Abstract
Nucleophosmin (NPM) exon-12 mutations occur in 50% to 60% of adult acute myeloid leukemia (AML) with normal karyotype and are predictors of favorable prognosis. We evaluated bone marrow or peripheral blood samples from 450 adult patients with AML of the GIMEMA (Gruppo Italiano Malattie Ematologiche Maligne dell'Adulto)/AML12 EORTC (European Organization for Research and Treatment of Cancer) trial to (1) search for new exon-12 NPM mutations; (2) determine whether NPM immunostaining on paraffin-embedded biopsies predicts NPM mutations; and (3) investigate altered nucleocytoplasmic NPM traffic in primary AML cells. Fourteen NPM mutations, including 8 new variants, were identified. All 200 AML cases expressing cytoplasmic NPM (NPMc+ AML) carried NPM mutations. None of the 250 cases with nucleus-restricted NPM (NPMc– AML) was mutated. At the C-terminus, NPM leukemic mutants carried mutations of only tryptophan 290 or of both tryptophans 288 and 290 and a new nuclear export signal (NES) motif, which appear to underlie their nuclear export. The specific Crm1/exportin-1 inhibitor leptomycin-B relocated NPM mutants from cytoplasm to nucleus of primary NPMc+ AML cells, demonstrating that nuclear export is NES dependent. NPM mutants bound and recruited wild-type NPM into leukemic cell cytoplasm. Because alterations at C-terminus of leukemic NPM mutants are similar, immunohistochemistry detects all exon-12 NPM mutations and is a valuable, inexpensive tool in the diagnostic-prognostic work-up of patients with AML with normal karyotype.
Introduction
Acute myeloid leukemia (AML) is a clinically and molecularly heterogeneous disease. Cytogenetic and/or molecular studies are used to assign 30% to 40% of AML cases carrying specific genetic lesions (eg, t(15;17), t(8;21) or Inv(16)) to different prognostic subgroups in order to monitor minimal residual disease and to select patients who could benefit from targeted therapies.1 However, they cannot be applied to 40% to 50% of patients with AML who at conventional cytogenetics exhibit a normal karyotype.1,2 We recently found exon-12 nucleophosmin (NPM) gene mutations in approximately 60% of AMLs with normal karyotype (about one third of all adult AML) that are characterized by distinct morphologic, phenotypic, and molecular features,3 as well as distinct gene expression profile signature.4 NPM mutations predict good response to induction therapy and favorable prognosis,5-8 and could serve to monitor minimal residual disease.9 Thus, analysis of NPM mutations emerges as a major new step in the diagnostic/prognostic work-up of patients with AML with a normal karyotype.
Reverse-transcriptase–polymerase chain reaction (RT-PCR), mutational analysis, or gene expression profiling is usually used for detecting genetic abnormalities or specifically overexpressed genes with diagnostic/prognostic significance. However, demand is growing for simple, inexpensive, and specific immunologic tests10 that can serve as a surrogate to, or used in integration with, molecular studies. Examples include immunohistochemical detection of proteins such as anaplastic lymphoma kinase (ALK) in CD30+ anaplastic large cell lymphoma,10 Zap-70 in chronic lymphocytic leukemia,11 annexin A1 in hairy cell leukemia,12 myeloid leukemia factor-1 (MLF1) in myelodysplasia/AML with t(3;5),13 and promyelocytic leukemia (PML) in acute promyelocytic leukemia carrying the t(15;17).14 In a small group of patients with AML, we showed that aberrant cytoplasmic expression of NPM, a multifunctional15-20 nucleocytoplasmic shuttling protein21 with restricted nucleolar localization,10,22 closely correlates with exon-12 NPM mutations.3 To validate these observations, the present study determined whether immunostaining for NPM in paraffin-embedded biopsies reliably predicts all exon-12 NPM mutations in a large cohort of adult patients with AML (n = 450) from the GIMEMA (Gruppo Italiano Malattie Ematologiche Maligne dell'Adulto)/AML12 EORTC (European Organization for Research and Treatment of Cancer) trial. In a few cases, we investigated the mechanism of altered nucleocytoplasmic traffic of wild-type and mutated NPM in primary AML cells.
Patients, materials, and methods
Case population
We selected bone marrow or peripheral blood samples from 450 patients with AML (ages 15-60 years) who were enrolled in the GIMEMA LAM99P/AML12 EORTC trial. Seventy of the 450 patients were previously reported by the authors.3 Inclusion criteria were provision of full information on subcellular expression of the NPM protein and NPM gene mutational status. Written informed consent to examine leukemic samples was obtained at each participating center, in accordance with the Declaration of Helsinki. Study protocol was approved by the ethics committee of each center participating in the trial.
Analysis of NPM mutations in leukemic samples
Antibodies
The value of immunohistochemistry in predicting exon-12 NPM mutations was assessed through detection of aberrant NPM cytoplasmic expression in leukemic cells. We immunostained 450 AML samples with mouse anti-NPM monoclonal antibodies (clones 322 and 376) that are suitable for detecting NPM in paraffin-embedded sections.3 These antibodies recognize both the wild-type NPM and the NPM leukemic mutants.
In selected NPMc+ AML cases, we used specifically directed antibodies to investigate by Western blot and coimmunoprecipitation the subcellular distribution of NPM leukemic mutants without interference of wild-type NPM (and vice versa) and to assess whether mutant and wild-type NPM proteins physically interact. NPM leukemic mutants were detected using a rabbit affinity-purified polyclonal antibody (Sil-C) generated against the synthetic peptide NHCOCH3-CLAVEEVSLRK-COOH (Inbio, Tallin, Estonia) corresponding to NPM mutant A.24 Wild-type NPM was detected using a mouse monoclonal antibody (clone FC-61991) recognizing wild-type, but not mutated, NPM (Invitrogen, Carlsbad, CA). Monoclonal antibodies against nucleolin (C23) and fibrillarin, nucleolar proteins used as controls, were purchased from Santa Cruz Biotechnology (Heidelberg, Germany) and Abcam (Cambridge, United Kingdom), respectively.
Immunohistochemical studies
All immunohistochemical procedures were performed blindly on paraffin sections cut from B5-fixed/EDTA-decalcified bone marrow biopsies (438 AML cases) or pellets of Ficoll-separated mononuclear cells (12 patients with AML presenting with high peripheral blood cell counts), as previously described.3 Sections were subjected to microwaving/antigen retrieval in 0.1 mM EDTA, pH 8.0, and immunostained with anti-NPM monoclonal antibodies (clones 376 and 322) using the immuno–alkaline phosphatase (APAAP) technique.3 The distinction between NPM cytoplasmic–positive (NPMc+) and NPM cytoplasmic–negative (NPMc–) cases was clear-cut, since most NPMc+ AML cases displayed a strong cytoplasmic expression of NPM in the majority of leukemic cells, with exception of M5b, in which NPMc+ blast cells ranged between 30% and 75%. Therefore, no grading system for the intensity of immunohistochemical staining was adopted. Accordingly, the AML cases were separated just as NPMc+ or NPMc–, as we previously reported.3 Paraffin sections from all cases were stained in parallel for nucleolin (C23), another nucleolar protein that, in NPMc+ AML cases, is nucleus restricted.3 Nucleus-restricted versus cytoplasmic NPM localization was correlated with the presence/absence of exon-12 NPM mutations.
Analysis of nucleocytoplasmic traffic of NPM in primary leukemic cells
Immunofluorescence analysis of NPM nucleocytoplasmic traffic. Studies were carried out on Ficoll-separated primary leukemic cells from 5 patients with AML carrying cytoplasmic/mutated NPM and presenting with high peripheral blood counts. Positive control was the human AML cell line OCI/AML3 carrying exon-12 NPM mutation A.25 Leukemic cells from the 5 NPMc+ AMLs and OCI/AML3 cells were seeded in growth medium (106 cells/mL in 24-well plates) and incubated at 37°C with 5% CO2 for 5 hours. After overnight incubation with leptomycin B (LMB, 20 ng/mL), cells were washed in PBS, centrifuged, fixed in B5, and embedded in paraffin for immunostaining. NPM was detected with anti-NPM monoclonal antibody (clone 376) followed by Alexa 488–conjugated secondary goat antimouse antibody (Molecular Probes, Eugene, OR); nuclei were counterstained with propidium iodide. Images were collected with a Zeiss LSM 510 confocal microscope (Carl Zeiss, Jena, Germany) using 488-nm (for Alexa 488) and 543-nm (for Texas Red and propidium iodide) laser lines for excitation. AOTF-controlled tuning of laser lines, pinhole diameters, and light collection configuration were optimized to obtain best signal-to-noise ratio and to avoid fluorescence crossover. LSM 510 software (Carl Zeiss) regulated the microscope, and collected images were transferred to an SGI Octane workstation (Silicon Graphics, Mountain View, CA) for further processing. Slices were reconstructed in 3D using the shadow technique or isosurface analysis, with Imaris software (Bitplane, Zurich, Switzerland).
Western blot and coimmunoprecipitation studies. Cells for biochemical studies were available in 3 of 5 NPMc+ patients with AML. For Western blotting, leukemic cells were incubated at 37°C with or without LMB (20 ng/mL) for 18 hours and washed with PBS. Dry cell pellets were snap-frozen in liquid nitrogen. Frozen pellets were thawed and lysed in hypotonic buffer using the slightly modified method of Schreiber et al.26 The supernatant was saved as cytoplasmic fraction. The pellet fraction, containing nuclei, was washed with hypotonic buffer and dissolved in hypertonic buffer. After centrifugation, the supernatant was recovered as nuclear extract; the pellet was saved as insoluble fraction. Equivalent dilutions (representing equal number of cells) of cytoplasmic, nuclear, or insoluble fractions were prepared for Western blot analysis of endogenous NPM mutant protein distribution using Sil-C, the specific anti-NPM mutant antibody.
To test whether wild-type and mutant NPM proteins interact with each other and with Crm1/exportin-1, coimmunoprecipitation experiments were performed on leukemic cell lysates from NPMc+ and NPMc– patients with AML, with OCI/AML3 cell line as control. Cell lysates were prepared as previously described,24 incubated with 2 μg of either control IgG, specific anti-NPM wild-type monoclonal antibody (clone FC-61991), or anti–mutated NPM rabbit polyclonal antibody (Sil-C), followed by 30 μL Protein A/G Plus-agarose beads (Santa Cruz Biotechnology) rocking at 4°C for 3 hours. Beads were washed with buffer containing 0.1% NP-40, 150 mM NaCl, 25 mM Tris, pH 7.5, 1 mM EDTA, and inhibitors. Proteins were separated by SDS–polyacrylamide gel electrophoresis, transferred to polyvinylidene difluoride (PVDF; Millipore, Billerica, MA), and probed with rabbit polyclonal antibody anti–mutated NPM (Sil-C), mouse monoclonal anti-NPM wild-type antibody (clone FC-61991), or mouse monoclonal anti-Crm1 antibody (BD Transduction Laboratories, Milan, Italy), followed by horseradish peroxidase–conjugated secondary antibodies. Polypeptides
Results
Exon-12 NPM mutations cause similar alterations at C-terminus of NPM mutants
NPM mutations were detected in 200 (44.4%) of 450 AML cases (Table 1). The frequency was higher than the expected 35% in adult AML as detected by cytoplasmic dislocation of NPM,3 because of case selection bias. In fact, DNA or RNA for NPM mutational analysis was more frequently available in patients with normal karyotype (often associated with NPM mutations) than in those carrying other genetic abnormalities.
Correlation between immunohistochemistry and exon-12 NPM mutations in 450 patients with AML
NPM location* . | No. patients with AML . | NPM mutations (exon-12) . |
|---|---|---|
| Cytoplasmic | 200 | 200 |
| Nuclear | 250 | 0 |
NPM location* . | No. patients with AML . | NPM mutations (exon-12) . |
|---|---|---|
| Cytoplasmic | 200 | 200 |
| Nuclear | 250 | 0 |
N = 450.
Detected by immunostaining of paraffin sections.
All NPM-mutated AMLs were heterozygous and retained a wild-type allele. Fourteen NPM mutations were identified (Table 2). Besides the original mutations A to F,3 we detected 8 new exon-12 NPM mutation variants, named J to Q (Table 2). Seven (J to P) are reported here for the first time. Mutation Q corresponds to mutation Mm, as described by Schnittger et al.6 Confirming our original findings,3 type A mutation, a TCTG insertion/duplication between position nt 960 and nt 961, was the most common variant (80.5% of cases) followed by mutation B (7.5%) and mutation D (5.5%). Other NPM mutations occurred rarely.
Immunohistochemical prediction of exon-12 NPM mutations in 200 adult NPMc+ AML patients
Mutation . | Nucleotide sequence . | . | . | . | . | Protein sequence . | Frequency of mutations, % . | IH . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| WT* | gaccaagaggctattcaagatctct | g | gcag | t | ggaggaagtctctttaagaaaatag | 286-DLWQWRKSL | Nucl | |||||
| A | gaccaagaggctattcaagatctct | gTCTG | gcag | t | ggaggaagtctctttaagaaaatag | 286-DLCLAVEEVSLRK | 80.5 | Cyt | ||||
| B | gaccaagaggctattcaagatctct | gCATG | gcag | t | ggaggaagtctctttaagaaaatag | 286-DLCMAVEEVSLRK | 7.5 | Cyt | ||||
| C | gaccaagaggctattcaagatctct | gCGTG | gcag | t | ggaggaagtctctttaagaaaatag | 286-DLCVAVEEVSLRK | < 1 | Cyt | ||||
| D | gaccaagaggctattcaagatctct | gCCTG | gcag | t | ggaggaagtctctttaagaaaatag | 286-DLCLAVEEVSLRK | 5.5 | Cyt | ||||
| E | gaccaagaggctattcaagatctct | g | gcag | tCTCTTGCCC | aagtctctttaagaaaatag | 286-DLWQSLAQVSLRK | < 1 | Cyt | ||||
| F | gaccaagaggctattcaagatctct | g | gcag | tCCCTGGAGA | aagtctctttaagaaaatag | 286-DLWQSLEKVSLRK | < 1 | Cyt | ||||
| J | gaccaagaggctattcaagatctct | g | gcag | tCTCTTTCTA | aagtctctttaagaaaatag | 286-DLWQSLSKVSLRK | < 1 | Cyt | ||||
| L | gaccaagaggctattcaagatctctCCCGg | gcag | t | aagtctctttaagaaaatag | 286-DLSRAVEEVSLRK | < 1 | Cyt | |||||
| K | gaccaagaggctattcaagatctct | g | gcag | tCCCTTTCCA | aagtctctttaagaaaatag | 286-DLWQSLSKVSLRK | < 1 | Cyt | ||||
| M | gaccaagaggctattcaagatctct | gTAGC | gcag | t | ggaggaagtctctttaagaaaatag | 286-DLCTAVEEVSLRK | < 1 | Cyt | ||||
| N | gaccaagaggctattcaagatctct | gCCAC | gcag | t | ggaggaagtctctttaagaaaatag | 286-DLCHAVEEVSLRK | < 1 | Cyt | ||||
| O | gaccaagaggctattcaagatctct | g | gcagCGTTTCC | aggaagtctctttaagaaaatag | 286-DLWQRFQEVSLRK | < 1 | Cyt | |||||
| P | gaccaagaggctattcaagatctct | gTACCTTCC | t | ggaggaagtctctttaagaaaatag | 286-DLCTFLEEVSLRK | < 1 | Cyt | |||||
| Q | gaccaagaggctattcaagatctct | g | gcagAGGA | t | ggaggaagtctctttaagaaaatag | 286-DLWQRMEEVSLRK | < 1 | Cyt | ||||
Mutation . | Nucleotide sequence . | . | . | . | . | Protein sequence . | Frequency of mutations, % . | IH . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| WT* | gaccaagaggctattcaagatctct | g | gcag | t | ggaggaagtctctttaagaaaatag | 286-DLWQWRKSL | Nucl | |||||
| A | gaccaagaggctattcaagatctct | gTCTG | gcag | t | ggaggaagtctctttaagaaaatag | 286-DLCLAVEEVSLRK | 80.5 | Cyt | ||||
| B | gaccaagaggctattcaagatctct | gCATG | gcag | t | ggaggaagtctctttaagaaaatag | 286-DLCMAVEEVSLRK | 7.5 | Cyt | ||||
| C | gaccaagaggctattcaagatctct | gCGTG | gcag | t | ggaggaagtctctttaagaaaatag | 286-DLCVAVEEVSLRK | < 1 | Cyt | ||||
| D | gaccaagaggctattcaagatctct | gCCTG | gcag | t | ggaggaagtctctttaagaaaatag | 286-DLCLAVEEVSLRK | 5.5 | Cyt | ||||
| E | gaccaagaggctattcaagatctct | g | gcag | tCTCTTGCCC | aagtctctttaagaaaatag | 286-DLWQSLAQVSLRK | < 1 | Cyt | ||||
| F | gaccaagaggctattcaagatctct | g | gcag | tCCCTGGAGA | aagtctctttaagaaaatag | 286-DLWQSLEKVSLRK | < 1 | Cyt | ||||
| J | gaccaagaggctattcaagatctct | g | gcag | tCTCTTTCTA | aagtctctttaagaaaatag | 286-DLWQSLSKVSLRK | < 1 | Cyt | ||||
| L | gaccaagaggctattcaagatctctCCCGg | gcag | t | aagtctctttaagaaaatag | 286-DLSRAVEEVSLRK | < 1 | Cyt | |||||
| K | gaccaagaggctattcaagatctct | g | gcag | tCCCTTTCCA | aagtctctttaagaaaatag | 286-DLWQSLSKVSLRK | < 1 | Cyt | ||||
| M | gaccaagaggctattcaagatctct | gTAGC | gcag | t | ggaggaagtctctttaagaaaatag | 286-DLCTAVEEVSLRK | < 1 | Cyt | ||||
| N | gaccaagaggctattcaagatctct | gCCAC | gcag | t | ggaggaagtctctttaagaaaatag | 286-DLCHAVEEVSLRK | < 1 | Cyt | ||||
| O | gaccaagaggctattcaagatctct | g | gcagCGTTTCC | aggaagtctctttaagaaaatag | 286-DLWQRFQEVSLRK | < 1 | Cyt | |||||
| P | gaccaagaggctattcaagatctct | gTACCTTCC | t | ggaggaagtctctttaagaaaatag | 286-DLCTFLEEVSLRK | < 1 | Cyt | |||||
| Q | gaccaagaggctattcaagatctct | g | gcagAGGA | t | ggaggaagtctctttaagaaaatag | 286-DLWQRMEEVSLRK | < 1 | Cyt | ||||
Mutations A to F correspond to those previously described by Falini et al3 ; mutation Q corresponds to mutation Mm described by Schnittger et al.6 Lowercase letters in nucleotide sequences indicate wild-type sequence residues; capital letters, mutated nucleotides. Underlined text indicates leucine-rich NES motif; italics, tryptophan (W) residues.
IH indicates immunohistochemistry; WT, wild type; Nucl, nuclear; Cyt, cytoplasmic.
NM_002520.
Despite DNA heterogeneity, all NPM mutations resulted in similar alterations at the C-terminus of the predicted mutant proteins (ie, a new leucine-rich nuclear export signal [NES] motif27 [Table 2] and mutated tryptophans 288 and 290 [or tryptophan 290 alone]).24 The most frequent NES motif (L-xxx-V-xx-V-x-L) was present in 7 (50%) of 14 NPM leukemic mutants. Other mutants carried a NES motif with another hydrophobic amino acid replacing valine at the second position (Table 2). In particular, 5 (35.7%) of 14 mutants carried the L-xxx-L-xx-V-x-L motif; 1 (7.1%) of 14 mutants (Table 2, mutation O) displayed the L-xxx-F-xx-V-x-L motif; and 1 (7.1%) of 14 mutants (Table 2, mutation Q) had the L-xxx-M-xx-V-x-L motif. All 14 NPM mutant proteins carried a mutated tryptophan 290, while tryptophan 288 was retained in 6 (42.9%) of 14 mutants (Table 2). In keeping with our previous report,24 the most frequent NES motif (L-xxx-V-xx-V-x-L) was associated with mutations of both tryptophans 288 and 290. Tryptophan 288 was retained only in NPM mutants carrying variants of the most common NES motif (ie, leucine, phenylalanine, or methionine instead of valine at the second position).
Immunohistochemistry predicts all exon-12 NPM gene mutation variants
The results of the immunohistochemical studies are shown in Table 1. Expression of NPM was cytoplasmic (NPMc+) in 200 cases, all carrying NPM exon-12 mutations. NPM immunoreactivity in bone marrow trephines and cell pellets was comparable. Cytoplasmic NPM was predominantly observed in M4 and M5 categories. NPM expression was nucleus restricted (NPMc–) in 250 AML cases, all carrying wild-type NPM gene. All 450 cases were characterized by nucleus-restricted expression of C23 (nucleolin).
One-hundred ninety-five of 200 NPMc+ cases were easily recognized since they exhibited strong cytoplasmic expression of NPM in most leukemic cells. Five cases showing equivocal weak NPM cytoplasmic positivity (most likely due to suboptimal fixation) were reviewed by 2 independent observers and correctly assigned to the NPMc+ category (as proved by the results of mutational analysis) based on the comparison with the C23/nucleolin staining that showed a clear nucleus-restricted reactivity (the staining pattern that would be expected for unmutated NPM). Thus, despite molecular heterogeneity, immunostaining for NPM detected correctly all 14 NPM mutation variants.
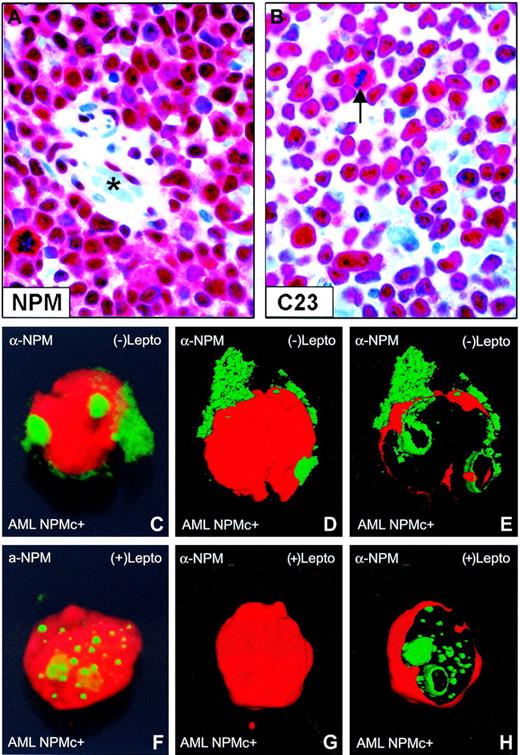
Nuclear relocation of NPM by leptomycin B in primary NPMc+ AML cells. (A-B) Bone marrow from an NPMc+ patient with AML bearing NPM mutation A. (A) Leukemic cells show cytoplasmic (in addition to nuclear) expression of NPM; endothelial cells of a vessel (lumen indicated with an asterisk) show nucleus-restricted NPM (Olympus 100×/1.3 NA UPlanFl oil objective). (B) Leukemic cells, with the exception of a mitotic figure (arrow), show the expected nucleus-restricted expression of C23/nucleolin (Olympus 100×/1.3 NA UPlanFl oil objective). (C-E) Purified leukemic cells from the patient were immunostained with a monoclonal anti-NPM antibody (clone 376) and observed at the confocal laser microscope (100×/1.4 NA PlanApochromat oil objective). Images were reconstructed in 3 dimensions, and an electronic cut of the nuclear surface was performed to better analyze the localization of NPM. The antibody 376, which cannot distinguish between the wild-type and NPM mutant A proteins (both contained in the leukemic cells), labels both nucleoli and cytoplasm in the absence of LMB. (F-H) Incubation with LMB results in nucleoplasmic relocation of cytoplasmic NPM.
Nuclear relocation of NPM by leptomycin B in primary NPMc+ AML cells. (A-B) Bone marrow from an NPMc+ patient with AML bearing NPM mutation A. (A) Leukemic cells show cytoplasmic (in addition to nuclear) expression of NPM; endothelial cells of a vessel (lumen indicated with an asterisk) show nucleus-restricted NPM (Olympus 100×/1.3 NA UPlanFl oil objective). (B) Leukemic cells, with the exception of a mitotic figure (arrow), show the expected nucleus-restricted expression of C23/nucleolin (Olympus 100×/1.3 NA UPlanFl oil objective). (C-E) Purified leukemic cells from the patient were immunostained with a monoclonal anti-NPM antibody (clone 376) and observed at the confocal laser microscope (100×/1.4 NA PlanApochromat oil objective). Images were reconstructed in 3 dimensions, and an electronic cut of the nuclear surface was performed to better analyze the localization of NPM. The antibody 376, which cannot distinguish between the wild-type and NPM mutant A proteins (both contained in the leukemic cells), labels both nucleoli and cytoplasm in the absence of LMB. (F-H) Incubation with LMB results in nucleoplasmic relocation of cytoplasmic NPM.
NPM cytoplasmic accumulation in primary NPMc+ AML is NES dependent
In transfected cells, cytoplasmic accumulation of NPM mutants depends both on a new NES motif and mutated tryptophan(s) at the C-terminus of leukemic mutants.24 To assess whether NES-dependent nuclear export of NPM operates in primary AML cells, we evaluated the effect of LMB, a specific inhibitor of Crm1/exportin-1 (the receptor for protein nuclear export signal in yeast and mammalian cells28 ), on NPM nucleocytoplasmic traffic in leukemic cells from 5 patients with AML carrying cytoplasmic/mutated NPM (Figure 1A-B). After immunostaining with anti-NPM monoclonal antibodies, confocal microscopy analysis of primary leukemic cells showed that cytoplasmic NPM (Figure 1C-E) relocated into nucleoplasm after incubation with LMB (Figure 1F-H).
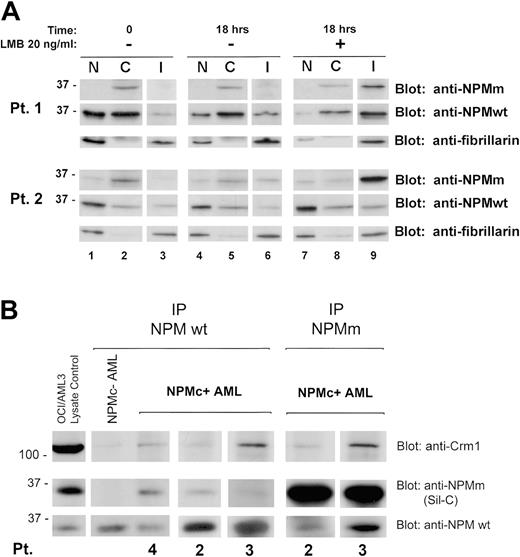
Western blotting confirmed these data in the 2 cases investigated. LMB treatment induced endogenous NPM mutant accumulation in the insoluble fraction (Figure 2A) containing most of the nucleolar compartment (see blot for the nucleolar marker fibrillarin) and nucleoplasmic insoluble structures, possibly appearing as nonnucleolar dots at immunofluorescence (for comparison, see Figure 1F-H). These findings provide evidence that nuclear export of NPM mutants in primary leukemic cells is NES dependent.
NPM mutants bind and recruit wild-type NPM in the cytoplasm of primary NPMc+ AML cells
We investigated whether NPM leukemic mutants physically interact with wild-type NPM and perturb its nucleocytoplasmic traffic. Immunohistochemical staining of the patients' cell pellets (not shown) and Western blot analysis of subcellular protein extracts with antibodies specifically directed against wild-type and mutant NPM (Figure 2A, Pt 1 and Pt 2, time 0, upper and middle panels) showed both forms delocalized into leukemic cell cytoplasm, suggesting mutant NPM bound and recruited wild-type NPM from nucleus to cytoplasm.
Coimmunoprecipitation experiments showed mutant NPM (recognized by Sil-C antibody) interacts with wild-type NPM protein (recognized by the specific anti-NPM wild-type antibody) (Figure 2B), suggesting cytoplasmic dislocation of wild-type NPM may occur through formation of NPM mutant/NPM wild-type heterodimers inside leukemic cells. Crm1/exportin-1 protein coprecipitated more with heterodimers containing mutant NPM than with wild-type NPM homodimers. These findings are consistent with the presence of a physiological NES motif in wild-type NPM29 and point to an additive effect of the acquired C-terminus NES motif for Crm1-mediated nuclear export of NPM mutants. These results confirm in primary leukemic cells our previous observations in transfected cells.24 Incubation with LMB relocated cytoplasmic wild-type NPM into the insoluble nuclear fraction, reducing the protein in the other compartments (Figure 2A, compare lanes 7-9 with lanes 1-3).
NES-dependent altered nucleocytoplasmic traffic of NPM in NPMC+ AML. (A) Subcellular distribution of wild-type and mutant NPM in NPMc+ AML cells. Data of experiments on 2 NPMc+ AML patients are shown. The anti-NPMm blotting (both patients, top panels) shows accumulation, upon LMB treatment, of NPM mutant protein in the insoluble fraction (I) (both patients, lane 9, top panels). Relocation, upon LMB treatment, of NPM wild-type protein in the same fraction was demonstrated by blotting the membrane with an anti-NPMwt–specific antibody (both patients, lane 9, middle panels). The insoluble fraction represents mainly subcellular fraction containing most of the nucleolar compartment, as assessed by stripping the membrane and reblotting for the nucleolar marker fibrillarin (both patients, bottom panels, lanes 3, 6, and 9), and possibly other insoluble nuclear structures. NPMm indicates mutated NPM; NPMwt, wild-type NPM. (B) Interaction between wild-type NPM, leukemic NPM mutants, and Crm1 in NPMc+ AML cells. Cell lysates from either NPMc– or NPMc+ AML patients were subjected to immunoprecipitation (IP) with either control IgG (not shown), mouse monoclonal anti-NPMwt antibody from Invitrogen (Pts 4, 2, and 3), or rabbit polyclonal antibody anti–mutated NPM (anti-NPMm, Sil-C) (Pts 2 and 3), as indicated. Coprecipitation of either NPM mutant protein or wild-type NPM, in the reciprocal experiments, was detected by Western blot with the anti-NPMm–specific antibody Sil-C (middle panels) and anti-NPMwt–specific antibody (bottom panels), respectively. Stechiometry of the NPMwt/NPMm protein complex changes with patients. Western blot analysis with a rabbit polyclonal antibody anti-Crm1 shows a much stronger signal in the NPMc+ than NPMc– AML sample (top panels). OCI/AML3 cell lysate was included as control for antibody activity. NPMm indicates mutated NPM; NPMwt, wild-type NPM.
NES-dependent altered nucleocytoplasmic traffic of NPM in NPMC+ AML. (A) Subcellular distribution of wild-type and mutant NPM in NPMc+ AML cells. Data of experiments on 2 NPMc+ AML patients are shown. The anti-NPMm blotting (both patients, top panels) shows accumulation, upon LMB treatment, of NPM mutant protein in the insoluble fraction (I) (both patients, lane 9, top panels). Relocation, upon LMB treatment, of NPM wild-type protein in the same fraction was demonstrated by blotting the membrane with an anti-NPMwt–specific antibody (both patients, lane 9, middle panels). The insoluble fraction represents mainly subcellular fraction containing most of the nucleolar compartment, as assessed by stripping the membrane and reblotting for the nucleolar marker fibrillarin (both patients, bottom panels, lanes 3, 6, and 9), and possibly other insoluble nuclear structures. NPMm indicates mutated NPM; NPMwt, wild-type NPM. (B) Interaction between wild-type NPM, leukemic NPM mutants, and Crm1 in NPMc+ AML cells. Cell lysates from either NPMc– or NPMc+ AML patients were subjected to immunoprecipitation (IP) with either control IgG (not shown), mouse monoclonal anti-NPMwt antibody from Invitrogen (Pts 4, 2, and 3), or rabbit polyclonal antibody anti–mutated NPM (anti-NPMm, Sil-C) (Pts 2 and 3), as indicated. Coprecipitation of either NPM mutant protein or wild-type NPM, in the reciprocal experiments, was detected by Western blot with the anti-NPMm–specific antibody Sil-C (middle panels) and anti-NPMwt–specific antibody (bottom panels), respectively. Stechiometry of the NPMwt/NPMm protein complex changes with patients. Western blot analysis with a rabbit polyclonal antibody anti-Crm1 shows a much stronger signal in the NPMc+ than NPMc– AML sample (top panels). OCI/AML3 cell lysate was included as control for antibody activity. NPMm indicates mutated NPM; NPMwt, wild-type NPM.
These findings demonstrate that wild-type NPM protein physically interacts with NPM mutants in NPMc+ AML patient cells. This interaction is likely responsible for the aberrant cytoplasmic accumulation of wild-type NPM and its relocation, together with the NPM mutant, to the nuclear compartment upon Crm1 inhibition by LMB.
Discussion
This paper describes 8 sporadic new exon-12 NPM mutation variants and, in a large series of AML cases, demonstrates that immunohistochemistry serves as a highly specific predictor of all exon-12 NPM mutations. It also shows that cytoplasmic accumulation of NPM mutants in cells from NPMc+ patients with AML is mediated by the NES motif, and provides biochemical evidence that NPM mutants physically interact with wild-type NPM to dislocate it aberrantly into primary leukemic cell cytoplasm.
Although 8 new NPM mutations, besides 29 so far identified,3,5-7,30,31 confirm that mutations at exon-12 of the NPM gene are heterogeneous, it remains a mystery why these mutations occur at such a high frequency in, and are so specific to, AML.
Here we report a perfect correlation between cytoplasmic expression of NPM and the presence of exon-12 NPM mutations. This immunohistochemical finding is consistent with the presence at the C-terminus of all 14 NPM mutant proteins of a new NES motif27 and mutated tryptophans 288 and 290 (or tryptophan 290 alone). In fact, in transfected cells, both of these molecular alterations are crucial for aberrant NPM mutant export from nucleus to cytoplasm.24 In primary AML cells, the role of carboxyterminus–mutated tryptophan(s) in NPM cytoplasmic accumulation is difficult to demonstrate experimentally. However, the role of the NES motif in aberrant NPM nuclear export can be addressed using LMB, a specific inhibitor of Crm1, the receptor for protein nuclear export signal in yeast and mammalian cells.28 In the present study, 5 NPMc+ AMLs were investigated by immunohistochemistry and biochemical techniques. In all cases, cytoplasmic NPM was completely relocated into the nucleus following incubation with LMB. Finally, coimmunoprecipitation experiments, carried out in 3 NPMc+ AML cases, showed that NPM leukemic mutants directly bind to wild-type NPM protein, recruiting it into the leukemic cell cytoplasm.
In addition to the mutations described in this study, NPM cytoplasmic dislocation has been also reported by Thiede et al8 in association with exon-12 NPM mutations termed DD-1, DD-3, DD-4, and DD-9. Unfortunately, no immunohistochemical data on NPM subcellular localization are available for the other exon-12 NPM mutation variants that have been reported to date.5-7,30-32 However, all leukemic NPM mutants in those reports carried a new NES and mutated tryptophan(s) at their C-terminus. Thus, it is reasonable to assume that, as in the present study, immunohistochemistry would detect NPM mutant proteins as cytoplasmic-dislocated and serve as a highly specific predictor of exon-12 NPM mutations.
Immunostaining for NPM is another example of how a leukemia-specific genetic lesion may be identified by investigating localization of altered gene products. A similar approach in acute promyelocytic leukemia is the immunocytologic detection of t(15;17) through visualization of wild-type PML protein delocalization from nuclear bodies to microspeckles, mediated by the chimeric PML-RARα protein.10,14
As NPM protein is not reliably detected in smears and cytospins,3,22 we recommend immunostaining bone marrow biopsy samples, although paraffin-embedded bone marrow clots33 or leukemic cell pellets are valid alternatives. Given its low cost and high specificity, the NPM immunohistochemical test seems ideal for centers that are not equipped for molecular screening and do not have highly specialized personnel.
In state-of-the-art centers, it might configure as the first step in the differential characterization of AML by molecular and genetic studies. A proposal of how the immunohistochemical test could be used for this purpose is shown in Figure 3. In NPMc– AML, molecular/cytogenetics analyses are mandatory for detecting major recurrent changes (eg, t(15;17), t(8;21), inv(16), t(6;9), and 11q23/MLL rearrangements). In NPMc+ AML, they can be avoided since they do not concur with cytoplasmic NPM.3 In approximately 14% of NPMc+ AMLs with minor chromosomal abnormalities,3 cytogenetics help pick up rare changes with potential prognostic significance. NPM mutational analysis is unnecessary in NPMc– AML but should be performed in samples of NPMc+ AML (Figure 3) since it can serve as a marker for monitoring minimal residual disease in patients with AML with a normal karyotype.9 FLT3 gene mutations34 should be sought since their correlation with cytoplasmic/mutated NPM helps identify new prognostic subgroups in AML with normal karyotype.5-8 Validation of the integrated NPM-based immunohistochemical-cytogenetic-molecular approach to AML characterization in large multicenter trials is warranted.
NPM-immunostaining–based approach to molecular-cytogenetic study of AML. AML cases can be simply subdivided by immunohistochemistry in 2 groups (ie, expressing nucleus-restricted NPM [NPMc–] or cytoplasmic NPM [NPMc+]; both figures were captured with an Olympus 100×/1.3 UPlanFl oil objective). The arrow in the NPMc– panel indicates the expected cytoplasmic positivity of a mitotic figure. Molecular studies for recurrent genetic abnormalities can be avoided in NPMc+ AML since the 2 events are mutually exclusive. For the same reason, mutational analysis of NPM is unnecessary in patients with NPMc– AML. Cytogenetics and other molecular studies should be performed in both NPMc+ and NPMc– patients with AML for diagnostic and investigational purposes.
NPM-immunostaining–based approach to molecular-cytogenetic study of AML. AML cases can be simply subdivided by immunohistochemistry in 2 groups (ie, expressing nucleus-restricted NPM [NPMc–] or cytoplasmic NPM [NPMc+]; both figures were captured with an Olympus 100×/1.3 UPlanFl oil objective). The arrow in the NPMc– panel indicates the expected cytoplasmic positivity of a mitotic figure. Molecular studies for recurrent genetic abnormalities can be avoided in NPMc+ AML since the 2 events are mutually exclusive. For the same reason, mutational analysis of NPM is unnecessary in patients with NPMc– AML. Cytogenetics and other molecular studies should be performed in both NPMc+ and NPMc– patients with AML for diagnostic and investigational purposes.
In routine diagnostics, when no fresh tissue is available for cytogenetic/molecular studies (eg, marrow punctio sicca or small biopsies such as in skin involvement by myeloid sarcoma35 ), the NPM-based immunohistochemical test is at present the only tool for diagnosing AML carrying NPM exon-12 mutations (NPMc+ AML). When cytogenetics fails because of lack of mitoses, as in 20% to 40% of AML in multicenter trials, the NPM-based immunohistochemical test can serve to diagnose normal karyotype and absence of recurrent genetic rearrangements. In retrospective screening for NPM mutations in paraffin sections of archived biopsy material from patients with AML, the immunohistochemical test can replace molecular biology procedures. Moreover, as immunohistochemistry correlates cytoplasmic expression of NPM (and therefore NPM mutations) with leukemic cell morphology, it may be a valuable tool for studying hemopoietic lineage involvement in NPMc+ AML. Detecting NPM mutations by immunohistochemistry may also impact on the current AML classification because cytoplasmic NPM identifies a type of leukemia with distinct clinicobiologic features,3 which, in the World Health Organization (WHO) classification, would be grouped (given their normal karyotype) under the term “acute myeloid leukemia not otherwise categorized.”36 Immunohistochemical detection of NPM mutations is of prognostic value since they appear to be predictors of good response to induction therapy3,6 and long-term prognosis5-8 in AML with normal karyotype. In the future, specific anti–mutant NPM antibodies are expected to play a role in monitoring minimal residual disease.
In conclusion, inserting NPM immunostaining in the diagnostic-prognostic work-up of AML offers significant advantages in terms of costs and effectiveness.
Appendix
The centers and investigators contributing to the GIMEMA Study were as follows (listed in order of the number of cases provided): Istituto di Ematologia, Università La Sapienza, Rome (G. Meloni); Divisione di Ematologia, Ospedale V. Cervello, Palermo (F. Fabbiano); Cattedra di Ematologia, Bari (V. Liso); Divisione di Ematologia, Azienda USL di Pescara, Pescara (M. Sborgia); Ospedale Ferrarotto S. Bambino, Catania (F. Di Raimondo); Divisione di Ematologia, Ospedale S. Eugenio, Rome (A. Venditti); Divisione di Ematologia e Oncologia Clinica, Catanzaro (D. Magro); Dipartimento di Emato-Oncologia, Azienda Ospedaliera Bianchi-Melacrino-Morelli, Reggio Calabria (F. Nobile); Istituto di Ematologia, Università Federico II, Naples (B. Rotoli); Azienda Ospedaliera S.G. Moscati, Avellino (N. Cantore); Divisione di Ematologia, Ospedale Casa Sollievo della Sofferenza, S. Giovanni Rotondo (L. Melillo); Istituto di Ematologia, Ospedale A. Businco, Cagliari (E. Angelucci); Istituto di Ematologia, Policlinico Monteluce, Perugia (A. Tabilio); Cattedra di Ematologia, Ospedale S. Chiara, Pisa (M. Petrini); Policlinico Gemelli, Rome (S. Sica); Università di Ancona, Ancona (P. Leoni); Sezione di Medicina Interna, Oncologia ed Ematologia, Dipartimento Scienze Mediche, Oncologiche e Radiologiche, Modena (G. Torelli); Divisione di Ematologia, Ospedale SS. Antonio e Biagio, Alessandria (A. Levis); Divisione di Ematologia, Fondazione Centro S. Raffaele del Monte Tabor, Milan (L. Camba); Divisione di Ematologia, Ospedale S. Carlo, Potenza (F. Ricciuti); Divisione di Ematologia, Ospedale S. Giovanni Bosco, Naples (E. Miraglia); Azienda Ospedaliera A. Di Summa, Brindisi (G. Quarta); Divisione di Ematologia, Ospedale S. Francesco, Nuoro (A. Gabbas); Divisione di Ematologia con Trapianto Midollo Osseo, Università di Palermo, Palermo (M.E. Mitra); Cattedra di Ematologia-Centro Trapianto Midollo Osseo, Università di Parma, Parma (V. Rizzoli); Divisione Ematologica di Muraglia, Ospedale S. Salvatore, Pesaro (G. Sparaventi); Divisione di Ematologia, Azienda Ospedaliera Cremona, Cremona (S. Moranti); Divisione di Ematologia, Ospedale S. Croce, Cuneo (A. Gallamini); Divisione di Medicina Interna, Ospedale S. Luigi, Orbassano (A. Serra); Sezione Ematologia, Dipartimento Scienze Biomediche, Arcispedale S. Anna, Ferrara (P.-L. Castaldi); Divisione di Ematologia, Università di Sassari, Sassari (F. Dore); Divisione di Medicina, Azienda Ospedaliera E. Morelli, Sondalo (E. Epis); Divisione Medicina 1, Ospedale S. Antonio Abate, Gallarate (R. Mozzana); Divisione Medica, Ospedale Maggiore, Lodi (G. Nalli); Azienda Sanitaria Locale Salerno 1, Medicina Interna, Ematologia-Oncologia, Nocera Inferiore (A.M. D'Arco); Divisione di Ematologia, Policlinico Careggi, Florence (P.-L. Rossi Ferrini); Unità Operativa Ematologia, Ospedale di Foggia, Foggia (M. Monaco); Divisione di Ematologia, Ospedale di Messina, Messina (M. Brugiatelli); and S. Vincenzo, Ospedale di Taormina, Divisione di Ematologia, Taormina (M. Russo)—all in Italy.
Prepublished online as Blood First Edition Paper, May 23, 2006; DOI 10.1182/blood-2006-03-007013.
The membership list for the GIMEMAAcute Leukemia Working Party is listed in the “Appendix.”
Supported by the Italian Association for Cancer Research (AIRC). N.B. is a recipient of a fellowship from the Italian Federation for Cancer Research (FIRC). B.F. and C.M. have applied for a patent on clinical use of NPM mutants.
B.F. had the original idea for the study; N.B. and A.L. were involved in the functional studies of NPM nucleocytoplasmic traffic; M.P.M, M.T.P. and B.V.G. were involved in biochemical studies; I.N. and R.M. were involved in the confocal microscopic analysis of cells; R.P., A.T., and E.T. were involved in the immunohistochemical study of AML samples; C.M. was involved in the identification of NPM mutations; R.B., D.D., G.R., R.R., M.A., L.L., S.V., L.B., E.G., A.G., G.S., F.P., F.L.-C., and P.-G.P. were involved in the analysis of NPM gene mutations in the GIMEMA-EORTC trial; and F.M., S.A., G.S., and M.F.M. were involved in organizing the clinical trial.
M.P.M. and N.B. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Dr Geraldine Boyd for editing the paper. We acknowledge Valentina Pettirossi for her help in coimmunoprecipitation studies. We thank Federica Frenguelli, Barbara Bigerna, and Alessandra Pucciarini for the helpful technical assistance, and Claudia Tibidò for secretarial assistance.

![Figure 3. NPM-immunostaining–based approach to molecular-cytogenetic study of AML. AML cases can be simply subdivided by immunohistochemistry in 2 groups (ie, expressing nucleus-restricted NPM [NPMc–] or cytoplasmic NPM [NPMc+]; both figures were captured with an Olympus 100×/1.3 UPlanFl oil objective). The arrow in the NPMc– panel indicates the expected cytoplasmic positivity of a mitotic figure. Molecular studies for recurrent genetic abnormalities can be avoided in NPMc+ AML since the 2 events are mutually exclusive. For the same reason, mutational analysis of NPM is unnecessary in patients with NPMc– AML. Cytogenetics and other molecular studies should be performed in both NPMc+ and NPMc– patients with AML for diagnostic and investigational purposes.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/108/6/10.1182_blood-2006-03-007013/4/m_zh80180601250003.jpeg?Expires=1770969394&Signature=b4RCXZHUQLVHaSBfoJntEsVADIDWfZ-j9QDWtKU8Yp9fheSfVoGHlzPdELfeRyYrAn13A-yrtH4YalvlrxzJItADxx7UxTaT94gzDQkYB9cgzqQaYNpQLRz~LLhPXzxqmg4SwC~jRPCi50VkX~tywQXBxMB3mPu~dr8h5UdDyoM5hL1n7LeauuyqJROnK6RZbbdR~B-jFe~AqJejug4rixyWsMDPIHFkIp6dOBy7ljxQ5NVFnqVRCAvKtINtfQ8IXNRr9TAgZeb0Ghe3QpXTGKC58aqs-~5reCJ6GPbQbnh4QSFqVGzYcxpQTN0UyBcnwhXcUbpM2lxu1MUEI2vsvw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal