59,63 Reactivation of KSHV infection in a non-Hodgkin lymphoma patient undergoing autologous peripheral stem cell transplantation resulted in fever and bone marrow aplasia with plasmacytosis.59 Suppression of hematopoiesis in CFA as a result of KSHV infection of HPCs may reflect documented cases of bone marrow failure resulting from KSHV primary infection and reactivation in immunosuppressed patients.59,60,63
The predominant localization of KSHV and rKSHV.219 infection in CFU-GM in vitro and in CD14+ and CD19+ cells from NOD/SCID-hu mice suggests that viral infection may be preferentially maintained in progenitor cells that differentiate into these hematopoietic lineages. We speculate that KSHV infection of HPCs also may have a deleterious effect on the development of BFU-E. The inability to detect KSHV genomic DNA in BFU-E presumably accounts for the suppressive effect on clonogenic colony formation by KSHV- and rKSHV.219-infected HPCs in vitro and could suggest that KSHV infection is detrimental for the development of erythroid progenitor cells. Indeed, detection of lytic viral gene expression following infection of CD34+ HPCs suggests that viral replication may induce cytolysis in a subset of CD34+ cells. Although KSHV infection and persistence in CD34+ HPCs is predominantly latent, the concurrent latent and lytic gene expression demonstrated by KSHV and rKSHV.219-infected CD34+ HPCs in tissue culture, and in infected cells from rKSHV.219-infected NOD/SCID-hu mouse bone marrow and spleen, is intriguing. These observations are consistent with recent reports demonstrating the concurrent expression of KSHV lytic and latent genes in primary endothelial and fibroblast cells52 and in epithelial cells differentiated from rKSHV.219-infected keratinocytes.53 It is possible that RFP detection in rKSHV.219-infected cells is the result of a low basal level of gene expression from the PAN promoter. The PAN promoter is directly activated by Rta64 and is indicative of early lytic KSHV gene expression. A recent report of rKSHV.219 infection of epithelial raft cultures demonstrates that not all cells expressing RFP progress to late gene expression and that lytic gene expression occurs in differentiated epithelial cells.53 Similarly, differentiation and maturation of KSHV-infected CD34+ HPCs may result in the induction of lytic viral gene expression. Notably, primary KSHV infection and lytic viral reactivation have been documented in posttransplantation individuals.59,60,63 Our observation of latent and lytic co-expression by rKSHV.219 in the NOD/SCID-hu mouse suggests the possibility of concomitant viral latency and lytic reactivation in an in vivo immunosuppressive setting.
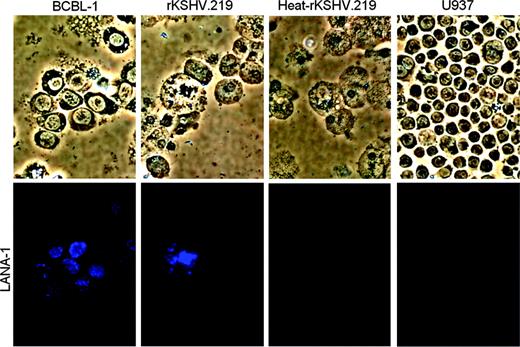
Viral gene expression in spleen and bone marrow cells recovered from NOD/SCID-hu mice reconstituted with rKSHV.219-infected CD34+HPCs. Bone marrow cells recovered from rKSHV.219 mouse D6 were cultured ex vivo for 33 days in complete IMDM. Bone marrow cells were analyzed by IFA using a rat anti-LANA-1 primary antibody and an AMCA antirat secondary antibody. Bone marrow cells recovered from a NOD/SCID mouse inoculated with heat-inactivated rKSHV.219-infected HPCs also were analyzed. BCBL-1, a KSHV-positive primary effusion lymphoma cell line, was used as a positive control for LANA-1 IFA. U937, a myeloid leukemia cell line negative for KSHV infection, was used as a negative control. Cells were visualized under a 20 ×.0.45 Plan-Fluor extra-long working distance (ELWD) dark medium (DM) objective lens (Nikon, Tokyo, Japan); total magnification, ×/200.
Viral gene expression in spleen and bone marrow cells recovered from NOD/SCID-hu mice reconstituted with rKSHV.219-infected CD34+HPCs. Bone marrow cells recovered from rKSHV.219 mouse D6 were cultured ex vivo for 33 days in complete IMDM. Bone marrow cells were analyzed by IFA using a rat anti-LANA-1 primary antibody and an AMCA antirat secondary antibody. Bone marrow cells recovered from a NOD/SCID mouse inoculated with heat-inactivated rKSHV.219-infected HPCs also were analyzed. BCBL-1, a KSHV-positive primary effusion lymphoma cell line, was used as a positive control for LANA-1 IFA. U937, a myeloid leukemia cell line negative for KSHV infection, was used as a negative control. Cells were visualized under a 20 ×.0.45 Plan-Fluor extra-long working distance (ELWD) dark medium (DM) objective lens (Nikon, Tokyo, Japan); total magnification, ×/200.
The presence of GFP+ lymphocytes in the spleens of NOD/SCID-hu mice demonstrates that rKSHV.219 infection and gene expression is maintained in human B and pre-B cells. Notably, Dittmer et al previously demonstrated KSHV infection of the SCID-hu Thy/Liv mouse involved a biphasic infection with early lytic replication accompanied by sustained latency.65 KSHV was primarily detected in CD19+ B lymphocytes in this model, reflecting the natural tropism of the virus. Additional experiments will be required to fully characterize infected cell lineages and the role of KSHV infection on human B lymphopoiesis in vivo. The detection of CD14+/GFP+ cells in the bone marrow and spleens of NOD/SCID-hu mice inoculated with rKSHV.219-infected CD34+ HPCs also suggests that monocyte/macrophages may participate in viral maintenance and dissemination in vivo. Monocyte-macrophages previously have been proposed as cellular reservoirs for KSHV,7 and these cell types presumably play an important role in the development of Kaposi sarcoma.7,8,66-68 It is feasible that infected monocytes transport KSHV to tissues such as the skin, spread viral infection to neighboring cells, and differentiate into latently infected spindlelike endothelial macrophages found in KS lesions.8,69-72 Finally, it will be important to further characterize the extent and nature of pleural effusions that arise with a low frequency in rKSHV.219-infected NOD/SCID-hu mice and determine if infection with KSHV also induces a similar phenotype. The consistent detection of rKSHV.219-infected human monocyte/macrophages (CD14+/GFP+) in pleural effusions allows speculation that virally infected monocytes/macrophages may contribute to lymphomagenesis. Notably, injection of PEL cells or PEL-associated macrophages previously has been reported to induce murine lymphomas in SCID mice,73 and it has been hypothesized that KSHV-infected human macrophages are central in a novel model of initiation and progression of oncogenesis termed “sequential neoplasia.”73,74 This suggests that a similar mechanism of virally induced lymphomagenesis may be involved in the induction of malignant murine B cells in rKSHV.219-infected NOD/SCID-hu mice. rKSHV.219 infection of NOD/SCID-hu mice clearly provides a compelling animal model to study KSHV infection, dissemination, and pathogenesis in an in vivo model.
Prepublished online as Blood First Edition Paper, March 16, 2006; DOI 10.1182/blood-2005-04-1697.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Vincent Lau and Joseph Domachowske for providing guidance and equipment for IFA, Rebecca Greenblatt for help with confocal microscopy, and Charles and Ying Hwang for assistance with fluorescence microscopy.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal