Abstract
Neutrophil-specific granule deficiency (SGD) is a rare congenital disorder marked by recurrent bacterial infections. Neutrophils from SGD patients lack secondary and tertiary granules and their content proteins and lack normal neutrophil functions. Gene-inactivating mutations in the C/EBPϵ gene have been identified in 2 SGD patients. Our studies on a third SGD patient revealed a heterozygous mutation in the C/EBPϵ gene. However, we demonstrate elevated levels of C/EBPϵ and PU.1 proteins in the patient's peripheral blood neutrophils. The expression of the transcription factor growth factor independence-1 (Gfi-1), however, was found to be markedly reduced in our SGD patient despite the absence of an obvious mutation in this gene. This may explain the elevated levels of both C/EBPϵ and PU.1, which are targets of Gfi-1 transcriptional repression. We have generated a growth factor–dependent EML cell line from the bone marrow of Gfi-1+/− and Gfi-1+/+ mice as a model for Gfi-1–deficient SGD, and demonstrate that lower levels of Gfi-1 expression in the Gfi-1+/− EML cells is associated with reduced levels of secondary granule protein (SGP) gene expression. Furthermore, we demonstrate a positive role for Gfi-1 in SGP expression, in that Gfi-1 binds to and up-regulates the promoter of neutrophil collagenase (an SGP gene), in cooperation with wild-type but not with mutant C/EBPϵ. We hypothesize that decreased Gfi-1 levels in our SGD patient, together with the mutant C/EBPϵ, block SGP expression, thereby contributing to the underlying etiology of the disease in our patient.
Introduction
The generation of mature neutrophils in the peripheral blood of adult mammals is a highly regulated process that is initiated in the bone marrow (reviewed in Bellantuono1 ). As the developing neutrophil makes the transition from the promyelocyte to the myelocyte stage, it acquires secondary (“specific”) granules and their content proteins (reviewed in Berliner2 ; Borregaard et al3 ; and Borregaard et al4 ), a process that is dependent on the interplay of a number of lineage-specific transcription factors (reviewed in Rosmarin et al5 ; Shivdasani and Orkin6 ; Skalnik7 ; and Friedman8 ). Specific granules contain 4 major proteins, namely, transcobalamin 1 (TC1), lactoferrin (LF), human neutrophil collagenase (HNC), and human neutrophil gelatinase (HNG), and their acquisition provides a unique marker of commitment to terminal neutrophil differentiation (reviewed in Berliner2 ; and Borregaard et al3 ). Several lines of evidence from our laboratory have established that the expression of the secondary granule protein (SGP) genes, which are functionally diverse and physically unlinked, is coordinately regulated at the level of mRNA transcription (reviewed in Berliner2 ; and Borregaard et al3 ). Absence of SGP expression is a consistent abnormality in acute myelogenous leukemia (AML), myelodysplastic syndrome (MDS), and neutrophil-specific granule deficiency (SGD), all disorders associated with disruption of the more distal events of the neutrophil maturation program.
SGD is a rare congenital disorder thought to be inherited in an autosomal recessive manner. Neutrophils from affected individuals display atypical pseudo–Pelger-Huet–type bilobed nuclei, and lack secondary and tertiary granules and their content proteins. Additional defects in neutrophil function including abnormal chemotaxis, respiratory burst, and receptor up-regulation are also observed. As a result of these combined abnormalities, SGD patients are prone to bacterial infections.9,10
Mutations in the CCAAT enhancer-binding protein epsilon (C/EBPϵ) gene that result in absent C/EBPϵ protein expression have been found in 2 cases of SGD, and are thought to be responsible for the disease in those patients.11,12 C/EBPϵ belongs to a family of basic region leucine zipper (b-Zip) proteins that bind the consensus DNA sequence 5′TKNNGYAAK3′ (Y = C or T, K = T or G) within the regulatory regions of target genes as homodimers or heterodimers (reviewed in Lekstrom-Himes and Xanthopoulos13 ). Of interest, neutrophils from C/EBPϵ−/− mice have morphologic and biochemical features very similar to those observed in patients with SGD, including absent neutrophil and eosinophil secondary and tertiary granules and their content proteins, defects in respiratory burst, chemotaxis and bactericidal activity, and atypical bilobed nuclei.14 Sequence analysis of the C/EBPϵ locus of our SGD patient was recently performed in our laboratory. We have identified a heterozygous valine to alanine mutation at amino acid 218 in the basic region of the C/EBPϵ gene. However, this mutation does not diminish the transactivation capability of the mutant C/EBPϵ in transient transfection assays. Our observations suggest that this structural abnormality in the C/EBPϵ gene likely contributes to the lesion underlying SGD since it is incapable of synergistically activating SGP gene expression in concert with the Gfi-1 transcription factor, which itself is essential for SGP expression.
Patient, materials, and methods
IRB approval was obtained from both the University of Michigan and Yale University for collecting blood and bone marrow on this patient.
Patient information and isolation of bone marrow
The SGD patient has been previously described.9,15 Briefly, the patient is a 47-year-old man with no evidence of infection at the time of this study. Following informed consent in accordance with the Declaration of Helsinki, bone marrow was isolated from the patient, and showed normal cellularity comparable with his previous marrows.9,15 Normal bone marrow was obtained from anonymous discarded samples.
Isolation of peripheral blood neutrophils from healthy individuals and the SGD patient
Whole blood samples were obtained from the SGD patient and from healthy volunteers following guidelines for informed consent set by the University of Michigan School of Medicine Committee on the Use of Human Subjects in Research. Neutrophils were isolated as previously described.16 Since the SGD polymorphonuclear leukocytes (PMNs) lack secondary granules, the buoyant density of the diseased PMNs is altered. Therefore, to isolate neutrophils efficiently, the specific gravity of the Ficoll-Paque had to be adjusted to 1.066.
PCR amplification and sequencing of the C/EBPϵ gene from the SGD patient
Genomic DNA was obtained from the peripheral blood neutrophils of the SGD patient and from healthy donors using the TriZol method (Invitrogen, Frederick, MD). Genomic DNA was polymerase chain reaction (PCR) amplified using Platinum Taq DNA polymerase (Life Technologies, Bethesda, MD) per the manufacturer's instructions and cycled as follows: 96°C for 12 minutes, followed by a 3-step cycle 94°C for 30 seconds, 60°C for 30 seconds, and 72°C for 2 minutes for 35 to 40 cycles. PCR products were purified and recovered using PCR purification columns (Qiagen, Valencia, CA). The products were sequenced with an ABI Prism Dye Terminator Cycle Sequencing Ready Reaction Kit (Perkin-Elmer, Shelton, CT). Primers were based on published sequence available from EMBL/GenBank/DDBJ under accession no. U48865, as previously described.17 A mutation identified at aa 218 converting a valine to an alanine residue was introduced into pCDNA3.1 C/EBPϵ using the QuickChange protocol (Stratagene, Cedar Creek, TX), using the oligomers: sense, 5′CGCAACAACATCGCCGCGCGCAAGAGCCGAC3′ and antisense, 5′GTCGGCTCTTGCGCGGGGCGATGTTGTTGCG3′.
Tissue culture, transient transfection, and luciferase assays
HEK293T (293T) cells were a gift from Dr Ghosh (Yale University School of Medicine, New Haven, CT), 32Dwt18 cells were a gift from Dr Daniel Link (Washington University, St Louis, MO), mouse promyelocytic (MPRO) cells were obtained from Dr Schickwann Tsai (University of Utah, Salt Lake City, UT), and NB4 cells were all grown as previously described.18–20 All cells were maintained at 37°C in a humidified 5% CO2 incubator. 32Dwt18 cells were transiently transfected as previously described, using 10 μg wild type or Gfi-1 mutant, generated by Quik-Change (Stratagene) (resulting in a mutant Gfi-1 site 5′TAGCAGC3′) reporter HNC plasmid and 2 μg pCMVβgal (Clontech, Palo Alto, CA), an internal control plasmid used to monitor transfection efficiency as previously described.21 Transfected cells were incubated at 37°C in 5% CO2 for 16 to 20 hours. 293T cells were transfected using FuGene (Roche Applied Sciences, Indianapolis, IN) according to the manufacturer's protocol. Luciferase activity was then determined using an assay kit from Promega Biotech (Madison, WI) as per the manufacturer's instructions. Cotransfection experiments included 10 μg pCMV-C/EBPϵ32 or a Gfi-1 expression plasmid (a gift from Dr M. Horwitz, University of Washington, Seattle, WA). Luciferase expression levels were normalized to the levels of β-galactosidase expression.21
Generation of erythroid myeloid lymphoid (EML) cell line from Gfi-1+/− and Gfi-1+/+ bone marrow
An EML cell line was generated from the bone marrow of mice deficient for the Gfi-1 gene using methodology previously described.22 Briefly, Gfi-1+/− mice and wild-type littermates were injected with 5-fluorouracil (5-FU, 100 mg/kg). Whole bone marrow was harvested 3 to 5 days later and cultured for 48 hours in IMDM supplemented with 20% (vol/vol) horse serum, murine GM-CSF (2.5 ng/mL; Immunex, Thousand Oaks, CA), human IL-6 (20 ng/mL; Peprotech, Rocky Hill, NJ), and murine IL-1b (10 ng/mL; Peprotech). The cells were transduced with a truncated retinoic acid receptor alpha (RARα403) by spinfection using supernatants derived from a stable GP + E86 producer line. Spinfections were performed over 2 hours with the addition of polybrene (4 μg/mL). Following spinfection, the cells were transferred to IMDM supplemented with 20% horse serum, rat SCF (200 ng/mL; Peprotech), Wehi-3B conditioned medium (0.25%) as a source of IL-3 (2.5-5 ng/mL; Peprotech), and human erythropoietin (8 U/mL; Amgen, Thousand Oaks, CA). Transduced cells were passaged every 2 to 3 days in IMDM medium supplemented with 20% (vol/vol) horse serum and 15% BHK-MKL conditioned medium as a source of SCF. The cells were exposed to G418 for an initial period of 10 days. Within 1 to 2 months, rapidly proliferating cell lines were generated. The genotype of the resulting Gfi-1+/− EML and Gfi-1+/+ cells was confirmed by PCR analysis using previously described oligonucleotide primers23 (data not shown).
Induction of Gfi-1+/− and Gfi-1+/+ EML cells
EML cells were induced sequentially to mature neutrophils via the promyelocytic stage (EPRO cells) using 10 μM ATRA as previously described.22,24 Cultures were harvested at the indicated times for RNA and whole cell protein extract preparation. Additionally, cells were cytospun using a Cytospin 3 instrument (Thermo-Shandon, Cheshire, United Kingdom), Wright-Giemsa–stained, and analyzed at original magnification, × 500, using a Nikon Eclipse E400 light microscope equipped with a Nikon 50 ×/0.90 oil-immersion objective lens and a Nikon PDX-35 camera (Nikon, Melville, NY).
Flow cytometry
Gfi-1+/− and Gfi-1+/+ EML cells were stained and analyzed using a FACSvantage flow cytometer and CellQuest software (BD Biosciences, San Jose, CA). Nonspecific antibody binding was inhibited by preincubating cells in staining medium (PBS + 0.5% BSA + 1% rat serum + 1% mouse serum) for 10 minutes at 4°C. Cells (1.5 × 105) were then labeled with conjugated antibodies for 20 minutes at 4°C. Propidium iodide (PI, 1 μg/mL; Molecular Probes, Eugene, OR) was used to eliminate dead cells during analysis. All antibodies (B220, Sca-1, CD34, c-kit, Mac-1, Gr-1, and Ter119) used for flow cytometry including matched isotype controls were conjugated to phycoerythrin (PE) and purchased from eBiosciences (Boston, MA). For each sample, 10 000 to 20 000 viable (PI negative) cells were acquired on the FACSvantage flow cytometer.
Preparation of nuclear extracts
All nuclear extracts were prepared as described previously.21
Oligonucleotide pull-down assay
This assay was conducted as previously documented25 using 20 μg biotinylated C/EBP/Gfi-1 oligomers: 5′CATTATAAATTGCAAAACTGTCAGAGTG3′. The DNA-protein complexes were then collected by low-speed centrifugation and resolved in a 4% to 12% gradient NuPage gel (Invitrogen).
Western blot analysis
Western blotting and detection were performed as previously described.25 Antibodies used were as follows: C/EBPϵ (sc-158, 1:3000; Santa Cruz Biotech, Santa Cruz, CA), Gfi-1 (sc-8558,1:500; Santa Cruz Biotech), PU.1 (sc-352, 1:500; Santa Cruz Biotech), MMP8 (neutrophil collagenase, sc-8848, 1:1000; Santa Cruz Biotech), and lactoferrin (antilactotransferrin, 1:10 000; Upstate Biotech, Lake Placid, NY).
Microarray analysis
Total RNA isolated from SGD and normal bone marrow was purified using an RNAeasy kit (Qiagen). High-quality total RNA (20 μg) prepared from each sample was submitted to the Keck Affymetrix Resource (http://info.med.yale.edu/wmkeck/affymetrix/), where cRNA labeling, hybridization, and data analysis were carried out. The Affymetrix GeneChip analysis (Santa Clara, CA) of gene expression used the human U133 set (900370) representing 22 000 genes or ORFs and 22 000 ESTs. Each gene or EST is represented on these chips by at least 11 × 25-mer oligonucleotides. The array images were compared following the guidelines provided by Affymetrix. Genes were considered up- or down-regulated if the relative expression changed 2-fold or more above the baseline control as per guidelines described at http://info.med.yale.edu/wmkeck/biostats/overview.htm#gene. Finally, expression of some differentially regulated genes of interest was confirmed by real-time PCR and or Western blot analysis.
Real-time PCR analysis
Total RNA (0.1 μg) from SGD and controls was used to generate first-strand cDNA using random hexamer primers with the Superscript II reverse transcription kit (Invitrogen). Of each cDNA, 5 ng was analyzed by real-time PCR in duplicate for each primer pair, and the corresponding threshold cycle (Ct) values were determined. All reactions were performed in a 25-μL volume using 50 ng each primer and the Platinum SYBR green qPCR Supermix UGD kit from Invitrogen as per the manufacturer's instructions using default cycling parameters in an ABI Prism 7200 Sequence detector. Transcript levels of each mRNA was normalized to that of β-actin and expressed as a percentage of the signal observed in the normal sample. The following primer pairs were used: Gfi-1 sense, 5′GACACCATGCCGCGCTCATTTC3′ and antisense, 5′ GTGCTCATTTGAGCCCATGCTG3′; PU.1 sense, 5′GAGAGCCATAGCGACCATTACT3′ and antisense, 5′GTACAGGCGGATCTTCTTCTTG3′; C/EBPϵ sense, 5′GACCTACTATGAGTGCGAGCCT3′ and antisense, 5′ACACCCTTGATGAGGGTAGCAG3′; M-CSF-R sense, 5′GTCACCCACAGACACCTCCT3′ and antisense, 5′AAGCTGGAAATCCCCCTAAA3′.
Results
A heterozygous mutation in the basic region of the C/EBPϵ gene in the SGD patient does not alter the transactivation ability of C/EBPϵ
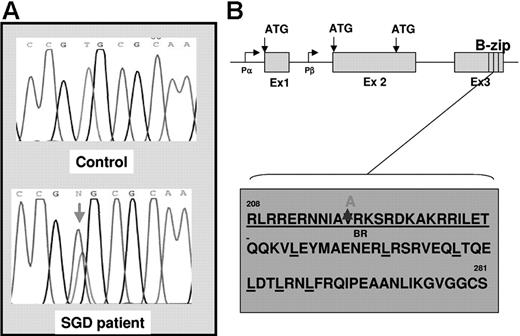
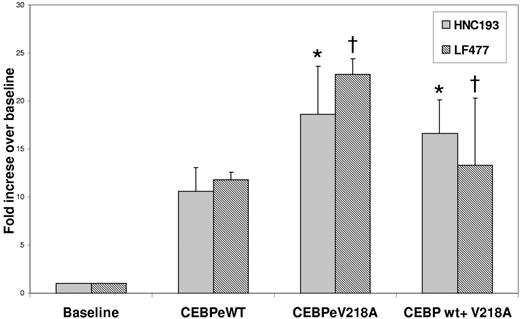
Since gene-inactivating mutations in the C/EBPϵ gene have been shown to underlie the etiology of SGD in 2 previously documented cases (reviewed in Lekstrom-Himes26 ), we looked for similar mutations in our SGD patient. Using oligomers designed to span the coding regions of the C/EBPϵ locus (Figure 1), we PCR amplified and sequenced genomic DNA from the SGD patient and from healthy controls. A heterozygous valine to alanine substitution was observed in the SGD but not in the normal DNA (Figure 1A). To rule out a naturally occurring polymorphism, we sequenced 108 normal DNA samples in this region of the C/EBPϵ gene and did not find the valine to alanine substitution (data not shown). This V218A mutation appears to disrupt the highly conserved basic or DNA-binding region of C/EBPϵ that is critical for association of this gene with C/EBP cis-elements located in its target genes (Figure 1C). In order to determine whether the V218A mutation in C/EBPϵ has functional repercussions, we introduced the mutation into full-length human C/EBPϵ cDNA and examined the ability of the mutant to transactivate SGP (LF and HNC) gene reporter constructs in cotransfection experiments. Figure 2 illustrates an experiment in which 2 SGP promoter reporter constructs (HNC193 and LF477) were cotransfected with either wild-type (wt) or mutant (V218A) C/EBPϵ expression plasmids in 293 cells. Both HNC193 and LF477 (SGP) reporter gene activity was up-regulated in the presence of wt C/EBPϵ (HNC193, 10.6-fold; LF477, 11.8-fold) when compared with the SGP reporter plasmids alone (Figure 2 baseline). Of interest, the mutant V128A C/EBPϵ plasmid transactivated the SGP reporter plasmids with higher efficiency (HNC193, 18.6-fold; LF477, 22.8-fold) when compared with the wt C/EBPϵ plasmid. This suggests that the mutant V218A C/EBPϵ protein is capable of binding to and transactivating a C/EBP reporter plasmid. An equivalent valine to alanine substitution in the C/EBPα gene also demonstrated a modest increase in reporter gene activity of a 2X C/EBP-Luciferase plasmid.27 To rule out a dominant-negative effect of the V218A mutant, we cotransfected both wt and V218A C/EBPϵ expression plasmids together with the SGP reporter plasmids. As is evident in Figure 2, there was no statistical difference between the transactivation activity of theV218A mutant versus the wt C/EBPϵ (compare wt vs wt + V218A). These experiments suggest that the V218A substitution in the SGD C/EBPϵ gene does not adversely affect SGP gene expression.
DNA sequence analysis of the C/EBPϵ gene in the SGD patient. (A) Genomic DNA isolated from a healthy individual (top panel, control) and from the SGD patient (bottom panel) was PCR amplified and sequenced using oligomers designed to capture the C/EBPϵ exons and intron-exon boundaries. The red arrow in the bottom panel indicates the heterozygous mutation in the SGD patient. (B) The genomic organization of the C/EBPϵ locus is indicated showing 3 exons (Ex 1, 2, and 3), 2 promoters (Pα and Pβ), 3 alternate translational start sites (ATG), and the highly conserved basic leucine zipper region (B-zip). (C) Amino acid sequence of the C/EBPϵ B-zip region. The heterozygous valine to alanine substitution in the SGD patient is indicated at position 218 in the basic region, which is underlined. The relevant leucine moieties are also underlined.
DNA sequence analysis of the C/EBPϵ gene in the SGD patient. (A) Genomic DNA isolated from a healthy individual (top panel, control) and from the SGD patient (bottom panel) was PCR amplified and sequenced using oligomers designed to capture the C/EBPϵ exons and intron-exon boundaries. The red arrow in the bottom panel indicates the heterozygous mutation in the SGD patient. (B) The genomic organization of the C/EBPϵ locus is indicated showing 3 exons (Ex 1, 2, and 3), 2 promoters (Pα and Pβ), 3 alternate translational start sites (ATG), and the highly conserved basic leucine zipper region (B-zip). (C) Amino acid sequence of the C/EBPϵ B-zip region. The heterozygous valine to alanine substitution in the SGD patient is indicated at position 218 in the basic region, which is underlined. The relevant leucine moieties are also underlined.
Transient cotransfection analysis of SGP promoter plasmids (HNC and LF) with wild-type and mutant C/EBPϵ expression plasmids in 32Dwt18 cells. 32Dwt18 cells were transiently cotransfected with HNC193 and LF477 (SGP) reporter plasmids (see “Patients, materials, and methods”) with 10 μg expression plasmids for wt C/EBPϵ (e) and mutant C/EBPϵ (V218A) either separately or together. Normalized luciferase values have been represented as a ratio of enzyme activity of SGP promoter plasmids plus C/EBPϵ expression plasmids to that of the SGP promoter plasmid without expression plasmids. The figure represents normalized mean ± SE obtained from 3 independent experiments, each performed in duplicate. Statistical analysis performed by Student t test revealed no statistical significance in the transactivation ability of wt and mutant plasmids. *P = .15; †P = .22.
Transient cotransfection analysis of SGP promoter plasmids (HNC and LF) with wild-type and mutant C/EBPϵ expression plasmids in 32Dwt18 cells. 32Dwt18 cells were transiently cotransfected with HNC193 and LF477 (SGP) reporter plasmids (see “Patients, materials, and methods”) with 10 μg expression plasmids for wt C/EBPϵ (e) and mutant C/EBPϵ (V218A) either separately or together. Normalized luciferase values have been represented as a ratio of enzyme activity of SGP promoter plasmids plus C/EBPϵ expression plasmids to that of the SGP promoter plasmid without expression plasmids. The figure represents normalized mean ± SE obtained from 3 independent experiments, each performed in duplicate. Statistical analysis performed by Student t test revealed no statistical significance in the transactivation ability of wt and mutant plasmids. *P = .15; †P = .22.
C/EBPϵ is expressed in the bone marrow and peripheral blood neutrophils of our SGD patient
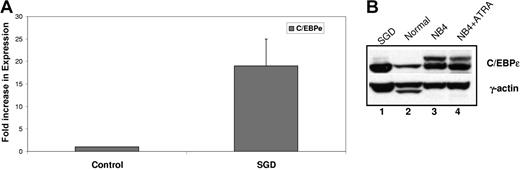
Since the heterozygous V218A mutation in the C/EBPϵ gene from the SGD patient did not impair transactivation of SGP reporter genes, we examined whether this substitution had an effect on C/EBPϵ expression at the RNA or protein level. We evaluated C/EBPϵ expression by both real-time PCR and Western blot analysis. Real-time PCR using cDNA prepared from SGD and normal control PMN RNA was performed using oligomers specific for C/EBPϵ. The expression of C/EBPϵ was normalized to that of β-actin and expressed as a fold increase over the normal control. As shown in Figure 3A, the C/EBPϵ transcript was expressed at 19-fold higher levels compared with the normal control. Western blot analysis revealed that this increase in expression was also reflected at the protein level, with higher levels of C/EBPϵ protein in the SGD PMNs (Figure 3B lane 1) compared with the normal PMNs (lane 2). The human acute promyelocytic leukemic NB4 cells, both uninduced (Figure 3B lane 3) and ATRA induced (lane 4) served as a positive control. These observations confirm that the V218A C/EBPϵ mutation does not cause the C/EBPϵ protein to be unstable. In fact, we cannot rule out the possibility that it may actually be more stable than its wild-type counterpart.
Expression of C/EBPϵ in the SGD patient. (A) Real-time PCR analysis of C/EBPϵ expression in a normal sample and in the SGD patient was carried out in triplicate. Transcript levels were normalized to that of β-actin and expressed as a ratio of the signal observed in the normal sample (1). Error bars indicate standard error of the mean (SEM). (B) Western blot analysis of normal and SGD PMNs. NB4 and NB4 cells treated with ATRA serve as an additional control for C/EBPϵ expression.
Expression of C/EBPϵ in the SGD patient. (A) Real-time PCR analysis of C/EBPϵ expression in a normal sample and in the SGD patient was carried out in triplicate. Transcript levels were normalized to that of β-actin and expressed as a ratio of the signal observed in the normal sample (1). Error bars indicate standard error of the mean (SEM). (B) Western blot analysis of normal and SGD PMNs. NB4 and NB4 cells treated with ATRA serve as an additional control for C/EBPϵ expression.
Microarray analysis of SGD versus normal bone marrow reveals the up-regulation of the transcription factor PU.1 and its target genes
Since the V218A C/EBPϵ mutation did not appear to account for the SGD phenotype in our patient, we sought to identify other potential candidate factors to explain this disease. We performed microarray analysis on RNA isolated from the bone marrow of the patient and from a healthy volunteer using the human Affymetrix U133 set (900370) gene chip representing 22 000 genes or ORFs and 22 000 ESTs. A total of 4393 differentially regulated genes were identified. We identified several genes on the list that have been previously demonstrated to be involved in neutrophil development and/or function (Table 1).
A partial list of genes up-/down-regulated in SGD versus normal bone marrow by Affymetrix Chip analysis
| . | Fold change . | Abbreviation . | Genbank accession no. . |
|---|---|---|---|
| Defensin alpha 1 | −3.7 | DEFA1 | NM_004084.2 |
| Defensin alpha 4 | −5.4 | DEFA4 | NM_001925.1 |
| Proteinase 3* | 4.6 | PRTN3 | NM_002777.2 |
| BPI* | 4.3 | BPI | NM_001725.1 |
| Secondary granule proteins | |||
| Neutrophil collagenase | −5.4 | MMP8 | NM_002424.1 |
| Neutrophil gelatinase | −3.5 | MMP9 | NM_004994.1 |
| Transcobalamin 1 | −4.5 | TCN1 | NM_001062.1 |
| Specific granule protein 28 | −9 | SGP28 | NM_006061.1 |
| Neutrophil function | |||
| gp47 phox* | 9.1 | NCF1 | NM_000265.1 |
| gp91 phox* | 6 | CYBB | NM_000397.1 |
| IL-8 | −2 | IL8 | AF043337.1 |
| Leukocyte IgG receptor, FcgR | 4.7 | FCGR | J04162.1 |
| Proteoglycan recognition protein | 13.8 | PGLYRP | NM_005091.1 |
| Transcription factors | |||
| CKLF | 5.6 | CKLF | AF132818 |
| KLF4 | 3.6 | KLF4 | NM_004235 |
| C/EBPδ | 2.1 | C/EBPd | AV655640 |
| C/EBPϵ | 1.5 | C/EBPe | NM_001805 |
| C/EBPα | 1.5 | C/EBPa | NM_004364.1 |
| Hox A9 | 4.3 | HoxA9 | U41813.1 |
| Fox B | 4.1 | FOSB | NM_006732.1 |
| PU.1 | 3.2 | PU.1 | NM_003120 |
| Egr-1 | 1.5 | Egr-1 | NM_001964 |
| Other genes | |||
| Eosinophil cataionic protein | −4.8 | RNASE3 | NM_002935.1 |
| Cathepsin W | −2.5 | CTSW | NM_001335.1 |
| Cathepsin D | 3.2 | CTSD | NM_001909.1 |
| CD11b* | 5 | ITGAM | NM_000632.2 |
| CD18* | 4.1 | Mac-1B | NM_000211.1 |
| GM-CSF receptor* | 2.7 | CSF2RA | NM_006140.1 |
| TGFb1 | 3.4 | TGFB1 | NM_000358.1 |
| Calpain-3 | 3.1 | p94 | BC003169.1 |
| . | Fold change . | Abbreviation . | Genbank accession no. . |
|---|---|---|---|
| Defensin alpha 1 | −3.7 | DEFA1 | NM_004084.2 |
| Defensin alpha 4 | −5.4 | DEFA4 | NM_001925.1 |
| Proteinase 3* | 4.6 | PRTN3 | NM_002777.2 |
| BPI* | 4.3 | BPI | NM_001725.1 |
| Secondary granule proteins | |||
| Neutrophil collagenase | −5.4 | MMP8 | NM_002424.1 |
| Neutrophil gelatinase | −3.5 | MMP9 | NM_004994.1 |
| Transcobalamin 1 | −4.5 | TCN1 | NM_001062.1 |
| Specific granule protein 28 | −9 | SGP28 | NM_006061.1 |
| Neutrophil function | |||
| gp47 phox* | 9.1 | NCF1 | NM_000265.1 |
| gp91 phox* | 6 | CYBB | NM_000397.1 |
| IL-8 | −2 | IL8 | AF043337.1 |
| Leukocyte IgG receptor, FcgR | 4.7 | FCGR | J04162.1 |
| Proteoglycan recognition protein | 13.8 | PGLYRP | NM_005091.1 |
| Transcription factors | |||
| CKLF | 5.6 | CKLF | AF132818 |
| KLF4 | 3.6 | KLF4 | NM_004235 |
| C/EBPδ | 2.1 | C/EBPd | AV655640 |
| C/EBPϵ | 1.5 | C/EBPe | NM_001805 |
| C/EBPα | 1.5 | C/EBPa | NM_004364.1 |
| Hox A9 | 4.3 | HoxA9 | U41813.1 |
| Fox B | 4.1 | FOSB | NM_006732.1 |
| PU.1 | 3.2 | PU.1 | NM_003120 |
| Egr-1 | 1.5 | Egr-1 | NM_001964 |
| Other genes | |||
| Eosinophil cataionic protein | −4.8 | RNASE3 | NM_002935.1 |
| Cathepsin W | −2.5 | CTSW | NM_001335.1 |
| Cathepsin D | 3.2 | CTSD | NM_001909.1 |
| CD11b* | 5 | ITGAM | NM_000632.2 |
| CD18* | 4.1 | Mac-1B | NM_000211.1 |
| GM-CSF receptor* | 2.7 | CSF2RA | NM_006140.1 |
| TGFb1 | 3.4 | TGFB1 | NM_000358.1 |
| Calpain-3 | 3.1 | p94 | BC003169.1 |
Known PU.1 regulated genes.
Our laboratory has previously reported reduced expression of the primary granule proteins defensin alpha1 and alpha4 as well as the secondary granule proteins neutrophil collagenase (HNC), neutrophil gelatinase (HNG), and transcobalamin 1 (TC1) in this SGD patient at the RNA level.15 Our microarray analysis (Table 1) corroborates this observation. Furthermore, the presence of a down-regulated eosinophil-specific transcript (eosinophil cationic protein, −4.8) was also confirmatory, since SGD patients lack both neutrophil and eosinophil secondary granules (reviewed in Gombart and Koeffler28 ). Additionally, a number of proteins involved in neutrophil function were also found to be dysregulated in SGD (Table 1).
The SGD bone marrow also showed abnormal expression of a number of transcription factors (Table 1). Of interest, several members of the C/EBP family, including C/EBPα, C/EBPδ, and C/EBPϵ, were all expressed at slightly higher levels (1.5- to 2.1-fold) in the SGD bone marrow compared with the control. Additionally, levels of PU.1 were up-regulated (3.2-fold) in the SGD bone marrow. PU.1 belongs to the Ets family of transcription factors and plays a vital role in B-cell, macrophage, and late neutrophil development.29,30 PU.1−/− mice have markedly impaired neutrophil production, with an abnormal pattern of neutrophil-specific gene expression, absence of secondary and tertiary granules, and functional impairment with defects in respiratory burst and phagocytosis.31,32 In the face of its altered expression in the SGD patient, we examined the PU.1 gene for mutations. Thorough examination of all exons and exon-intron boundaries of the PU.1 gene by direct sequencing of PCR products revealed no mutations (data not shown). However, we observed that expression of a number of PU.1 regulated genes was also up-regulated in the SGD bone marrow (Table 1 asterisk). These include early growth response 1 (Egr1), a transcription factor that plays an important role in macrophage differentiation (Laslo et al33 ); CD11b (integrin αM) and CD18 (mac-1 β subunit), both members of the integrin family of cell-surface markers; proteinase 3 and bactericidal permeability-increasing protein (BPI), both primary granule proteins; and gp91phox, also known as cytochrome b-245 b polypeptide (CYBB) and gp47phox or NCF1, both of which contribute to the NADPH-reductase complex associated with the respiratory burst activity of normal neutrophils (Skalnik et al7 and references within).
PU.1 RNA and protein levels are up-regulated in SGD PMNs and bear a reciprocal relationship to that of growth factor independence-1 (Gfi-1)
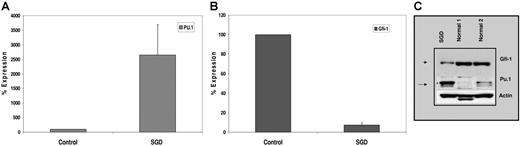
We confirmed increased expression of PU.1 in SGD PMNs by real-time PCR using cDNA prepared from SGD and normal control PMN RNAs. The expression of PU.1 was normalized to that of β-actin and expressed as a percentage of control (100%). As shown in Figure 4A, PU.1 expression in SGD PMNs was 25-fold higher than in normal PMNs, confirming an even higher level of up-regulation than was suggested by microarray analysis (Table 2). Increased PU.1 mRNA suggests transcriptional up-regulation of the PU.1 promoter, perhaps through disrupted binding of a negative regulator. Such a negative regulator has recently been described. In a recent study, Hock et al described mice nullizygous for the zinc finger–containing transcription factor Gfi-1,23 which bear striking similarities to the C/EBPϵ knock-out mice and to patients with SGD. Gfi-1−/− mice lack secondary and tertiary granule protein gene expression despite C/EBPϵ expression.23 Furthermore, these mice express elevated levels of PU.1 and its downstream target genes including the M-CSF receptor.23 Gfi-1 has been described as a negative regulator of gene expression in T lymphocytes (reviewed in Moroy34 ; Duan and Horwitz35 ; Hock and Orkin36 ). Since the expression of Gfi-1 and PU.1 has been reported to have a reciprocal relationship, we examined levels of Gfi-1 in SGD PMNs by real-time detection PCR. Significantly decreased levels of Gfi-1 RNA were observed in our SGD patient (7%) compared with the healthy control (100%) (Figure 4A).
Expression of PU.1 and Gfi-1 in the SGD patient. Real-time PCR analysis of (A) PU.1 and (B) Gfi-1 expression in normal versus SGD PMNs. Transcript levels of each mRNA were assessed in triplicate and normalized to that of β-actin and expressed as a percentage of the signal observed in the normal sample (100%). Due to the paucity of RNA, this experiment was performed only once in triplicate. Error bars indicate SEM. (C) Western blot analysis of PMNs from SGD and normal samples. Nuclear extracts prepared from peripheral blood neutrophils (PMNs) of 2 healthy volunteers and our SGD patient were subjected to Western blot analysis. Equal concentrations of protein were loaded in each lane. The blot was sequentially probed with antibodies for Gfi-1, PU.1, and β-actin.
Expression of PU.1 and Gfi-1 in the SGD patient. Real-time PCR analysis of (A) PU.1 and (B) Gfi-1 expression in normal versus SGD PMNs. Transcript levels of each mRNA were assessed in triplicate and normalized to that of β-actin and expressed as a percentage of the signal observed in the normal sample (100%). Due to the paucity of RNA, this experiment was performed only once in triplicate. Error bars indicate SEM. (C) Western blot analysis of PMNs from SGD and normal samples. Nuclear extracts prepared from peripheral blood neutrophils (PMNs) of 2 healthy volunteers and our SGD patient were subjected to Western blot analysis. Equal concentrations of protein were loaded in each lane. The blot was sequentially probed with antibodies for Gfi-1, PU.1, and β-actin.
We next examined Gfi-1 and PU.1 protein levels by Western blot analysis. Figure 4B demonstrates markedly reduced levels of Gfi-1 protein in the SGD PMN sample compared with 2 normal controls. In contrast, PU.1 levels were significantly higher in the SGD patient than in the 2 control PMN samples. Equal loading of protein in each lane was confirmed by stripping the blot and probing with a β-actin antibody.
Gfi-1+/− EML cell line shares morphologic and biochemical features of the Gfi-1–null and SGD patient bone marrow
SGD is extremely rare, and obtaining adequate patient samples is difficult. Therefore, we established a cell line model in which levels of Gfi-1 are significantly reduced while C/EBPϵ levels remain normal to high. We harvested bone marrow from 5-FU–treated Gfi-1+/− mice and age-matched wild-type littermates, and transduced them with RARα403 as previously described.22,24 Within 1 to 2 months, rapidly proliferating factor-dependent cell lines emerged. We confirmed the genotype of the Gfi-1+/− and Gfi-1+/+ EML cells using a previously described PCR approach (data not shown).
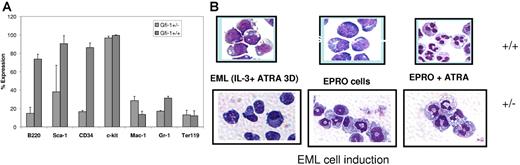
Analysis of the cell-surface markers on the EML Gfi-1+/+ and Gfi-1+/− cells revealed clear differences (Figure 5A). The expression of Sca-1 (hematopoietic stem-cell [HSC] marker), B220 (B-cell marker), and CD34 were significantly lower in Gfi-1+/− cells than in Gfi-1+/+ EML cells. No change was observed with respect to the expression of c-kit (HSC marker) and Ter119 (erythroid marker). High-level expression of c-kit in both cell lines was predictable as EML cells were selected to be SCF dependent. Of interest, the level of Mac-1 expression (myeloid marker) was 2-fold higher in Gfi-1+/− cells than in Gfi-1+/+ cells, while that of Gr-1 was about 2-fold lower in the Gfi-1+/− EML cells. With the exception of c-kit, this general expression pattern followed the same trend as that observed for bone marrow myeloid progenitors of Gfi-1–null mice.23 Furthermore, induction of EML Gfi-1+/+ and Gfi-1+/− cells with ATRA and IL-3 (3 days) followed by GM-CSF induction gave rise to EML-derived promyelocyte (EPRO) cells. Wright-Giemsa staining (Figure 5B) of Gfi-1+/− cells revealed abnormal ring-shaped EPRO cells, while EML Gfi-1+/+ cells yielded promyelocytes with normal morphology. These abnormal cells have been described previously in the bone marrow of Gfi-1−/− mice.23 Further maturation of the EPRO cells with ATRA gave rise to neutrophils with fewer nuclear lobes and a more monocyte/macrophage-like cytoplasm compared with the equivalent wild-type cells (Figure 5B). Of interest, we failed in 2 attempts to isolate EML cells from Gfi-1−/− bone marrow. In 2 recent studies, Gfi-1 has been shown to restrict proliferation and maintain quiescence of hematopoietic stem cells (reviewed in Hock and Orkin36 ). We hypothesize that complete loss of Gfi-1 activity may increase cycling of the stem-cell progenitors, which in turn may compromise the ability to transduce an early progenitor cell, an essential step in the process of generating EML cells.
A flow cytometric analysis of Gfi-1+/− and Gfi-1+/+ EML cells. Gfi-1+/− and Gfi-1+/+ EML cells were stained and analyzed using a FACSvantage flow cytometer and CellQuest software. (A) Cells (1.5 × 105) were then labeled with conjugated antibodies for 20 minutes at 4°C. All antibodies (B220, Sca-1, CD34, c-kit, Mac-1, Gr-1, and Ter119) used for flow cytometry including matched isotype controls were conjugated to phycoerythrin (PE) and purchased from eBiosciences. Error bars indicate SEM. (B) Wright-Giemsa staining of EML+/− and EML+/+ cells following terminal neutrophil maturation. Myeloid differentiation of EML cells was conducted in IMDM medium containing 20% horse serum, IL-3, SCF, and 10 μM ATRA for 3 days. The cells were then transferred to IMDM medium containing 20% horse serum and GM-CSF. Within a week, GM-CSF–dependent EPRO cells emerged. These cells were terminally differentiated by the addition of 10 μM ATRA for 3 days. Cells were cytospun on the days indicated and subjected to Wright-Giemsa staining.
A flow cytometric analysis of Gfi-1+/− and Gfi-1+/+ EML cells. Gfi-1+/− and Gfi-1+/+ EML cells were stained and analyzed using a FACSvantage flow cytometer and CellQuest software. (A) Cells (1.5 × 105) were then labeled with conjugated antibodies for 20 minutes at 4°C. All antibodies (B220, Sca-1, CD34, c-kit, Mac-1, Gr-1, and Ter119) used for flow cytometry including matched isotype controls were conjugated to phycoerythrin (PE) and purchased from eBiosciences. Error bars indicate SEM. (B) Wright-Giemsa staining of EML+/− and EML+/+ cells following terminal neutrophil maturation. Myeloid differentiation of EML cells was conducted in IMDM medium containing 20% horse serum, IL-3, SCF, and 10 μM ATRA for 3 days. The cells were then transferred to IMDM medium containing 20% horse serum and GM-CSF. Within a week, GM-CSF–dependent EPRO cells emerged. These cells were terminally differentiated by the addition of 10 μM ATRA for 3 days. Cells were cytospun on the days indicated and subjected to Wright-Giemsa staining.
Gfi-1+/− EPRO cells express higher levels of C/EBPϵ and PU.1 and reduced levels of SGP genes
Gfi-1+/− and Gfi-1+/+ EPRO cells were induced to terminally differentiate by the addition of ATRA. RNA and total cellular proteins were harvested at 24-hour intervals. The RNA was analyzed by real-time PCR. The data in Figure 6A are represented as a ratio of the expression in the Gfi-1+/− cells to that in the wild-type cells. At 24 hours, Gfi-1 expression in the Gfi-1+/− cells was about a third (0.36-fold) of that observed in the wild-type cells. This fraction fell even further to about one tenth (0.14-fold) after 48 hours. In contrast, the levels of both PU.1 (3.4-fold) and C/EBPϵ (1.9-fold) after 24 hours of induction were higher in the Gfi-1+/− cells than in the Gfi-1+/+ cells. While the relative expression of C/EBPϵ continued to increase upon ATRA induction (3.6-fold), the expression of PU.1 seemed to taper off to wild-type levels (0.9-fold) at 48 hours after ATRA induction. The protein expression pattern for both C/EBPϵ and PU.1 was identical to the RNA expression pattern in ATRA-induced EPRO cells (data not shown). We also examined expression of M-CSF-R, a known direct target of PU.1 in the monocyte/macrophage lineage,30,37 in ATRA-induced Gfi-1+/− and Gfi-1+/+ EPRO cells (Figure 6A). While mRNA expression levels of M-CSF-R were similar following 24-hour ATRA induction (0.6-fold), a 5.4-fold increase in expression was noted in 48-hour ATRA-induced Gfi-1+/− EPRO cells (Figure 6A) when compared with the equivalent wild-type cells. (Figure 6A). This suggests that high levels of PU.1 at 24 hours are sufficient to elevate M-CSF-R expression at 48 hours after ATRA treatment. It has been previously demonstrated that M-CSF expression is dependent not only on PU.1 but also on the synergistic activity of C/EBPs and AML-1.38 This may explain why sustained PU.1 expression is not required to up-regulate M-CSF-R mRNA in the 48-hour ATRA-treated Gfi-1+/− cells (Figure 6A). A similar increase in the levels of the M-CSF-R was noted in the SGD patient compared with a healthy control (Table 1).
Expression pattern of Gfi-1, PU.1, C/EBPϵ, and their downstream targets in ATRA-induced Gfi-1+/+ and Gfi-1+/– EPRO cells. (A) Real-time PCR analysis of ATRA-induced Gfi-1+/+ and Gfi-1+/− EPRO cells. EPRO cells were induced to terminally differentiate in the presence of ATRA. RNA samples were collected at the times indicated and subjected to real-time PCR analysis. Transcript levels of each mRNA were normalized to that of 18S rRNA and expressed as a percentage of the signal observed in the uninduced EPRO+/+ cells. This experiment was performed 3 times in triplicate. (B) Western blot analysis of ATRA-induced Gfi-1+/+ and Gfi-1+/− EPRO cells. Whole cell extracts prepared from ATRA-induced EPRO cells at the time indicated were subjected to Western blot analysis. Equal concentrations of protein were loaded in each lane. The blots were sequentially probed with antibodies for LF and β-actin, and NC and β-actin.
Expression pattern of Gfi-1, PU.1, C/EBPϵ, and their downstream targets in ATRA-induced Gfi-1+/+ and Gfi-1+/– EPRO cells. (A) Real-time PCR analysis of ATRA-induced Gfi-1+/+ and Gfi-1+/− EPRO cells. EPRO cells were induced to terminally differentiate in the presence of ATRA. RNA samples were collected at the times indicated and subjected to real-time PCR analysis. Transcript levels of each mRNA were normalized to that of 18S rRNA and expressed as a percentage of the signal observed in the uninduced EPRO+/+ cells. This experiment was performed 3 times in triplicate. (B) Western blot analysis of ATRA-induced Gfi-1+/+ and Gfi-1+/− EPRO cells. Whole cell extracts prepared from ATRA-induced EPRO cells at the time indicated were subjected to Western blot analysis. Equal concentrations of protein were loaded in each lane. The blots were sequentially probed with antibodies for LF and β-actin, and NC and β-actin.
The expression of 2 SGP genes, LF and neutrophil collagenase (NC, MMP-8), was next assessed in the Gfi-1 EPRO cells. While protein levels of both LF and NC were up-regulated upon ATRA induction in the Gfi-1+/+ EPRO cells (Figure 6B), up-regulation was markedly diminished in the ATRA-treated Gfi-1+/− EPRO cells (Figure 6B). This observation is consistent with the observation that expression of LF and NC was completely absent in the bone marrow of Gfi-1−/− mice.23
Gfi-1 cooperates with wt C/EBPϵ but not mutant C/EBPϵ to up-regulate the expression of the neutrophil collagenase promoter, a secondary granule protein gene
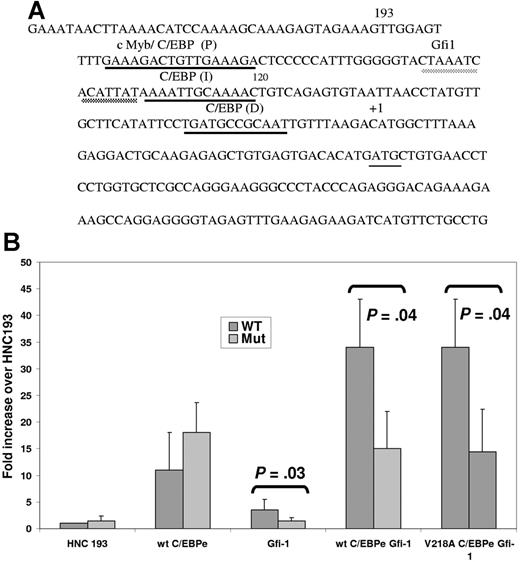
Despite adequate levels of both C/EBPϵ (Figure 3) and PU.1 (Figure 4A-B) in the SGD PMNs, both essential factors for neutrophil secondary and tertiary granule protein gene expression, no expression of the SGP genes was observed in the SGD patient.15 We hypothesized that as in the Gfi-1+/− EPRO cells, decreased Gfi-1 in this patient may contribute to decreased secondary granule protein gene expression. To test this hypothesis, we examined the sequence of the human neutrophil collagenase promoter in which we have demonstrated 3 functional C/EBP-binding sites termed proximal (P), intermediate (I), and distal (D) (underlined in black in Figure 7A). 25 A putative Gfi-1–binding site was identified just upstream of the C/EBP I site in the first 193 bp of the HNC promoter (underlined in gray in Figure 7A). To demonstrate the functional relevance of the putative Gfi-1 site, we cotransfected a previously described plasmid containing either a wild-type (wt) or Gfi-1 mutant (Mut) 193 bp of the minimal HNC promoter cloned upstream of the promoterless pGL3 basic plasmid25 with expression plasmids for wt C/EBPϵ and V218A C/EBPϵ with and without Gfi-1 into 32Dwt18 myeloid cells.25 As expected, cotransfection of both the wt and Gfi-1 mutant HNC193 plasmids with a C/EBPϵ expression plasmid resulted in an 11- to 18-fold increase in luciferase activity compared with HNC193 alone (Figure 7B). Cotransfection of the HNC promoter plasmids with a Gfi-1 expression plasmid also resulted in transactivation, but only of the wild-type reporter plasmid (3.4-fold above HNC193 alone) and not the Gfi-1 mutant HNC193 plasmid (Mut). Gfi-1–mediated transactivation was considerably lower than that observed with the C/EBPϵ expression plasmid. This modest but statistically significant increase (P = .03) in reporter gene activity in response to Gfi-1 has not been previously reported in a myeloid context. A 34-fold increase in reporter gene activity was observed when C/EBPϵ and Gfi-1 were cotransfected with the HNC193 plasmid. This synergistic (and statistically significant; P = .04) effect is suggestive of a cooperative interaction of these 2 transcription factors in activating the HNC promoter in myeloid cells. In contrast, no synergy was observed either when the Gfi-1 site in the HNC promoter was mutated or when the V218A C/EBPϵ mutant protein was used (Figure 7B). A similar experiment conducted in nonmyeloid NIH3T3 cells yielded similar but less dramatic results (data not shown). In an equivalent experiment using a LF promoter reporter plasmid harboring Gfi-1– and C/EBP-binding sites and expression plasmids for Gfi-1 and C/EBPϵ, a similar synergistic effect was observed (Figure S1, available on the Blood website; see the Supplemental Figure link at the top of the online article). These observations suggest that an interaction between C/EBPϵ and Gfi-1 is vital to SGP gene expression, and disruption of this synergy may contribute to the SGD phenotype in our patient.
Transient transfection analysis of an SGP promoter (NC) harboring a Gfi-1 and a C/EBP site. (A) DNA sequence of the promoter of an SGP gene, neutrophil collagenase (NC). The first 193 bp of the human (HNC193) neutrophil collagenase promoter sequences representing the minimal promoter is illustrated. The 3 C/EBP sites (P, I, and D) are underlined in black and the conserved Gfi-1 sequence, in gray. The transcription start site is marked by a +1 sign and the translation start site ATG is indicated. (B) 32Dwt18 cells were transiently cotransfected with wild-type HNC193 (wt) or Gfi-1 mutant HNC193 (“Patients, materials, and methods”) and 10 μg expression plasmids for Gfi-1 and wt C/EBPϵ (e) separately or together. HNC193 (wt) was also cotransfected with the Gfi-1 expression plasmid and wt or V218A C/EBPϵ plasmids. Normalized luciferase values have been represented as a ratio of enzyme activity of HNC193 promoter plasmid plus C/EBP and/or Gfi-1 expression plasmids to that of HNC193 promoter plasmid without expression plasmids. The figure represents normalized mean ± SE obtained from 3 independent experiments, each performed in duplicate. Statistical significance was determined using Student t test, and the P values have been indicated.
Transient transfection analysis of an SGP promoter (NC) harboring a Gfi-1 and a C/EBP site. (A) DNA sequence of the promoter of an SGP gene, neutrophil collagenase (NC). The first 193 bp of the human (HNC193) neutrophil collagenase promoter sequences representing the minimal promoter is illustrated. The 3 C/EBP sites (P, I, and D) are underlined in black and the conserved Gfi-1 sequence, in gray. The transcription start site is marked by a +1 sign and the translation start site ATG is indicated. (B) 32Dwt18 cells were transiently cotransfected with wild-type HNC193 (wt) or Gfi-1 mutant HNC193 (“Patients, materials, and methods”) and 10 μg expression plasmids for Gfi-1 and wt C/EBPϵ (e) separately or together. HNC193 (wt) was also cotransfected with the Gfi-1 expression plasmid and wt or V218A C/EBPϵ plasmids. Normalized luciferase values have been represented as a ratio of enzyme activity of HNC193 promoter plasmid plus C/EBP and/or Gfi-1 expression plasmids to that of HNC193 promoter plasmid without expression plasmids. The figure represents normalized mean ± SE obtained from 3 independent experiments, each performed in duplicate. Statistical significance was determined using Student t test, and the P values have been indicated.
Gfi-1 and C/EBPϵ both bind the HNC promoter
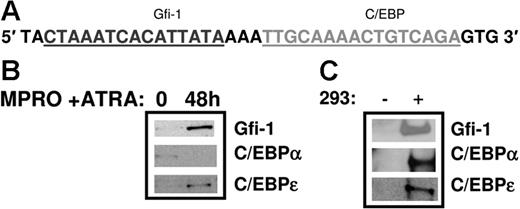
In order to validate the role of Gfi-1 in directly activating the HNC promoter, we performed an oligonucleotide pull-down assay to demonstrate binding of this factor to the Gfi-1 site in the HNC promoter. Nuclear extracts prepared from uninduced and ATRA-induced MPRO cells, a model for neutrophil maturation, were incubated with biotinylated oligomers representing the Gfi-1/C/EBP I site in the HNC promoter (Figure 8A). The DNA-protein complexes were recovered using streptavidin-agarose beads and the bound proteins resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and Western blot analysis. As shown in Figure 8B, Gfi-1 is bound at very low levels to the Gfi-1/C/EBP I probe in uninduced MPRO cells. Gfi-1 binding is significantly increased upon induction with ATRA. C/EBPϵ binds to this probe with the same binding pattern, with increased binding of C/EBPϵ in ATRA-induced MPRO cells (Figure 8B). C/EBPα binding to the same oligomers, however, appears to be higher in uninduced MPRO cells and decreases upon induction. Of interest, mutating the C/EBP site in the Gfi-1/C/EBP I prevents C/EBPα and C/EBPϵ binding to the probe in both uninduced and ATRA-induced MPRO cells, but Gfi-1 binding remains unaffected (data not shown). The Gfi-1/C/EBP I probe and Gfi-1–, C/EBPα-, and C/EBPϵ-overexpressing 293T nuclear extracts were used as positive controls in this experiment (Figure 8C).19,20
Oligonucleotide pull-down assay using the Gfi-1/C/EBP probe in the HNC promoter. Nuclear extracts prepared from uninduced and ATRA-induced MPRO cells (a murine myeloid cell line for neutrophil maturation) were incubated with the biotinylated Gfi-1/C/EBP probe from the HNC promoter (A). The DNA-protein complexes were recovered using streptavidin-agarose beads and the bound proteins resolved by SDS-PAGE and Western blot analysis. The blot was probed sequentially with Gfi-1, C/EBPα, and C/EBPϵ antibodies (B). Oligonucleotide pull-down assays using the same probe and 293 extracts overexpressing Gfi-1, C/EBPα, and C/EBPϵ served as a positive control (C). (− indicates untransfected 293 cells; +, transfected 293 cells.)
Oligonucleotide pull-down assay using the Gfi-1/C/EBP probe in the HNC promoter. Nuclear extracts prepared from uninduced and ATRA-induced MPRO cells (a murine myeloid cell line for neutrophil maturation) were incubated with the biotinylated Gfi-1/C/EBP probe from the HNC promoter (A). The DNA-protein complexes were recovered using streptavidin-agarose beads and the bound proteins resolved by SDS-PAGE and Western blot analysis. The blot was probed sequentially with Gfi-1, C/EBPα, and C/EBPϵ antibodies (B). Oligonucleotide pull-down assays using the same probe and 293 extracts overexpressing Gfi-1, C/EBPα, and C/EBPϵ served as a positive control (C). (− indicates untransfected 293 cells; +, transfected 293 cells.)
Discussion
We have described a patient with the rare congenital disorder SGD. SGD is typically associated with inactivating mutations in the C/EBPϵ gene. DNA sequence analysis in our patient revealed a heterozygous substitution at amino acid 218 (NM_001805) that lies within the highly conserved DNA-binding basic region of the C/EBPϵ gene. This results in the substitution of a valine to an alanine at this position. We introduced this mutation into wild-type C/EBPϵ and evaluated the ability of the mutant gene to transactivate SGP reporter genes (HNC and LF). We showed that the mutant C/EBPϵ transactivated the SGP promoters with equal efficiency as the wild type, suggesting that the mutation is unlikely to alter C/EBPϵ-mediated SGP gene expression in the SGD patient. Given that the mutation occurs in the basic region of C/EBPϵ, coinciding with the region harboring the nuclear localization signal of b-Zip proteins, we examined whether this substitution altered the ability of C/EBPϵ to localize to the nucleus. In preliminary transient transfection immunofluorescence studies, we showed that the V218AC/EBPϵ protein does not accumulate in the cytoplasm (A.K.-G., H.S., and N.B., unpublished observations, December 2005), thus suggesting that mislocalization of the mutant C/EBPϵ protein is unlikely to explain the SGD phenotype.
In a recent study, Miller et al mutated the equivalent conserved Val (296) to Ala in the basic region of the C/EBPα gene and found that it strongly enhanced C/EBPα binding to cAMP response element (CRE) sites while retaining affinity for C/EBP sites. This evolutionarily conserved Val (296) is thus thought to function primarily to restrict interactions with related sequences such as CRE sites rather than to specify binding to C/EBP sites.27 Of interest, in other b-Zip transcription factors such as AP-1 and CREB, Ala occupies the position analogous to Val218.27 While it is possible that the mutant C/EBPϵ (V218A) may promiscuously activate inappropriate downstream targets with C/EBP-like elements in their promoters, we have observed that unlike the mutant C/EBPα, the mutant C/EBPϵ does not preferentially transactivate a CRE reporter gene (A.K.-G., M.S., and N.B., unpublished observations, June 2005). We predict that the mutant protein binds canonical C/EBPϵ target gene promoters and directs their expression in a manner similar to wild-type C/EBPϵ.
To gain a better understanding of the molecular basis of SGD in our patient, we performed microarray analysis using RNA isolated from the SGD patient compared with normal bone marrow. A number of genes contributing to normal neutrophil development and function were dysregulated in the bone marrow of the SGD patient. The sum of these gene expression changes undoubtedly contributes to the disease phenotype. Although limited material precluded us from performing exhaustive analysis of the gene expression profile of this patient, microarray analysis provided several candidate genes for further study.
Microarray analysis revealed dysregulated expression of several transcription factors, among which were C/EBPϵ and PU.1. Both these factors were up-regulated at the protein and RNA levels in the SGD bone marrow. The increased expression of C/EBPϵ and PU.1, both critical for normal development of neutrophils (reviewed in Skalnik7 ), strongly suggested decreased activity of a negative regulator of both factors. The Gfi-1 gene encodes a nuclear protein with 6 carboxy terminal zinc-finger domains and a novel, highly conserved N-terminal domain of 20 amino acids termed the SNAG domain, which is thought to confer negative regulatory properties on Gfi-1 (reviewed in Moroy34 ; and Jafar-Nejad and Bellen39 ). Gfi-1–null mice, as described by Hock et al,23 have defects similar to those in C/EBPϵ knock-out mice and patients with SGD. Mice lacking Gfi-1 are severely neutropenic and eventually succumb to bacterial infections. Much like our SGD patient in whom Gfi-1 levels are significantly reduced, Gfi-1–null mice lack secondary and tertiary granule protein gene expression despite C/EBPϵ expression.23
In an attempt to design an in vitro model for Gfi-1lo SGD, we used bone marrow from Gfi-1 wild-type and heterozygous mice to generate factor-dependent EML cell lines that can terminally differentiate into mature neutrophils. The Gfi-1 heterozygous mice were previously reported to have normal life spans and have therefore not been described in detail.23 Our ATRA-induced EML+/− cells express reduced levels of Gfi-1, increased levels of PU.1 and its downstream target M-CSF-R, increased levels of C/EBPϵ, and significantly reduced levels of the SGP genes LF and NC. In a recent study, Zhuang et al40 also demonstrated increased levels of C/EBPϵ in 32D myeloid cells programmed to express a mutant Gfi-1 gene. The expression pattern in the Gfi-1+/− EML cell line thus parallels the findings in our Gfi-1lo SGD patient. While both the SGD patient and the Gfi-1+/− EML cell line express decreased levels of Gfi-1, SGP expression is completely abrogated in the patient bone marrow. Given that complete loss of Gfi-1 expression in Gfi-1–null mice leads to loss of SGP expression,23 it is tempting to speculate that the defect in our SGD patient results in a hypomorphic Gfi-1 allele leading to lower than heterozygous levels of Gfi-1, which in turn contributes to loss of SGP expression. Why the levels of Gfi-1 are diminished in the SGD patient is a question currently under study in our laboratory.
Heterozygous dominant-negative mutations in the Gfi-1 gene have been described in 2 patients with severe congenital neutropenia (SCN),41 underscoring the role of Gfi-1 in the neutrophil maturation pathway. These observations led us to hypothesize that both C/EBPϵ and Gfi-1 are required for secondary and tertiary granule protein gene expression in the developing neutrophil. However, our SGD patient is not neutropenic, an observation that may be explained by the fact that Gfi-1 levels in this patient are significantly reduced, but not completely absent.
Gfi-1 is a negative regulator of gene expression in T lymphocytes and can affect target promoters differentially according to the cellular and promoter context (reviewed in Moroy34 ). Recent chromatin immunoprecipitation42 studies and analysis of the Gfi-1–null mice23 demonstrate that Gfi-1 negatively regulates PU.1 and C/EBPϵ expression in myeloid cells. Gfi-1 has not been frequently described as a transactivator. In one study, Gfi-1 and its paralog Gfi-1B activated transcription of a promoter containing 4 copies of their consensus-binding site in an erythroid cell line.43 We demonstrate for the first time that Gfi-1 acts to up-regulate neutrophil collagenase (MMP-8) promoter activity in myeloid cells. Additionally, we show that together with wt C/EBPϵ, Gfi-1 synergistically activates the HNC promoter during myeloid differentiation.
We hypothesize that decreased levels of Gfi-1 in the SGD bone marrow prevent the up-regulation of the SGP genes by 2 mechanisms. First, decreased levels of Gfi-1 are insufficient to provide synergistic cooperation with mutant C/EBPϵ to transactivate the SGP gene promoters. Second, in light of a recent report by Laslo et al,33 increased PU.1 levels result in the up-regulation of the zinc-finger–containing Egr-1 transcription factor (also up-regulated in our SGD patient, Table 1), which in turn binds to the Gfi-1 promoter and blocks its expression. Diminished Gfi-1 levels then contribute to decreased SGP expression in the SGD patient.
It is possible that the heterozygous C/EBPϵ gene mutation observed in our SGD patient may in fact be responsible for the observed decrease in Gfi-1 levels in this patient. This possibility is currently being tested in our laboratory.
In summary, we have demonstrated that in an SGD patient with a V218A heterozygous mutation in C/EBPϵ, an imbalance in the expression of myeloid-specific transcription factors leads to developmental and functional neutrophil abnormalities. Efforts are currently under way to delineate the mechanism underlying the disruption of the neutrophil development pathway in this C/EBPϵ-positive Gfi-1lo SGD patient.
Authorship
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Nancy Berliner, Section of Hematology, WWW 428, Yale University School of Medicine, 333 Cedar St, New Haven, CT 06510; e-mail: nancy.berliner@yale.edu.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
This project has been funded with federal funds from the National Heart, Lung, and Blood Institute, National Institutes of Health, under contract no. N01-8V-28186 and award P01-HL63357.
We thank Drs Stuart Orkin, Hanno Hock, and Melanie Hamblen for making the Gfi-1–null mice available to us. We also thank Dr Stephanie Halene for providing us with viral supernatants used to make the Gfi-1+/− and Gfi-1+/+ cell lines, Dr Peter Gaines for helpful advice on generating the EML cell lines, and Sharon Lin for critically reviewing the paper.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal