Abstract
Ferroportin disease is caused by mutation of one allele of the iron exporter ferroportin (Fpn/IREG1/Slc40a1/MTP1). All reported human mutations are missense mutations and heterozygous null mutations in mouse Fpn do not recapitulate the human disease. Here we describe the flatiron (ffe) mouse with a missense mutation (H32R) in Fpn that affects its localization and iron export activity. Similar to human patients with classic ferroportin disease, heterozygous ffe/+ mice present with iron loading of Kupffer cells, high serum ferritin, and low transferrin saturation. In macrophages isolated from ffe/+ heterozygous mice and through the use of Fpn plasmids with the ffe mutation, we show that Fpnffe acts as a dominant negative, preventing wild-type Fpn from localizing on the cell surface and transporting iron. These results demonstrate that mutations in Fpn resulting in protein mislocalization act in a dominant-negative fashion to cause disease, and the Fpnffe mouse represents the first mouse model of ferroportin disease.
Introduction
Hereditary hemochromatosis is a common disorder in humans, characterized by iron overload resulting in tissue injury and ultimately organ failure. Typically, hemochromatosis exhibits an autosomal-recessive pattern of inheritance and is associated with mutations in HFE, hemojuvelin, hepcidin, or transferrin receptor 2.1,2 Targeted deletion of these genes in the mouse results in hemochromatosis, providing mouse models for most forms of the disease. Hemochromatosis type IV, also referred to as ferroportin (Fpn) disease, results from mutations in the iron transporter ferroportin. Fpn is the only known iron exporter in mammalian cells and is present on the surface of macrophages, intestinal enterocytes, hepatocytes, and placental cells.3–5 The level of cell surface Fpn is regulated by its interaction with hepcidin, a peptide secreted by the liver in response to iron stores and inflammation. Hepcidin binds to Fpn, inducing its internalization and degradation, thus regulating the export of iron from cells to plasma.6
Mutations in Fpn lead to iron-overload disease but, in contrast to other forms of hemochromatasis, ferroportin disease exhibits an autosomal-dominant pattern of inheritance.7 The disorder has different presentations depending on the Fpn mutation. Mutations leading to Fpn that is not internalized by hepcidin result in iron accumulation in hepatocytes and high transferrin saturation.8,9 Mutations leading to Fpn that is not appropriately targeted to the cell surface result in iron accumulation in Kupffer cells and low transferrin saturation.9–11 The mechanism by which the disease mutations exert a dominant effect is unclear. Some groups that study the disease suggest that it results from haploinsufficiency,10,12 whereas others suggest that the disorder results from a dominant-negative effect of the mutant allele.9,13 Importantly, all human mutations are missense mutations and mice that are heterozygous for a targeted deletion of Fpn do not show the disease.14 Treatment for hemochromatosis aims to decrease iron load by repeated phlebotomy and this treatment works well for most patients. Many patients with ferroportin disease, however, become anemic with phlebotomy, highlighting the need for a mouse model to develop better treatments.
We report here on a missense mutation in mouse Fpn that results in a disorder that is identical to classic human ferroportin disease. We show that macrophages isolated from mutant mice have no Fpn on their cell surface and that expression of Fpn constructs containing the missense mutation (H32R) affects the behavior of wild-type Fpn. These results show that Fpn disease is due to a dominant-negative effect of the mutant allele and provide the first mouse model for this disorder.
Materials and methods
Generation of mutant mice and identification of Fpn mutation
The ffe mouse line was identified in a screen for recessive ethylnitrosourea (ENU)–induced mutations that cause morphologic abnormalities at embryonic day (E) 12.5.15–17 The ffe mutation was generated on a C57BL/6J genetic background and backcrossed to C3H/HeJ or 129/SvJ. In a mapping cross of 1078 opportunities for recombination, ffe was mapped between Massachusetts Institute of Technology (mit) simple sequence-length polymorphism (SSLP) markers D1mit213 and D1mit528. For high-resolution mapping, additional polymorphic DNA markers were generated based on nucleotide repeat sequences. D1ski4-L: CCTCTACCACTGCCTATTCTGT; D1ski4-R: ACAGGCTGGACCTGAGCA; D1ski6-L: GTTACCGAGCAACAGCGAAG; D1ski6-R: TGAATGTGCACCTGTCTATGG; D1ski7-L: TTTGGCGATGAATCTTCTGA; D1ski7-R: TGAAAAGATGGCCAATTGCT; D1ski12-L: GGGTTAGAACCAAAAGGGTGA; D1ski12-R: CCAAGAAGCAGGAGTGGGTA; D1ski13-L: GCCTGATGTCTTTGTCATGC; D1ski13-R: ATGGAACTTCAGGGTTGACG; D1ski14-L: TGGACCTGAAAATCATATTATTACA; D1ski14-R: CTGGGTCCCCTCCTTCTTAC. The entire Fpn transcript was sequenced by reverse transcriptase–polymerase chain reaction (RT-PCR; Superscript One-Step RT-PCR; Invitrogen, Carlsbad, CA) using RNA isolated from E10.5 ffe/ffe and C57BL/6 control embryos. Sequencing was confirmed using DNA isolated from 10 additional ffe/ffe mutant embryos.
Prussian Blue staining
Livers were fixed overnight in 4% paraformaldehyde, cryopreserved in 30% sucrose, embedded in OCT compound (Tissue-Tek; Electron Microscopy Science, Hatfield, PA) and sectioned at 10 mM. Fixed frozen sections were incubated in hydrochloric acid (10%) and potassium ferrocyanide (5%) as described18 and counterstained with eosin.
Constructs and cells
The cloning and expression of mouse Fpn in a cytomegalovirus (CMV)–containing vector (pEGFP-N1 [Clontech, Mountain View, CA] or pCMV-Tag4 [FLAG; Stratagene, La Jolla, CA]) was described previously.9 pEGFP-FpnH32R was generated in pFpn-EGFP-N1 by using the QuikChange Site-Directed Mutagenesis Kit (Stratagene), amplified in Escherichia coli and sequence verified. HEK293T cells were maintained in Dulbecco minimal essential media (DMEM) with 10% fetal bovine serum and were transfected with pFpn-GFP and pFpn(H32R)-GFP or pFpn-FLAG using Nucleofector technology (Amaxa, Gaithersburg, MD) according to the manufacturer's directions. Mouse bone marrow macrophages were harvested from femurs and maintained as described previously.19
Erythrophagocytosis
Erythrophagocytosis was performed as described.3 Macrophages were incubated with IgG-coated red blood cells (RBCs) at a ratio of 20 RBCs/macrophage for 90 minutes. The cultures were then washed free of extracellular RBCs and incubated for 18 hours before measurement of ferritin. Wild-type and ffe/+ macrophages phagocytosed an average of 10 RBCs/macrophage with more than 95% of the cells containing RBCs. There was no difference between wild-type and ffe/+ macrophages in the average number of RBCs/cell or the percent of cells with ingested RBCs.
siRNA transfection
siRNA oligonucleotide pools, nonspecific and mouse Fpn specific, were obtained from Dharmacon (Lafayette, CO). Mouse bone marrow macrophages were transfected with siRNAs at concentrations up to 100 nM using Oligofectamine reagent (Invitrogen) as described.4 At 24 hours after transfection, the cells were trypsinized and plated onto 35-mm plates with or without ferric ammonium citrate (FAC) (10 μM Fe). Cells were grown for 18 hours and then processed for Western blot and ferritin analysis.
Other procedures
Immunofluorescence was performed as described previously.9 Fluorescent images were captured on an Olympus BX51 microscope (Olympus, Tokyo, Japan) using an Olympus U-CMAD-Z camera and a 60×/1.30 NA oil-immersion objective lens. Images were acquired using Picture Frame 2.5 software (Olympus America, East Muskogee, OK). Captured images were combined in Adobe Photoshop 6.0 (Adobe Systems, San Jose, CA) to generate multiple-image figures for publication. Fpn was detected using a rabbit anti-Fpn antibody (1:100; a generous gift from Dr David Haile). Transferrin receptor 1 was detected using a mouse anti–human transferrin receptor 1 antibody (CD81 1:100; RDI, Concord, MA) followed by Alexa 594–conjugated goat anti–rabbit IgG (1:750; Molecular Probes, Eugene, OR), Alexa 594–conjugated goat anti–mouse IgG (1:750; Molecular Probes). Mouse anti-FLAG antibody M2 (Sigma, St Louis, MO) was used to detect Fpn-FLAG by immunofluorescence (1:750) followed by Alexa 594–conjugated goat anti–mouse IgG (1:750; Molecular Probes). Cellular protein was extracted with 150 mM NaCl, 10 mM EDTA, 10 mM Tris (pH 7.4), 1% Triton X-100, and a protease inhibitor cocktail (Roche, Indianapolis, IN). Western analysis was performed as described previously9 using rabbit anti-Fpn (1:500) and mouse anti–human α-tubulin (1:5000) followed by either peroxidase-conjugated goat anti–rabbit IgG (1:12 500, Jackson ImmunoResearch Labs, West Grove, PA) or peroxidase-conjugated goat anti–mouse IgG (1:12 500, Jackson ImmunoResearch Labs). Mice were bled retro-orbitally and peripheral blood smears were stained with Wright-Giemsa. Ferritin analysis was performed as described.6 Transferrin saturation was determined by means of a ferrozine-based iron and total iron-binding capacity assay (Teco Diagnostic, Anaheim, CA). Protein concentration was measured using the BCA assay (Pierce, Rockford, IL). All experiments were repeated a minimum of 3 times.
Results
The flatiron (ffe) mutation is in the Fpn gene
In a screen for recessive mutations induced by ethylnitrosurea that affect development of the mouse embryo, we identified ffe as a recessive mutation that causes developmental defects on a mixed C3H/HeJ X C57BL/6 background, including neural tube closure defects, microphthalmia, forebrain truncations, generalized edema, severe anemia, and mid-gestation lethality (data not shown). When the ffe mutation was crossed onto the 129/SvJ inbred strain, homozygous mutant embryos showed only severe anemia and mid-gestation lethality (Figure 1A-B). Using a positional cloning strategy, the ffe mutation was mapped to a 2.6-megabase interval on mouse chromosome 1 that contains 16 transcripts, including the Slc40a1 gene encoding the Fpn protein (Figure 1C). Fpn is the only exporter of cellular iron in mammalian cells5,20,21 and is expressed on placental syncytiotrophoblast cells where it is required for iron transport from the maternal circulation to the fetus.14 Sequencing of the cDNA encoding Fpn from ffe mutant embryos revealed a c.95A>G nucleotide substitution. The deduced amino acid substitution is H32R in the first putative transmembrane domain (Figure 1D). Most Fpnnull/null embryos die by E7.514 because of defects in iron transfer across the visceral endoderm, indicating that Fpnffe is a hypomorphic allele of Fpn. It is likely that the more complex phenotype seen in the original mutant was due to the background strain, as the complex phenotype was lost when the mice were bred onto different backgrounds.
The ffe mouse mutation is in the gene encoding Fpn. (A, B) Wild-type and ffe/ffe mutant embryos dissected at E13.5 on a 129/SvJ inbred mouse background. ffe mutants exhibit severe anemia. (C) Genetic map of ffe interval on mouse chromosome 1. The number of recombination events over number of opportunities for recombination is indicated for each polymorphic marker. Markers D1ski7, D1mit236, and D1ski12 never separated from the ffe phenotype. Within this interval are 7 known protein-coding transcription units: Gulp1 (GULP, engulfment adaptor PTB domain containing 1), Col3a1 (procollagen, type III, alpha 1), Col5a2 (procollagen, type V, alpha 2), Wdr75 (WD repeat domain 75), Slc40a1 (Fpn, solute carrier family 40 member 1), Dnahc7 (dynein, axonemal, heavy chain 7), and Slc39a10 (solute carrier family 39 [zinc transporter], member 10), and 9 novel predicted transcripts. (D) The ffe ENU-induced mutation results in an A-to-G transition at position 95 (green and black arrows) in the coding sequence of Fpn, changing a histidine to an arginine in the first putative transmembrane domain. Image was taken using a Leica M2FLIII fluorescence stereomicroscope with a 10× eyepiece and a 1.0 × planachromic objective with a 0.125 numerical aperture set on 1× (Leica Microsystems Gmbh, Wetzlar, Germany). Image was captured using a Qimaging RETGA 1200 digital CCD camera (Qimaging Corporation, Surrey, BC, Canada) and IPLab Advanced Image Analysis Software Version 3.9.5r3 (BD Biosciences, Rockville, MD). Subsequent image processing was done using Adobe Photoshop 7.0 software (Adobe Systems, San Jose, CA).
The ffe mouse mutation is in the gene encoding Fpn. (A, B) Wild-type and ffe/ffe mutant embryos dissected at E13.5 on a 129/SvJ inbred mouse background. ffe mutants exhibit severe anemia. (C) Genetic map of ffe interval on mouse chromosome 1. The number of recombination events over number of opportunities for recombination is indicated for each polymorphic marker. Markers D1ski7, D1mit236, and D1ski12 never separated from the ffe phenotype. Within this interval are 7 known protein-coding transcription units: Gulp1 (GULP, engulfment adaptor PTB domain containing 1), Col3a1 (procollagen, type III, alpha 1), Col5a2 (procollagen, type V, alpha 2), Wdr75 (WD repeat domain 75), Slc40a1 (Fpn, solute carrier family 40 member 1), Dnahc7 (dynein, axonemal, heavy chain 7), and Slc39a10 (solute carrier family 39 [zinc transporter], member 10), and 9 novel predicted transcripts. (D) The ffe ENU-induced mutation results in an A-to-G transition at position 95 (green and black arrows) in the coding sequence of Fpn, changing a histidine to an arginine in the first putative transmembrane domain. Image was taken using a Leica M2FLIII fluorescence stereomicroscope with a 10× eyepiece and a 1.0 × planachromic objective with a 0.125 numerical aperture set on 1× (Leica Microsystems Gmbh, Wetzlar, Germany). Image was captured using a Qimaging RETGA 1200 digital CCD camera (Qimaging Corporation, Surrey, BC, Canada) and IPLab Advanced Image Analysis Software Version 3.9.5r3 (BD Biosciences, Rockville, MD). Subsequent image processing was done using Adobe Photoshop 7.0 software (Adobe Systems, San Jose, CA).
The ffe mutation results in mislocalization of Fpn
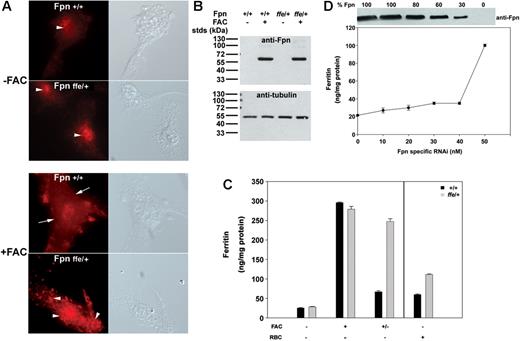
The cell surface localization of Fpn is essential for its function,6 and the ffe mutation could result in mislocalization of Fpn, thus inhibiting iron transport into the fetal circulation. To determine if the Fpnffe mutation results in its mislocalization, this mutation (H32R) was introduced into a plasmid containing a green fluorescent protein (GFP)–tagged Fpn. As shown in Figure 2A, Fpn-GFP expressed in HEK293T cells is localized to the plasma membrane. In contrast, Fpn(H32R)-GFP is predominantly intracellular. To determine if Fpn(H32R)-GFP has iron-exporting activity, cells expressing Fpn-GFP or Fpn(H32R)-GFP were iron loaded by incubation with ferric ammonium citrate (FAC), inducing the expression of the iron storage protein ferritin (Figure 2B). Expression of Fpn-GFP leads to iron export, resulting in decreased ferritin levels,6 whereas expression of Fpn(H32R)-GFP results in only a minor decrease in ferritin, indicating that the Fpnffe allele has significantly decreased iron-exporting activity.
The ffe mutation affects the localization of transfected Fpn and iron-exporting activity. (A) HEK293T cells were transiently transfected with plasmids containing either Fpn-GFP or Fpn(H32R)-GFP. Localization of Fpn was assessed by epifluorescence microscopy 12 to 18 hours after transfection. The arrows denote plasma membrane localization. The arrowheads denote intracellular localization. (B) Cells, treated as described in panel A, were incubated with FAC (10 μM Fe) for 18 hours and ferritin levels were determined by enzyme-linked immunosorbent assay (ELISA). (C) HEK293T cells were transiently cotransfected with plasmids containing Fpn-GFP and Fpn-FLAG or Fpn(H32R)-GFP and Fpn-FLAG. Fpn or transferrin receptor 1 localization was determined by immunofluorescence microscopy 18 hours after transfection. (D) Cells, treated as in panel C, were incubated with FAC (10 μM Fe) for 18 hours and ferritin levels were determined by ELISA. The error bars represent the standard deviation.
The ffe mutation affects the localization of transfected Fpn and iron-exporting activity. (A) HEK293T cells were transiently transfected with plasmids containing either Fpn-GFP or Fpn(H32R)-GFP. Localization of Fpn was assessed by epifluorescence microscopy 12 to 18 hours after transfection. The arrows denote plasma membrane localization. The arrowheads denote intracellular localization. (B) Cells, treated as described in panel A, were incubated with FAC (10 μM Fe) for 18 hours and ferritin levels were determined by enzyme-linked immunosorbent assay (ELISA). (C) HEK293T cells were transiently cotransfected with plasmids containing Fpn-GFP and Fpn-FLAG or Fpn(H32R)-GFP and Fpn-FLAG. Fpn or transferrin receptor 1 localization was determined by immunofluorescence microscopy 18 hours after transfection. (D) Cells, treated as in panel C, were incubated with FAC (10 μM Fe) for 18 hours and ferritin levels were determined by ELISA. The error bars represent the standard deviation.
We have shown that Fpn is a multimer and mutations in Fpn that affect cell surface localization also result in mislocalization of wild-type Fpn.9,13 To determine if expression of Fpn(H32R)-GFP can change the localization of the wild-type protein, Fpn-FLAG was coexpressed with Fpn-GFP or mutant Fpn(H32R)-GFP and localization of the tagged proteins was determined by immunofluorescence. Coexpression of Fpn-GFP with Fpn-FLAG resulted in plasma membrane localization of both proteins (Figure 2C). In contrast, coexpression of Fpn-GFP(H32R) with Fpn-FLAG resulted in the intracellular localization of GFP and FLAG-tagged proteins, indicating that mutant Fpn can change the localization of wild-type protein. This is specific to Fpn, as expression of Fpn(H32R)-GFP did not affect localization of transferrin receptor 1. To determine if mutant Fpn can affect the iron-exporting activity of wild-type Fpn, Fpn-FLAG was coexpressed with Fpn-GFP or Fpn(H32R)-GFP in iron-loaded HEK293T cells. Coexpression of wild-type proteins resulted in export of cellular iron and decreased ferritin levels (Figure 2D). Cells coexpressing Fpn(H32R)-GFP with Fpn-FLAG had elevated levels of ferritin, indicating that the Fpnffe allele (FpnH32R) can act as a dominant negative by changing the localization of wild-type Fpn and its iron-exporting activity.
Mutant Fpnffe acts as a dominant negative in macrophages isolated from ffe/+ mice
To determine if endogenous levels of Fpnffe also act as a dominant negative in vivo, macrophages were isolated from Fpnffe/+ heterozygous mice and localization of endogenous Fpn was examined. Fpn expression in macrophages is induced in response to high levels of cellular iron.3 Little Fpn is expressed in untreated primary cultures of macrophages isolated from wild-type or Fpnffe/+ animals (Figure 3A-B). Upon iron loading, high levels of Fpn are detected at the cell surface of wild-type macrophages. Iron loading of macrophages from Fpnffe/+ heterozygous mice did not result in expression of Fpn on the cell surface, although equivalent levels of Fpn were detected in wild-type and Fpnffe/+ macrophages (Figure 3B). Since Fpnffe/+ mice express both a wild-type and mutant allele of Fpn, these results indicate that Fpnffe inhibits the cell surface localization of wild-type Fpn.
Localization of Fpn and iron transport activity in macrophages from ffe/+ mice. (A,B) Bone marrow macrophages isolated from wild-type mice or ffe/+ mice were incubated in the presence or absence of FAC (10 μM Fe) for 24 hours. The localization of Fpn was analyzed by immunofluorescence using an antibody to Fpn and the amount of Fpn was determined by Western analysis using an antibody to α-tubulin as a loading control. The arrows denote plasma membrane localization. The arrowheads denote intracellular localization. (C) Cells were incubated with or without FAC (10 μM Fe) for 24 hours or with IgG-coated RBCs for 90 minutes. Cells were washed and ferritin levels measured by ELISA after 16 hours (+/− refers to cells incubated with FAC and then incubated in the absence of FAC; black bars represent wild type; gray bars represent ffe/+). The error bars represent the standard deviation. (D) Bone marrow macrophages from wild-type mice were transfected with either nonspecific oligonucleotide pools or with different concentrations of oligonucleotide pools specific for mouse Fpn. Cells were incubated with FAC (10 μM Fe) for 24 hours and then in the absence of FAC for 16 hours. Cell extracts were isolated and assayed for ferritin by ELISA or for Fpn by Western blot analysis. The amount of Fpn in cells incubated with nonspecific oligonucleotides was identical to that of cells not transfected and was taken as 100%. The Western blots were analyzed by densitometry and Fpn was normalized to α-tubulin. The amount of ferritin in cells incubated with 50 nM to 100 nM Fpn-specific oligonucleotide pools was equivalent to that of untransfected cells. The error bars represent the standard deviation.
Localization of Fpn and iron transport activity in macrophages from ffe/+ mice. (A,B) Bone marrow macrophages isolated from wild-type mice or ffe/+ mice were incubated in the presence or absence of FAC (10 μM Fe) for 24 hours. The localization of Fpn was analyzed by immunofluorescence using an antibody to Fpn and the amount of Fpn was determined by Western analysis using an antibody to α-tubulin as a loading control. The arrows denote plasma membrane localization. The arrowheads denote intracellular localization. (C) Cells were incubated with or without FAC (10 μM Fe) for 24 hours or with IgG-coated RBCs for 90 minutes. Cells were washed and ferritin levels measured by ELISA after 16 hours (+/− refers to cells incubated with FAC and then incubated in the absence of FAC; black bars represent wild type; gray bars represent ffe/+). The error bars represent the standard deviation. (D) Bone marrow macrophages from wild-type mice were transfected with either nonspecific oligonucleotide pools or with different concentrations of oligonucleotide pools specific for mouse Fpn. Cells were incubated with FAC (10 μM Fe) for 24 hours and then in the absence of FAC for 16 hours. Cell extracts were isolated and assayed for ferritin by ELISA or for Fpn by Western blot analysis. The amount of Fpn in cells incubated with nonspecific oligonucleotides was identical to that of cells not transfected and was taken as 100%. The Western blots were analyzed by densitometry and Fpn was normalized to α-tubulin. The amount of ferritin in cells incubated with 50 nM to 100 nM Fpn-specific oligonucleotide pools was equivalent to that of untransfected cells. The error bars represent the standard deviation.
To determine if macrophages from Fpnffe/+ mice are able to export iron, macrophages from wild-type or Fpnffe/+ mice were loaded with iron and cellular ferritin levels were measured. Wild-type and Fpnffe/+ cells expressed equivalent amounts of ferritin in the absence or presence of FAC. Wild-type macrophages exported cellular iron upon removal of FAC, as indicated by decreased ferritin levels (Figure 3C, black bars); however, macrophages isolated from Fpnffe/+ mice were not able to efficiently export iron, and ferritin levels remained high (Figure 3C, gray bars). Similar results were obtained when macrophages were loaded with iron by engulfing RBCs, although the absolute level of ferritin was lower in RBC-fed macrophages (Figure 3C). Together these results indicate that Fpnffe acts as a dominant negative in vivo by associating with wild-type Fpn, causing its mislocalization and thus preventing its ability to export iron.
It has been suggested that mutations in Fpn might result in cellular iron overload due to haploinsufficiency10,12 ; however, mice that are heterozygous for a targeted gene deletion of Fpn do not show evidence of iron-overload disease.14 To further explore the possibility that haploinsufficiency could explain decreased iron export, we used RNAi to silence mouse Fpn in macrophages. Cells were transfected with nonspecific oligonucleotides or different concentrations of oligonucleotides specific to mouse Fpn. After 24 hours, cells were loaded with iron and levels of Fpn and cellular ferritin were determined 48 hours after transfection. Macrophages showed almost normal levels of iron export even when Fpn protein levels were decreased by 50% (Figure 3D). Reduction in iron-exporting activity (higher levels of ferritin) did not occur until Fpn levels decreased to below 30% of wild-type levels. These results indicate that half normal levels of Fpn, which would be expected in haploinsufficiency, cannot explain the defect in Fpn disease.
ffe/+ mice have ferroportin disease
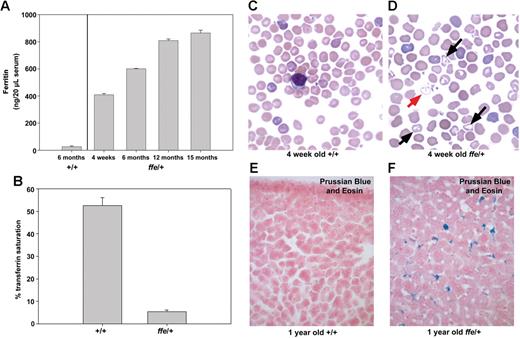
We sought to determine if expression of the dominant-negative Fpnffe allele in Fpnffe/+ heterozygous mice can mimic the human disease. In human patients, Fpn-linked hemochromatosis has a heterogeneous presentation.22 Some patients exhibit the classic symptoms of hemochromatosis, such as high transferrin saturation and iron accumulation in parenchymal cells. This presentation is associated with mutations in Fpn that affect its response to hepcidin.8,9 Other patients present with an early rise in ferritin levels, low to normal transferrin saturation, and iron accumulation primarily in Kupffer cells. This presentation is associated with Fpn mutations that affect its plasma membrane localization or iron-exporting activity.8,9 Serum ferritin levels in Fpnffe/+ heterozygous animals were increased 10-fold over 6-month-old control animals and increased with age (Figure 4A). In addition, transferrin saturation in serum from 6-month-old heterozygotes was much lower than that of control mice (Figure 4B). Blood smears from 4-week-old wild-type animals showed normochromic and normocytic RBCs that have a uniform size and shape and no target cells (Figure 4C). Blood smears from 4-week-old Fpnffe/+ heterozygous animals revealed some hypochromic red blood cells with variable size and some target cells (Figure 4D). These results show that Fpnffe/+ animals have a mild anemia, which is consistent with the extremely low transferrin saturation. To determine if heterozygous animals also present with iron accumulation in Kupffer cells, Prussian Blue staining was used to visualize accumulated ferric iron. Livers from wild-type mice do not accumulate iron (Figure 4E) whereas livers from aged Fpnffe/+ animals accumulate high levels of iron in Kupffer cells (Figure 4F). Together, these results indicate that expression of the Fpnffe allele affects the ability of wild-type Fpn to export iron by inhibiting the localization and activity of wild-type Fpn. Furthermore, the close similarities between the clinical phenotypes of Fpnffe/+ mice and human patients demonstrates the utility of the Fpnffe/+ mouse as a model for this disease.
Serum ferritin and iron accumulation in ffe/+ mice. (A) Ferritin levels were measured in serum obtained from 3 6-month-old control mice and from 2 to 3 ffe/+ mice of the specified age. (B) Serum transferrin saturation was measured in 2 6-month-old control mice and 4 6-month-old ffe/+ mice. Samples from each mouse were measured in triplicate and the data from mice of similar ages were pooled. The error bars represent standard error of the mean. (C,D) Blood smears from 4-week-old +/+ mice (C) and ffe/+ mice were stained with Wright-Giemsa. Red arrow indicates hypochronic red blood cells; black arrows, target cells. (E,F) Sections of liver from 1-year-old wild-type (+/+) and ffe/+ mice stained with Prussian Blue to visualize accumulated iron. Image was taken using a Zeiss Axiophot2 using a 40× Plan Neofluar air objective with a 0.74 numerical aperture (CarlZeiss, Thornwood, NY). Image was captured using a Photometrics CoolSNAPfx camera (Image Processing Solutions, North Reading, MA) and IPLab Advanced Image Analysis Software Version 3.9.5r3 (BD Biosciences, Rockville, MD). Subsequent image processing was done using Adobe Photoshop 7.0 software (Adobe Systems, San Jose, CA).
Serum ferritin and iron accumulation in ffe/+ mice. (A) Ferritin levels were measured in serum obtained from 3 6-month-old control mice and from 2 to 3 ffe/+ mice of the specified age. (B) Serum transferrin saturation was measured in 2 6-month-old control mice and 4 6-month-old ffe/+ mice. Samples from each mouse were measured in triplicate and the data from mice of similar ages were pooled. The error bars represent standard error of the mean. (C,D) Blood smears from 4-week-old +/+ mice (C) and ffe/+ mice were stained with Wright-Giemsa. Red arrow indicates hypochronic red blood cells; black arrows, target cells. (E,F) Sections of liver from 1-year-old wild-type (+/+) and ffe/+ mice stained with Prussian Blue to visualize accumulated iron. Image was taken using a Zeiss Axiophot2 using a 40× Plan Neofluar air objective with a 0.74 numerical aperture (CarlZeiss, Thornwood, NY). Image was captured using a Photometrics CoolSNAPfx camera (Image Processing Solutions, North Reading, MA) and IPLab Advanced Image Analysis Software Version 3.9.5r3 (BD Biosciences, Rockville, MD). Subsequent image processing was done using Adobe Photoshop 7.0 software (Adobe Systems, San Jose, CA).
Discussion
Mutations in Fpn result in an autosomal-dominant disorder with 2 different patient presentations. Mutations that render Fpn insensitive to down-regulation by hepcidin result in iron overload in hepatocytes.8,11,23 In this form of the disorder there is no regulation of Fpn, leading to high transferrin saturation and iron deposition in hepatocytes. The iron burden of hepatocytes can be reduced by phlebotomy, as liver iron stores are capable of being mobilized through Fpn. A second class of Fpn mutations prevents localization to the cell surface or permits its localization to the cell surface but affects the ability of Fpn to transport iron.8,11,23 This form of the disorder, referred to as classic Fpn disease, results in low transferrin saturation, high serum ferritin, and excessive iron deposits in Kupffer cells, not hepatocytes. Patients with these types of Fpn mutations develop severe anemia upon repeated phlebotomy and iron stores in liver are not mobilized because mutant Fpn cannot export the excess iron. It has been a source of debate whether haploinsufficiency would explain Fpn disease or whether the disease results from a dominant-negative effect. While studies have supported the view that Fpn is a multimer9,13,24 and that the mutant allele can affect the behavior of the wild-type allele, other studies have suggested that Fpn is monomeric.10,12,25 All human Fpn mutations are missense mutations. If haploinsufficiency was the explanation for Fpn disease, then nonsense mutations should also result in the disorder; however, none have been found. Additionally, a targeted gene deletion in the murine Fpn gene has little effect in heterozygous animals.14 While these data suggest that haploinsufficiency cannot explain the disorder it is formally possible that Fpn disease in mice cannot recapitulate the human disease.
The discovery of the ffe mutation shows that mice can have Fpn disease, as mice heterozygous for the mutation exhibit all of the features of the human disorder: low transferrin saturation, high serum ferritin, and excessive iron in Kupffer cells. Our studies show that macrophages from ffe/+ mice have no detectable Fpn on their cell surface and cannot export iron. Additionally, generation of the H32R mutation in constructs of Fpn-GFP demonstrates that mutant Fpn can exert a dominant-negative effect and prevent the surface localization of wild-type Fpn-FLAG. Finally, using siRNA oligonucleotides to decrease the expression of Fpn by 50%, as would be expected for haploinosufficiency, has no effect on macrophage iron export. Iron export is only compromised when Fpn levels decrease below 30% of wild type. Together, these data show that Fpn disease does not result from haploinsufficiency but rather from a dominant-negative effect of the mutant allele.
The available clinical data,2 supported by the studies reported here, show that Fpn mutations that lead to Kupffer cell iron loading result in low transferrin saturation. The low transferrin saturation may lead to iron-limited anemia during periods of active growth or in the face of phlebotomy. In is unclear, however, whether these Fpn mutations have adverse clinical affects independent of the low transferrin saturation.26 The Fpnffe/+ mouse will be a useful system to explore the pathophysiological consequences of Kupffer cell iron loading.
Authorship
Contribution: I.E.Z. performed the positional cloning, preparation of mouse samples for further analysis, histologic analysis, and wrote the manuscript; I.D.D. generated the Fpn(H32R)-GFP plasmid, performed the immunofluorescence, macrophage culturing, ferritin analysis, and transferrin saturation analysis, and wrote the manuscript; A.P. performed the positional cloning, sequencing of the ffe mutation, and histologic analysis; D.M.W. performed the immunofluorescence and assisted in the writing and preparation of the manuscript; A.J.S. performed the positional cloning; J.F.G. and X.L. analyzed the peripheral blood smears; and L.N. and J.K. guided these studies and assisted in manuscript preparation.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
I.E.Z. and I.D.D. contributed equally to this work.
Correspondence: Jerry Kaplan, Department of Pathology, School of Medicine, University of Utah, Salt Lake City, UT 84132; e-mail: jerry.kaplan@path.utah.edu.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors wish to express their appreciation to Dr Richard Ajioka for help in measuring transferrin saturation; Kathryn V. Anderson and members of the Anderson and Niswander laboratories for performing the mutagenesis screen; Lori Bulwith for technical assistance; and Christopher Porter for help with blood smears. The current address for I.E.Z. is Center for Neuroscience Research, Children's Research Institute, Children's National Medical Center, Washington, DC.
This work was supported by National Institutes of Health (NIH) grants DK070 947 (J.K.) and F32-HD08 605 (I.E.Z.). J.F.G. is supported by an NIH training grant (T32CA8608604). L.N. is a Howard Hughes Medical Institute (HHMI) investigator.

![Figure 1. The ffe mouse mutation is in the gene encoding Fpn. (A, B) Wild-type and ffe/ffe mutant embryos dissected at E13.5 on a 129/SvJ inbred mouse background. ffe mutants exhibit severe anemia. (C) Genetic map of ffe interval on mouse chromosome 1. The number of recombination events over number of opportunities for recombination is indicated for each polymorphic marker. Markers D1ski7, D1mit236, and D1ski12 never separated from the ffe phenotype. Within this interval are 7 known protein-coding transcription units: Gulp1 (GULP, engulfment adaptor PTB domain containing 1), Col3a1 (procollagen, type III, alpha 1), Col5a2 (procollagen, type V, alpha 2), Wdr75 (WD repeat domain 75), Slc40a1 (Fpn, solute carrier family 40 member 1), Dnahc7 (dynein, axonemal, heavy chain 7), and Slc39a10 (solute carrier family 39 [zinc transporter], member 10), and 9 novel predicted transcripts. (D) The ffe ENU-induced mutation results in an A-to-G transition at position 95 (green and black arrows) in the coding sequence of Fpn, changing a histidine to an arginine in the first putative transmembrane domain. Image was taken using a Leica M2FLIII fluorescence stereomicroscope with a 10× eyepiece and a 1.0 × planachromic objective with a 0.125 numerical aperture set on 1× (Leica Microsystems Gmbh, Wetzlar, Germany). Image was captured using a Qimaging RETGA 1200 digital CCD camera (Qimaging Corporation, Surrey, BC, Canada) and IPLab Advanced Image Analysis Software Version 3.9.5r3 (BD Biosciences, Rockville, MD). Subsequent image processing was done using Adobe Photoshop 7.0 software (Adobe Systems, San Jose, CA).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/109/10/10.1182_blood-2007-01-066068/4/m_zh80100700910001.jpeg?Expires=1764990774&Signature=o1E0vv2WWyO~zRPDHgY1ijrkvHBibE75KAJiJ0qnvatiNmCGiiB-KGTKNTyvyDvOW2xrTxGaUfb7ul-jaDNAP6TETLg~mvoV6kx1lN5wYj1gz8ATbrXhgvLLoFWApStm8~azMB60ID110teHrrc2JIHegJEBw75jU4qx9uVwkXEM1vsVVeQFT5H0EnI4K~KmMNSVZ9EgANsmEmZ2MC3-QRGGcsYSoJDDzz-zyHGli4gST5ygXxEoO~v7HdtXHkuxiPL-H~WE1RX6vgQJ~kubfwq0aZHghASE5T7WWagcQpUUbapB1RhCcX6~PI4g0eOKUuuAyh6OZTyj8vb11xJe0g__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal