Erythropoietin (Epo) and its cognate receptor (EpoR) are required for maintaining adequate levels of circulating erythrocytes during embryogenesis and adulthood. Here, we report the functional characterization of the zebrafish epo and epor genes. The expression of epo and epor was evaluated by quantitative reverse transcriptase–polymerase chain reaction (RT-PCR) and whole-mount in situ hybridization, revealing marked parallels between zebrafish and mammalian gene expression patterns. Examination of the hypochromic mutant, weissherbst, and adult hypoxia-treated hearts indicate that zebrafish epo expression is induced by anemia and hypoxia. Overexpression of epo mRNA resulted in severe polycythemia, characterized by a striking increase in the number of cells expressing scl, c-myb, gata1, ikaros, epor, and βe1-globin, suggesting that both the erythroid progenitor and mature erythrocyte compartments respond to epo. Morpholino-mediated knockdown of the epor caused a slight decrease in primitive and complete block of definitive erythropoiesis. Abrogation of STAT5 blocked the erythropoietic expansion by epo mRNA, consistent with a requirement for STAT5 in epo signaling. Together, the characterization of zebrafish epo and epor demonstrates the conservation of an ancient program that ensures proper red blood cell numbers during normal homeostasis and under hypoxic conditions.
Introduction
The glycoprotein erythropoietin (Epo) is essential for definitive erythropoiesis during ontogeny and for maintaining appropriate numbers of circulating erythrocytes in the adult.1,2 Epo binds the erythropoietin receptor (EpoR) on erythroid progenitors and stimulates an intracellular signaling cascade initiated by autophosphorylation of the receptor-associated Janus kinase 2 (Jak2) and subsequent tyrosine phosphorylation of EpoR (reviewed in Richmond et al3 ). Proteins with Src homology 2 (SH2) domains, such as STAT5 (signal transducer and activator of transcription factor 5) and phosphoinositide 3-kinase (PI3K), associate with the EpoR and are activated by Jak2 phosphorylation.4 One primary action of Epo is to inhibit apoptosis,5 which is mediated by STAT5 induction of the antiapoptotic B-cell lymphoma-XL (bcl-XL) response pathway and activation of Akt by PI3K.6 The Epo-EpoR interaction also activates the Ras–mitogen-activated protein kinase (MAPK) pathways7 and nuclear factor-κB (NFκB)–dependent transcription.8
In mammals, Epo is produced by hepatocytes during development and by interstitial peritubular cells in the adult kidney.9,–11 In response to chronic hypoxia, extrarenal Epo is expressed by the adult liver and spleen. In contrast, EpoR is expressed by primitive and definitive erythroid progenitors, endothelial cells, neural cells, and at low levels in cardiomyocytes.12,,,–16 Mice lacking Epo or EpoR have fewer primitive erythrocytes in the yolk sac blood islands and die between embryonic day 13 and embryonic day 15 due to severe anemia.17,–19 Analysis of these mice revealed that neither Epo nor EpoR is required for erythroid lineage commitment, or for the proliferation and differentiation of burst-forming unit–erythroid (BFU-E) progenitors into colony-forming unit–erythroid (CFU-E) progenitors. Epo is essential for the survival, proliferation, and terminal differentiation of late CFU-E progenitors, resulting in the formation of mature circulating red blood cells.17
The maintenance of red cell volume is critical for the supply of oxygen to all tissues, and Epo expression is tightly regulated to ensure appropriate levels of circulating erythrocytes. Hypoxia, due to congenital or acquired anemia, causes Epo levels in the plasma to increase exponentially, thereby stimulating erythropoiesis and raising the oxygen carrying capacity of the blood (reviewed in Fandrey and Fisher2,20 ). While decreased Epo causes anemia, excess Epo results in secondary erythrocytosis (reviewed in McMullin and Percy21 ). Increased red blood cell number is associated with greater blood viscosity and thereby with increased hypertension, heart failure, myocardial infarction, seizures, and thromboses.22 Epo signaling is therefore essential for balancing the dynamics of erythropoiesis.
As in other vertebrates, zebrafish hematopoiesis occurs in several anatomic locations during ontogeny (reviewed in Davidson and Zon23 ). Primitive hematopoietic cells originate from bilateral stripes in the posterior lateral plate mesoderm and migrate medially to form the intermediate cell mass (ICM), a structure analogous to the mammalian yolk sac blood islands. Primitive erythroid cells enter circulation concomitant with the onset of heart contractions at 24 hours after fertilization (hpf). The transition to definitive hematopoiesis occurs around 36 hpf and is characterized by the formation of self-renewing hematopoietic stem cells (HSCs) from the ventral wall of the dorsal aorta, corresponding to the mammalian aorta-gonad-mesonephros (AGM) region. By 4 days after fertilization (4 dpf), hematopoietic cells are found in the kidney primordium, the mammalian bone marrow equivalent and the site of adult hematopoiesis.24
To date, Epo signaling has been primarily investigated in mammalian systems. Here, we report the characterization of cDNAs for zebrafish epo and epor. Our analysis reveals moderate sequence identity of zebrafish epo and epor to their mammalian orthologs, yet the zebrafish genes do share respective tissue-specific expression, with the exception of zebrafish epo expression in the adult heart. Furthermore, epo was up-regulated in response to hypoxia and anemia, epo overexpression induced polycythemia, morpholino (MO)–mediated knock-down of epor caused erythropoietic defects, and abrogation of STAT5 blocked epo-induced erythrocytosis. Taken together, our data establishes zebrafish epo and epor as essential components of a conserved signaling program that regulates vertebrate erythropoiesis.
Materials and methods
Fish maintenance
Genetic mapping
The epo and epor genes were genetically mapped onto the Goodfellow RH panel by the Children's Hospital Genome Initiative group using the following primer sets: epo: forward (5′-TAATGAAATGGTCGAGGACGC-3′), reverse (5′-ATGAGTGCAGGCTTGCCAGATCTT-3′); and epor: forward (5′-CCCGCTGAAGGTGTTATCTGA-3′), reverse (5′-GGAGCGGGAGCGTTTC-3′).
Comparison of predicted peptide sequences
The deduced amino acid sequence of zebrafish epo was aligned with the predicted translation of Takifugu rubripes (fugu) epo29 and to Mus musculus and Homo sapiens Epo protein sequences using the CLUSTALW alignment algorithm30 of the MacVector 8.0 software package (Accelrys Software, San Diego, CA). An identical comparison was performed between the predicted translation of zebrafish epor to Xenopus laevis, X tropicalis, T rubripes, Tetraodon nigroviridis, M musculus, and H sapiens EpoR proteins.
Adult hypoxia treatment
Adult zebrafish were incubated in a modular incubator chamber (Billups-Rothenberg, Del Mar, CA) equilibrated with 5% O2 balanced with N2 for 6.5 hours at 25°C. Hearts were removed following treatment and placed in Trizol (Invitrogen, Carlsbad, CA); untreated hearts were used as a control. After homogenization (Tekmar Tissumizer, Tekmar Cincinnati, OH), total RNA was extracted, cDNA was generated, and gene expression was assessed by quantitative real-time polymerase chain reaction (PCR) as described in “Quantitative PCR analysis.”
RNA isolation and cDNA preparation
Zebrafish embryos (3 pools of 50 embryos each) and dissected adult tissues (3 pools of 10 organs each) were placed in Trizol and homogenized (Tekmar Tissumizer). Total RNA was isolated and treated with DNase I (Roche Applied Science, Indianapolis, IN) to remove genomic DNA. cDNA was synthesized from DNA-free total RNA using the BD Sprint PowerScript Double PrePrimed 96 plate with oligo(dT)18 and random hexamer primers (BD Sciences, Palo Alto, CA). Total RNA was extracted and cDNA generated from weh embryos as described.31
Quantitative PCR analysis
Quantitative real-time PCR was performed on an iCycler iQ5 Real-Time PCR detection system (Bio-Rad, Hercules, CA) using SYBR Green Supermix (Bio-Rad), and fold change was determined using the Gene Expression Analysis for iCycler iQ Macro (Bio-Rad).32 PCR primers were designed using Beacon Designer software (PREMIER Biosoft International, Palo Alto, CA). epo: forward (5′-AGGAGGCAGGATATGGACTATTAC-3′), reverse (5′-ACAGTTGGAGGTGCTTGAGG-3′); epor: forward (5′- GTCAGATACGCTGTGGAGGA-3′), reverse (5′- TCACAGGACTGGTCCAAGAA-3′); and beta-actin: forward (5′-GCTGTTTTCCCCTCCATTGTT-3′), reverse (5′-TCCCATGCCAACCATCACT-3′). Reaction mixtures were denatured at 95°C for 3 minutes, followed by 40 cycles of 95°C for 15 seconds, 53°C for 20 seconds, and 60°C for 45 seconds. cDNA starting material was normalized to beta-actin expression and the mean threshold cycle (Ct) was used to determine relative expression.
Whole-mount in situ hybridization and o-dianisidine staining
Digoxigenin-labeled RNA probes were transcribed from linear cDNA constructs (Roche Applied Science, Indianapolis, IN). Whole-mount in situ hybridization was performed as described.33 Embryos that had developed melanocytes were bleached.33 Staining for hemoglobinized red cells was achieved by incubating embryos in a solution containing o-dianisidine (Sigma, St Louis, MO) as described.33
epo overexpression
epo cDNAs were subcloned into pCS2+ for mRNA synthesis using mMESSAGE mMACHINE High Yield Capped RNA Transcription Kit (Ambion, Austin, TX). A total of 1 nL transcribed epo mRNA was injected into 1- to 4-cell embryos at 20 ng μL−1.
MO-mediated knockdown of epor and STAT5
Antisense morpholino oligos (Gene Tools, Corvalis, OR) for epor (5′-AA-CTGGGCCACTGAACAATCAAATT-3′; 5′-TATGCGATTGACTTACATGTATGTA-3′) and STAT5 (5′-AATCCACACGGCCATGATCACTCT-3′; 5′-CTTGTGACTTACCAGAGTTGTCCC-3′) hybridize to the start site and predicted exon/intron junctions. Control 4-bp mismatch morpholinos (bold letters) were also created for the epor (5′-AACAGGGCCAGTGAAGAATCTAATT-3′; 5′-TATGCCATTGAGTTACATCTATCTA-3′) and STAT5 MOs (5′-AATGCACACCGCCATGTTCACACT-3′; 5′-CTTGAGACTAACCAGACTTGTGCC-3′). MOs were resuspended in Danieau solution and 1 nL was microinjected at the 1- to 4-cell stage at a concentration of 0.8 mM (epor) or 0.2 mM (STAT5) for each MO. A subset of embryos was coinjected with epor MO (0.8 mM) or STAT5 MO (0.2 mM) and epo mRNA (10 ng μL−1).
Microscopy
Visible light imaging was performed using a Leica MZ16 stereomicroscope (Leica, Northvale, NJ) with 10×/21 B eyepiece and rotary objective 6.3-8.0× using a Nikon E995 digital camera (Nikon, Melville, NY). Embryos were photographed in 100% glycerol and images were prepared using Adobe Photoshop CS version 8.0 (Adobe, San Jose, CA). gata1DsRed embryos were imaged with a Hamamatsu Orca-ER digital camera (Hamamatsu Photonics, Bridgewater, NJ) and Openlab software (Improvision, Lexington, MA).
Results
Isolation of epo and epor cDNAs
Efforts to isolate zebrafish homologs of mammalian cytokines and cytokine receptors from zebrafish sequence databases have had little success, most likely due to a high degree of evolution since the divergence of the mammalian and fish lineages 450 million years ago.34 A cDNA clone for zebrafish epo was isolated from a zebrafish cardiac library in a screen to identify transcripts up-regulated under hypoxic conditions (C. Yang and R. H., unpublished results, July 2001). Radiation hybrid mapping of zebrafish epo placed it on linkage group 7 (LG7). A cDNA for epor was recovered by synteny and homology searches between murine EpoR and zebrafish sequences,35 resulting in the identification of a conserved domain. epor-directed oligonucleotides were used to probe a kidney cDNA library, yielding a partial clone; the full-length cDNA was cloned from 24 hpf zebrafish embryos and RH mapped to LG3.
Structure of predicted zebrafish Epo and EpoR proteins
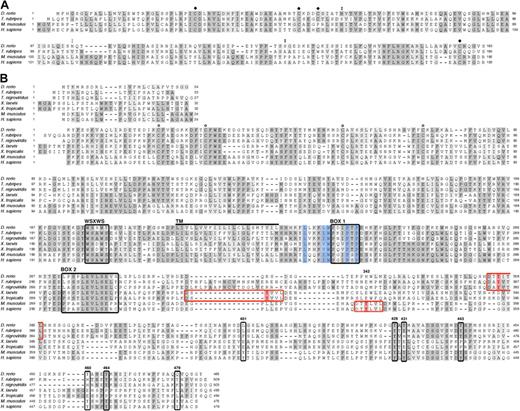
Alignment of zebrafish epo and epor protein sequences to other vertebrate orthologs revealed a moderate level of sequence conservation (Figure 1A). Zebrafish Epo shares 55% identity with epo cDNA from T rubripes,29 yet is 35% and 33% identical to mouse and human, respectively. While the sequences are largely dissimilar, residues known to be required for folding and glycosylation in mammals are conserved in zebrafish Epo. The tertiary structure of mammalian Epo contains an antiparallel bundle of 4 α-helices with 4 cysteine residues that form 2 disulfide bridges.36 The main bridge, between Cys7 and Cys161, links the amino and carboxyl terminal ends and is required for stability and function.37 The zebrafish Epo protein sequence was predicted to be a member of the 4-helical cytokine superfamily using the Superfamily HMM Library and Genome Assignments Server (data not shown).38 Structural modeling using the Swiss-Prot program (Geneva, Switzerland) shows that zebrafish Epo forms a bundle of 4 alpha-helices with conserved disulfide bridges (data not shown).39,40 Thus, despite dissimilarity between mammalian and zebrafish Epo sequences, the structural homology of the predicted tertiary structure suggests functional conservation.
Alignment of vertebrate Epo and EpoR Sequences highlights conserved functional residues. Amino acid sequence comparison of Epo (A) and EpoR (B) from various vertebrate species (see “Results” for percentage of identity and similarity). (A) ● marks cysteines known to be required for the formation of disulphide bridges in mammals; the cysteines near the N- and C- termini are essential for stability and function. Double black circles indicate conserved N- and O-linked glycosylation sites; double gray circles indicate a predicted N-linked glycosylation site. (B) Conserved cysteines in the extracellular domain, required for ligand binding, are indicated by ○; the WSXWS domain, box 1, and box 2 are demarcated by black boxes; and the transmembrane domain (240-262) is marked by a thick black line. The 5 residues that comprise the hydrophobic patch are shaded in blue. Murine phosphotyrosines (with the exception of Y343) are boxed in black and labeled according to the mouse EpoR sequence. Murine Y343 is shaded in red within a red box, believed to be a consensus site for STAT5 binding; putative STAT5 binding sites in fish and frog are likewise marked. Identical amino acids are shaded in dark gray; similar residues, in light gray. The signal peptide is depicted above the mature EpoR.
Alignment of vertebrate Epo and EpoR Sequences highlights conserved functional residues. Amino acid sequence comparison of Epo (A) and EpoR (B) from various vertebrate species (see “Results” for percentage of identity and similarity). (A) ● marks cysteines known to be required for the formation of disulphide bridges in mammals; the cysteines near the N- and C- termini are essential for stability and function. Double black circles indicate conserved N- and O-linked glycosylation sites; double gray circles indicate a predicted N-linked glycosylation site. (B) Conserved cysteines in the extracellular domain, required for ligand binding, are indicated by ○; the WSXWS domain, box 1, and box 2 are demarcated by black boxes; and the transmembrane domain (240-262) is marked by a thick black line. The 5 residues that comprise the hydrophobic patch are shaded in blue. Murine phosphotyrosines (with the exception of Y343) are boxed in black and labeled according to the mouse EpoR sequence. Murine Y343 is shaded in red within a red box, believed to be a consensus site for STAT5 binding; putative STAT5 binding sites in fish and frog are likewise marked. Identical amino acids are shaded in dark gray; similar residues, in light gray. The signal peptide is depicted above the mature EpoR.
The glycosylation of Epo is essential for in vivo function; nearly 40% of its molecular weight is attributed to carbohydrate moieties, which are required for synthesis and secretion.41 Sialylation of the carbohydrates both reduces the affinity of Epo to its receptor and extends its half-life.42,43 Human Epo contains 3 N-linked sugar chains (Asn24, Asn38, and Asn83) and 1 O-linked sugar chain (Ser126).37 The N-linked (N-X-S/T) and O-linked glycosylation sites are conserved in mammals except for rodents, which lack the O-glycosylation site.36 Zebrafish Epo is predicted to maintain 2 N-linked glycosylation sites, at Asn38 and proximal to Asn83, and 1 O-linked glycosylation site at Ser126 (Figure 1A). fugu Epo lacks all 3 N-linked glycosylation sites, yet contains the O-linked glycosylation site.29 Addition of glycan moieties to Epo in mammals and teleosts underscores the importance of posttranslational modifications for in vivo function, yet the localization and number of glycosylation sites appear to have diverged.
EpoR belongs to the type I superfamily of single-transmembrane cytokine receptors.21 This family shares conserved extracellular ligand–binding regions and a cytoplasmic box 1 motif, which selectively binds Jaks, conferring kinase activity to the receptor. Sequence identity between zebrafish EpoR and other vertebrates is relatively low: T rubripes, 44%; T nigroviridis, 41%; X laevis, 22%; X tropicalis, 22%; M musculus, 26%; and H sapiens, 27%. Alignment of fish, frog, and mammalian proteins, however, reveals key features of single-transmembrane cytokine receptors that have been maintained in EpoR throughout evolution (Figure 1B). These include 4 cysteines in the extracellular domain that are required for homodimerization and for transmitting a conformational change through the transmembrane domain upon ligand binding, a WSXWS domain, a hydrophobic patch, and a box 1 domain, both of which are essential for binding and activation of Jak2, as well as a box 2 domain. Also conserved are 5 of 8 tyrosines in the distal cytoplasmic domain (corresponding to murine P-Y429, P-Y431, P-Y443, P-Y460, and P-Y464) that are phosphorylated after Epo stimulation and act as docking sites for downstream signaling molecules (Figure 1B). Of the 3 nonconserved phosphotyrosines, 2 (P-Y343 and P-Y401) are recognized by STAT5 and 1 (P-Y479) is recognized by PI3K.3 Upon further sequence analysis, we observed 6 residues flanking P-Y343 (DT[b]YLVL), which appear to be similarly conserved in fish (DT[b]YITL) and frogs (DS[b]YVVL), raising the possibility that zebrafish STAT5 uses these binding sites. The maintenance of these functionally relevant motifs throughout evolution highlights their importance and is suggestive of functional conservation between zebrafish and mammalian Epo signaling.
Expression of epo and epor in the zebrafish
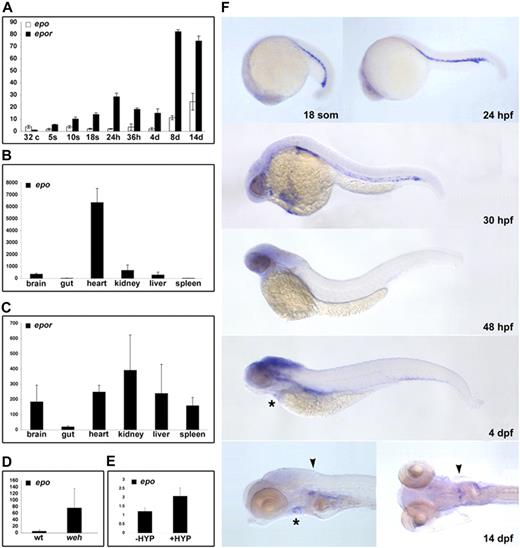
The expression of epo and epor was evaluated by quantitative reverse transcriptase (RT)–PCR using cDNA isolated from a progressive developmental series of zebrafish embryos and from adult tissues. Maternal transcripts for zebrafish epo are detected at low levels from prezygotic transcription (32-cell stage) to 24 hpf (Figure 2A). A subtle peak in expression occurs at 36 hpf, coinciding with AGM hematopoiesis, after which expression levels are constant until 8 to 14 dpf. In adults, epo transcripts are detectable at high levels in the heart, and at lower levels in the brain, liver, and kidney, but are not expressed in the spleen or gut (Figure 2B).
Conservation of zebrafish epo and epor expression. Detection of transcripts for epo and epor during ontogeny (A) and in select adult tissues (B,C) by quantitative RT-PCR. Expression of epo peaks slightly at 36 hpf, followed by an increase between 4 and 8 dpf. A total of 2 waves of epor expression are detected, 1 peak at 24 hpf, and another between 4 and 8 dpf. Expression of epo is induced in the hypochromic mutant weh, with respect to wild-type siblings (D) as well as in adult hypoxia-treated hearts, with respect to untreated controls (E). The average transcript abundance is represented by the bar graphs with standard deviations. (F) Spatiotemporal expression of epor was evaluated during embryonic development by whole-mount in situ hybridization. Transcripts were detected in the ICM from 18 somites until the onset of circulation at 24 hpf. Circulating erythroid cells maintain expression of epor until 32 hpf. At 4 dpf, epor is weakly expressed in the heart (*), and by 14 dpf, epor is localized to the heart (*) and is strongly expressed in the head kidney (▾). Lateral views, anterior to the left, a dorsal view of the 14-dpf embryo is also shown.
Conservation of zebrafish epo and epor expression. Detection of transcripts for epo and epor during ontogeny (A) and in select adult tissues (B,C) by quantitative RT-PCR. Expression of epo peaks slightly at 36 hpf, followed by an increase between 4 and 8 dpf. A total of 2 waves of epor expression are detected, 1 peak at 24 hpf, and another between 4 and 8 dpf. Expression of epo is induced in the hypochromic mutant weh, with respect to wild-type siblings (D) as well as in adult hypoxia-treated hearts, with respect to untreated controls (E). The average transcript abundance is represented by the bar graphs with standard deviations. (F) Spatiotemporal expression of epor was evaluated during embryonic development by whole-mount in situ hybridization. Transcripts were detected in the ICM from 18 somites until the onset of circulation at 24 hpf. Circulating erythroid cells maintain expression of epor until 32 hpf. At 4 dpf, epor is weakly expressed in the heart (*), and by 14 dpf, epor is localized to the heart (*) and is strongly expressed in the head kidney (▾). Lateral views, anterior to the left, a dorsal view of the 14-dpf embryo is also shown.
The first wave of epor expression begins at the 5-somite stage (12 hpf) and peaks at 24 hpf (Figure 2A). A second wave is apparent between 4 and 8 dpf, corresponding to the observed expansion of hematopoietic cells in the kidney primordium.24 In the adult, epor transcripts are highly expressed in the kidney, and are also found in brain, heart, liver, and spleen (Figure 2C). With the exception of epo transcripts in the adult heart, the expression profiles of zebrafish epo and epor are consistent with their mammalian counterparts.
The localization of epor expression was determined by whole-mount in situ hybridization. Transcripts for zebrafish epor are first detected in presumptive hematopoietic precursors in bilateral stripes of lateral plate mesoderm at 11 somites (data not shown). These cells express epor as they migrate to the midline, ultimately residing in the ICM at 18 somites to 24 hpf (Figure 2F). Circulating erythrocytes express epor until 32 hpf, and by 48 hpf epor is only observed in the brain. By 4 dpf, epor transcripts are restricted to the brain and heart. Transcripts for epor are localized to heart and kidney primordia at 14 dpf, consistent with the hematopoietic nature of the kidney in the zebrafish adult. epo expression was not detected by whole-mount in situ hybridization in embryonic stages, which is likely due to the low expression level of this potent cytokine.
Zebrafish epo is regulated by anemia and hypoxia
The induction of Epo expression in response to anemia and hypoxia is fundamental to Epo function in regulating erythrocyte number in mammals, and we sought to ascertain whether zebrafish epo is similarly controlled. Quantitative RT-PCR was used to compare transcript levels between 3-dpf hypochromic weh mutant embryos,27 which exhibit severe anemia by 48 hpf, and their wild-type siblings. This analysis revealed a 70-fold increase in epo transcripts in response to anemia (Figure 2D). In a similar analysis, adult zebrafish were incubated at 5% O2 for 6.5 hours, resulting in a 2-fold increase in epo expression in hypoxia-treated hearts compared with untreated controls (Figure 2E). This modest induction may be an underestimate of the hypoxic response as exposures of longer duration had a high mortality rate. These findings highlight the conservation of epo signaling in response to oxygen tension, which is integral to its function.
Exogenous zebrafish epo expands erythroid populations
In mammals, excess Epo production causes polycythemia. Microinjection of zebrafish epo mRNA into 1-cell–stage gata1DsRed embryos caused an increase in the number of circulating gata1+ cells at 4.5 dpf (Videos S1–S2, available on the Blood website; see the Supplemental Videos link at the top of the online article), providing strong evidence that the isolated epo clone was functionally analogous to mammalian Epo. This polycythemia continued, such that by 7 dpf, embryos exhibited increased circulation viscosity associated with cardiac regurgitation, inhibited blood flow, localized vascular stasis, and an enlarged posterior tail-vein lumen (Video S3Video 4. -epo7dpfclose.QT (MOV, 1.31 MB)Video 5. +epo7dpf.QT (MOV, 983 KB)–S6). The lack of blood flow caused widespread necrosis, and all embryos died between 9 and 12 dpf with no other morphologic abnormalities.
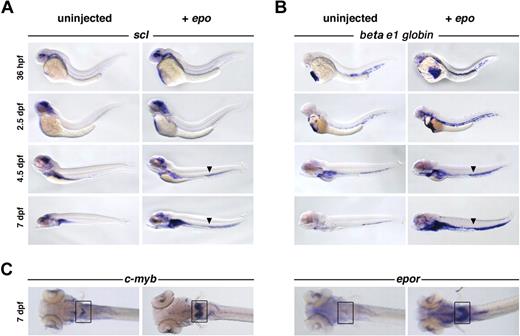
To characterize the circulating cells, hematopoietic gene expression was evaluated by whole-mount in situ hybridization. epo-injected embryos exhibited increased numbers of circulating scl+ and beta e1-globin+ cells at 36 hpf, compared with their uninjected siblings (Figure 3A-B). The expansion of scl, a transcription factor expressed in hematopoietic stem cells and erythroid precursors,23 and beta e1-globin, a gene found in mature erythrocytes,23 demonstrates an increase in early and late erythroid populations. This expansion was observed in circulation, as well as in the kidney primordium at 4.5 dpf, and continued until 7 dpf. While both populations increased, the number of beta e1-globin+ cells exceeded the number of scl+ cells at all stages, suggesting that erythroblasts proliferated in response to epo. Exogenous epo also increased the expression of primitive erythroid progenitor markers c-myb, epor (Figure 3C), ikaros (data not shown), and gata1. Expression of hematopoietic precursor markers gata2 and runx1, myeloid markers pu.1 and mpo, lymphoid markers rag1 and lck, and the vascular marker fli1 was unaffected (data not shown). The expansion, proliferation, and differentiation of erythroid progenitors into mature erythrocytes in response to exogenous epo indicates that zebrafish and mammalian Epo signaling are functionally analogous. There appears to be no cross-reactivity between mammalian Epo and zebrafish EpoR, as 24- and 48-hpf zebrafish embryos injected with human EPO (103 U/mL) did not display an erythroid expansion (data not shown)
Overexpression of epo expands erythroid populations. Capped epo mRNA was injected into 1-cell–stage zebrafish embryos and evaluated by whole-mount in situ hybridization. A clear increase in (A) scl+ and (B) beta e1-globin+ cell number in response to exogenous epo is evident in circulation at all stages examined, as well as in the kidney primordium beginning at 4.5 dpf. The arrowhead marks the enlarged vein lumen caused by excess cells in circulation. Lateral views, anterior to the left. (C) An expansion of c-myb+ and epor+ cells can be observed in the kidney primordium (boxed) at 7 dpf, dorsal views, anterior to the left.
Overexpression of epo expands erythroid populations. Capped epo mRNA was injected into 1-cell–stage zebrafish embryos and evaluated by whole-mount in situ hybridization. A clear increase in (A) scl+ and (B) beta e1-globin+ cell number in response to exogenous epo is evident in circulation at all stages examined, as well as in the kidney primordium beginning at 4.5 dpf. The arrowhead marks the enlarged vein lumen caused by excess cells in circulation. Lateral views, anterior to the left. (C) An expansion of c-myb+ and epor+ cells can be observed in the kidney primordium (boxed) at 7 dpf, dorsal views, anterior to the left.
epor morphants exhibit erythropoietic defects
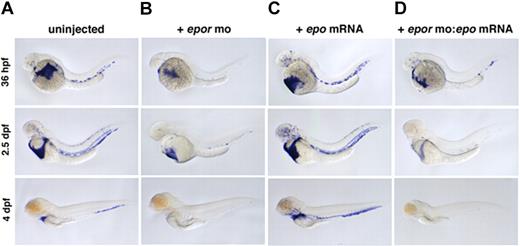
In mammals, loss of Epo or EpoR causes a slight decrease in primitive and a complete block in definitive erythropoiesis.17 To examine the function of zebrafish epor, 2 MOs were designed against its exon-intron boundaries, and resulting morphants were evaluated by whole-mount in situ hybridization for beta e1-globin expression. As in mammals, loss of zebrafish EpoR caused a modest decrease in primitive erythrocyte number at 36 hpf (Figure 4A,B). A more significant reduction was apparent at 2.5 dpf, and by 4 dpf, only a few cells remained (Figure 4C; Video S7, S8). The absence of erythrocytes at 4 dpf suggests a complete block to definitive erythropoiesis, consistent with the mouse EpoR knockout data. No other morphologic differences were detected between morphants and their uninjected siblings. MO-mediated knock-downs are transient; the morphants recovered normal levels of blood by 6 or 7 dpf. Embryos coinjected with 2 4-bp mismatch controls exhibited normal erythropoiesis (data not shown). To examine whether zebrafish epo and epor are a functional ligand-receptor pair, epor morphants were coinjected with epo mRNA. Injection of epo mRNA alone induces erythropoiesis, but coinjection of epor MO with epo mRNA blocks this erythroid expansion (Figure 4C,D). These data clearly demonstrate that epor is functioning downstream of epo.
Loss of EpoR decreases primitive and blocks definitive erythropoiesis. beta e1-globin expression in (A) uninjected, (B) epor MO–injected, (C) epo mRNA–injected, and (D) epor MO:epo mRNA–injected embryos at 36 hpf, 2.5 dpf, and 4 dpf. Embryos injected with epor MO show a slight decrease in erythropoiesis beginning at 36 hpf, and by 4 dpf there is a complete absence of circulating erythrocytes. Injection of epo mRNA alone causes a progressive increase in the number of beta e1-globin+ cells in circulation throughout ontogeny. To demonstrate that epo and the epor are a functional ligand-receptor pair, epor MO was coinjected with epo mRNA. Loss of EpoR effectively blocks the expansion of erythrocytes by epo overexpression as shown by a decrease in beta e1-globin+ cell numbers. Lateral views, anterior to the left.
Loss of EpoR decreases primitive and blocks definitive erythropoiesis. beta e1-globin expression in (A) uninjected, (B) epor MO–injected, (C) epo mRNA–injected, and (D) epor MO:epo mRNA–injected embryos at 36 hpf, 2.5 dpf, and 4 dpf. Embryos injected with epor MO show a slight decrease in erythropoiesis beginning at 36 hpf, and by 4 dpf there is a complete absence of circulating erythrocytes. Injection of epo mRNA alone causes a progressive increase in the number of beta e1-globin+ cells in circulation throughout ontogeny. To demonstrate that epo and the epor are a functional ligand-receptor pair, epor MO was coinjected with epo mRNA. Loss of EpoR effectively blocks the expansion of erythrocytes by epo overexpression as shown by a decrease in beta e1-globin+ cell numbers. Lateral views, anterior to the left.
epo signaling uses STAT5
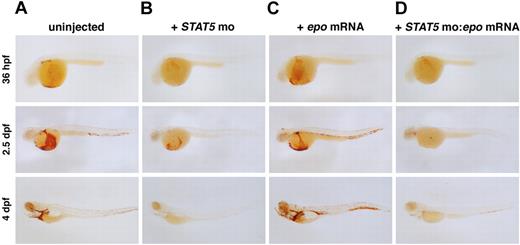
EpoR lacks intrinsic kinase activity and recruits Jak2 to mediate downstream signaling upon Epo binding. This leads to STAT5 phosphorylation, which translocates to the nucleus as a transcription factor. Zebrafish have 2 STAT5 genes, STAT5.1, which is most homologous to mammalian STAT5a and STAT5b, and STAT5.2, which has no apparent mammalian ortholog.44 To ascertain whether zebrafish EpoR uses STAT5 similarly to mammals, we tested the function of zebrafish STAT5.1, hereafter referred to as STAT5, using MOs targeted to the start site and a splice junction. Loss of STAT5 caused anemia, as assessed by o-dianisidine staining (Figure 5A,B) at 36 hpf. The anemia persisted until 4 dpf, when only a few erythrocytes remained in circulation. The STAT5 morphants appeared otherwise morphologically normal. Embryos coinjected with 4-bp mismatch controls were unaffected (data not shown). To evaluate whether STAT5 is required for Epo signaling, we overexpressed epo mRNA in the STAT5 morphants. The STAT5 MO blocked the epo-induced erythrocyte expansion, indicating that STAT5 is required for signaling (Figure 5C,D). This finding demonstrates that Epo signaling is conserved in zebrafish and opens new avenues for investigating signaling pathways resulting from EpoR activation.
Abrogation of STAT5 blocks epo-induced polycythemia.o-dianisidine expression in (A) uninjected, (B) STAT5 MO–injected, (C) epo mRNA–injected, and (D) STAT5 MO:epo mRNA–injected embryos at 36 hpf, 2.5 dpf, and 4 dpf. Embryos injected with STAT5 MO show a slight decrease in erythropoiesis beginning at 36 hpf, and by 4 dpf there are very few erythrocytes in circulation. Injection of epo mRNA alone causes a progressive increase in the number of o-dianisidine+ cells in circulation. To demonstrate that STAT5 is required for signaling through the EpoR, STAT5 MO was coinjected with epo mRNA. Loss of STAT5 effectively blocks the expansion of erythrocytes by epo overexpression as shown by a decrease in o-dianisidine+ cell numbers. Lateral views, anterior to the left.
Abrogation of STAT5 blocks epo-induced polycythemia.o-dianisidine expression in (A) uninjected, (B) STAT5 MO–injected, (C) epo mRNA–injected, and (D) STAT5 MO:epo mRNA–injected embryos at 36 hpf, 2.5 dpf, and 4 dpf. Embryos injected with STAT5 MO show a slight decrease in erythropoiesis beginning at 36 hpf, and by 4 dpf there are very few erythrocytes in circulation. Injection of epo mRNA alone causes a progressive increase in the number of o-dianisidine+ cells in circulation. To demonstrate that STAT5 is required for signaling through the EpoR, STAT5 MO was coinjected with epo mRNA. Loss of STAT5 effectively blocks the expansion of erythrocytes by epo overexpression as shown by a decrease in o-dianisidine+ cell numbers. Lateral views, anterior to the left.
Discussion
Here we describe the characterization of epo and epor genes in zebrafish, and our evaluation demonstrates that their hematopoietic function remains highly conserved with respect to mammals. Several lines of evidence, namely the conservation of functionally critical residues in Epo and EpoR, the regulation of epo by anemia and hypoxia, the resulting polycythemia upon epo overexpression, and the erythropoietic defect in epor morphants, indicate that these genes are the zebrafish homologs of their mammalian counterparts.
Structure and expression of zebrafish epo is conserved
Alignment of the predicted amino acid sequence of zebrafish and fugu Epo with mammalian Epo proteins revealed only a moderate degree of sequence identity. The preservation of residues essential for N-/O-linked glycosylation and formation of key disulfide bridges, as well as the prediction that zebrafish Epo forms a bundle of 4 α-helices, suggests that the structure and posttranslational modifications are largely conserved. Differences in glycosylation and sialylation between mammalian and zebrafish Epo proteins may be relevant to hormone function. Biochemically, deglycosylated Epo can bind EpoR, yet, in vivo, it is rapidly metabolised.45 The isolation and description of additional divergent Epo proteins may shed light on the importance of these posttranslational modifications, which is relevant toward developing therapeutic proteins with improved molecular potency and therefore advantages over their natural counterparts.
Transcripts for mammalian epo have been notoriously difficult to detect due to its low level of gene expression. In adult mammals, epo is predominately expressed in the kidney, at lower levels in the liver, and at barely measurable levels in the brain and spleen.36,46 Zebrafish epo expression was not detectable by in situ hybridization, yet quantitative RT-PCR revealed epo expression in the adult heart, brain, liver, and kidney. The abundant localization of epo transcripts in adult zebrafish hearts was unexpected given that there is no evidence of heart-specific expression in mammals, yet our finding is consistent with epo expression in adult fugu cardiac tissue.29
Differences in epo expression between fish and mammals may be explained in several ways. The fugu report postulates that the heart was the original source of epo prior to the split between the common mammalian and teleost ancestor. The heart-specific expression is believed to have been lost during mammalian evolution and instead localized to the kidney.29 The expression of epo in the adult zebrafish kidney is in contrast to fugu, in which kidney-expressed epo was not detected. Determining the localization of epo expression in additional nonmammalian vertebrates should resolve this contradiction.
Zebrafish epo is induced by anemia and hypoxia
The regulation of epo by oxygen tension is central to its function as a modulator of erythroid cell mass, guaranteeing adequate oxygen supply to all tissues. Our results demonstrate that, as in mammals, zebrafish epo expression is induced in hypochromic weh mutants and in adult hypoxia-treated hearts. In contrast, a recent study tested the promoter and 3′ flanking region of fugu epo using a luciferase reporter in a zebrafish hepatic cell line and discovered that fugu epo is not hypoxia inducible.29 The discrepancy in results may be attributable to the nature of the in vitro fugu experiments, as opposed to our in vivo studies. Specifically, it is unclear if the cell line used in the fugu study was competent to initiate a hypoxic response. The induction of zebrafish epo by hypoxia in vivo demonstrates the conservation of a critical response pathway for erythroid homeostasis.
The modest 2-fold increase in zebrafish epo expression in response to hypoxia was surprising given that, in mammals, EPO induction can be exponential.2,20 Our finding may underestimate the actual hypoxia induction because of the nature of our treatment, which stressed the fish considerably. Future studies will be designed to further dissect the zebrafish hypoxic response.
Conserved signal transduction in zebrafish EpoR
The comparison of zebrafish and mammalian EpoR sequences highlights significant residues that have been maintained throughout evolution. Alignment of multiple cytokine receptor family members reveals conserved hydrophobic residues proximal to the transmembrane domain.47 In the murine EpoR, this cytosolic juxtamembrane domain contains a precisely oriented hydrophobic motif that couples ligand-induced conformational changes in the receptor to intracellular activation of Jak2.47 A total of 3 of the hydrophobic residues in this hydrophobic patch, L253, I257, and W258, are required for Jak2 activity. The zebrafish EpoR juxtamembrane domain and the hydrophobic patch are conserved, suggesting that, as with mammals, this region is critical for associating with Jak2, enabling proper EpoR folding, presentation to the cell membrane, and intracellular activation of signaling upon binding of Epo.48 The conservation of this interaction is of particular interest given that patients with myeloproliferative disorders maintain a mutation in Jak2,49,,–52 which may confer an altered orientation with respect to the EpoR, and places the zebrafish in a unique position to investigate the etiology of myeloproliferative diseases.
The type I receptor superfamily contains cytoplasmic tyrosine residues that are phosphorylated by Jak2 and function as docking sites for signal transduction factors containing SH2 domains.53 For a few cytokine receptors, such as EpoR, the contributing roles of certain phosphotyrosine sites have been postulated.54 In the zebrafish EpoR, 5 of 8 tyrosines are maintained, some of which are required for cell proliferation and differentiation in mammals. For instance, the adapter Grb2 is believed to associate with EpoR at the consensus site P-Y464ENS and can couple to mSOS/Ras/Raf, leading to MAPK activation, an effector of mitogenesis.55,56 The interaction between tyrosine kinase Lyn with P-Y464 is important for differentiation in response to Epo signaling, but not for viability.57 Negative EpoR regulation is executed by the phosphatase SHP-1, which binds to P-Y429 and P-Y431, resulting in the dephosphorylation and inactivation of Jak2.58
Among the Jak2 substrates is STAT5, and the role of STAT5 activation in erythroid proliferation and differentiation has been well examined. Mice deficient in both isoforms of STAT5 exhibit decreased numbers of peripheral T cells, reduced bone marrow colonies, and extramedullary hematopoiesis.59 Subsequent studies demonstrated fetal anemia in vivo as well as defects in Epo-dependent production and survival of fetal liver hematopoietic colonies.60 Loss of zebrafish STAT5 blocked the expansion of erythrocytes by exogenous epo expression, suggesting a conserved role for STAT5 in zebrafish and implicating STAT5 in the proliferation of erythroid cells in response to epo.
The phosphotyrosine recruitment sites for murine STAT5 (P-Y343 and P-Y401) and PI3K (P-Y479) are notably absent in zebrafish EpoR. The 6 residues surrounding murine P-Y343 are similarly present in fish and frog sequences, suggesting that SH2 binding sites have shifted throughout evolution. Our finding that zebrafish STAT5 is required for EpoR signaling corroborates this hypothesis. Furthermore, a putative p85 binding site was predicted to occur in the fish at P-Y444 (YSPM), corresponding to murine P-Y443, using Scansite 2.061 (data not shown). The isolation of the zebrafish EpoR may reveal the functional significance of phosphotyrosine sites in steady-state and disease hematopoiesis. Future studies will address the function of these sites and the involvement of downstream mediators in zebrafish EpoR signaling.
Conserved function of zebrafish epo and epor
The exogenous expression of epo caused an increase in the number of circulating cells. Based on hematopoietic marker expression, we conclude that the expanded population includes proerythroblasts and fully differentiated cells. This expansion is functionally analogous to the induction of polycythemia in mammals in response to increased Epo levels.22,62
While injection of zebrafish epo induced a strong response, injection of human EPO failed to stimulate zebrafish erythropoiesis. The low degree of sequence identity between the human and zebrafish EpoR may prevent binding of human EPO. With the exception of the 4 conserved cysteines, the extracellular domain of EpoR is dissimilar between the 2 species. Furthermore, the importance of sugar residues in ligand-receptor interactions has been well established, and it is plausible that zebrafish and mammalian carbohydrate chains are divergent enough to preclude sufficient recognition. The binding specificity between Epo and EpoR may have evolved as a ligand-receptor pair within each species. Future studies will determine the degree of cross-reactivity of Epo and EpoR between other nonmammalian and mammalian species.
Gene ablation studies in the mouse have demonstrated that the Epo-EpoR signaling pathway is crucial for definitive hematopoiesis.17 Evaluation of zebrafish epor morphants demonstrates a similar erythropoietic phenotype, with a slight defect in primitive erythropoiesis and a complete block in definitive erythropoiesis. The mouse knockout also maintains defects in angiogenesis, cardiac development, and neural development, yet zebrafish epor morphants do not exhibit nonhematopoietic abnormalities. These findings are consistent with recent work in which epor was expressed under the gata1 promoter to rescue the hematopoietic and nonhematopoietic defects in epor-null mice, demonstrating that Epo signaling is not required in nonhematopoietic tissues during development.63
In summary, we present the characterization of epo and epor in the zebrafish, demonstrating both marked parallels to their mammalian homologs as well as interesting divergences that have occurred over the course of evolution. The introduction of these critical factors in the zebrafish system creates innovative avenues for studying the Epo signaling pathway. For instance, the zebrafish is amenable to genetic and chemical screens, facilitating the identification of novel factors and further elucidating important pathways. Epo is used for diverse applications in the clinic, stressing the need to clarify the function of Epo signaling in nonhematopoietic tissues and in diseased states. The zebrafish system provides another model to accelerate the analysis of Epo signaling.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this manuscript contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We acknowledge Chun Yang for his work on cloning and sequencing the zebrafish epo cDNA and for contributing unpublished results. We are grateful to Jenna Galloway, Trista North, Michael Dovey, and Gerhard Weber for critical reading of this manuscript. We thank Alan Davidson and Caroline Burns for helpful discussions and members of the zebrafish community for donated cDNAs.
This work was supported by a grant from the National Institutes of Health (R01 HL48801-13).
National Institutes of Health
Authorship
Contribution: N.P.L. designed and performed research, analyzed data, and wrote paper; N.H., P.G.F., B.P., I.L., B.B., and N.B. performed research; J.C., R.H., and L.I.Z. designed research.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Leonard I. Zon, Children's Hospital and Dana-Farber Cancer Institute, Howard Hughes Medical Institute, Stem Cell Program and Division of Hematology/ Oncology, Karp 7, 1 Blackfan Circle, Boston, MA 02115; e-mail:zon@enders.tch.harvard.edu.