Abstract
The Smad-signaling pathway downstream of the transforming growth factor–β superfamily of ligands is an evolutionarily conserved signaling circuitry with critical functions in a wide variety of biologic processes. To investigate the role of this pathway in the regulation of hematopoietic stem cells (HSCs), we have blocked Smad signaling by retroviral gene transfer of the inhibitory Smad7 to murine HSCs. We report here that the self-renewal capacity of HSCs is promoted in vivo upon blocking of the entire Smad pathway, as shown by both primary and secondary bone marrow (BM) transplantations. Importantly, HSCs overexpressing Smad7 have an unperturbed differentiation capacity as evidenced by normal contribution to both lymphoid and myeloid cell lineages, suggesting that the Smad pathway regulates self-renewal independently of differentiation. Moreover, phosphorylation of Smads was inhibited in response to ligand stimulation in BM cells, thus verifying impairment of the Smad-signaling cascade in Smad7-overexpressing cells. Taken together, these data reveal an important and previously unappreciated role for the Smad-signaling pathway in the regulation of self-renewal of HSCs in vivo.
Introduction
Hematopoiesis, the process of blood-cell formation, is sustained throughout life by a rare population of multipotent hematopoietic stem cells (HSCs), which reside in the bone marrow (BM) of adult vertebrates.1,2 HSCs have the capacity to both self-renew, thereby maintaining stem-cell numbers intact over many years, and to give rise to differentiated progeny of all mature blood-cell lineages.3 HSC fate decisions such as self-renewal and differentiation are kept under strict control by a combination of cell-intrinsic and external factors present in the HSC niche.4 However, the precise molecular mechanisms that govern HSC fate options are poorly defined.
The transforming growth factor–β (TGF-β) superfamily of ligands, including the TGF-βs, activins, and bone morphogenetic proteins (BMPs), represents an important group of growth factors that have been shown to regulate a wide array of processes including apoptosis, cell proliferation, and differentiation.5-8 TGF-β family members bind and signal through 2 types of serine/threonine kinase receptors, type I and type II, both of which are necessary for signal transduction. Upon ligand binding and receptor activation, the intracellular mediators of TGF-β superfamily signaling, the Smad proteins, are activated through phosphorylation by type I receptors.5,6 Three groups of Smads have been identified: receptor-activated Smads (R-Smads), common-partner Smads (Co-Smads), and inhibitory Smads (I-Smads). In general, TGF-β and activin signal via R-Smad2 and 3, whereas BMP signals are transduced through R-Smad1, 5, and 8. Phosphorylated R-Smads subsequently associate with the Co-Smad Smad4, creating a complex that carries the signal to the nucleus, modifying target gene transcription. In contrast to R- and Co-Smads, the inhibitory Smad7 has been shown to inhibit both TGF-β/activin signaling and BMP signaling by associating with activated type I receptors, thus preventing phosphorylation of R-Smads.9-11 Smad7 is up-regulated in response to TGF-β, activin, and BMP, indicating its involvement in a negative feedback loop in response to stimulation by TGF-β and related ligands.9,12,13
In hematopoiesis, BMP-4 has been shown to modify proliferation, differentiation, and survival of primitive human hematopoietic cells in a concentration-dependent manner.14 Additionally, during embryogenesis, BMP-4 acts as an inducer of ventral mesoderm, the tissue from which hematopoiesis originates.15 The role of TGF-β in hematopoiesis has been studied extensively over the years and it has been identified as one of the most potent inhibitors of cell proliferation in primitive hematopoietic cells in vitro.16-24 However, studies performed using TGF-β type I receptor–conditional knockout mice suggest that TGF-β is dispensable for normal mouse hematopoiesis in vivo under both steady-state and stressed conditions.25,26 Whether activin plays a role in the regulation of HSCs in vivo has so far not been explored in detail.27
Since the Smad-signaling pathway is shared by numerous ligands, this system is inherently redundant. Therefore, to investigate whether simultaneous blocking of both Smad-signaling branches in HSCs might affect fate decisions, such as self-renewal and differentiation, we have overexpressed the inhibitory Smad7 in murine HSCs by a retroviral gene transfer approach. We report here that blocking of the entire Smad pathway in murine HSCs causes increased self-renewal in vivo, as assessed by both phenotypic and functional assays in primary and secondary recipients. Importantly, Smad7-overexpressing HSCs could give rise to both myeloid and lymphoid cell compartments at normal distributions, providing evidence for an unperturbed differentiation capacity. Furthermore, gene-expression analysis of purified HSCs from BM of recipient mice revealed decreased levels of p21 in parallel with an increase of Bmi-1, thus suggesting a plausible mechanism downstream of Smad7. This is, to our knowledge, the first report describing a model where the entire Smad network has been blocked in murine HSCs using overexpression of Smad7, revealing an important role for Smad signaling in the self-renewal of HSCs in vivo.
Materials and methods
Retroviral vector production
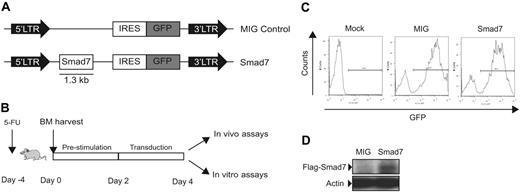
The 1.3-kb BamHI-XhoI murine Flag-tagged Smad7 cDNA fragment (a gift from P. ten Dijke) was blunted and subsequently subcloned into the blunted EcoRI-XhoI sites of the MSCV-based vector (previously described by Hawley et al28 ) upstream of the internal ribosomal entry site (IRES) and GFP reporter gene.
The Smad7 and MIG control vector plasmids were transfected into amphotropic Phoenix cells, and supernatant was harvested. Stable retroviral packaging cell lines were obtained by multiple infections of GP+E86 ecotropic cells with supernatant from the transient infection. Retroviral GP+E86 packaging cells produced stable titers of retroviral particles, as measured by infecting NIH3T3 cells and subsequent detection of GFP+ cells by fluorescence-activated cell sorter (FACS). Both Smad7- and MIG-producing cell lines reached titers above 1 × 106 infectious particles/mL.
Retroviral transduction
BM cells were extracted from femora and tibiae of wild-type (WT) C57Bl/6 mice treated with 5-fluorouracil (5-FU; 150 mg/kg) 4 days prior to harvest. The cell suspension was filtrated through a 70-μm cell strainer (BD Biosciences, Franklin Lakes, NJ) and subsequently cultured at a density of 0.5 × 106 cells/mL for 48 hours in X-Vivo medium (BioWhittaker, Walkersville, MD) supplemented with 1% BSA (Stem Cell Technologies, Vancouver, BC, Canada), 10–4 M 2-ME (Sigma-Aldrich, St Louis, MO), 2 mM L-glutamine (Gibco, Paisley, United Kingdom), 100 U/mL penicillin, 100 μg/mL streptomycin (Gibco), 50 ng/mL murine stem-cell factor (mSCF; R&D Systems, Minneapolis, MN), 10 ng/mL murine IL-3 (mIL-3; R&D Systems), and 50 ng/mL human IL-6 (hIL-6). After stimulation, cells were harvested and subjected to cocultivation on mitomycin C–treated (5 μg/mL for 2 h; Sigma-Aldrich) GP+E86 viral producer cells for another 48 hours in IMDM supplemented with 20% FCS (Gibco), 10–4 M 2-ME, 2 mM L-glutamine, 100 U/mL penicillin, 100 μg/mL streptomycin, and the above cytokines, with the addition of 6 μg/mL of protamine sulfate (Sigma-Aldrich). After the infection procedure, nonadherent cells were harvested and used for further experiments.
Mice
C57Bl/6 (Ly5.2), congenic Bl6SJL (Ly5.1), and C57Bl/6 × Bl6SJL (Ly5.1/5.2) as well as the MxCre Smad4 conditional knockout (KO) mice were bred in the barrier facility at the biomedical center, Lund University. Smad4-conditional KO mice have been described previously29 and were backcrossed for 6 generations onto C57Bl/6 mice carrying the Cre recombinase under the Mx1-inducible promoter. pIpC was administered to conditional KO and littermate controls 1 week prior to 5-FU administration, as previously described.25 Mice were kept in ventilated racks and given autoclaved food and water. All animal experiments were approved by Lund University's ethical committee.
Transplantations
A total of 4 separate transduction experiments were performed. After retroviral transduction, 2.5 × 105 to 5 × 105 unsorted BM cells (Ly5.1/5.2 or Ly5.2) were transplanted together with 2 × 105 congenic support cells (Ly5.1) into 12 lethally irradiated (950 cGy) primary recipient mice (Ly5.1) for each vector, giving a total of 24 recipients. Primary mice were killed 16 to 17 weeks after BM transplantation and the equivalent of half a femur was transplanted to each lethally irradiated (950 cGy) secondary recipient mouse. Three to 4 secondary recipients received transplants per primary donor animal.
Cell preparations
Peripheral blood (PB) was collected from the tail veins and kept in heparin (2000 IU per/mL; Leo Pharma, Ajax, ON, Canada) until further analysis. When necessary, red blood cells (RBCs) were lysed with ammonium chloride (NH4Cl; Stem Cell Technologies). BM was harvested as described above and kept in PBS (Gibco-BRL) supplemented with 2% FCS. Lineage depletion was performed as previously described.25
Flow cytometry
Rat antibodies against murine CD45.1 (Ly5.1), CD45.2 (Ly5.2), Mac1, Gr1, B220, CD3ϵ, Sca1, and c-kit were obtained from Pharmingen (Franklin Lakes, NJ). Antibodies were conjugated with PE, allophycocyanin (APC), or biotin. Biotinylated antibodies were detected by streptavidin APC (Pharmingen). Unconjugated antibodies were detected with tricolor conjugated goat F(ab′)2 anti–rat IgG (H+L) (Caltag Lab, Burlingame, CA). Dead cells were excluded by staining with 7-aminoactinomycin D (7-AAD; Sigma-Aldrich). Cells were sorted on a FACSVantage Cell Sorter (BD Biosciences) or analyzed on a FACSCalibur (BD Biosciences), and results were subsequently analyzed by using CellQuest software (BD Biosciences).
Western blot analysis
Purified GFP+ BM cells from Smad7 recipients or unsorted BM cells from control mice were treated for 1 hour with 10 ng/mL human TGF-β1 (PeproTech, London, United Kingdom), 100 ng/mL human activin A, or 50 ng/mL human BMP-4 (R&D Systems). Protein extraction, separation, transfer, and detection were performed as previously described.30 The following antibodies were used: anti-FLAG (Sigma, St Louis, MO) for detection of Smad7; anti-Smad1 and antiphosphorylated Smad2 (Cell Signaling Technology, Beverly, MA); antiphosphorylated Smad1 (Upstate Biotechnology, Lake Placid, NY); and anti-Smad2 and antiactin (BD Transduction Laboratories, Franklin Lakes, NJ).
Quantitative reverse transcriptase–polymerase chain reaction (RT-PCR)
GFP+ lineage– Sca1+ c-kit+ (LSK) cells were purified from BM of primary recipients 16 weeks after transplantation. RNA from WT and Smad7-overexpressing GFP+ LSK cells was isolated (RNeasy; Qiagen, Hilden, Germany) and reverse transcribed (SuperScript III; Invitrogen, Carlsbad, CA) in the presence of random hexamers. Quantitative PCRs (Taqman; Applied Biosystems, Foster City, CA) were performed in an ABI Prism 7700 (Applied Biosystems) according to the manufacturer's protocol using gene-specific primers (Applied Biosystems). Each assay was performed in duplicate and the results were normalized to Hprt levels.
In vitro culture and colony assays
Following transduction, cells were sorted for GFP and subsequently cultured in serum-free X-Vivo media containing 1% BSA, 10–4 M 2-ME, 2 mM L-glutamine, 100 U/mL penicillin, 100 μg/mL streptomycin (Gibco), 50 ng/mL mSCF, 10 ng/mL mIL-3, and 50 ng/mL hIL-6. Cells were plated at a concentration of 0.2 × 106 cells/mL. Every third day, cells were counted and replated at 0.2 × 106 cells/mL. For growth inhibition assays, 1, 10, or 50 ng/mL of active TGF-β1 (PeproTech) was added to media, and cells were cultured for 6 days, counted, and replated every second day. For colony assays, 1000 to 5000 cultured cells/mL were plated in 35-mm Petri dishes in methylcellulose medium (M3236; Stem Cell Technologies) containing 50 ng/mL mSCF, 10 ng/mL mIL-3, and 10 ng/mL hIL-6. Colonies were scored after 7 days of culture.
Stromal coculture
The mouse fibroblast cell line sl/sl was cultured in DMEM supplemented with 15% FCS, 100 U/mL penicillin, and 100 μg/mL streptomycin. Prior to coculture, sl/sl cells were treated with 5 μg/mL mitomycin C for 3 hours and subsequently replated at a density of 0.5 × 106 cells/collagen-coated well (Collagen I–coated 6-well plates; BD Labware, Franklin Lakes, NJ). Transduced BM cells were purified for GFP and seeded onto the sl/sl cells at a density of 0.1 × 106 cells/mL in serum-free X-vivo media as described in the previous section. Cells were counted and replated every third day onto fresh stromal cells.
Statistical analysis
Data were analyzed using Student t test unless otherwise stated. P values less than .05 were considered significant.
Results
Smad7-overexpressing HSCs can reconstitute irradiated recipients long-term and show a normal differentiation capacity
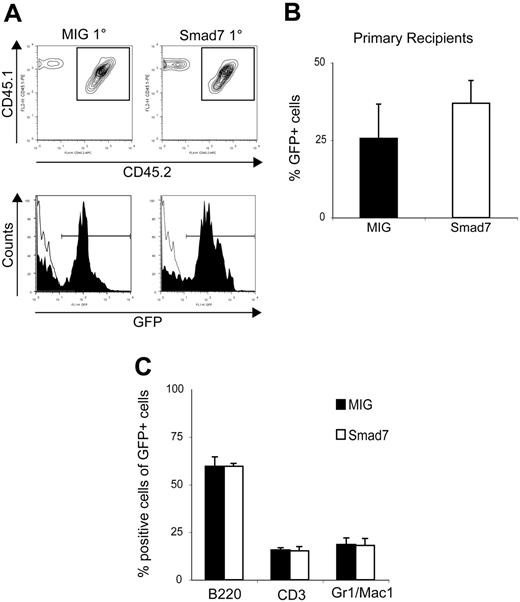
To investigate the effects of simultaneous blocking of multiple Smad pathways in HSCs in vivo, 5-FU–treated BM cells from WT mice were transduced with Smad7 and MIG control vectors (Figure 1A-B). Flow-cytometric analysis revealed similar transduction efficiencies between MIG and Smad7 vectors (Figure 1C), and Western blot analysis confirmed Smad7 protein expression in transduced BM cells (Figure 1D). Following transduction, equal numbers of unsorted cells (Ly5.1/5.2) from MIG and Smad7 transductions were transplanted in conjunction with 2 × 105 freshly isolated congenic Ly5.1 WT BM support cells into lethally irradiated recipient mice (Ly5.1). Both MIG- and Smad7-transduced HSCs were able to reconstitute lethally irradiated recipients long-term, as determined by FACS analysis of PB at 16 to 18 weeks after BM transplantation (Figure 2A-B; total GFP contribution in PB averaged to 25.6% ± 11.1% for MIG vs 36.9% ± 7.35% for Smad7; n = 4, P > .05). The slightly increased repopulative capacity suggested that Smad7-overexpressing HSCs might have an advantage over MIG-transduced HSCs. This could have resulted from an increased homing capacity of Smad7-overexpressing cells. To investigate this possibility, homing experiments were performed. However, intravenous injection of cells from coculture transductions into lethally irradiated recipients revealed no differences in the number of GFP+ cells that homed to the BM 24 hours later (n = 3, P > .05; data not shown).
Experimental design and evidence for Smad7 expression in transduced BM cells. (A) MSCV control (MIG) and Smad7 retroviral constructs. The Flag-tagged Smad7 cDNA was subcloned into the control vector backbone upstream of IRES and GFP sequences. LTR indicates long terminal repeat. (B) Experimental outline. (C) Transduction efficiencies of BM cells were determined 3 days after transduction and shown to be between 60% and 80% for MIG and 50% and 80% for Smad7. Mock indicates untransduced cultured cells. The figure shows a representative experiment. (D) Western blot analysis showing Smad7 protein expression in BM cells using an anti-Flag antibody. Actin was used as loading control.
Experimental design and evidence for Smad7 expression in transduced BM cells. (A) MSCV control (MIG) and Smad7 retroviral constructs. The Flag-tagged Smad7 cDNA was subcloned into the control vector backbone upstream of IRES and GFP sequences. LTR indicates long terminal repeat. (B) Experimental outline. (C) Transduction efficiencies of BM cells were determined 3 days after transduction and shown to be between 60% and 80% for MIG and 50% and 80% for Smad7. Mock indicates untransduced cultured cells. The figure shows a representative experiment. (D) Western blot analysis showing Smad7 protein expression in BM cells using an anti-Flag antibody. Actin was used as loading control.
Smad7-overexpressing cells can reconstitute lethally irradiated recipients long-term and generate a normal lineage distribution. (A) FACS analysis of PB from primary recipients at 16 to 18 weeks after BM transplantation. The top panel shows Ly5.1/Ly5.2 analysis and the bottom panel shows the GFP contribution within the Ly5.1/5.2 population. (B) Average percentage of total GFP+ cells in PB of primary recipients 16 to 18 weeks after BM transplantation: MIG, 25.6% ± 11.1%; Smad7, 36.9 ± 7.35%. (C) Lineage distribution within the GFP+ population in PB of primary recipients. B220, CD3, and Gr1/Mac1 indicate B cells, T cells, and myeloid cells, respectively. B220+ cells were represented on average 59.6% ± 4.95% for MIG and 59.6% ± 1.50% for Smad7, CD3+ cells 15.7% ± 1.15% for MIG and 15.2% ± 2.20% for Smad7, Gr1+/Mac1+ cells 18.5% ± 3.50% for MIG versus 18.0% ± 3.70% for Smad7. Data are pooled from 4 separate transduction experiments and a total of 24 recipients. Error bars indicate standard error of the mean (SEM).
Smad7-overexpressing cells can reconstitute lethally irradiated recipients long-term and generate a normal lineage distribution. (A) FACS analysis of PB from primary recipients at 16 to 18 weeks after BM transplantation. The top panel shows Ly5.1/Ly5.2 analysis and the bottom panel shows the GFP contribution within the Ly5.1/5.2 population. (B) Average percentage of total GFP+ cells in PB of primary recipients 16 to 18 weeks after BM transplantation: MIG, 25.6% ± 11.1%; Smad7, 36.9 ± 7.35%. (C) Lineage distribution within the GFP+ population in PB of primary recipients. B220, CD3, and Gr1/Mac1 indicate B cells, T cells, and myeloid cells, respectively. B220+ cells were represented on average 59.6% ± 4.95% for MIG and 59.6% ± 1.50% for Smad7, CD3+ cells 15.7% ± 1.15% for MIG and 15.2% ± 2.20% for Smad7, Gr1+/Mac1+ cells 18.5% ± 3.50% for MIG versus 18.0% ± 3.70% for Smad7. Data are pooled from 4 separate transduction experiments and a total of 24 recipients. Error bars indicate standard error of the mean (SEM).
Importantly, Smad7-overexpressing cells could give rise to both myeloid and lymphoid cell compartments at normal distributions, indicating an unperturbed differentiation capacity of Smad7-overexpressing HSCs (Figure 2C). Thus, our data revealed that Smad7-overexpressing HSCs could reconstitute lethally irradiated recipients long-term while maintaining a normal differentiation capacity.
Smad7 increases the stem-cell pool in vivo
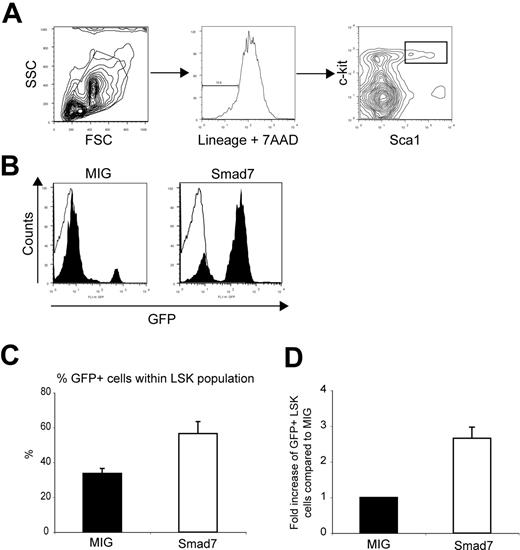
To investigate the impact of blocked Smad signaling on primitive hematopoietic cells, flow-cytometric analysis was used at 16 to 18 weeks after transplantation to measure the proportions and absolute numbers of GFP+ cells within the HSC-enriched LSK population of BM cells from animals that received a primary transplant (Figure 3A-B). FACS analysis revealed both significantly higher proportions and absolute numbers of GFP+ LSK cells overexpressing Smad7 compared with GFP+ control cells. The percentage of GFP+ cells within the LSK population was 33.7% ± 2.85% for MIG versus 56.5% ± 6.85% for Smad7 (Figure 3C; P = .03, n = 4). The absolute number of GFP+ LSK cells per femur was increased on average 2.7-fold in animals that received transplants of Smad7-transduced BM compared with animals that received MIG transplants (Figure 3D; 2.7 ± 0.31, P = .04). Consistent with the slight increase in reconstitution activity by Smad7-overexpressing cells, these data suggested that overexpression of Smad7 expanded the HSC pool in vivo.
Smad7 increases HSC self-renewal in vivo
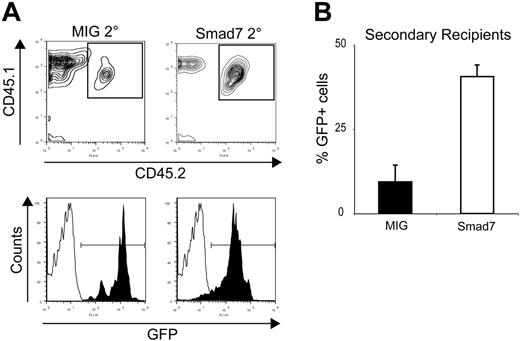
To evaluate whether the increase in GFP+ LSK cell numbers would functionally translate into an enhanced regenerative ability of the hematopoietic system upon reconstitution, secondary transplantations were carried out. Sixteen to 18 weeks after the first transplantation, BM from primary mice was transplanted into 3 to 4 lethally irradiated recipients per donor, and engraftment of secondary recipients was followed for another 12 to 14 weeks. Interestingly, the GFP contribution from Smad7-transduced cells in secondary recipients was significantly higher in PB compared with MIG-transduced cells, as assessed by FACS analysis (Figure 4A-B; total percentage GFP in PB was 9.40% ± 4.95% for MIG vs 40.6% ± 3.45% for Smad7; P = .003). Importantly, the total GFP contribution in PB was maintained, or even slightly increased, between primary and secondary recipients of Smad7-transduced cells, functionally suggesting increased self-renewal activity of Smad7-overexpressing HSCs. In contrast, the GFP contribution from MIG-transduced cells dropped between primary and secondary recipients, indicative of exhaustion of the stem-cell pool. Furthermore, no significant alterations were detected in the lineage distribution of Smad7-overexpressing cells 12 to 14 weeks after transplantation in PB, as determined by FACS analysis (Table 1). In addition, BM from some secondary recipients was transplanted to lethally irradiated tertiary recipients, as a proof of principle for sustained self-renewal. Persistent multi-lineage engraftment was observed in tertiary recipients receiving Smad7-overexpressing BM cells but not control BM, indicating that self-renewal was maintained and the HSC compartment was not exhausted (data not shown). These data were in agreement with the increased pool of GFP+ LSK cells observed in primary recipients and moreover confirmed that this population contained functional HSCs. Together, these findings suggested that upon inhibition of Smad-mediated signals, the self-renewal capacity of HSCs was promoted in vivo whereas the differentiation potential was left unaffected.
Lineage distribution within the GFP+ population of PB in secondary recipients at 12 weeks after BM transplantation
. | No. experiments . | B220+, % . | CD3+, % . | Gr1+/Mac1+, % . |
|---|---|---|---|---|
| MIG | 4 | 22.8 ± 3.77 | 26.2 ± 6.10 | 43.7 ± 7.75 |
| Smad7 | 4 | 30.3 ± 5.45 | 31.1 ± 1.53 | 28.8 ± 5.95 |
. | No. experiments . | B220+, % . | CD3+, % . | Gr1+/Mac1+, % . |
|---|---|---|---|---|
| MIG | 4 | 22.8 ± 3.77 | 26.2 ± 6.10 | 43.7 ± 7.75 |
| Smad7 | 4 | 30.3 ± 5.45 | 31.1 ± 1.53 | 28.8 ± 5.95 |
Increased numbers of GFP+ LSK cells in primary recipients of Smad7-transduced cells. (A) FACS analysis of BM from primary recipients at 16 to 18 weeks after BM transplantation. Lineage- and 7AAD-positive cells were excluded and Sca1/c-kit-positive cells selected by gating. SSC indicates side scatter; FSC, forward scatter. (B) Histogram showing the GFP contribution within the LSK population from representative recipients of MIG and Smad7 cells, respectively. Filled bars indicate MIG; open bars, Smad7. (C) Average percentage of GFP+ cells within the LSK population: MIG, 33.7% ± 2.85%; Smad7, 56.5% ± 6.85 (P = .03 as analyzed by paired t test). (D) Fold increase of absolute numbers of GFP+ LSK cells/femur compared with MIG. Fold increase is 2.7 (P = .04 as analyzed by paired t test). Data represent mean values from 4 independent experiments ± SEM.
Increased numbers of GFP+ LSK cells in primary recipients of Smad7-transduced cells. (A) FACS analysis of BM from primary recipients at 16 to 18 weeks after BM transplantation. Lineage- and 7AAD-positive cells were excluded and Sca1/c-kit-positive cells selected by gating. SSC indicates side scatter; FSC, forward scatter. (B) Histogram showing the GFP contribution within the LSK population from representative recipients of MIG and Smad7 cells, respectively. Filled bars indicate MIG; open bars, Smad7. (C) Average percentage of GFP+ cells within the LSK population: MIG, 33.7% ± 2.85%; Smad7, 56.5% ± 6.85 (P = .03 as analyzed by paired t test). (D) Fold increase of absolute numbers of GFP+ LSK cells/femur compared with MIG. Fold increase is 2.7 (P = .04 as analyzed by paired t test). Data represent mean values from 4 independent experiments ± SEM.
Smad7-overexpressing cells show increased repopulative capacity upon secondary transplantation. (A) FACS analysis of PB from secondary recipients 12 to 14 weeks after BM transplantation. The top panel shows Ly5.1/Ly5.2 analysis and the bottom panel shows GFP within the Ly5.1/5.2 population. Filled bars indicate MIG; open bars, Smad7. (B) Average percentage of total GFP+ cells in PB of secondary recipients 12 to 14 weeks after BM transplantation: MIG, 9.40% ± 4.95%; Smad7, 40.6% ± 3.45% (P = .003). All mean values are from 4 independent transductions ± SEM; a total of 78 secondary recipients were used.
Smad7-overexpressing cells show increased repopulative capacity upon secondary transplantation. (A) FACS analysis of PB from secondary recipients 12 to 14 weeks after BM transplantation. The top panel shows Ly5.1/Ly5.2 analysis and the bottom panel shows GFP within the Ly5.1/5.2 population. Filled bars indicate MIG; open bars, Smad7. (B) Average percentage of total GFP+ cells in PB of secondary recipients 12 to 14 weeks after BM transplantation: MIG, 9.40% ± 4.95%; Smad7, 40.6% ± 3.45% (P = .003). All mean values are from 4 independent transductions ± SEM; a total of 78 secondary recipients were used.
Forced expression of Smad7 results in down-regulation of p21 and up-regulation of Bmi1
To study changes in gene expression provoked by inhibition of Smad signaling in HSCs, GFP+ LSK cells were purified from BM of primary recipients. Quantitative RT-PCR was performed to investigate the relative expression levels of known TGF-β target genes and other genes previously identified as regulators of HSC self-renewal. Although not significant, a number of negative regulators of self-renewal were found to be down-regulated upon forced expression of Smad7, including p21 and lnk (Table 2). In addition, Bmi1, a crucial component of the self-renewal machinery in HSCs, was slightly but significantly up-regulated (Table 2). Among other positive regulators tested, c-mpl was found to be up-modulated. These data suggest that the increase in self-renewal promoted by forced expression of Smad7 is likely a combinatorial effect of decreased levels of negative factors and increased amounts of genes positively regulating self-renewal.
Gene expression analysis of GFP+ LSK cells
Gene . | MIG, AU . | Smad7, AU . | Fold difference Smad7 vs MIG . |
|---|---|---|---|
| p18 | 0.33 ± 0.015 | 0.41 ± 0.054 | 1.22 |
| p21* | 0.47 ± 0.008 | 0.25 ± 0.005 | - 1.88 |
| p27 | 0.46 ± 0.025 | 0.41 ± 0.007 | - 1.13 |
| p57 | ND | ND | ND |
| id2 | 0.009 ± 0.001 | 0.01 ± 0.0009 | 1.11 |
| c-myc | 7.25 ± 0.181 | 8.86 ± 1.21 | 1.22 |
| gata2 | 1.18 ± 0.142 | 1.43 ± 0.061 | 1.21 |
| hoxB4 | ND | ND | ND |
| bmi1 | 0.15 ± 0.004 | 0.18 ± 0.003 | 1.21† |
| gfi1 | 0.03 ± 0.006 | 0.05 ± 0.009 | 1.42 |
| hes1 | ND | ND | ND |
| notch1 | 0.19 ± 0.03 | 0.24 ± 0.016 | 1.24 |
| C/EBPa | 0.29 ± 0.011 | 0.38 ± 0.007 | 1.31 |
| c-mpl* | 1.41 ± 0.237 | 2.18 ± 0.464 | 1.56 |
| Ink* | 0.012 ± 0.005 | 0.007 ± 0.0006 | - 1.65 |
| flt3 | 0.47 ± 0.046 | 0.62 ± 0.040 | 1.32 |
| spry2 | 0.16 ± 0.014 | 0.19 ± 0.022 | 1.18 |
| smad7‡ | .055‡ | 6.85‡ | 124 |
Gene . | MIG, AU . | Smad7, AU . | Fold difference Smad7 vs MIG . |
|---|---|---|---|
| p18 | 0.33 ± 0.015 | 0.41 ± 0.054 | 1.22 |
| p21* | 0.47 ± 0.008 | 0.25 ± 0.005 | - 1.88 |
| p27 | 0.46 ± 0.025 | 0.41 ± 0.007 | - 1.13 |
| p57 | ND | ND | ND |
| id2 | 0.009 ± 0.001 | 0.01 ± 0.0009 | 1.11 |
| c-myc | 7.25 ± 0.181 | 8.86 ± 1.21 | 1.22 |
| gata2 | 1.18 ± 0.142 | 1.43 ± 0.061 | 1.21 |
| hoxB4 | ND | ND | ND |
| bmi1 | 0.15 ± 0.004 | 0.18 ± 0.003 | 1.21† |
| gfi1 | 0.03 ± 0.006 | 0.05 ± 0.009 | 1.42 |
| hes1 | ND | ND | ND |
| notch1 | 0.19 ± 0.03 | 0.24 ± 0.016 | 1.24 |
| C/EBPa | 0.29 ± 0.011 | 0.38 ± 0.007 | 1.31 |
| c-mpl* | 1.41 ± 0.237 | 2.18 ± 0.464 | 1.56 |
| Ink* | 0.012 ± 0.005 | 0.007 ± 0.0006 | - 1.65 |
| flt3 | 0.47 ± 0.046 | 0.62 ± 0.040 | 1.32 |
| spry2 | 0.16 ± 0.014 | 0.19 ± 0.022 | 1.18 |
| smad7‡ | .055‡ | 6.85‡ | 124 |
AUs indicate arbitrary units; and ND, not detectable.
Fold difference > 1.5.
P = .003.
Representative experiment.
Smad7 overexpression blocks phosphorylation of R-Smads and eliminates the growth inhibitory effect of TGF-β
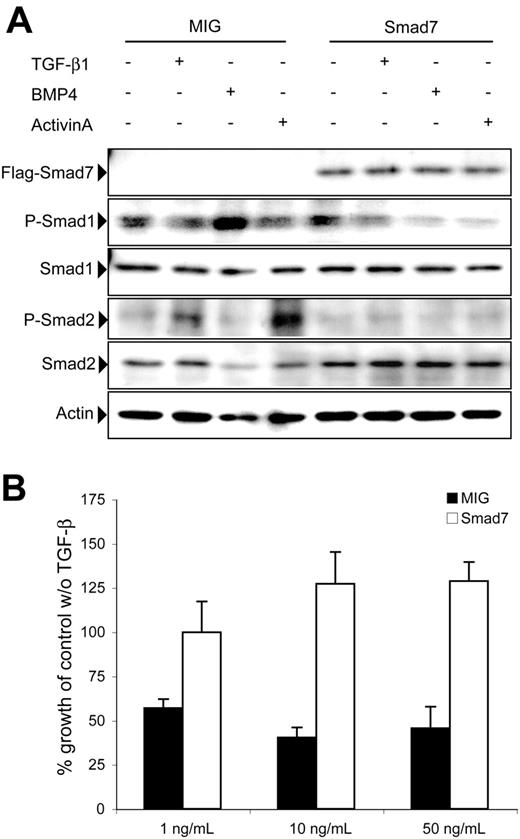
To confirm that the Smad7 protein was expressed and functionally active in the BM, GFP+ cells were purified from BM of recipients that received Smad7-transduced cells. Western blot analysis revealed strong expression of the Smad7 protein in GFP+ BM cells from Smad7-transduced donor cells (Figure 5A). In addition, cells were stimulated with TGF-β1, activin A, or BMP-4, and subsequently the phosphorylation status of R-Smads was examined by Western blot analysis. As expected, control BM cells showed robust induction of phosphorylation of Smad1 and Smad2 in response to BMP-4 and TGF-β1 or activin A, respectively (Figure 5A). In contrast, BM cells overexpressing Smad7 showed no increased phosphorylation of Smad1 or Smad2 in response to BMP-4 or TGF-β1 and activin A stimulation, respectively. To investigate whether forced expression of Smad7 resulted in functional resistance to TGF-β–mediated growth inhibition in vitro, GFP+ cells were sorted after transduction and cultured under serum-free conditions for 6 days in the presence or absence of increasing concentrations of TGF-β1. As expected, MIG cells were growth inhibited at all concentrations of TGF-β1 tested (Figure 5B). In contrast, the growth inhibitory effect conveyed by TGF-β1 was completely abrogated upon overexpression of Smad7, irrespective of the concentration of TGF-β1 (Figure 5B). Collectively, these data demonstrated that robust expression of the Smad7 protein in BM cells impeded R-Smad phosphorylation and yielded resistance to the growth inhibitory effects of TGF-β in vitro.
Smad7 reduces the proliferative and colony-forming capacity of cells in vitro
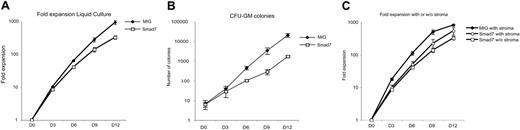
To investigate the growth characteristics of transduced cells in vitro, GFP+ BM cells from Smad7 and MIG cocultures were sorted immediately after transduction and subsequently plated in serum-free liquid cultures supplemented with hematopoietic cytokines. Cells were kept in culture for up to 12 days after transduction. Every third day, the cells were counted and replated in fresh media. In contrast to the increased repopulative capacity conferred by Smad7 in vivo, Smad7-overexpressing cells had a significantly reduced proliferative capacity compared with MIG-transduced cells in vitro. Significant differences in growth were observed from day 6 and onwards (Figure 6A; P < .01). MIG cells expanded on average 931-fold over 12 days in culture, whereas Smad7-overexpressing cells expanded on average 327-fold over the same time. Thus, at day 12 of culture, the fold expansion of Smad7-overexpressing cells was only 35% of that of MIG cells (Figure 6A). To investigate the colony-forming capacity of cultured cells, methylcellulose supporting CFU-GM growth was seeded at the same time intervals. Interestingly, Smad7-overexpressing cells formed fewer CFU-GM colonies at all time points of culture compared with control cells. At day 12 of culture, the colony numbers of Smad7-overexpressing cells were only 11.8% of that of MIG cells (Figure 6B). Similarly, when cells were grown in serum-containing media, Smad7-overexpressing cells showed decreased proliferative capacity of similar magnitude as observed for serum-free cultures (data not shown). To ask whether stem-cell self-renewal might be favored over differentiation in vitro, cells from days 6 and 12 of liquid culture (in serum-containing media) were transplanted into lethally irradiated recipients. However, when the GFP contribution was analyzed 12 weeks later, no differences or GFP contribution were found from day 6 and day 12 cultures, respectively, suggesting that the stem-cell activity was equally decreased over time in MIG- and Smad7-overexpressing cultures (data not shown). Thus, overexpression of Smad7 resulted in decreased proliferative capacity in vitro, suggesting that the increased regenerative capacity imparted by Smad7 in vivo was dependent on the BM microenvironment.
Overexpression of Smad7 inhibits R-Smad phosphorylation and abrogates TGF-β–mediated growth inhibition. (A) Western blot analysis performed on BM cells from recipient mice. Protein was extracted from BM cells stimulated with the indicated ligands and subsequently used for Western blot analysis. Actin was used as control for loading. P-Smad indicates phosphorylated. (B) GFP+ cells were sorted after transduction and cultured for 6 days under serum-free conditions with or without increasing concentrations of active TGF-β1 as indicated. The growth was compared with cultures without exogenous TGF-β1 and calculated as percentage of values. Data represent mean values ± SEM from 3 independent experiments on day 6 of culture.
Overexpression of Smad7 inhibits R-Smad phosphorylation and abrogates TGF-β–mediated growth inhibition. (A) Western blot analysis performed on BM cells from recipient mice. Protein was extracted from BM cells stimulated with the indicated ligands and subsequently used for Western blot analysis. Actin was used as control for loading. P-Smad indicates phosphorylated. (B) GFP+ cells were sorted after transduction and cultured for 6 days under serum-free conditions with or without increasing concentrations of active TGF-β1 as indicated. The growth was compared with cultures without exogenous TGF-β1 and calculated as percentage of values. Data represent mean values ± SEM from 3 independent experiments on day 6 of culture.
Smad7-overexpressing BM cells exhibit reduced proliferative and clonogenic capacity in vitro. (A) BM cells were sorted for GFP directly after transduction and plated in serum-free media. Cells were counted and replated every third day. Day 0 indicates day of sorting. Data represent fold increase in number of cells ± SEM from 4 separate experiments (P ≤ .03 for days 6, 9, and 12). (B) Cells from the liquid culture were plated at indicated days into methylcellulose supporting growth of CFU-GM colonies. Data represent number of colonies/1000 input cells on day 0. Mean values ± SEM (P = .02 day 12). (C) BM cells were sorted for GFP directly after transduction (day 0) and plated onto preseeded sl/sl stromal cells in serum-free media. BM cells were counted and replated every third day for up to 12 days after transduction. Data represent mean values of fold expansion of number of cells ± SEM (n = 3). No significant differences in growth were detected between MIG and Smad7 on sl/sl stroma on days 9 and 12 (P > .05). Also included in the graph is Smad7-overexpressing cells grown under stroma-free conditions (n = 4).
Smad7-overexpressing BM cells exhibit reduced proliferative and clonogenic capacity in vitro. (A) BM cells were sorted for GFP directly after transduction and plated in serum-free media. Cells were counted and replated every third day. Day 0 indicates day of sorting. Data represent fold increase in number of cells ± SEM from 4 separate experiments (P ≤ .03 for days 6, 9, and 12). (B) Cells from the liquid culture were plated at indicated days into methylcellulose supporting growth of CFU-GM colonies. Data represent number of colonies/1000 input cells on day 0. Mean values ± SEM (P = .02 day 12). (C) BM cells were sorted for GFP directly after transduction (day 0) and plated onto preseeded sl/sl stromal cells in serum-free media. BM cells were counted and replated every third day for up to 12 days after transduction. Data represent mean values of fold expansion of number of cells ± SEM (n = 3). No significant differences in growth were detected between MIG and Smad7 on sl/sl stroma on days 9 and 12 (P > .05). Also included in the graph is Smad7-overexpressing cells grown under stroma-free conditions (n = 4).
Stromal cocultures partially restore the proliferative capacity of Smad7-overexpressing cells in vitro
In an effort to provide evidence for a microenvironmental-dependent mechanism of expansion of Smad7-overexpressing cells, GFP+ cells were purified after transduction and plated onto the preseeded stromal cell line sl/sl. Cells were cultured for up to 12 days after transduction in serum-free media containing hematopoietic cytokines and counted and replated in fresh media every third day. MIG cells grown on stroma expanded on average 818-fold over 12 days, whereas Smad7-overexpressing cells expanded on average 542-fold over the same time (Figure 6C). Moreover, no significant differences in growth between MIG- and Smad7-overexpressing cells were observed at days 9 and 12 of culture. Thus, the proliferative capacity of Smad7-overexpressing cells increased in the presence of stroma cells compared with stroma-free cultures. Although the proliferative capacity of Smad7-overexpressing cells in the presence of stroma was not statistically increased over Smad7 cultures without stroma cells, the average growth on day 12 was increased by 66% (Figure 6C). Taken together, these data suggested that the proliferative capacity of Smad7-overexpressing cells could be modulated and increased in the presence of additional signals provided by stroma cells, thus supporting a niche-dependent mechanism.
The in vivo effect of Smad7 overexpression on HSCs is dependent on Smad4
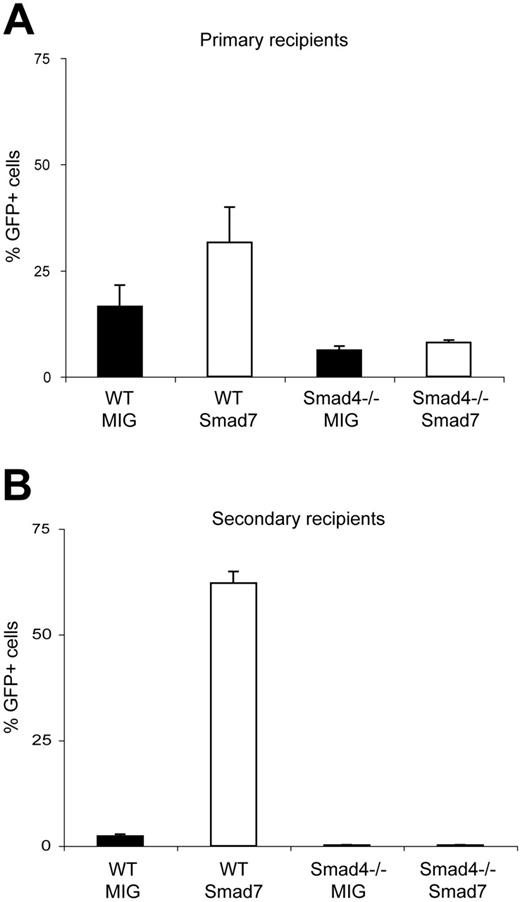
So far our data suggested that simultaneous blocking of R-Smad pathways in HSCs caused increased self-renewal in vivo. However, a simultaneous study in our laboratory suggested that complete disruption of the Smad pathway in HSCs by targeted deletion of Smad4 had a negative effect on self-renewal (G.K., U.B., J.L.M., Mats Ehinger, S.S., Chu-Xia Deng, S.K., manuscript submitted). This prompted us to investigate whether the in vivo effect observed in Smad7-overexpressing cells was in fact dependent on the presence of Smad4 or whether ectopic expression of Smad7 resulted in a novel function. To dissect between these possibilities, we overexpressed Smad7 in HSCs where Smad4 had been conditionally deleted. Smad4 null and WT littermate BM cells were transduced with both MIG control and Smad7 vectors. Following transduction, 2.5 × 105 Ly5.2 unsorted cells from the respective transductions were transplanted in conjunction with 2 × 105 Ly5.1 fresh support cells into lethally irradiated Ly5.1 recipients. At 13 weeks after BM transplantation, the mice were killed and the reconstitution capacity in PB was analyzed by flow-cytometric analysis. In agreement with our previous in vivo data, recipients of WT cells transduced with the Smad7 vector showed the highest GFP contribution (31.6% ± 8.26%), whereas MIG-transduced WT cells gave a GFP contribution of 16.5% ± 5.0%. Interestingly, the increase in repopulative capacity imparted by Smad7 in WT BM cells was lost in cells lacking Smad4 (31.6% ± 8.26% and 7.97% ± 0.6% respectively; P < .05; Figure 7A). This became increasingly apparent upon secondary transplantation where the GFP contribution of Smad7-transduced WT cells was increased to 62.2% ± 2.7% and the Smad7-transduced Smad4–/– cells dropped to 0.2% ± 0.05% (P < .05; Figure 7B). Since forced expression of Smad7 in Smad4 null HSCs failed to recapitulate the phenotype observed in the WT genetic background, it was unlikely that ectopic expression of Smad7 had effects unrelated to blocking the Smad pathway. Rather, these data provided evidence for a Smad4-dependent mechanism, suggesting that the increased repopulative capacity conveyed by Smad7 in vivo requires Smad4 either as a transducer of low residual Smad signals or alternatively as a mediator of cross-talk signals from other signaling pathways.
Discussion
Smad7 promotes HSC self-renewal in vivo
A thorough understanding of the molecular machinery that regulates HSC fate decisions in vivo, such as self-renewal and differentiation, is germane to our understanding and subsequent development of in vitro protocols for HSC expansion. Numerous members of the TGF-β superfamily of ligands have been shown to be involved in hematopoietic regulation at various stages. In an effort to decipher the role of Smad signaling in HSCs, we have overexpressed the inhibitory Smad7 in HSCs by a retroviral gene transfer approach. Here, we report for the first time that blocking of the entire Smad-signaling network downstream of TGF-β, activin, and BMPs promotes self-renewal of HSCs in vivo. Smad7-overexpressing cells could repopulate lethally irradiated recipients long-term and gave rise to a completely normal lineage distribution. Interestingly, when primitive hematopoietic cell subsets were analyzed phenotypically by flow cytometry, primary recipients of Smad7-overexpressing HSCs showed both increased absolute numbers and proportions of GFP+ LSK cells in BM. Importantly, the total numbers of LSK cells (GFP+ and GFP–) were not significantly different between control mice and mice that received Smad7-transduced cells, suggesting that Smad7-overexpressing HSCs do not expand in an uncontrollable manner but are kept under control by niche-specific signals. Interestingly, when self-renewal was functionally tested by performing secondary transplantations, Smad7-overexpressing cells gave rise to a significantly higher GFP contribution in PB compared with MIG-transduced cells. Of particular importance, the GFP contribution from control transduced cells dropped between primary and secondary recipients, whereas the Smad7-overexpressing cell contribution was maintained or even increased between primary and secondary transplantations. These data provide evidence that when Smad signaling is blocked in HSCs, self-renewal is promoted in vivo, whereas the hematopoietic stem-cell pool becomes exhausted in control animals. Given the normal lineage distribution and sustained engraftment even upon tertiary BM transfer, manipulation of the Smad-signaling pathway may be of considerable importance for regenerative medicine in the future.
The in vivo phenotype conferred by Smad7 is dependent on Smad4. (A) GFP contribution in PB of primary recipients as assessed by FACS analysis at 13 weeks after BM transplantation: WT cells transduced with the MIG vector, 16.5% ± 5.0%; WT cells transduced with Smad7 vector, 31.6% ± 8.26%; Smad4–/– cells transduced with MIG vector, 6.19% ± 0.96%; Smad4–/– cells transduced with Smad7 vector, 7.97% ± 0.6%. (B) The percentage of GFP+ cells in PB of secondary recipients at 12 weeks after BM transplantation: WT cells transduced with MIG vector, 2.3% ± 0.51%; WT cells transduced with Smad7 vector, 62.2% ± 2.7%; Smad4–/– cells transduced with MIG vector, 0.2% ± 0.06%; Smad4–/– cells transduced with Smad7 vector, 0.2% ± 0.05%. Data are from 1 representative experiment and calculated as mean values ± SEM (P < .05 between WT/Smad7 and Smad4–/–/Smad7). A total of 20 primary and 16 secondary recipients were used.
The in vivo phenotype conferred by Smad7 is dependent on Smad4. (A) GFP contribution in PB of primary recipients as assessed by FACS analysis at 13 weeks after BM transplantation: WT cells transduced with the MIG vector, 16.5% ± 5.0%; WT cells transduced with Smad7 vector, 31.6% ± 8.26%; Smad4–/– cells transduced with MIG vector, 6.19% ± 0.96%; Smad4–/– cells transduced with Smad7 vector, 7.97% ± 0.6%. (B) The percentage of GFP+ cells in PB of secondary recipients at 12 weeks after BM transplantation: WT cells transduced with MIG vector, 2.3% ± 0.51%; WT cells transduced with Smad7 vector, 62.2% ± 2.7%; Smad4–/– cells transduced with MIG vector, 0.2% ± 0.06%; Smad4–/– cells transduced with Smad7 vector, 0.2% ± 0.05%. Data are from 1 representative experiment and calculated as mean values ± SEM (P < .05 between WT/Smad7 and Smad4–/–/Smad7). A total of 20 primary and 16 secondary recipients were used.
It was recently reported by Chadwick et al31 that overexpression of Smad7 in human severe combined immunodeficiency (SCID)–repopulating cells (SRCs) from cord-blood sources caused a shift from a B-lymphoid–dominant graft toward a graft with higher frequency of myeloid progeny within the murine BM. However, we did not find any abnormalities in lineage distribution in our mice that received transplants. Species-related and/or ontogenic differences between human and mouse might account for the discrepancies. Alternatively, the expression level of the Smad7 protein might vary between studies, causing a more or less efficient block in R-Smad phosphorylation. In our study, the RNA level of Smad7 within the transduced LSK population was elevated 124-fold, whereas Chadwick et al31 noted an approximately 3-fold increase in the level of Smad7 in transduced CD34+ cells.
Forced expression of Smad7 results in decreased proliferative capacity in vitro
When cultured under serum-free conditions in vitro, Smad7-overexpressing cells proliferated less efficiently compared with control transduced cells. Similarly, when progenitor-cell function was analyzed using methylcellulose colony-forming cell (CFC) assays, Smad7-overexpressing cells formed significantly fewer colonies compared with control transduced cells. There is evidence in the literature showing that CFC capacity in vitro may paradoxically be reduced at the expense of increased self-renewal as assayed in vivo.32,33 Accordingly, our data support an in vivo niche-dependent mechanism of increased self-renewal of Smad7-overexpressing HSCs. This hypothesis is further supported by the fact that Smad7-overexpressing cells proliferate more efficiently in the presence of stroma cells in vitro compared with stroma-free cultures under serum-free conditions. The inability of the stromal cell line to completely mimic the in vivo HSC niche could provide an explanation why the proliferative capacity was not statistically increased over stromal-free cultures. The stromal cell line used here is of fibroblastic origin, whereas osteoblasts have been shown to be an important constituent of the HSC niche.34-37 Taken together, our data emphasize the importance of the niche in the context of blocked Smad signaling.
Differential gene expression
The exact molecular mechanism behind these data is likely to be multifaceted. It is possible that the phenotype observed here stems from a lack of TGF-β signaling in Smad7-overexpressing HSCs. Indeed, a large body of evidence exists showing that TGF-β inhibits proliferation of primitive hematopoietic cells of both human and mouse origins in vitro.18,38,39 However, contrary to this, in vivo data of TGF-β signaling-deficient HSCs have shown that TGF-β is dispensable for normal repopulative potential and self-renewal capacity in vivo.25,26 In the context of the present study, it is appealing to speculate that compensatory mechanisms due to the highly promiscuous and redundant nature of the TGF-β superfamily might have contributed to the negative findings previously reported.25,26 In support of this, we found that the cell-cycle regulator and equally known TGF-β target gene p21 had a 2-fold decreased expression level in GFP+ LSK cells overexpressing Smad7 compared with control GFP+ LSK cells. p21 has previously been shown to regulate quiescence of murine HSCs, and p21 null mice exhibit premature exhaustion of the HSC pool upon serial transplantations.40 However, experiments with human cells demonstrated that antisense-mediated repression of p21 led to increased engraftment capacity of nonobese diabetic (NOD)–SCID mice.41 Since Smad7-overexpressing HSCs did not show any evidence of premature exhaustion, it is possible that decreased expression levels of p21 might have profoundly different effects than complete lack of p21. In addition, we found that Bmi1, previously recognized as a central player in HSC self-renewal,42,43 was modestly but consistently up-regulated in Smad7-overexpressing LSK cells. Thus, our data imply that down-modulation of negative regulators coupled with elevated levels of positive factors altogether favors self-renewal upon overexpression of Smad7.
Smad7-mediated effects are dependent on Smad4 in vivo
Intriguingly, our data revealed that the effect of Smad7 overexpression on HSCs was dependent on the presence of Smad4, since overexpression of Smad7 in cells deficient in Smad4 failed to phenocopy WT cells overexpressing Smad7. This discovery was unexpected, since overexpression of Smad7 would be predicted to yield a similar hematopoietic phenotype to deletion of Smad4. Nonetheless, this finding is important because it shows that the level at which Smad signaling is disrupted holds great biologic importance for HSCs. Although deletion of Smad4 is likely to yield a more complete block in Smad signaling as opposed to overexpression of Smad7, our data suggest that forced expression of Smad7 efficiently impedes Smad phosphorylation, resulting in robust resistance to TGF-β–mediated growth inhibition in vitro. Thus, low levels of residual Smad signals are not likely to explain the differences between the 2 models. An alternate explanation is that deletion of Smad4 affects non-Smad–signaling pathways in addition to disrupting the Smad pathway. For example, Smad4 has been shown to physically interact with β-catenin and Lef1/Tcf during Xenopus development and in embryonic stem (ES) cells.44,45 Cooperation between the Notch and Smad pathways has also been observed in a variety of model systems.46-48 Indeed, both Wnt and Notch pathways have been implicated in the regulation of self-renewal of HSCs,32,49-52 thus these pathways may represent possible cross-talk candidates. Consequently, disruption of the Smad pathway at the level of Smad4 is more likely to affect cross-talk mechanisms than the overexpression of Smad7. Further characterization of cross-talk signals between the Smad pathway and other signaling pathways in HSCs will certainly be an important mission in the future.
In summary, our data reveal that inhibition of multiple Smad-signaling pathways simultaneously in HSCs by overexpression of Smad7 promotes their self-renewal in vivo, uncovering an important role for the Smad-signaling circuitry in the regulation of HSC self-renewal.
Authorship
Contribution: U.B. designed and performed research, analyzed data, and wrote the paper. G.K. designed and performed research, analyzed data, and contributed to writing the paper. J.L.M. performed research and contributed to writing the paper. T.U., M.M., and S.S. performed research. J.L. designed research and contributed to writing the paper. S.K. directed the research program, designed research, and contributed to writing the paper.
Conflict of interest disclosure: The authors declare no competing financial interests.
Prepublished online as Blood First Edition Paper, 8/17/2006; DOI 10.1182/blood-2006-02-005611.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
This work was supported by grants from The Swedish Cancer Society (S.K.), The European Commission (INHERINET and CONSERT; S.K.), The Swedish Gene Therapy Program (S.K.), The Swedish Medical Research Council (S.K.), The Swedish Children Cancer Foundation (S.K.), a Clinical Research Award from Lund University Hospital (S.K.), The Joint Program on Stem Cell Research supported by The Juvenile Diabetes Research Foundation and The Swedish Medical Research Council (S.K.), and Kungliga Fysiografiska Sällskapet (U.B.). The Lund Stem Cell Center is supported by a Center of Excellence grant in life sciences from the Swedish Foundation for Strategic Research.
We thank Dr C.-X. Deng for the Smad4 conditional mice; Dr P. ten Dijke for the Smad7 cDNA; Drs M.-J. Goumans and R. Månsson for valuable advice; and E. Gynnstam, A. Fossum, and Z. Ma for expert technical assistance.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal