Abstract
Tumor necrosis factor (TNF)–related apoptosis-inducing ligand (TRAIL) induces apoptosis in many transformed cells; however, not all human tumors respond to TRAIL, potentially limiting its therapeutic utility. Although there is substantial evidence that cytotoxic drugs can augment sensitivity to TRAIL, it has become important to know what kinds of nontoxic drugs can be used together with TRAIL. We thus screened several natural compounds that can overcome resistance to TRAIL and found that a cycloanthranilylproline derivative, Fuligocandin B (FCB), an extract of myxomycete Fuligo candida, exhibited significant synergism with TRAIL. Treatment of the TRAIL-resistant cell line KOB with FCB and TRAIL resulted in apparent apoptosis, which was not induced by either agent alone. FCB increased the production of 15-deoxy-Δ12,14 prostaglandin J2 (15d-PGJ2), an endogenous PPARγ ligand, through activation of cyclooxygenase-2 (COX-2). This unique mechanism highlighted the fact that 15d-PGJ2 directly enhanced sensitivity to TRAIL by inhibiting multiple antiapoptotic factors. More importantly, similar effects were observed in other leukemia cell lines irrespective of their origin. The enhancement was observed regardless of PPARγ expression and was not blocked even by peroxisome proliferator-activated receptor-γ (PPARγ) siRNA. These results indicate that 15d-PGJ2 sensitizes TRAIL-resistant cells to TRAIL in a PPARγ-independent manner and that the use of 15d-PGJ2 or its inducers, such as FCB, is a new strategy for cancer therapy.
Introduction
Tumor necrosis factor (TNF)–related apoptosis-inducing ligand (TRAIL), a member of the TNF superfamily, selectively induces apoptosis in various cancer cells with little toxicity toward normal cells.1-3 This unique property of TRAIL has prompted attempts to develop drugs that are effective against malignant diseases. Recombinant TRAIL and agonistic antibodies that bind to the TRAIL death receptor (DR) has been used in phase I-II human clinical trials.4,5
TRAIL can bind to 2 DRs, DR4 (TRAIL-R1) and DR5 (TRAIL-R2), each containing a cytoplasmic death domain, and to 2 decoy receptors (DcR), DcR1 (TRAIL-R3) and DcR2 (TRAIL-R4), each lacking a functional death domain.6 DcRs compete with DRs for ligands and therefore suppress apoptotic signals. There are at least 2 apoptotic pathways: the extrinsic pathway and the intrinsic pathway.7 TRAIL triggers apoptosis by binding to DRs, which engage the extrinsic pathway. Once bound to DRs, TRAIL forms a death-inducing signaling complex, resulting in the proteolytic activation of caspase-8 and, consequently, the activation of effector caspases such as caspase-3. Proteolytic caspase-8 further activates the Bcl-2 family member Bid, which in turn translocates to mitochondria and activates the intrinsic pathway.8 In the intrinsic pathway, death signals lead to changes in mitochondrial outer membrane permeability, subsequently releasing cytochrome c, which forms an apoptosome with Apaf-1 and caspase-9 and finally activates effector caspases.9 Although the TRAIL-resistant mechanisms differ among cancer cells and are poorly understood, DcRs, FLICE/caspase-8-inhibitory protein (FLIP), antiapoptotic Bcl-2 family members, and the inhibitor of apoptosis (IAP) family are all important antiapoptotic effectors.10
Adult T-cell leukemia/lymphoma (ATLL) is a neoplasm of T lymphocyte origin etiologically associated with human T-lymphotropic virus type I (HTLV-I) and is known to be resistant to standard anticancer therapies.11 In a previous study, we showed that although most ATLL cells express TRAIL-DRs, they are resistant to TRAIL.12 Although previous studies have suggested that multiple myeloma cells might be susceptible to TRAIL-induced apoptosis, most hematologic malignancies, especially primary leukemic cells are resistant to TRAIL.13,14 A majority of solid tumor cells are also relatively resistant to TRAIL-induced apoptosis. Nevertheless, several studies have shown that resistance to TRAIL can be overcome by the combined use of chemotherapeutic drugs and irradiation.15
Using a TRAIL-resistant ATLL cell line, we have been searching for natural compounds that have strong synergism with TRAIL but minimal toxicity to normal cells, as a new tool for cancer therapy.16 Natural products have played a highly significant role over the years in the discovery of new drugs. This is particularly evident in the treatment of cancers and infectious diseases, where more than 60% and 75% of drugs, respectively, are of natural origin.17 In this study, we found a natural compound that exhibits antitumor activity with a brand new mechanism. The cycloanthranilylproline derivative Fuligocandin B (FCB), an extract of myxomycete Fuligo candida, enhanced the sensitivity to TRAIL in leukemia cells by increasing the expression of cyclooxygenase-2 (COX-2) and 15-deoxy-Δ12,14 prostaglandin J2 (15d-PGJ2). Although FCB increased the expression of peroxisome proliferation activated receptor γ (PPARγ), the cytotoxic activity was not negated by a PPARγ antagonist or PPARγ siRNA, indicating an unidentified PPARγ-independent antitumor mechanism.
Materials and methods
Cell preparations
The ATLL cell lines KOB, KK1, ST1, and SO4 were established in our laboratory from respective patients with ATLL.18 These cell lines are dependent on exogenously added IL-2 and were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS) and 0.5 U/mL IL-2 (kindly provided by Takeda Pharmaceutical Company, Osaka, Japan). We also used the human T-cell leukemia cell lines Jurkat and MOLT-4, acute myeloid leukemia cell line HL-60, myeloma cell line HS-Sultan, erythromyeloblastic cell line K562, Burkitt lymphoma cell line Ramos, and monocytic leukemia cell line THP-1. These cell lines were maintained in RPMI 1640 medium supplemented with 10% FBS. Primary leukemia cells from 6 patients with acute-type ATLL were also analyzed. The diagnosis of ATLL was established hematocytologically, and the integration of the monoclonal HTLV-I provirus into the genome was confirmed by Southern blot hybridization in all cases (data not shown). Peripheral blood mononuclear cells (PBMCs) from patients with ATLL and healthy donors were purified by density gradient centrifugation using Lymphoprep (Axis-Shield PoC AS, Oslo, Norway). Each patient sample contained more than 90% leukemia cells at the time of analysis. After approval by the Ethics Committee at Nagasaki University Hospital of Medicine and Dentistry, Nagasaki, Japan, all materials from patients and normal healthy donors were obtained with informed consent in accordance with the Declaration of Helsinki.
Chemicals, cell-proliferation assay, apoptosis assay, and statistical analysis
Chemicals used in this study were the COX-2 inhibitor NS398 (Alexis, San Diego, CA), PPARγ agonists 15d-PGJ2, troglitazone, ciglitazone, and MCC-555, and PPARγ antagonist GW-9662 (Cayman Chemical, Ann Arbor, MI). In the cell-proliferation assay and apoptosis assay, cells were incubated with various concentrations of natural compounds from our libraries, or chemicals, and/or recombinant human soluble TRAIL (BIOMOL Research Laboratories, Plymouth Meeting, PA). Cell proliferation (percentage of control cells) was determined with the 3-(4,5 dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl-2-(4-sulfophenyl)-2H-tetrazolium (MTS) assay using a Cell Titer 96 AQueous Cell Proliferation Assay kit (Promega, Madison, WI) in accordance with the manufacturer's protocols. Determination of a synergistic, additive, or antagonistic effect of the combined treatment with TRAIL and sFCB was achieved with an isobolographic analysis as described previously.19 To evaluate apoptotic changes, cells were stained simultaneously with fluorescein isothiocyanate (FITC)–conjugated Annexin-V and the nonvital dye propidium iodide (PI) (Bender Medsystems, Vienna, Austria) to enable the discrimination of intact cells (Annexin-V− PI−), early apoptotic cells (Annexin-V+ PI−) and late apoptotic or necrotic cells (Annexin-V+ PI+). Cells were harvested after the treatment, and 104 cells per sample were analyzed by examined by flow cytometry (FCM) and were analyzed using a FACSCalibur flow cytometer and Cellquest software (BD Biosciences Immunocytometry Systems, San Jose, CA). All experiments were performed in triplicate, and the results were expressed as the mean plus or minus SD.
Chemical synthesis of FCB
The isolation and structural elucidation of cycloanthranilylproline and its derivatives, extracts of Myxomycete Fuligo candida, were described previously.20 We named cycloanthranilylproline derivatives 2 and 3 Fuligocandin A and Fuligocandin B, respectively. FCB was synthesized by aldol condensation of Fuligocandin A with 1-(triisopropylsilyl) indole-3-carboxaldehyde in the presence of 3 Eq of lithium diisopropylamide, followed by deprotection of the triisopropylsilyl group by acid hydrolysis in good yield.21
Surface expression of TRAIL receptors
The cell-surface expression of TRAIL receptors was examined by FCM. In brief, cells were incubated for 30 minutes on ice with mouse IgG1 anti-TRAIL-R1, -R2, -R3, or -R4 monoclonal antibodies (Alexis). Mouse IgG1 (Dako Japan, Kyoto, Japan) was used as a negative control. Cells were washed 3 times in phosphate-buffered saline and then incubated with FITC-conjugated goat anti–mouse IgG1 (DAKO Japan) as the secondary antibody. After a wash, 104 cells were analyzed.
Flow cytometric analysis of caspase-9 activity and Bax conformational changes
Caspase-9 activity was determined using a fluorescein active caspase-9 staining kit (MBL, Nagoya, Japan) according to the manufacturer's instructions. In brief, cells were incubated with FITC-LEHD-FMK for 1 hour at 37° C and analyzed using FCM. Comparison of the fluorescence-treated sample with an untreated control allows determination of the fold-increase in caspase-9 activity. The Bax conformational changes were investigated by using permeabilized samples. A FIX & PERM cell permeabilization kit from Invitrogen (Carlsbad, CA) was used according to the manufacturer's directions. Cells were then incubated with an N-terminal polyclonal antibody against Bax (Upstate Biotechnology, Lake Placid, NY) and isotype-matched control antibody (DAKO Japan). Next, the cells were stained with Alexafluor 488-labeled anti–rabbit IgG (Invitrogen). After washing steps, conformational changes of Bax were examined using FCM.
Measurement of mitochondrial transmembrane potential
Variations in mitochondrial transmembrane potential (Δψm) during the induction of apoptosis were examined with 3,3′-dihexyloxacarbocyanine iodide (DiOC6(3)) (Lambda Fluoreszenztechnologie, Graz, Austria). Cells were harvested after treatment, and DiOC6(3) was added at a final concentration of 40 nM. After 20 minutes of incubation at 37°C, the cells were washed and analyzed using FCM, and the percentage of cells with low mitochondrial potential was then calculated. For each sample, 104 cells were investigated, and all experiments were performed in triplicate.
Western blot analysis and antibodies
Cells were harvested after treatment, washed, and homogenized at 4°C in lysis buffer (0.1% sodium dodecyl sulfate [SDS], 1% Igepal CA-630, and 0.5% sodium deoxycholate) and a protease inhibitor cocktail (Sigma, St Louis, MO). Cell lysates (20 μg-50 μg) were resolved by electrophoresis on a 12.5% or 15% polyacrylamide gel and transferred to a polyvinylidine difluoride membrane. After blocking of the membrane in 10% FBS and 0.1% Tween 20 in Tris-buffered saline for 1 hour at room temperature, the blots were hybridized overnight at 4°C with primary antibodies. After hybridization with secondary antibodies conjugated with horseradish peroxidase, the immunocomplexes were visualized using an ECL Western blotting detection system (GE Healthcare, Chalfont St. Giles, United Kingdom). The cytosolic fraction was prepared using a mitochondrial/cytosol kit in accordance with the manufacturer's instructions (BioVision, Mountain View, CA) and 5 μg of cell lysate was used. The analysis was performed using antibodies to caspase-8, -9, and -3, Bid, Bax, Bak, Bcl-xL, XIAP, survivin, cytochrome c, phospho-AKT ser473, AKT, phospho-extracellular signal-regulated kinase (ERK) 1/2, ERK 1/2, p53, phospho-p53-ser15, and p21 (Cell Signaling Technology, Danvers, MA), TRAIL-R1 (Abcam, Cambridge, MA), TRAIL-R2 and COX-2 (Cayman Chemical), COX-1 and PPARγ (Santa Cruz Biotechnology, Santa Cruz, CA), p53-up-regulated modulator of apoptosis (PUMA) and neutrophil NADPH oxidase factor (Noxa) (Calbiochem, La Jolla, CA), FLIP (Dave-2) (Alexis), Bcl-2 (MBL), cIAP-1 and cIAP-2 (R&D Systems, Minneapolis, MN), and the monoclonal anti β-actin antibody AC-15 (Sigma).
NFκB transcription factor assay
Nuclear extracts from cells were obtained using a nuclear/cytosol fractionation kit (BioVision) according to the manufacturer's protocol. Activities of NFκB p50 and p65 were investigated using an NFκB transcription factor assay kit (Chemicon, Temecula, CA) according to the manufacturer's directions. In short, nuclear extract (10 μg/sample) from untreated or treated cells was added to the capture probe, a double-stranded biotinylated oligonucleotide containing the consensus sequence for binding NFκB. After incubation, the sample was transferred to a streptavidin-coated 96-well plate. After washing, the bound NFκB transcription factor subunit was detected with a primary antibody. The plate was incubated with a secondary antibody, a chromogenic substrate was added to the cells, and then the absorbance of each sample was read using a microplate reader.
Measurement of prostaglandins
The amount of each prostaglandin (PG) released into the medium was measured using an enzyme immunoassay (EIA) kit. The kits used were a PGE2 Express EIA Kit, a PGD2-MOX Express EIA Kit (Cayman Chemical), and a 15d-PGJ2 Correlate-EIA immunoassay kit (Assay Designs, Ann Arbor, MI). Assays were performed according to the manufacturer's directions.
PPARγ transcription factor assay
Activities of PPARγ were investigated using a PPARγ transcription factor assay kit (Cayman Chemical) according to the manufacturer's directions. A specific double stranded DNA sequence containing the peroxisome proliferator response element (PPRE) was immobilized onto the bottom of a 96-well plate and nuclear extracts (10 μg/sample) from untreated or treated cells were added. PPARs in samples then bind to the PPRE. Next, a specific primary antibody directed against PPARγ was added. After washing, the plate was incubated with a secondary antibody, and the absorbance of each sample was measured using a microplate reader at 450 nm.
Transfection of small interfering RNA
Transfection was performed with a cell line nucleofector kit V and the Nucleofector system (Amaxa Biosystems, Cologne, Germany). Cell preparations were adjusted so that the counts of viable cells were the same in each cell line after 24 hours of transfection. The transfection programs for KOB and K562 cells were T-20 and T-16, respectively. The siRNA of PPARγ (silencer-validated siRNA 5821) and control siRNA (silencer negative control 1) were purchased from Ambion (Austin, TX). Each siRNA was transfected at a final concentration of 100 nM. At 24 hours after transfection, cells were used for experimentation. Each siRNA experiment was performed in triplicate.
Results
Screening of natural compounds
We explored a variety of natural compounds to find extracts that enhance sensitivity to TRAIL using the TRAIL-resistant ATLL cell line KOB. KOB cells express TRAIL-R1 and TRAIL-R2 but lack DcRs. Jurkat cells, known to be sensitive to TRAIL, express TRAIL-R2 but not DcRs (Figure 1A). Although Jurkat cells exhibited remarkably reduced proliferation at 20 ng/mL of TRAIL, KOB cells were resistant (IC50 was 20.5 mg/mL) and showed only a 20% reduction even at a high TRAIL concentration, 2000 ng/mL (Figure 1C). In the screening assay, KOB cells were sequentially treated with various natural compounds and 500 ng/mL of TRAIL for 48 hours, and cell proliferation was assessed by MTS assay. Among more than 200 natural compounds in our library, we found that an extract of Fuligo candida, a kind of myxomycete (true slime mold), enhanced sensitivity to TRAIL in KOB cells (data not shown). We identified this compound as a cycloanthranilylproline derivative (11-[4-(1H-indol-3-yl)-2-oxo-but-3-enylidene]-1,2,3,10,11,11a-hexahydro-benzo[e]pyrrolo[1,2-a][1,4]diazepin-5-one) and named it Fuligocandin B (Figure 1B). Next, we chemically synthesized a synthetic FCB (sFCB). KOB cells showed resistance to sFCB alone at 0.05 to 5 μg/mL (IC50 was 11.1 μg/mL); however, when the cells were treated with a combination of TRAIL and sFCB, cell proliferation was significantly decreased, and a synergistic effect was observed by isobolographic analysis (Figure 1C,D). As shown in Figure 1E, a combination of 500 ng/mL TRAIL and 2.5 μg/mL sFCB increased the proportion of Annexin-V-positive cells to 69%. In contrast, that of Annexin-V-positive cells on treatment with TRAIL or sFCB alone was 13.4% or 6.8%, respectively. We further investigated whether sFCB induces this synergism in other ATLL cell lines and primary ATLL cells. Using Annexin-V staining, we confirmed that all cell lines examined showed increased apoptotic changes on combined treatment (Figure 1F). Four primary ATLL samples also showed a similar enhancement in the MTS assay, but normal peripheral blood mononuclear cells (PBMCs) were not affected at all by these conditions (Figure 1G).
Screening of natural compounds using the TRAIL-resistant cell line KOB. (A) TRAIL receptor expression was analyzed by FCM. Shaded and unshaded peaks correspond to specific and control staining, respectively. RFI (the ratio of mean fluorescence intensity for specific staining to that for control staining) is indicated in each panel. (B) The structure of the cycloanthranilylproline-derivative, Fuligocandin B (FCB). (C) Inhibition of cell proliferation. KOB cells (3.5 × 105/mL) were cultured for 48 hours with TRAIL and/or sFCB and cell proliferation was evaluated by MTS assay. The sensitivity of Jurkat cells to TRAIL is also indicated. (D) Isobolographic analysis. The fractional inhibitory concentrations were determined by using the IC50 of either agent alone or in combination. Sums of less than 1, 1, and greater than 1 indicate synergy, additivity, and antagonism, respectively. Four experimental points were found to be significantly below the theoretical additive line (dotted line), indicating a synergistic effect. (E-G) Combined treatment of ATLL cell lines, primary ATLL cells, and normal PBMCs. Cells (3.5-5.0 × 105/mL) were incubated for 48 hours with 500 ng/mL of TRAIL, 2.5 μg/mL of sFCB, sFCB→TRAIL (TRAIL was added after incubation with sFCB for 24 hours), or without reagents. (E) Annexin-V/PI staining of KOB. Percentages of intact cells, and early and late apoptotic cells are indicated in the bottom panels. (F) Annexin-V-positive cells in various ATLL cell lines are indicated. (G) Results of MTS assays of primary ATLL cells and normal PBMCs. Values are expressed as mean plus or minus SD.
Screening of natural compounds using the TRAIL-resistant cell line KOB. (A) TRAIL receptor expression was analyzed by FCM. Shaded and unshaded peaks correspond to specific and control staining, respectively. RFI (the ratio of mean fluorescence intensity for specific staining to that for control staining) is indicated in each panel. (B) The structure of the cycloanthranilylproline-derivative, Fuligocandin B (FCB). (C) Inhibition of cell proliferation. KOB cells (3.5 × 105/mL) were cultured for 48 hours with TRAIL and/or sFCB and cell proliferation was evaluated by MTS assay. The sensitivity of Jurkat cells to TRAIL is also indicated. (D) Isobolographic analysis. The fractional inhibitory concentrations were determined by using the IC50 of either agent alone or in combination. Sums of less than 1, 1, and greater than 1 indicate synergy, additivity, and antagonism, respectively. Four experimental points were found to be significantly below the theoretical additive line (dotted line), indicating a synergistic effect. (E-G) Combined treatment of ATLL cell lines, primary ATLL cells, and normal PBMCs. Cells (3.5-5.0 × 105/mL) were incubated for 48 hours with 500 ng/mL of TRAIL, 2.5 μg/mL of sFCB, sFCB→TRAIL (TRAIL was added after incubation with sFCB for 24 hours), or without reagents. (E) Annexin-V/PI staining of KOB. Percentages of intact cells, and early and late apoptotic cells are indicated in the bottom panels. (F) Annexin-V-positive cells in various ATLL cell lines are indicated. (G) Results of MTS assays of primary ATLL cells and normal PBMCs. Values are expressed as mean plus or minus SD.
sFCB and TRAIL synergistically activated both extrinsic and intrinsic pathways
To evaluate the synergism, we examined a series of apoptotic signals from caspase-8 to caspase-3. The band densities of the full-length caspase-8 and -3 decreased after treatment and bands for cleaved caspase-8 and -3 appeared in both concurrently treated and sequentially treated KOB cells but were stronger in the latter (Figure 2A). In contrast, there was no change in cells treated with sFCB alone, although cells treated with TRAIL showed faint activation. The decrease in the amount of full-length Bid implies a truncation of Bid and the transference of apoptotic signals from the extrinsic pathway to mitochondria.8 As shown, these changes and the appearance of cleaved Bid were noticed in cells cotreated with sFCB and TRAIL (Figure 2A). Apoptotic events in mitochondria were evaluated by measuring Δψm using FCM. A marked reduction in Δψm was observed in the sequentially treated cells but not in the cells treated with sFCB or TRAIL alone (Figure 2B). Likewise, appearance of cleaved caspase-9 (Figure 2A), increase of caspase-9 activities (Figure 2C), release of cytochrome c into the cytosol (Figure 2A), and conformational changes of Bax (Figure 2D) were elicited after combined treatment. These results suggest that the combined treatment activated the intrinsic pathway through engagement of the extrinsic pathway.
Analysis of caspase activation after treatment with sFCB and TRAIL. 3.5 × 105 KOB cells/mL were incubated for 48 hours with 2.5 μg/mL of sFCB [F], 500 ng/mL of TRAIL [T], simultaneous addition [F + T], or sequential [F→T], or without reagents [C]. (A) Western blot analysis. Using 30 μg of the cell extract and 5 μg of the cytosolic fraction, caspases, and cytochrome c were detected, respectively. (B) Mitochondrial membrane potential (Δψm). After treatment, cells were evaluated using DiOC6(3) by FCM and loss of Δψm (%) is indicated in each panel. (C) Caspase-9 activity. After treatment, cells were evaluated using FITC-LEHD-fluoromethyl ketone by FCM. Shaded and unshaded peaks correspond to treated and untreated cells, respectively. RFI is indicated in each panel. (D) Detection of Bax activation. After treatment, cells were evaluated using a conformation-specific Bax antibody by FCM. Shaded and unshaded peaks correspond to specific and isotype-matched control staining, respectively. RFI is indicated in each panel.
Analysis of caspase activation after treatment with sFCB and TRAIL. 3.5 × 105 KOB cells/mL were incubated for 48 hours with 2.5 μg/mL of sFCB [F], 500 ng/mL of TRAIL [T], simultaneous addition [F + T], or sequential [F→T], or without reagents [C]. (A) Western blot analysis. Using 30 μg of the cell extract and 5 μg of the cytosolic fraction, caspases, and cytochrome c were detected, respectively. (B) Mitochondrial membrane potential (Δψm). After treatment, cells were evaluated using DiOC6(3) by FCM and loss of Δψm (%) is indicated in each panel. (C) Caspase-9 activity. After treatment, cells were evaluated using FITC-LEHD-fluoromethyl ketone by FCM. Shaded and unshaded peaks correspond to treated and untreated cells, respectively. RFI is indicated in each panel. (D) Detection of Bax activation. After treatment, cells were evaluated using a conformation-specific Bax antibody by FCM. Shaded and unshaded peaks correspond to specific and isotype-matched control staining, respectively. RFI is indicated in each panel.
Analysis of modulators of extrinsic and intrinsic pathways
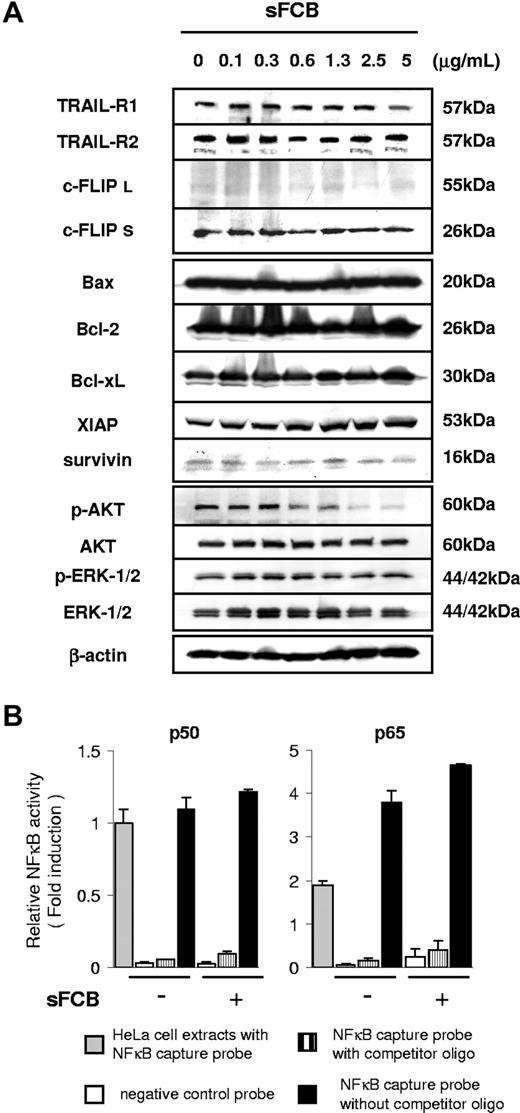
The up-regulation of DRs is known to be one of the most important events in the enhancement of TRAIL-induced apoptosis.7,22 However, we found no significant change in TRAIL receptor expression in sFCB-treated cells by either Western blot analysis (Figure 3A) or FCM (data not shown). FLIP is often expressed in long (c-FLIPL) and short (c-FLIPS) forms as a result of differential splicing and is considered a key regulator of DR signaling.23,24 Although the strong expression of c-FLIPS suggests its involvement in resistance to TRAIL, the expression was not influenced by sFCB. In our previous study, KOB cells had been demonstrated to be resistant to TRAIL because of the blocking of death signal at the mitochondrial level.12 However, the levels of Bax, Bak, Bcl-2, Bcl-xL, XIAP, cIAP-1, cIAP-2, and survivin were not influenced by the sFCB treatment (Figure 3A and data not shown).
Analysis of modulators of TRAIL sensitivity after treatment with sFCB. (A) 3.5 × 105 KOB cells/mL were incubated for 24 hours with sFCB. Cells were harvested and Western blot analysis was performed using 30 μg of the cell extract. (B) Activities of NFκB transcription factors. Cells were incubated for 24 hours with or without 2.5 μg/mL of sFCB, nuclear extracts were prepared, and activities of p50 and p65 were evaluated. HeLa cells were used as a positive control and the fold activation was obtained by setting the value for the positive control cells of p50 as 1.0. Values are expressed as mean plus or minus SD.
Analysis of modulators of TRAIL sensitivity after treatment with sFCB. (A) 3.5 × 105 KOB cells/mL were incubated for 24 hours with sFCB. Cells were harvested and Western blot analysis was performed using 30 μg of the cell extract. (B) Activities of NFκB transcription factors. Cells were incubated for 24 hours with or without 2.5 μg/mL of sFCB, nuclear extracts were prepared, and activities of p50 and p65 were evaluated. HeLa cells were used as a positive control and the fold activation was obtained by setting the value for the positive control cells of p50 as 1.0. Values are expressed as mean plus or minus SD.
AKT and ERK 1/2 might affect sensitivity to TRAIL.25,26 Phospho-AKT was observed in untreated cells and was diminished by sFCB treatment (Figure 3A). Phospho-ERK 1/2 was also observed but showed no change. The constitutive activation of NFκB is thought to be an important factor in antiapoptotic effects on cancer cells including ATLL.27,28 The levels of NFκB p50 and p65 subunit activities were higher in untreated KOB cells than in HeLa cells used as a positive control but showed no change or rather increased after sFCB treatment (Figure 3B). These results suggest that sFCB has a novel way of overriding resistance to TRAIL.
Increase of COX-2 expression by sFCB treatment
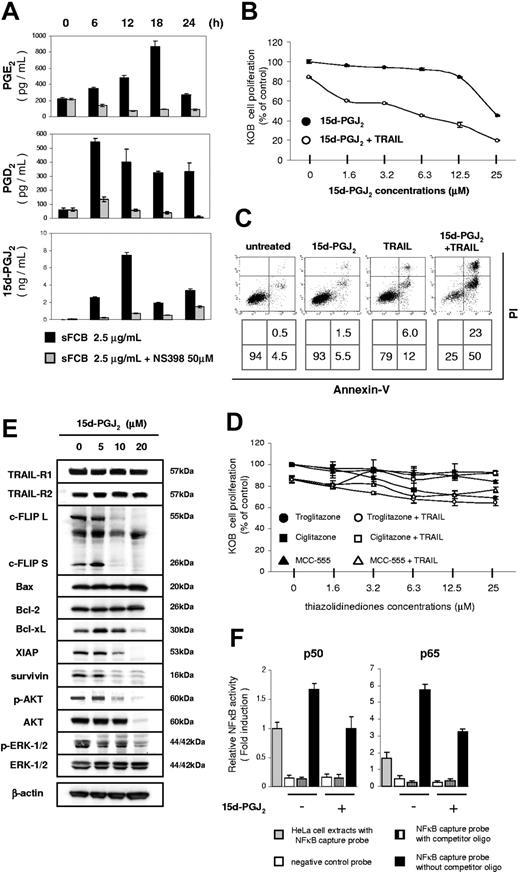
COX inhibitors have been demonstrated as TRAIL enhancers in cancer cells.29 The anthranilic compound mefenamic acid is well known to be a COX inhibitor. Because sFCB has an anthranilic structure, we expected sFCB to be a potential COX inhibitor. We thus performed Western blot analysis to determine whether COX expression is suppressed by sFCB treatment. The level of COX-2 expression in KOB cells was very low before treatment but was increased dramatically by 2.5 μg/mL of sFCB; it is noteworthy that this effect was not observed at a higher concentration, 10 μg/mL (Figure 4A). COX-2 expression was activated within 6 hours and continued for 24 hours. In contrast, COX-1 expression showed no change after sFCB treatment in either dose or time course experiments. These results suggest that low concentrations of sFCB selectively activate COX-2 expression and enhance sensitivity to TRAIL. We then used a selective COX-2 inhibitor, NS398. At 1.5 μM to 50 μM, NS398, either as a single agent or in combination with TRAIL, did not affect cell proliferation (Figure 4B). We confirmed that the expression of COX-2 caused by sFCB was repressed by NS398 treatment without any affect on COX-1 expression (Figure 4C). It is noteworthy that the synergistic effect of TRAIL and sFCB was clearly inhibited by NS398 in a dose-dependent manner (Figure 4D). These results indicate that COX-2 is a key factor influencing sensitivity to TRAIL.
Increase of COX-2 expression by sFCB treatment. (A,C,E) Western blot analysis. 3.5 × 105 KOB cells/mL were incubated for 24 hours with sFCB, NS398, or both at the concentrations and for the time course indicated. Using 30 μg of the cell extract, proteins were detected with the antibodies described in “Materials and methods, Western blot analysis and antibodies.” (B) Effects of TRAIL and NS398. Cells were incubated for 48 hours with NS398 and/or 500 ng/mL of TRAIL and cell proliferation was assessed by MTS assay. (D) Inhibition of synergism. Cells were treated as described in Figure 1E. Indicated concentrations of NS398 were added and cell proliferation was assessed by MTS assay. Values are expressed as mean plus or minus SD.
Increase of COX-2 expression by sFCB treatment. (A,C,E) Western blot analysis. 3.5 × 105 KOB cells/mL were incubated for 24 hours with sFCB, NS398, or both at the concentrations and for the time course indicated. Using 30 μg of the cell extract, proteins were detected with the antibodies described in “Materials and methods, Western blot analysis and antibodies.” (B) Effects of TRAIL and NS398. Cells were incubated for 48 hours with NS398 and/or 500 ng/mL of TRAIL and cell proliferation was assessed by MTS assay. (D) Inhibition of synergism. Cells were treated as described in Figure 1E. Indicated concentrations of NS398 were added and cell proliferation was assessed by MTS assay. Values are expressed as mean plus or minus SD.
Although COX-2 is thought to have antiapoptotic effects in cancer cells, there is some evidence that many of the signals that activate COX-2 also induce expression of the tumor suppressor p53.30 We therefore examined p53 and p53-dependent key molecules by Western blot analysis (Figure 4E). As shown, the expression of p53, phospho-p53 (ser15), p21, PUMA, and Noxa was not affected by sFCB. These results suggest that another pathway, which is triggered by COX-2 activation, is involved in the synergism.
15d-PGJ2, a final metabolite of the arachidonic-acid cascade, enhanced sensitivity to TRAIL
It has been recognized that metabolites of the arachidonic-acid cascade are synthesized by COXs.31 We first analyzed PGE2 expression, which is often investigated to assesses this cascade. When KOB cells were treated with sFCB, the PGE2 concentration increased 4-fold at 18 hours after incubation (Figure 5A top), which is consistent with the increase in COX-2 expression shown in Figure 4A. Among the metabolites and receptors in the arachidonic-acid cascade, we focused on PPARγ rather than other PG-receptors because of its proapoptotic properties.32 Although a specific 15d-PGJ2 synthase has not yet been identified, 15d-PGJ2 is a derivative of PGD2.33 After the treatment with sFCB, the PGD2 concentration had increased almost 10-fold at 6 hours, an activation faster and stronger than that of PGE2 (Figure 5A middle). The concentration of 15d-PGJ2 also increased, and was more than 10-fold higher at 12 hours (Figure 5A bottom). It is noteworthy that the activation of each PG caused by sFCB was mostly inhibited by 50 μM NS398 (Figure 5A).
Increase of 15d-PGJ2 expression by sFCB treatment. (A) Measurement of PGs. 3.5 × 105 KOB cells/mL were incubated for 24 hours with 2.5 μg/mL of sFCB and/or 50 μM NS398. Activation of PGE2, PGD2, and 15d-PGJ2 was detected by EIA. (B) 15d-PGJ2 enhances sensitivity to TRAIL. Cells were incubated for 48 hours with the indicated concentrations of 15d-PGJ2 and/or 500 ng/mL of TRAIL. Cell proliferation was assessed by MTS assay. (C) Evaluation of apoptosis. KOB cells were incubated for 48 hours with 10 μM 15d-PGJ2 and/or 500 ng/mL of TRAIL and analyzed as described in Figure 1E. (D) Combination use of synthetic PPARγ agonists and TRAIL. 3.5 × 105 KOB cells/mL were incubated for 48 hours with the indicated concentrations of TZDs and/or 500 ng/mL of TRAIL and cell proliferation was assessed by MTS assay. (E) Western blot analysis. 3.5 × 105 KOB cells/mL were incubated for 24 hours with 15d-PGJ2. 30-50 μg of the cell extract was used and were detected with the antibodies described in “Materials and methods, Western blot analysis and antibodies.” (F) Activities of NFκB. Cells were incubated for 24 hours with or without 10 μM 15d-PGJ2. The activities of p50 and p65 were evaluated as described in Figure 3B. Values are expressed as mean plus or minus SD.
Increase of 15d-PGJ2 expression by sFCB treatment. (A) Measurement of PGs. 3.5 × 105 KOB cells/mL were incubated for 24 hours with 2.5 μg/mL of sFCB and/or 50 μM NS398. Activation of PGE2, PGD2, and 15d-PGJ2 was detected by EIA. (B) 15d-PGJ2 enhances sensitivity to TRAIL. Cells were incubated for 48 hours with the indicated concentrations of 15d-PGJ2 and/or 500 ng/mL of TRAIL. Cell proliferation was assessed by MTS assay. (C) Evaluation of apoptosis. KOB cells were incubated for 48 hours with 10 μM 15d-PGJ2 and/or 500 ng/mL of TRAIL and analyzed as described in Figure 1E. (D) Combination use of synthetic PPARγ agonists and TRAIL. 3.5 × 105 KOB cells/mL were incubated for 48 hours with the indicated concentrations of TZDs and/or 500 ng/mL of TRAIL and cell proliferation was assessed by MTS assay. (E) Western blot analysis. 3.5 × 105 KOB cells/mL were incubated for 24 hours with 15d-PGJ2. 30-50 μg of the cell extract was used and were detected with the antibodies described in “Materials and methods, Western blot analysis and antibodies.” (F) Activities of NFκB. Cells were incubated for 24 hours with or without 10 μM 15d-PGJ2. The activities of p50 and p65 were evaluated as described in Figure 3B. Values are expressed as mean plus or minus SD.
We next investigated whether 15d-PGJ2 contributes to the synergism with TRAIL in KOB cells. 15d-PGJ2 enhanced sensitivity to TRAIL in a dose-dependent manner, although 25 μM 15d-PGJ2 inhibited cell proliferation alone (Figure 5B). Apoptotic cells were identified using Annexin-V and PI staining as shown in Figure 5C. The treatment of KOB cells with 500 ng/mL of TRAIL and 10 μM 15d-PGJ2 resulted in apparent apoptosis, which was not observed with either agent alone. We investigated whether other PPARγ agonists have a similar effect.34,35 Synthetic PPARγ agonists such as thiazolidinediones (TZD), however, did not affect cell proliferation either alone or in combination with TRAIL (Figure 5D). We further examined what kind of modulators play a key role in the enhancement. We were surprised to find that most of the antiapoptotic factors (c-FLIPL, c-FLIPS, Bcl-xL, XIAP, survivin, phospho-AKT, phospho-ERK1/,2 and AKT) were all decreased by 10 μM 15d-PGJ2 (Figure 5E). Likewise, activities of the NFκB p50 and p65 subunits were also attenuated (Figure 5F). Together, these results indicate that the activation of COX-2 triggered by sFCB caused the increase in 15d-PGJ2, which enhanced susceptibility to TRAIL in KOB cells. Among typical PPARγ agonists, only 15d-PGJ2 showed enhancement with TRAIL and suppressed multiple antiapoptotic molecules.
15d-PGJ2 enhanced sensitivity to TRAIL in a PPARγ-independent manner
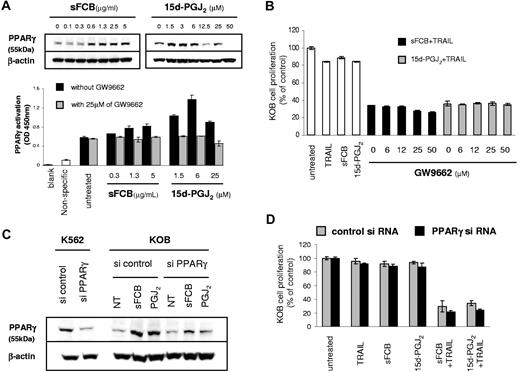
The activation of PPARγ by synthetic PPARγ agonists causes growth inhibition in a variety of cancer cells.32 Western blot analyses were performed to investigate whether sFCB participates in the activation of PPARγ (Figure 6A top). KOB cells showed faint PPARγ expression before treatment, which was up-regulated by sFCB; similarly, 15d-PGJ2 also up-regulated PPARγ expression. We further investigated the up-regulation using a PPARγ-specific transcription factor assay. As shown in Figure 6A bottom, PPARγ was activated by both sFCB and 15d-PGJ2, especially the latter, and suppressed by the specific PPARγ antagonist GW9662. These results strongly suggest that the activation of PPARγ contributes to the enhancement of sensitivity to TRAIL. To make clear this point, we performed an inhibition assay using GW9662. Contrary to expectation, the enhancement was not inhibited (Figure 6B). To further examine whether the enhancement requires PPARγ activation, we performed a knockdown experiment using PPARγ siRNA. As shown in Figure 6C, the expression of PPARγ protein in K562 cells that have high levels of PPARγ expression was clearly suppressed by PPARγ siRNA. Likewise, the up-regulated PPARγ expression by sFCB or 15d-PGJ2 in KOB cells was suppressed by PPARγ siRNA. However, PPARγ siRNA did not rescue cells from apoptosis, indicating that the enhancement by sFCB or 15d-PGJ2 occurred regardless of PPARγ expression (Figure 6D). These results confirmed that although sFCB or 15d-PGJ2 activate PPARγ in both transcriptional and protein levels, they contribute to the enhancement of sensitivity to TRAIL in a PPARγ-independent manner (Figure 7D scheme).
15d-PGJ2 enhances sensitivity to TRAIL in a PPAR γ-independent manner. (A) Western blot analysis and PPARγ transcription factor assay. KOB cells were treated for 24 hours with indicated concentrations of sFCB or 15d-PGJ2. 20 μg of the cell extract was used for the Western blot analysis and 10 μg of the nuclear extract was used for the transcription factor assay. (B) 3.5 × 105 KOB cells/mL were incubated for 48 hours with 2.5 μg/mL of sFCB, 10 μM 15d-PGJ2, 500 ng/mL of TRAIL, or a combination thereof. The indicated concentrations of GW9662 were added in the sFCB + TRAIL or 15d-PGJ2 + TRAIL setting, and cell proliferation was assessed by MTS assay. (C) Effect of siRNA on PPARγ expression. At 24 hours after transfection, cells were incubated with or without 2.5 μg/mL of sFCB and 10 μM 15d-PGJ2 for 24 hours, and Western blot analysis was performed using 20 μg of the cell extract. (D) Effect of siRNA on cell proliferation. At 24 hours after transfection, cells were treated for 48 hours with 2.5 μM of sFCB, 10 μmol/L 15d-PGJ2, 500 ng/mL of TRAIL, or a combination thereof. Cell proliferation was assessed by MTS assay. Values are expressed as mean plus or minus SD.
15d-PGJ2 enhances sensitivity to TRAIL in a PPAR γ-independent manner. (A) Western blot analysis and PPARγ transcription factor assay. KOB cells were treated for 24 hours with indicated concentrations of sFCB or 15d-PGJ2. 20 μg of the cell extract was used for the Western blot analysis and 10 μg of the nuclear extract was used for the transcription factor assay. (B) 3.5 × 105 KOB cells/mL were incubated for 48 hours with 2.5 μg/mL of sFCB, 10 μM 15d-PGJ2, 500 ng/mL of TRAIL, or a combination thereof. The indicated concentrations of GW9662 were added in the sFCB + TRAIL or 15d-PGJ2 + TRAIL setting, and cell proliferation was assessed by MTS assay. (C) Effect of siRNA on PPARγ expression. At 24 hours after transfection, cells were incubated with or without 2.5 μg/mL of sFCB and 10 μM 15d-PGJ2 for 24 hours, and Western blot analysis was performed using 20 μg of the cell extract. (D) Effect of siRNA on cell proliferation. At 24 hours after transfection, cells were treated for 48 hours with 2.5 μM of sFCB, 10 μmol/L 15d-PGJ2, 500 ng/mL of TRAIL, or a combination thereof. Cell proliferation was assessed by MTS assay. Values are expressed as mean plus or minus SD.
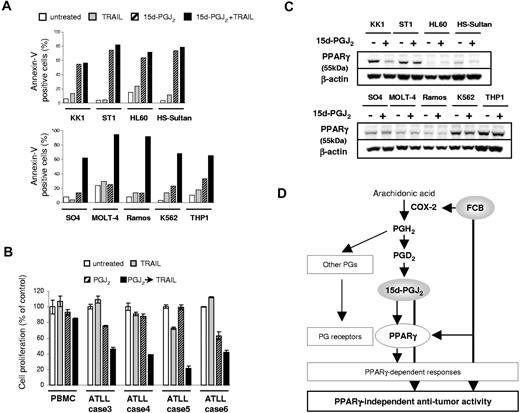
15d-PGJ2 enhance TRAIL sensitivity in various leukemia cell lines. (A) Annexin-V/PI staining; 3.5–5.0 × 105 cells/mL were incubated for 48 hours with 15d-PGJ2, TRAIL, or both. Percentages of Annexin-V-positive cells are indicated. Concentrations of each agent are as follows. KK1, ST1, and SO4: 10 μM 15d-PGJ2 and 500 ng/mL TRAIL. HL60, HS-sultan, Ramos, and K562: 10 μM 15d-PGJ2 and 125 ng/mL TRAIL. THP-1: 10 μM 15d-PGJ2 and 10 ng/mL TRAIL. MOLT-4: 2.5 μM 15d-PGJ2 and 125 ng/mL TRAIL. (B) 5.0 × 105 cells/mL cells were incubated for 48 hours with 500 ng/mL TRAIL, 10 μM 15d-PGJ2, both, or neither. Cell proliferation was assessed by MTS assay. (C) Effect of 15d-PGJ2 treatment on PPARγ expression. Cells were treated for 24 hours using the above described concentrations of 15d-PGJ2, and Western blot analysis was performed using 30 μg of the cell extract. (D) Proposed cascade of FCB-mediated sensitization to TRAIL-induced apoptosis. Values are expressed as mean plus or minus SD.
15d-PGJ2 enhance TRAIL sensitivity in various leukemia cell lines. (A) Annexin-V/PI staining; 3.5–5.0 × 105 cells/mL were incubated for 48 hours with 15d-PGJ2, TRAIL, or both. Percentages of Annexin-V-positive cells are indicated. Concentrations of each agent are as follows. KK1, ST1, and SO4: 10 μM 15d-PGJ2 and 500 ng/mL TRAIL. HL60, HS-sultan, Ramos, and K562: 10 μM 15d-PGJ2 and 125 ng/mL TRAIL. THP-1: 10 μM 15d-PGJ2 and 10 ng/mL TRAIL. MOLT-4: 2.5 μM 15d-PGJ2 and 125 ng/mL TRAIL. (B) 5.0 × 105 cells/mL cells were incubated for 48 hours with 500 ng/mL TRAIL, 10 μM 15d-PGJ2, both, or neither. Cell proliferation was assessed by MTS assay. (C) Effect of 15d-PGJ2 treatment on PPARγ expression. Cells were treated for 24 hours using the above described concentrations of 15d-PGJ2, and Western blot analysis was performed using 30 μg of the cell extract. (D) Proposed cascade of FCB-mediated sensitization to TRAIL-induced apoptosis. Values are expressed as mean plus or minus SD.
15d-PGJ2 enhanced sensitivity to TRAIL in a variety of leukemia cells
Finally, we investigated whether 15d-PGJ2 contributes to the enhancement of sensitivity to TRAIL in other leukemia cell lines, including ATLL and primary ATLL cells. Annexin-V and PI staining were performed, and we confirmed that KK1, ST1, HL-60, and HS-Sultan cells underwent apoptosis after 15d-PGJ2 treatment (Figure 7A top panel), whereas SO4, K562, Ramos, MOLT-4, and THP-1 cells underwent apoptosis in response to the combined use of 15d-PGJ2 and TRAIL (Figure 7A bottom panel). All primary ATLL samples also showed similar responses in the cell proliferation assay, but PBMCs were not affected by any treatment (Figure 7B). PPARγ protein expression showed various patterns depending on the cell lines (Figure 7C). 15d-PGJ2 actually decreased PPARγ expression in KK1 cells contrary to the case in KOB cells, and some cell lines showed faint or even no PPARγ protein expression irrespective of 15d-PGJ2 treatment. These results indicate that 15d-PGJ2 has potential as a new therapeutic agent for hematologic malignancies either alone or combination with TRAIL regardless of PPARγ protein expression (Figure 7D).
Discussion
The overexpression of COX-2 in cell and animal models is thought to be associated with tumor-cell proliferation, angiogenesis, invasiveness, and inhibition of apoptosis.36,37 The mechanisms of these tumor-promoting actions of COX-2 are not completely known but inhibition of COX-2 has been recognized as a promising strategy for cancer prevention or treatment.38,39 However, as shown in the present study, a new compound of natural origin, FCB, activated the arachidonic-acid cascade through COX-2 and enhanced sensitivity to TRAIL in TRAIL-resistant ATLL cells. There is some evidence that when overexpressed, COX-2 not only acts as an oncogene in tumor development but also functions as an inducer of apoptosis in some types of cancer cells.40-45 The explanation given in most of these reports is that COX-2 overexpression is induced by the activation of p53 in response to DNA damage. However, sFCB did not activate p53, suggesting that another pathway of the antitumor effect is caused by COX-2 activation.
COX-2 is the enzyme that catalyzes the rate-limiting step in the synthesis of PGs, converting arachidonic-acid into PGH2, which is then further metabolized to other PGs or eicosanoids in inflammatory cells, transformed cells, and malignant tumor cells.39 15d-PGJ2, the most recently discovered PG, is the end product of the dehydration of PGD2.33 We showed that sFCB increased production of PGE2, PGD2, and 15d-PGJ2 and these effects were clearly blocked by a selective COX-2 inhibitor. These results are consistent with a recent study reporting that 15d-PGJ2 is produced via the COX-2 pathway.46
15d-PGJ2 has 2 characteristic features: (1) it has the most potent antiproliferative properties of any PG,47,48 and (2) it is the most potent endogenous ligand for PPARγ.49 The use of synthetic PPARγ agonists, such as TZDs, has allowed, over the past several years, the recognition and extensive study of PPARγ-dependent antitumor properties.32 Although previous studies have paid attention to 15d-PGJ2 as a PPARγ agonist, the precise biologic activities of 15d-PGJ2, including its antitumor effect, have not been elucidated.50-52 It has been noted previously that 15d-PGJ2 inhibits NFκB activities, whose effects may help to overcome TRAIL resistance.53,54 In fact, inhibition of NFκB activities was observed on 15d-PGJ2 treatment in our study. A recent work showed that 15d-PGJ2 induced apoptosis in Burkitt lymphoma cell lines and myeloma cell lines and repressed NFκB activity.55 Consistent with our study, HS-Sultan cells were sensitive to 15d-PGJ2, whereas K562 cells were resistant. We found that combined use of 15d-PGJ2 and TRAIL caused significant apoptosis of 15d-PGJ2-resistant K562 cells. Thus, the concept of combined therapy using these compounds may extend to 15d-PGJ2-resistant cells as well as TRAIL-resistant cells. It is noteworthy that both sFCB and 15d-PGJ2 decreased the level of phospho-AKT. 15d-PGJ2 even repressed total AKT expression in KOB cells, indicating that 15d-PGJ2 has a potent inhibitory effect on the activation of AKT. It is also suggested that down-regulation of phospho-AKT is important for KOB cells to become sensitive to TRAIL.
PPARs are members of the steroid/retinoid superfamily, and activation of PPARγ decreases serum glucose level in diabetes, which led to the development of TZDs that are now in clinical use as antidiabetic drugs.34,56 Several antitumor properties of TZDs have also been reported, and TZDs are used in phase I-II human clinical trials as new anticancer agents.32 Moreover, in line with their proapoptotic function, TZDs reportedly enhance sensitivity to TRAIL in cancer cells.57-59 The mechanisms of enhancement addressed in these reports were of 2 types: apoptotic changes triggered by modifications of antiapoptotic factors such as FLIP and survivin, and a G1 phase cell-cycle arrest caused by cyclin D3 repression. In our study, 15d-PGJ2 caused apoptotic changes but not cell-cycle arrest (data not shown). In addition, many antiapoptotic factors as well as FLIP and survivin were repressed by 15d-PGJ2. Among the PPARγ agonists we examined, only 15d-PGJ2 enhanced sensitivity to TRAIL in KOB cells. Thus, the ability to enhance sensitivity to TRAIL may not be the same between TZDs and 15d-PGJ2. Several lines of evidence indicate that the antiproliferative effect of TZDs in cancer cell is independent of PPARγ activation.60-63 In this regard, Kim et al57 and Lu et al58 showed that TZDs augment TRAIL-induced apoptosis in a PPARγ-independent manner by using PPARγ dominant-negative mutant cells. In the present study, 15d-PGJ2 and sFCB clearly activated PPARγ expression. Nevertheless, the synergism with TRAIL was not negated by a specific PPARγ antagonist or by PPARγ siRNA, suggesting that these compounds contribute to sensitivity to TRAIL independent of PPARγ activation. It is noteworthy that there is persuasive evidence of PPARγ-independent functions of 15d-PGJ2, including inhibition of NFκB activity, inflammatory responses, and cytokine production.53,54,64,65 Together, our results indicate 2 independent functions of 15d-PGJ2: proapoptotic activities and as a natural ligand of PPARγ.
According to numerous studies, the combined use of conventional cytotoxic drugs or irradiation can augment TRAIL-induced apoptosis in tumor cells in vitro and in vivo.15 If possible, however, these agents should be nontoxic and should not undermine the tumor-specific proapoptotic effects of TRAIL. In addition to COX-2 inhibitors or TZDs, some existing natural compounds may be candidates for such agents; however, few studies have screened natural products for new agents that enhance sensitivity to TRAIL.66-69 Based on the proapoptotic mechanism that FCB harbors, we eventually proposed using 15d-PGJ2 as an antineoplastic PG (not as a PPARγ agonist). PGs are used clinically in the treatment of several diseases, including congenital heart disease and pulmonary arterial hypertension, and are generally effective and well tolerated.70-72 Thus, our results demonstrated 15d-PGJ2 to be an ideal TRAIL enhancer.
Consequently, we found a new natural compound, FCB, that up-regulates COX-2 expression through a p53-independent mechanism and sensitizes TRAIL-resistant cells to TRAIL. This unique mechanism highlighted a potential new effect of 15d-PGJ2. This is the first report to suggest a link between the up-regulation of COX-2 expression, the production of 15d-PGJ2, and enhancement of sensitivity to TRAIL via a PPARγ-independent mechanism. Furthermore, we showed that this strategy may suit hematologic malignancies of various origins.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This study was supported in part by a Grant-in-Aid for Scientific Research (18590510) from the Japan Society for the Promotion of Science.
Authorship
Contribution: H.H. performed research and wrote the manuscript; Y.Y. designed the research protocol; K.K. designed the research protocol and contributed to the investigation of natural products; M.H. screened and investigated the natural products; M.I. analyzed the structure of natural products; T.S. analyzed the structure and chemically synthesized the FCB; T.I. contributed to chemical synthesis of the FCB; K.S. and K. Tsuruda performed research and contributed to the analysis; M.M. and N.T. provided clinical samples; K. Tsukasaki and M.T. provided clinical samples; and S.K. contributed to the study design and summarized the research results.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Yasuaki Yamada, MD, PhD, Department of Laboratory Medicine, Nagasaki University Graduate School of Biomedical Sciences, 1-7-1 Sakamoto, Nagasaki City, 852-8501, Japan; e-mail: y-yamada@nagasaki-u.ac.jp.


![Figure 2. Analysis of caspase activation after treatment with sFCB and TRAIL. 3.5 × 105 KOB cells/mL were incubated for 48 hours with 2.5 μg/mL of sFCB [F], 500 ng/mL of TRAIL [T], simultaneous addition [F + T], or sequential [F→T], or without reagents [C]. (A) Western blot analysis. Using 30 μg of the cell extract and 5 μg of the cytosolic fraction, caspases, and cytochrome c were detected, respectively. (B) Mitochondrial membrane potential (Δψm). After treatment, cells were evaluated using DiOC6(3) by FCM and loss of Δψm (%) is indicated in each panel. (C) Caspase-9 activity. After treatment, cells were evaluated using FITC-LEHD-fluoromethyl ketone by FCM. Shaded and unshaded peaks correspond to treated and untreated cells, respectively. RFI is indicated in each panel. (D) Detection of Bax activation. After treatment, cells were evaluated using a conformation-specific Bax antibody by FCM. Shaded and unshaded peaks correspond to specific and isotype-matched control staining, respectively. RFI is indicated in each panel.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/110/5/10.1182_blood-2007-01-068981/6/m_zh80180707250002.jpeg?Expires=1766044392&Signature=HKukoApMSREP51CYnXgzZjZ1LDor-NM8BctfppKMT0PYgkkfHxov3OO1nNPE6sFpxBrh5LLJWGDEqk3WISgoNgBr1CGSf6TzgQoDTBclXCsk5Jih1VtD0vW9fH5fCObzQOAo-ArDl8edAwis7PBSxP-cklddJgs0P0HjBE7Sx1qMO80kGwrrscStWFE3H6UENDKyzX00tbJWQaxqn19LBu~CY9rrtYSM-a5Y5xptSkpp6A7f1~xK6hHKaiD0AcqoaLYpQUhhrvNKpCH67YdOzXqxZXdRGfgQbnPap6GVCSHFkVtyxEt-avyMNkmvIrfDHq-bL-NGoa7KrH6shDNghw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal