Abstract
Gene copy number variation (CNV) and single nucleotide polymorphisms (SNPs) count as important sources for interindividual differences, including differential responsiveness to infection or predisposition to autoimmune disease as a result of unbalanced immunity. By developing an FCGR-specific multiplex ligation-dependent probe amplification assay, we were able to study a notoriously complex and highly homologous region in the human genome and demonstrate extensive variation in the FCGR2 and FCGR3 gene clusters, including previously unrecognized CNV. As indicated by the prevalence of an open reading frame of FCGR2C, Fcγ receptor (FcγR) type IIc is expressed in 18% of healthy individuals and is strongly associated with the hematological autoimmune disease idiopathic thrombocytopenic purpura (ITP) (present in 34.4% of ITP patients; OR 2.4 (1.3-4.5), P < .009). FcγRIIc acts as an activating IgG receptor that exerts antibody-mediated cellular cytotoxicity by immune cells. Therefore, we propose that the activating FCGR2C-ORF genotype predisposes to ITP by altering the balance of activating and inhibitory FcγR on immune cells.
Introduction
Human Fcγ receptors are glycoproteins that bind the Fc portion of IgG. Fcγ receptors are encoded on chromosome 1q23-24. Depending on their expression on effector cells, FcγRs exert different effects.1
The low-affinity FcγRs have been implicated in immune-complex–mediated auto-inflammation and are encoded by FCGR2A, FCGR2B, FCGR2C, FCGR3A, and FCGR3B. The current paradigm in FcγR biology states that cell activation is balanced by activating and inhibitory FcγRs. Loss of inhibitory FcγRIIb expression, altered function due to single nucleotide polymorphisms (SNPs), or overrepresentation of activating FcγRs results in unbalanced immunity and subsequent auto-inflammation.
Genetic polymorphisms affecting IgG-subclass binding exist in FCGR2A2 , FCGR3A,3,4 and FCGR3B.5 A functional polymorphism has been identified in FcγRIIa.2 FcγRIIa-H131 binds human IgG2, whereas FcγRIIa-R131 does not.6 In FcγRIIIa, a polymorphism at amino acid 158, a valine or phenylalanine, has been identified. As a result, FcγRIIIa-V158 has a higher affinity for IgG1 and IgG3 than FcγRIIIa-F158.3,4 Allelic variation in FcγRIIIb is composed of differences in 4 amino acids, referred to as human neutrophil antigen 1 (HNA1a [NA1]) and HNA1b [NA2]).7 FcγRIIIb-HNA1a internalizes IgG1- or IgG3-opsonized particles more efficiently than does FcγRIIIb-HNA1b.5
Also, the negative signaling through FcγRIIb is subject to genetic variation. An SNP in exon 5 of FCGR2B, which changes isoleucine to threonine at residue 232 (I232T) in the transmembrane domain, alters receptor signaling through the inhibitory FcγRIIb.8-10 In addition, functional polymorphisms in the promoter region of the FCGR2B gene were shown to be linked to transcriptional activity.11 Close to FCGR2B, the FCGR2C gene is located, which is believed to be a pseudogene. Very likely, FCGR2C results from an unequal crossover event between the 5′ part of the genomic sequence of FCGR2B and the 3′ part of FCGR2A.12
Recent papers13,14 have described the identification of large deletions and duplications of DNA fragments when genomes of healthy individuals were compared. The identification and characterization of copy number variation (CNV) show that in addition to single nucleotide differences, genomes of unrelated individuals have large regions of thousands to millions of nucleotides that are different. From a functional perspective, gene copy number differences can contribute to variation in gene expression, at the transcript and/or protein level. Gene-expression studies have shown that subtle differences in expression levels of genes can have significant consequences, as reported for DEFB415 and CCL3L1.16
CNV in the human FcγR gene cluster is not a new phenomenon. Deletion and duplication of the FCGR3B gene has been described by our group,17-19 in linkage with a deletion/duplication of the FCGR2C gene.18 Recently, Aitman et al observed an association between low copy number of FCGR3B and glomerulonephritis in the autoimmune disease systemic lupus erythematosus (SLE).20
The contribution of Fcγ receptor genes to the genetic susceptibility of autoimmune/inflammatory diseases is very complex. Genetic variation, SNPs and CNV, within the Fcγ receptor cluster could collectively be responsible for a “susceptibility phenotype,” altering the balance between activating and inhibitory receptor signaling.
To study the genetic variation of this complex gene cluster, we developed a multiplex ligation-dependent probe amplification (MLPA) assay that enabled us to study the genetic variation in the low-affinity FcγR genes (FCGR2A, FCGR2B, FCGR2C, FCGR3A, and FCGR3B) in one assay. We used this MLPA assay to study a cohort of idiopathic thrombocytopenic purpura (ITP) patients in a case-control study.
ITP is a disease characterized by thrombocytopenia with otherwise normal cell lineages and no other explanation for the isolated thrombocytopenia. Destruction of autoantibody-sensitized platelets by FcγR-bearing phagocytic cells in the reticuloendothelial system plays an important role, although the exact pathophysiology of this autoimmune disorder is not precisely known.21 The role of Fc receptors is underscored by the fact that intravenous immunoglobulin (IVIg) treatment and splenectomy (removal of the platelet-destructing organ) are effective treatment options.21
Despite its proven beneficial effect, the exact mechanism of action of IVIg in ITP is not known. Several models have been proposed,22 including blockade of the Fc receptor for IgG,23,24 increased surface expression of FcγRIIB,25 and the acute interaction of IVIg with activating FcγRs on dendritic cells.26
In children, the disease is often self-limiting within a 6-month period. In about 20% of cases, the thrombocytopenia sustains.27,28 In adult ITP, the disease is more often chronic, and an association with a prior infection has not been observed.
Previous studies concerning polymorphisms in the FCGR gene cluster in ITP patients show conflicting results. While confirming the previously observed overrepresentation of the SNP in FCGR3A encoding the FcγRIIIa-158V variant in pediatric ITP, we demonstrate that the copy number of an open reading frame (ORF) form of the FCGR2C gene results in the expression of a functionally activating FcγRIIc. The activating FCGR2C-ORF allele is determined by an SNP in exon 3 that changes the common stop codon in the pseudogene FCGR2C into a glutamine. In this study, we show that FCGR2C gene variation represents a significant genetic parameter associated with ITP.
Methods
Subjects
DNA of 116 patients with ITP was available for analysis. Informed consent was obtained from all parents of patients younger than12 and from all patients older than 12. Diagnosis and treatment were according to the guidelines of the American Society of Hematology and the United Kingdom practice for management of acute childhood ITP.29-31 Healthy adult white volunteers (n = 100) served as controls. These donors were not known to have hematological disorders of any kind in the past or at present. The study was approved by the Medical Ethics Committee of the Academic Medical Center in Amsterdam and was performed in accordance with the Declaration of Helsinki.
Multiplex ligation-dependent probe amplification
MLPA was developed as an alternative for the Southern blot (RLFP) method to detect deletions and duplications of genes (or part of genes) but can be adapted to detect SNPs as well in one multiplex assay with 50 different targets (J. Schouten, MRC-Holland, Amsterdam, The Netherlands).
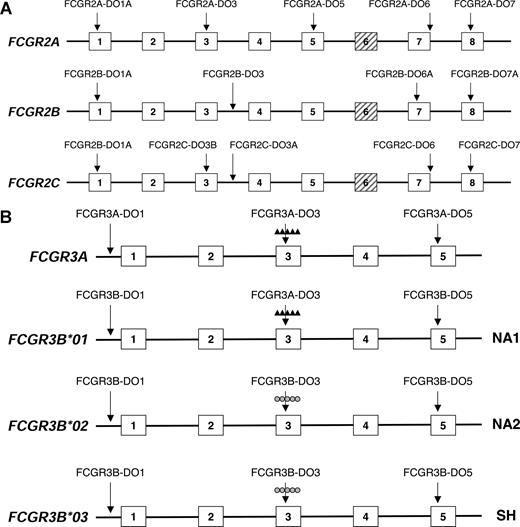
MLPA probes were designed specifically for the FCGR2A, FCGR2B, FCGR2C, FCGR3A, and FCGR3B genes. As the FCGR gene cluster is very homologous, this was not possible for all sites within a given gene. At least 3 probes per gene were designed to cover every single gene. In this way, CNV and partial insertions/deletions can be studied. Separate probes were designed to study known (functional) SNPs in FCGR2A (131H/R), FCGR2B (232I/T), promoter of FCGR2B and FCGR2C (−386 G/C), FCGR3A (158V/F), and FCGR3B (HNA1a/HNA1b/HNA1c). For an overview of the location and the specific target sequences of the probes, see Figure 1 and Table S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article). Several DNA samples of previously well-typed individuals were used for validation of the MLPA assay with respect to CNV and SNPs.17,18,27
Location of MLPA probes within the FCGR2 and FCGR3 gene clusters. (A) Location of MLPA probes within the FCGR2 gene cluster. Several probes within each separate gene, being either specific for FCGR2A, FCGR2B, or FCGR2C were used to determine CNV and SNPs. Shown are only the probes to determine CNV as indicated by the arrows. For a more detailed probe description, including the SNP probes, we refer to Table S1. (B) Location of MLPA probes within the FCGR3 gene cluster. Several probes within each separate gene were used to determine CNV and the SNPs. Note that the probe that recognizes FCGR3B-HNA1a is the same that recognizes FCGR3A. For a more detailed probe description, we refer to Table S1.
Location of MLPA probes within the FCGR2 and FCGR3 gene clusters. (A) Location of MLPA probes within the FCGR2 gene cluster. Several probes within each separate gene, being either specific for FCGR2A, FCGR2B, or FCGR2C were used to determine CNV and SNPs. Shown are only the probes to determine CNV as indicated by the arrows. For a more detailed probe description, including the SNP probes, we refer to Table S1. (B) Location of MLPA probes within the FCGR3 gene cluster. Several probes within each separate gene were used to determine CNV and the SNPs. Note that the probe that recognizes FCGR3B-HNA1a is the same that recognizes FCGR3A. For a more detailed probe description, we refer to Table S1.
Probes were constructed in collaboration with MRC-Holland and embedded in a standard MLPA kit. Because of the homology between certain probes, some probes had to be divided over 2 separate mixes to prevent competition. The MLPA assay was performed according to the first description by Schouten et al.32
In brief, 5 μL of DNA (20 ng/μL) was denatured at 98°C for 5 minutes and then cooled to 25°C in a thermal cycler with heated lid; 1.5 μL probe mix and 1.5 μL buffer were added to each sample and incubated for 1 minute at 95°C, followed by 16 hours at 60°C. Then, 32 μL of ligase-65 mix was added to each sample while at 54°C, followed by an incubation of 15 minutes at 54°C and 5 minutes at 98°C. This ligation mixture was diluted 4 times. Then, 10 μL polymerase mix was added, containing one single primer pair. Directly after adding the polymerase mix, the polymerase chain reaction (PCR) was started. PCR conditions were 36 cycles of 30 seconds at 95°C, 30 seconds at 60°C, and 60 seconds at 72°C, followed by 20 minutes at 72°C. After the PCR reaction, 1 μL of the PCR reaction was mixed with 0.5 μL CXR 60-400 (Promega, Madison, WI) internal size standards and 8.5 μL deionized formamide, and the mixture was incubated for 10 minutes at 90°C. The products were then separated by electrophoresis on an ABI-3130XL (Applied Biosystems, Foster City, CA).
The program Genemarker (version 1.30) was used to analyze the samples (Soft Genetics, State College, PA). Since MLPA is a relative quantification assay, one sample has to be assigned as reference sample. The normalized height (or the area) of the probe amplification product of the unknown sample was divided by the normalized height of the probe amplification product of the reference. A ratio between 0.8 and 1.2 was considered normal, below 0.8 was considered as loss in copy number, and above 1.2 as gain of copy number. For information concerning the control probes used in the standard MLPA, we refer to MRC-Holland (Amsterdam, The Netherlands).
FCGR2B/C promoter haplotypes
Due to the high homology between the promoter of the FCGR2B gene and that of the FCGR2C gene at position −386 G > C (rs3219018), it was not possible to design a specific MLPA probe exclusively for one of these genes. A gene-specific long-range PCR was performed on those samples carrying the uncommon FCGR2B/C variant −386C as detected by the MLPA assay. For this purpose, the expand long template PCR system (Roche Applied Science, Mannheim, Germany) was used according to the manufacturer's instructions. In brief, a 15-kb fragment was amplified with a nonspecific FCGR2B/C sense primer (5′-GCCATCCTGACATACCTCCT-3′), annealing in the promoter region and an FCGR2B-specific antisense primer (5′-CCCAACTTTGTCAGCCTCATC-3′) or the FCGR2C-specific antisense primer (5′-CTCAAATTGGGCAGCCTTCAC-3′), both annealing in exon 7. The PCR conditions were 94°C for 2 minutes, 10 cycles of 94°C for 10 seconds, 60°C for 30 seconds, 68°C for 12 seconds, followed by 20 cycles of 94°C for 15 seconds, 60 °C for 30 seconds, 68°C for 12 seconds, with an elongation of each cycle with 20 seconds at 68°C, followed by a final elongation at 72°C for 7 minutes. PCR products were purified by GFX PCR DNA and Gel Band Purification Kit (GE Healthcare, Amersham, United Kingdom) and sequenced with BigDye terminator cycle sequencing (v 1.1) (Applied Biosystems) with the sense primer used in the PCR reaction.
Real-time quantitative PCR
Intron-spanning primers specific for FcγRIIc were designed, forward primer 5′-ATCATTGTGGCTGTGGTCACTGG-3′ and reverse primer 5′-CTTTCTGATGGCAATCATTTGACG-3′, and β-glucuronidase was used as a reference gene. Amplification by PCR was performed on a LightCycler instrument (Roche, Almere, The Netherlands) and analyzed with software version 3.5. The reaction was performed with Lightcycler FastStart DNA MasterPLUS SYBR Green I (Roche Diagnostics, Indianapolis, IN), as has been previously described in detail.33
Monoclonal antibodies and reagents
The following monoclonal antibodies (mAbs) against human Fcγ receptors (FcγR) were used: CD16 (anti-FcγRIII, IgG1 isotype, clone 3G8; Sanquin, Amsterdam, The Netherlands); CD16-APC-Cy7 (anti-FcγRIII, IgG1 isotype, clone 3G8; BD Pharmingen, Alphen a/d Rijn, The Netherlands), CD32 (anti-FcγRII, IgG1 isotype, clone KB61; Sanbio, Uden, The Netherlands), and CD32-FITC (anti-FcγRII, IgG1 isotype, clone KB61; DakoCytomation, Glostrup, Denmark). For the detection of NK cells, CD56-APC (isotype IgG1, clone B159; BD Pharmingen) was used. Relevant isotype controls were IgG1-FITC (clone 203; Sanquin), IgG2b-FITC (clone GC198; Sanquin), IgG1-APC (clone MOPC-21; BD Pharmingen), and IgG1-APC-Cy7 (clone X40; BD Pharmingen). Expression of these markers was analyzed by tricolor flow cytometry on a FACS LSRII machine (BD Biosciences) using BD FACS Diva Software (version 5.0.1; BD Biosciences, San Jose, CA).
Cell lines
The FcγR-bearing P815 murine mastocytoma cells were a generous gift of Dr R. Mous (AMC, Amsterdam, The Netherlands) and were cultured in Iscove modified Dulbecco medium (IMDM; Biowhittaker Europe, Verviers, Belgium) supplemented with 10%, v/v, fetal calf serum (FCS; Bodinco, Alkmaar, The Netherlands), 100 U/mL penicillin (Gibco, Paisley, United Kingdom), and 100 μg/mL streptomycin (Gibco).
Isolation of neutrophils and PBMCs
Heparinized venous blood was collected from healthy donors and patients after obtaining informed consent and was separated over a Percoll gradient (GE Healthcare) into PBMCs as interphase and granulocytes in the pellet as described before.33 Purity and viability of both cell fractions were more than 95%, as determined by flow cytometry and trypan blue exclusion, respectively. PBL (peripheral blood lymphocytes) were obtained from PBMCs by allowing the monocytes to adhere for 1 hour on the plastic surface of a culture flask. In some experiments for mRNA isolation from selective cell types, further purification by MiniMACS (Miltenyi Biotec, Bergisch Gladbach, Germany) was performed with the relevant MoAbs for positive selection. In these cases, purity of cell fractions was more than 99%.
In vitro activation of neutrophils and PBMCs
Neutrophils and PBMCs were isolated as described above and cultured in 24-well plates at a density of 106 cells/mL. Wells contained medium alone (control cells), 50 U/mL rhGM-CSF (generous gift from Dr Lucien Aarden, Sanquin Research, Amsterdam, The Netherlands), 5 ng/mL TNFα (PeproTech EC, London, United Kingdom), or 20 ng/mL LPS (Sigma-Aldrich, St Louis, MO) supplemented with rhLBP (LPS-binding protein, Boehringer Ingelheim, Germany). Each condition was as applied in triplicate. After 4 hours, samples were taken for quantification of FcγRIIc mRNA.
In vitro activation of NK cells
PBLs were isolated as described above and cultured in 24-well plates at a density of 3.5 × 105 cells/mL. Wells contained either medium alone (control cells), 50 U/mL rhIL-2 (Strathmann Biotec, Hannover, Germany), or 10 ng/mL rhIL-15 (R&D Systems, Abingdon, United Kingdom). Each condition was as applied in duplicate. At days 0, 2, and 4, samples were taken for analysis by flow cytometry of their FcγR expression. Samples taken at day 2 were also assessed for cytotoxicity.
Redirected antibody-dependent cytotoxicity assay
The cytotoxicity of unstimulated and IL-12– or IL-15–stimulated NK cells was assessed by incubating NK cells in the presence of FcγR-bearing P815 cells as targets. Target cells were plated in 96-well round-bottom plates at 104/well. PBLs were mixed with target cells at E:T ratios of 50:1, 25:1, 12:1, and 6:1, in the presence or absence of either 5 μg/mL anti-FcγRII (clone KB61) or 5 μg/mL anti-FcγRIII (clone 3G8). After 4 hours of incubation at 37°C at 5% CO2, cells were harvested, and 7-amino-actinomycin (7-AAD; Invitrogen, Leiden, The Netherlands) was added to determine cell death by flow cytometry. The percentage of specific cell death was calculated by subtracting the percentage of spontaneous cell death from experimental samples.
Statistics
Genotyping data were analyzed by Fisher exact tests, a P value of less than .05 was considered statistically significant. Results of the in vitro data are expressed as mean plus or minus SEM. Where applicable, Student t test or one-way ANOVA was used. For comparison of the various time points, a 2-way ANOVA was performed.
Results
Distribution of FCGR gene CNV in healthy controls
Genetic variation in the FCGR gene cluster was studied with a newly developed MLPA assay (Figure 1 and Table S1). In total, 100 healthy white controls were evaluated. We observed no CNV for the FCGR2A and FCGR2B genes. In contrast, both the FCGR3A and the FCGR3B genes showed variation in copy number (Table 1). As has been reported previously by our group,17 CNV in FCGR3B is directly linked to similar variation in the FCGR2C gene number. Here, we confirm this linkage by use of MLPA.
Copy number variation in the FCGR2 and FCGR3 genes
| . | Controls . | ITP patients . | Adult ITP . | Children ITP . |
|---|---|---|---|---|
| FCGR2C | ||||
| 1 | 7 (7) | 6 (5.2) | 0 (0) | 6 (8.3) |
| 2 | 82 (82) | 103 (88.8) | 41 (93.1) | 62 (86.1) |
| 3 | 11 (11) | 7 (6.0) | 3 (6.8) | 4 (5.6) |
| FCGR3A | ||||
| 1 | 1 (1) | 4 (3.4) | 2 (4.5) | 2 (2.8) |
| 2 | 96 (96) | 108 (93.1) | 39 (88.6) | 69 (95.8) |
| 3 | 3 (3) | 4 (3.4) | 3 (6.8) | 1 (1.4) |
| FCGR3B | ||||
| 1 | 7 (7) | 6 (5.2) | 0 (0) | 6 (8.3) |
| 2 | 82 (82) | 103 (88.8) | 41 (93.1) | 62 (86.1) |
| 3 | 11 (11) | 7 (6.0) | 3 (6.8) | 4 (5.6) |
| . | Controls . | ITP patients . | Adult ITP . | Children ITP . |
|---|---|---|---|---|
| FCGR2C | ||||
| 1 | 7 (7) | 6 (5.2) | 0 (0) | 6 (8.3) |
| 2 | 82 (82) | 103 (88.8) | 41 (93.1) | 62 (86.1) |
| 3 | 11 (11) | 7 (6.0) | 3 (6.8) | 4 (5.6) |
| FCGR3A | ||||
| 1 | 1 (1) | 4 (3.4) | 2 (4.5) | 2 (2.8) |
| 2 | 96 (96) | 108 (93.1) | 39 (88.6) | 69 (95.8) |
| 3 | 3 (3) | 4 (3.4) | 3 (6.8) | 1 (1.4) |
| FCGR3B | ||||
| 1 | 7 (7) | 6 (5.2) | 0 (0) | 6 (8.3) |
| 2 | 82 (82) | 103 (88.8) | 41 (93.1) | 62 (86.1) |
| 3 | 11 (11) | 7 (6.0) | 3 (6.8) | 4 (5.6) |
Individuals with 1, 2, or 3 copies of a gene were observed. No significant difference existed in frequencies between controls and ITP patients. Data are no. (%).
We found a CNV in FCGR3A that was not linked to the gene copy number of the other FCGR genes studied here. Furthermore, the number of FCGR3A genes corresponded with the number of FcγRIIIa molecules on immune cells (manuscript in preparation), as has been described previously for FCGR3B gene copy variation.17 The CNV for the FCGR3A gene was observed in 3 individuals with 3 gene copies and in 1 individual with only a single FCGR3A gene copy (Figure 2).
CNV in FCGR3A. (A) MLPA electropherogram. Individual A with only 1 allele FCGR3A (gray line) compared with individual B with 2 alleles of FCGR3A (black line). FCGR3A-specific probes are indicated with an arrow. As an example, 2 of the control probes are indicated (C). One of the probes is specific for the X-chromosome (X). Individual A: Female, FCGR3B-Na1Na1, FCGR3A-158F. Individual B: Male, FCGR3B-Na2Na2, FCGR3A-158FF. (B) MLPA electropherogram. Individual C with 3 alleles FCGR3A (gray line) compared with individual D with 2 alleles of FCGR3A (black line). FCGR3A-specific probes are indicated with an arrow. As an example, 2 of the control probes are indicated (C). One of the probes is specific for the X-chromosome (X). Individual C: Female, FCGR3B-Na2Na2, FCGR3A-158VFF. Individual D: Female, FCGR3B-Na2Na2, FCGR3A-158VF. Because of the homology between certain probes, probes had to be divided over 2 separate mixes to prevent competition.
CNV in FCGR3A. (A) MLPA electropherogram. Individual A with only 1 allele FCGR3A (gray line) compared with individual B with 2 alleles of FCGR3A (black line). FCGR3A-specific probes are indicated with an arrow. As an example, 2 of the control probes are indicated (C). One of the probes is specific for the X-chromosome (X). Individual A: Female, FCGR3B-Na1Na1, FCGR3A-158F. Individual B: Male, FCGR3B-Na2Na2, FCGR3A-158FF. (B) MLPA electropherogram. Individual C with 3 alleles FCGR3A (gray line) compared with individual D with 2 alleles of FCGR3A (black line). FCGR3A-specific probes are indicated with an arrow. As an example, 2 of the control probes are indicated (C). One of the probes is specific for the X-chromosome (X). Individual C: Female, FCGR3B-Na2Na2, FCGR3A-158VFF. Individual D: Female, FCGR3B-Na2Na2, FCGR3A-158VF. Because of the homology between certain probes, probes had to be divided over 2 separate mixes to prevent competition.
Distribution of FCGR gene SNPs in healthy controls
Regarding the distribution of the most common SNPs within these genes, that is, FcγRIIa-131H/R, FcγRIIb-232I/T, FcγRIIIa-158V/F, and the FcγRIIIb HNA1a/HNA1b/HNA1c, we observed allele frequencies that were close to the frequencies reported before in the white population (Table 2).8,34,35
Genotyping results of SNPs in the FCGR2 and FCGR3 genes
| . | Controls . | All ITP . | Adult ITP . | Children ITP . |
|---|---|---|---|---|
| FCGR2A | ||||
| Genotype frequency | ||||
| 131RR | 20 (20) | 27 (23.3) | 8 (18.2) | 19 (26.3) |
| 131HR | 52 (52) | 54 (46.6) | 26 (59.1) | 28 (38.9) |
| 131HH | 28 (28) | 35 (30.2) | 10 (22.7) | 25 (34.7) |
| Allele frequency | ||||
| 131R | 92 (46) | 108 (46.6) | 42 (47.7) | 66 (45.8) |
| 131H | 108 (54) | 124 (53.4)* | 46 (52.3)* | 78 (54.2)* |
| FCGR2B | ||||
| Genotype frequency | ||||
| 232II | 81 (81) | 97 (83.6) | 37 (84.1) | 60 (83.3) |
| 232IT | 15 (15) | 15 (12.9) | 5 (11.4) | 10 (13.9) |
| 232TT | 4 (4) | 4 (3.4) | 2 (4.5) | 2 (2.8) |
| Allele frequency | ||||
| 232I | 177 (88.5) | 209 (90.1) | 79 (89.8) | 130 (90.2) |
| 232T | 23 (11.5) | 23 (9.9)* | 9 (10.2)* | 14 (9.7)* |
| FCGR3A | ||||
| Genotype frequency | ||||
| 158F | 0 (0) | 3 (2.6) | 2 (4.5) | 1 (1.4) |
| 158FF | 48 (48) | 30 (25.9) | 15 (34.1) | 15 (20.8) |
| 158FFF | 1 (1) | 2 (1.7) | 2 (4.5) | 0 (0) |
| 158V | 1 (1) | 1 (0.9) | 0 (0) | 1 (1.4) |
| 158VV | 7 (7) | 23 (19.8) | 8 (18.2) | 15 (20.8) |
| 158VVV | 1 (1) | 0 (0) | 0 (0) | 0 (0) |
| 158VF | 41 (41) | 55 (47.4) | 16 (36.4) | 39 (54.1) |
| 158VVF | 1 (1) | 0 (0) | 0 (0) | 0 (0) |
| 158VFF | 0 (0) | 2 (1.7) | 1 (2.3) | 1 (1.4) |
| Allele frequency | ||||
| 158F | 141 (69.5) | 128 (55.2) | 56 (62.9) | 72 (50.3) |
| 158V | 62 (30.5) | 104 (44.8)† | 33 (37.1)‡ | 71 (49.7)§ |
| FCGR3B | ||||
| Genotype frequency | ||||
| HNA1a | 4 (4) | 4 (3.4) | 0 (0) | 4 (5.6) |
| HNA1aHNA1a | 13 (13) | 13 (11.2) | 5 (11.4) | 8 (11.1) |
| HNA1aHNA1aHNA1 | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| HNA1b | 3 (3) | 2 (1.7) | 0 (0) | 2 (2.8) |
| HNA1bHNA1b | 34 (34) | 55 (47.4) | 22 (50.0) | 33 (45.8) |
| HNA1bHNA1bHNA1b | 1 (1) | 0 (0) | 0 (0) | 0 (0) |
| HNA1aHNA1b | 35 (35) | 35 (30.2) | 14 (31.8) | 21 (29.2) |
| HNA1aHNA1aHNA1b | 2 (2) | 1 (0.9) | 0 (0) | 1 (1.4) |
| HNA1aHNA1bHNA1b | 8 (8) | 6 (5.2) | 3 (6.8) | 3 (4.2) |
| Allele frequency | ||||
| HNA1a | 77 (37.7) | 73 (31.3) | 27 (29.7) | 46 (32.4) |
| HNA1b | 127 (62.3) | 160 (68.7)* | 64 (70.3)* | 96 (67.6)* |
| HNA1c (SH) | 4 (4) | 5 (4.3)* | 1 (2.3)* | 4 (5.6)* |
| . | Controls . | All ITP . | Adult ITP . | Children ITP . |
|---|---|---|---|---|
| FCGR2A | ||||
| Genotype frequency | ||||
| 131RR | 20 (20) | 27 (23.3) | 8 (18.2) | 19 (26.3) |
| 131HR | 52 (52) | 54 (46.6) | 26 (59.1) | 28 (38.9) |
| 131HH | 28 (28) | 35 (30.2) | 10 (22.7) | 25 (34.7) |
| Allele frequency | ||||
| 131R | 92 (46) | 108 (46.6) | 42 (47.7) | 66 (45.8) |
| 131H | 108 (54) | 124 (53.4)* | 46 (52.3)* | 78 (54.2)* |
| FCGR2B | ||||
| Genotype frequency | ||||
| 232II | 81 (81) | 97 (83.6) | 37 (84.1) | 60 (83.3) |
| 232IT | 15 (15) | 15 (12.9) | 5 (11.4) | 10 (13.9) |
| 232TT | 4 (4) | 4 (3.4) | 2 (4.5) | 2 (2.8) |
| Allele frequency | ||||
| 232I | 177 (88.5) | 209 (90.1) | 79 (89.8) | 130 (90.2) |
| 232T | 23 (11.5) | 23 (9.9)* | 9 (10.2)* | 14 (9.7)* |
| FCGR3A | ||||
| Genotype frequency | ||||
| 158F | 0 (0) | 3 (2.6) | 2 (4.5) | 1 (1.4) |
| 158FF | 48 (48) | 30 (25.9) | 15 (34.1) | 15 (20.8) |
| 158FFF | 1 (1) | 2 (1.7) | 2 (4.5) | 0 (0) |
| 158V | 1 (1) | 1 (0.9) | 0 (0) | 1 (1.4) |
| 158VV | 7 (7) | 23 (19.8) | 8 (18.2) | 15 (20.8) |
| 158VVV | 1 (1) | 0 (0) | 0 (0) | 0 (0) |
| 158VF | 41 (41) | 55 (47.4) | 16 (36.4) | 39 (54.1) |
| 158VVF | 1 (1) | 0 (0) | 0 (0) | 0 (0) |
| 158VFF | 0 (0) | 2 (1.7) | 1 (2.3) | 1 (1.4) |
| Allele frequency | ||||
| 158F | 141 (69.5) | 128 (55.2) | 56 (62.9) | 72 (50.3) |
| 158V | 62 (30.5) | 104 (44.8)† | 33 (37.1)‡ | 71 (49.7)§ |
| FCGR3B | ||||
| Genotype frequency | ||||
| HNA1a | 4 (4) | 4 (3.4) | 0 (0) | 4 (5.6) |
| HNA1aHNA1a | 13 (13) | 13 (11.2) | 5 (11.4) | 8 (11.1) |
| HNA1aHNA1aHNA1 | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| HNA1b | 3 (3) | 2 (1.7) | 0 (0) | 2 (2.8) |
| HNA1bHNA1b | 34 (34) | 55 (47.4) | 22 (50.0) | 33 (45.8) |
| HNA1bHNA1bHNA1b | 1 (1) | 0 (0) | 0 (0) | 0 (0) |
| HNA1aHNA1b | 35 (35) | 35 (30.2) | 14 (31.8) | 21 (29.2) |
| HNA1aHNA1aHNA1b | 2 (2) | 1 (0.9) | 0 (0) | 1 (1.4) |
| HNA1aHNA1bHNA1b | 8 (8) | 6 (5.2) | 3 (6.8) | 3 (4.2) |
| Allele frequency | ||||
| HNA1a | 77 (37.7) | 73 (31.3) | 27 (29.7) | 46 (32.4) |
| HNA1b | 127 (62.3) | 160 (68.7)* | 64 (70.3)* | 96 (67.6)* |
| HNA1c (SH) | 4 (4) | 5 (4.3)* | 1 (2.3)* | 4 (5.6)* |
Due to CNV, the 3-allelic variation was observed for FCGR3A-158VF and FCGR3B-HNA1aHNA1b. Data are no. (%). Significance levels are indicated by symbols.
P is not significant.
P is .003.
P is .3.
P is less than .001.
For the FcγRIIIb polymorphisms, we found that our MLPA assay is superior to tests previously described for genotyping. When conventional genotyping results of the FCGR3B*1 (HNA1a), FCGR3B*2 (HNA1b), FCGR3B*3 (HNA1c) alleles were compared with the results obtained with MLPA, all observed differences could be explained by variation in the copy number. For instance, an individual formerly predicted to be FCGR3B*1, FCGR3B*1 (HNA1a/HNA1a) could now be correctly typed as FCGR3B*1/− (HNA1a/null). The FCGR3B*3 (HNA1c) variant was observed in 4 (4%) of control individuals.
In the control group, 25% of the donors were heterozygous for the SNP at position −386 (G > C) in the promoter region of the FCGR2B/C genes (Table 3). In 100 donors, we did not observe the homozygous −386C variant. Gene-specific long-range PCR was performed on the DNA samples carrying the FCGR2B/C −386G/C variant to discriminate between the promoters of either the FCGR2B or FCGR2C gene, because it was not possible to design a discriminating gene-specific probe for our MLPA assay.
Frequency of nonspecific promoter polymorphism −386 G > C and frequency of stop codon versus ORF in exon 3 of FCGR2C
| . | Controls . | All ITP . | Adult ITP . | Children ITP . |
|---|---|---|---|---|
| Promotor 2B/C −386 | ||||
| GG | 75 (75) | 68 (58.6) | 25 (56.8) | 43 (59.7) |
| CG | 25 (25) | 47 (40.5) | 18 (40.9) | 29 (40.2) |
| CC | 0 (0) | 1 (0.9)*† | 1 (2.3)†‡ | 0 (0)†§ |
| FCGR2C exon 3 | ||||
| STOP | 6 (6) | 4 (3.4) | 0 (0) | 4 (5.6) |
| STOP/STOP | 66 (66) | 68 (58.6) | 28 (63.6) | 40 (55.6) |
| STOP/STOP/STOP | 10 (10) | 4 (3.4) | 1 (2.3) | 3 (4.2) |
| ORF | 1 (1) | 2 (1.7) | 0 (0) | 2 (2.8) |
| ORF/ORF | 0 (0) | 3 (2.6) | 2 (4.5) | 1 (1.4) |
| ORF/STOP | 16 (16) | 32 (27.6) | 11 (25) | 21 (29.2) |
| ORF/ORF/STOP | 0 (0) | 1 (0.9) | 1 (2.3) | 0 (0) |
| ORF/STOP/STOP | 1 (1) | 2 (1.7)¶‖ | 1 (2.3)§‖ | 1 (1.4)‖# |
| FCGR2C exon 3 (allele frequency) | ||||
| STOP | 186 (91.2) | 189 (81.1) | 73 (80.2) | 116 (81.7) |
| ORF | 18 (8.8) | 44 (18.9)** | 18 (19.8)* | 26 (18.3)* |
| . | Controls . | All ITP . | Adult ITP . | Children ITP . |
|---|---|---|---|---|
| Promotor 2B/C −386 | ||||
| GG | 75 (75) | 68 (58.6) | 25 (56.8) | 43 (59.7) |
| CG | 25 (25) | 47 (40.5) | 18 (40.9) | 29 (40.2) |
| CC | 0 (0) | 1 (0.9)*† | 1 (2.3)†‡ | 0 (0)†§ |
| FCGR2C exon 3 | ||||
| STOP | 6 (6) | 4 (3.4) | 0 (0) | 4 (5.6) |
| STOP/STOP | 66 (66) | 68 (58.6) | 28 (63.6) | 40 (55.6) |
| STOP/STOP/STOP | 10 (10) | 4 (3.4) | 1 (2.3) | 3 (4.2) |
| ORF | 1 (1) | 2 (1.7) | 0 (0) | 2 (2.8) |
| ORF/ORF | 0 (0) | 3 (2.6) | 2 (4.5) | 1 (1.4) |
| ORF/STOP | 16 (16) | 32 (27.6) | 11 (25) | 21 (29.2) |
| ORF/ORF/STOP | 0 (0) | 1 (0.9) | 1 (2.3) | 0 (0) |
| ORF/STOP/STOP | 1 (1) | 2 (1.7)¶‖ | 1 (2.3)§‖ | 1 (1.4)‖# |
| FCGR2C exon 3 (allele frequency) | ||||
| STOP | 186 (91.2) | 189 (81.1) | 73 (80.2) | 116 (81.7) |
| ORF | 18 (8.8) | 44 (18.9)** | 18 (19.8)* | 26 (18.3)* |
Data are no. (%).
P is .01.
GG vs CG + CC.
P is .03.
P is .05.
P is .009.
STOP + STOP/STOP + STOP/STOP/STOP vs ORF + ORF/ORF + ORF/STOP + ORF/ORF/STOP + ORF/STOP/STOP.
P is .02.
P is .004.
In the FCGR2C gene, an SNP in exon 3 converts a glutamine in the ORF to the most commonly found stop codon. In the control group, 82% of the individuals were homozygous for the STOP codon (FCGR2Cstop, FCGR2Cstop/stop, or FCGR2Cstop/stop/stop), which is close to what has been reported previously.36
Genetic variation in ITP
In total, 116 white ITP patients were evaluated, divided into ITP of childhood-onset (< 16 years at time of diagnosis; n = 72; male, 48.6%) and ITP of adult-onset (> 16 years at time of diagnosis; n = 44; male, 15.9%). Genetic analysis was performed by the same MLPA and PCR assays as used in controls.
First, variation in copy number was not associated with susceptibility to ITP in our cohort. As in the control group, we found no CNV in FCGR2A and FCGR2B. In the ITP group, 3 copies of the FCGR3B allele were found in 7 patients and 1 single FCGR3B allele in 6 patients, respectively. In all patients, these single or triple copy alleles were linked to an identical variation in the copy number of the FCGR2C gene. These results are similar to what was found in the control group. Hence, we did not observe an association between the CNV of FCGR3B and susceptibility to ITP (Table 1). In 4 patients, a loss of copy number for FCGR3A was observed, and also in 4 patients a gain in copy number was observed, resulting in 1* FCGR3A allele and 3* FCGR3A alleles, respectively (Figure 2).
Second, genotyping results of the FcγRIIa-131H/R and the FcγRIIb-232I/T SNPs also showed no significant differences in genotype or allele frequencies when ITP samples were compared with the control samples (Table 2). The allele frequencies of the FCGR3A SNP for the FcγRIIIa-158F and FcγRIIIa-158V variants were 55.2% and 44.8%, respectively, for the ITP patients. The allele frequencies differed significantly between ITP patients and healthy controls (P = .003; Table 2). When adult-onset and pediatric-onset ITP patients were separately analyzed, only for the children a significant overrepresentation of the FcγRIIIa-158V was observed (P < .001; Table 2), as shown before in pediatric ITP.27,28 Allele frequency in adult-onset ITP patients was comparable to the control samples. Any genotypic variation of the polymorphism FcγRIIIa-158V/F resulting from CNV was separately confirmed by quantitative PCR (LightCycler method; data not shown).
With MLPA, also a significant difference in genotype (P = .01) frequency for the FCGR2B/C promoter polymorphism at position −386 G/C was observed (Table 3). The genotype −386 CC was rare, as it was only observed in one patient.
Regarding the FCGR2C gene, a significant overrepresentation of the ORF in the ITP group was observed (P = .009; Table 3), with 82% being FCGR2Cstop, FCGR2Cstop/stop, or FCGR2Cstop/stop/stop in the control cohort versus 65.5% in the ITP group. Moreover, 3 ITP patients were found to carry the homozygous FCGR2CORF/ORF genotype. W observed that 86.2% of the individuals carrying an FCGR2C-ORF allele were also heterozygous for the FCGR2B/C −386 promoter polymorphism (data not shown). Individuals carrying an FCGR2C-ORF allele and a heterozygous FCGR2B/C −386 promoter polymorphism were all observed to have the promoter haplotype FCGR2C −386C/−120T, previously designated 2B.2,37 as was analyzed with specific long-range PCR (Table 4). When FcγRIIIa-158V data were combined with the FCGR2C-ORF allele and promoter haplotype FCGR2C −386C/−120T, we did not observe an absolute linkage (data not shown).
Correlation of FCGR2C-ORF with FcγRIIc expression on NK cells and with promoter haplotype FCGR2C 2B.2
| Sample . | Expression . | MLPA . | Gene-specific PCR . | |||||
|---|---|---|---|---|---|---|---|---|
| NK cells . | MFI . | FCGR2C stop/ORF . | 2B/C −386 . | 2B −386 . | 2B −120 . | 2C −386 . | 2C −120 . | |
| 1 | No | nd | stop/stop | W | — | — | — | — |
| 2 | No | nd | stop/stop | W | — | — | — | — |
| 3 | No | nd | stop/stop | W | — | — | — | — |
| 4 | No | nd | stop/stop | W | — | — | — | — |
| 5 | No | nd | stop/stop | W | — | — | — | — |
| 6 | No | nd | stop/stop | W | — | — | — | — |
| 7 | No | nd | stop/stop | W | — | — | — | — |
| 8 | No | nd | stop/stop | W | — | — | — | — |
| 9 | No | nd | stop/stop | W | — | — | — | — |
| 10 | No | nd | stop/stop | W | — | — | — | — |
| 11 | Yes | 590 | ORF/stop | He | GC | TA | GC | TT |
| 12 | Yes | 1188 | ORF/stop | He | GC | TA | GC | TT |
| 13 | Yes | 1648 | ORF/stop | He | GC | TA | GC | TT |
| 14 | Yes | 2602 | ORF/stop | He | GC | TA | GC | TT |
| 15 | Yes | 2123 | ORF/stop | He | GC | TA | GC | TT |
| 16 | Yes | 1109 | ORF/stop | He | GC | TA | GC | TT |
| 17 | Yes | 2380 | ORF/stop | He | GC | TA | GC | TT |
| 18 | Yes | 1243 | ORF/stop | He | GC | TA | GC | TT |
| 19 | Yes | 1394 | ORF/stop | He | CC | AA | GC | TT |
| 20 | Yes | 1629 | ORF/stop | He | GC | TA | GC | TT |
| Sample . | Expression . | MLPA . | Gene-specific PCR . | |||||
|---|---|---|---|---|---|---|---|---|
| NK cells . | MFI . | FCGR2C stop/ORF . | 2B/C −386 . | 2B −386 . | 2B −120 . | 2C −386 . | 2C −120 . | |
| 1 | No | nd | stop/stop | W | — | — | — | — |
| 2 | No | nd | stop/stop | W | — | — | — | — |
| 3 | No | nd | stop/stop | W | — | — | — | — |
| 4 | No | nd | stop/stop | W | — | — | — | — |
| 5 | No | nd | stop/stop | W | — | — | — | — |
| 6 | No | nd | stop/stop | W | — | — | — | — |
| 7 | No | nd | stop/stop | W | — | — | — | — |
| 8 | No | nd | stop/stop | W | — | — | — | — |
| 9 | No | nd | stop/stop | W | — | — | — | — |
| 10 | No | nd | stop/stop | W | — | — | — | — |
| 11 | Yes | 590 | ORF/stop | He | GC | TA | GC | TT |
| 12 | Yes | 1188 | ORF/stop | He | GC | TA | GC | TT |
| 13 | Yes | 1648 | ORF/stop | He | GC | TA | GC | TT |
| 14 | Yes | 2602 | ORF/stop | He | GC | TA | GC | TT |
| 15 | Yes | 2123 | ORF/stop | He | GC | TA | GC | TT |
| 16 | Yes | 1109 | ORF/stop | He | GC | TA | GC | TT |
| 17 | Yes | 2380 | ORF/stop | He | GC | TA | GC | TT |
| 18 | Yes | 1243 | ORF/stop | He | GC | TA | GC | TT |
| 19 | Yes | 1394 | ORF/stop | He | CC | AA | GC | TT |
| 20 | Yes | 1629 | ORF/stop | He | GC | TA | GC | TT |
MFI indicates mean fluorescence units; nd, not determined; W, wild-type; —, not tested because wild-type in MLPA; and He, heterozygous.
FCGR2C splice variants
To date, 5 splice variants of FcγRIIc have been reported, 2 of which result in a membrane-anchored receptor, that is, FcγRIIc1 encoded by the full transcript, and FcγRIIc3 reported to lack exon 7.38,39 We hypothesized that alternative splicing of the FcγRIIc transcript is restricted to the FCGR2C-stop allele. To investigate this, we cloned the FcγRIIc transcripts from a single individual with an FCGR2CORF/ORF genotype and from 3 individuals with an FCGR2CORF/stop genotype into a bacterial expression vector and obtained the sequence of at least 20 clones per individual. Upon sequencing of all these clones, we found that individuals genotyped as FCGR2CORF/ORF or FCGR2CORF/stop only express FcγRIIc1 (data not shown). Various studies have reported about a polymorphism at the splice donor site of the intron following the C2 exon (exon7).12,38,40 In general, we observed the nucleotides AT at the splice border of FCGR2Cstop/stop individuals and only in rare cases a heterozygous GT/AT (detectable with probe 2C-DO6 and 2A-DO6), which will, however, not be expressed due to the Stop codon. In case of an FCGR2C-ORF allele, the reverse was true. We mainly observed a GT at the splice border, but in some cases we did observe AT. This could imply that FcγRIIc1 may not be the only functional splice variant expressed. Further studies will have to be done to investigate the function of the transmembrane protein of transcripts with both the ORF and the AT splice border.
Distribution of FCGR2C expression
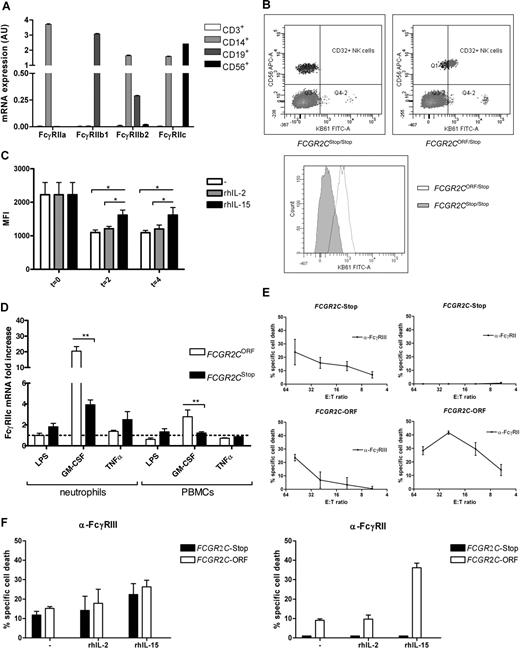
We explored the mRNA expression of FcγRII isoforms in neutrophils, monocytes, NK cells, T cells, and B cells from healthy volunteers. Transcripts for FcγRIIa, FcγRIIb2 (lacking exon 6 of FCGR2B), and FcγRIIc, but not FcγRIIb1 (containing exon 6 of FCGR2B), were found in both neutrophils and monocytes, while B cells contained only FcγRIIb1.33 T cells did not express any of the FcγRII isoforms, whereas NK cells solely expressed FcγRIIc. This is in line with previous findings (Figure 3A).41
FcγRIIc expression and function. (A) Distribution of FcγRIIc mRNA expression in leucocytes. T cells CD3+, monocytes, and neutrophils CD14+, B cells CD19+, NK cells CD 56+. (B) FcγRII expression on NK cells is limited to the FCGR2C-ORF genotype. Blood cells were incubated with CD56 and CD32. Lymphocytes were gated on the basis of their forward scatter/side scatter pattern. NK-cell population was determined as CD56-positive lymphocytes. (C) FcγRIIc expression on NK cells is modulated by IL-15 but not by IL-2. PBMCs were isolated and cultured as described in “In vitro activation of NK cells.” Expression of CD32 on CD56+ NK cells was measured by flow cytometry with CD32, at the indicated time-points. There was a significant higher expression on days 2 and 4 on the IL-15–stimulated NK cells (P = .01, n = 5). Data are expressed as mean plus or minus SEM. (D) FcγRIIc mRNA is strongly up-regulated by GM-CSF on cells of FCGR2C-ORF donors. Neutrophils and PBMCs were isolated and cultured for 4 hours with the indicated stimuli as described in “In vitro activation of neutrophils and PBMCs.” FcγRIIc mRNA was measured by quantitative RT-PCR. GM-CSF strongly up-regulated FcγRIIc mRNA in neutrophils and, to a lesser extent, in PBMCs of FCGR2C-ORF genotyped donors, but not in FCGR2C-Stop donors (P = .0001 and P = .01, respectively; n = 5-8). Data are expressed as mean plus or minus SEM. (E) rADCC. PBLs were isolated as described in “Isolation of neutrophils and PBMCs,” and FcγRIIc functionality was assessed by rADCC. Cells from both FCGR2C-Stop and FCGR2C-ORF genotyped donors killed anti-FcγRIII–coated targets with similar kinetics (left panels). In contrast, only cells from FCGR2C-ORF genotyped donors were capable of killing anti-FcγRII–coated targets (right panels) (n = 4). Data are expressed as mean plus or minus SEM. (F) rADCC with stimulated cells. PBLs were obtained and subsequently cultured for 2 days with or without IL-2 or IL-15. Thereafter, the cells were harvested and used in a rADCC. Cells from both FCGR2C-Stop and FCGR2C-ORF genotyped donors killed anti-FcγRIII–coated targets (left panel). In both cases IL-15 and, to a lesser extent, IL-2 enhanced specific lysis of the anti-FcγRIII–coated targets (n = 3). In contrast, only cells from FCGR2C-ORF genotyped donors were capable of killing anti-FcγRII–coated targets (right panel). Here again, IL-15 strongly enhanced the specific cell lysis (n = 3). Data are expressed as mean plus or minus SEM.
FcγRIIc expression and function. (A) Distribution of FcγRIIc mRNA expression in leucocytes. T cells CD3+, monocytes, and neutrophils CD14+, B cells CD19+, NK cells CD 56+. (B) FcγRII expression on NK cells is limited to the FCGR2C-ORF genotype. Blood cells were incubated with CD56 and CD32. Lymphocytes were gated on the basis of their forward scatter/side scatter pattern. NK-cell population was determined as CD56-positive lymphocytes. (C) FcγRIIc expression on NK cells is modulated by IL-15 but not by IL-2. PBMCs were isolated and cultured as described in “In vitro activation of NK cells.” Expression of CD32 on CD56+ NK cells was measured by flow cytometry with CD32, at the indicated time-points. There was a significant higher expression on days 2 and 4 on the IL-15–stimulated NK cells (P = .01, n = 5). Data are expressed as mean plus or minus SEM. (D) FcγRIIc mRNA is strongly up-regulated by GM-CSF on cells of FCGR2C-ORF donors. Neutrophils and PBMCs were isolated and cultured for 4 hours with the indicated stimuli as described in “In vitro activation of neutrophils and PBMCs.” FcγRIIc mRNA was measured by quantitative RT-PCR. GM-CSF strongly up-regulated FcγRIIc mRNA in neutrophils and, to a lesser extent, in PBMCs of FCGR2C-ORF genotyped donors, but not in FCGR2C-Stop donors (P = .0001 and P = .01, respectively; n = 5-8). Data are expressed as mean plus or minus SEM. (E) rADCC. PBLs were isolated as described in “Isolation of neutrophils and PBMCs,” and FcγRIIc functionality was assessed by rADCC. Cells from both FCGR2C-Stop and FCGR2C-ORF genotyped donors killed anti-FcγRIII–coated targets with similar kinetics (left panels). In contrast, only cells from FCGR2C-ORF genotyped donors were capable of killing anti-FcγRII–coated targets (right panels) (n = 4). Data are expressed as mean plus or minus SEM. (F) rADCC with stimulated cells. PBLs were obtained and subsequently cultured for 2 days with or without IL-2 or IL-15. Thereafter, the cells were harvested and used in a rADCC. Cells from both FCGR2C-Stop and FCGR2C-ORF genotyped donors killed anti-FcγRIII–coated targets (left panel). In both cases IL-15 and, to a lesser extent, IL-2 enhanced specific lysis of the anti-FcγRIII–coated targets (n = 3). In contrast, only cells from FCGR2C-ORF genotyped donors were capable of killing anti-FcγRII–coated targets (right panel). Here again, IL-15 strongly enhanced the specific cell lysis (n = 3). Data are expressed as mean plus or minus SEM.
Expression of FcγRIIc on NK cells
FCGR2A, FCGR2B, and FCGR2C have 92% to 96% sequence homology. There are no monoclonal antibodies available that can truly distinguish between FcγRIIa, FcγRIIb, and FcγRIIc, rendering it difficult to quantify the protein expression on cells. However, we found that NK cells only expressed mRNA of the FcγRIIc isoform. For this reason, we examined the expression of FcγRII on NK cells of individuals with the FCGR2CORF/stop genotype and the FCGR2Cstop/stop genotype. We found that the presence of an FCGR2C-ORF allele correlated with FcγRII expression on NK cells, whereas the absence of a functional allele corresponded with the total absence of any FcγRII on NK cells (Figure 3B and Table 4). Thus, FcγRIIc is present on NK cells.
In vitro activation of NK cells
The FcγRIIc expression on NK cells was measured 3 times at various time intervals (range, 2–5 months) in 4 individuals with the FCGR2CORF/stop genotype. Variable expression levels of FcγRIIc occurred, irrespective of the constant FCGR2B/C promoter haplotype 2B.237 in all individuals tested. Minor fluctuation was observed over time (MFI ± SEM: 1707 ± 211).
Subsequent testing in NK-cell cultures with the NK-cell activators IL-2 or IL-15 showed the ability to modulate the FcγR expression on NK cells. In line with earlier reports,42 we found that both IL-2 and IL-15 up-regulated the surface expression of FcγRIIIa on NK cells. In contrast, FcγRIIc expression was down-regulated during culture, although IL-15 significantly rescued the loss of FcγRIIc expression compared with either IL-2 or medium (Figure 3C).
Because surface expression of FcγRIIc on neutrophils and monocytes cannot be monitored, the regulation of FcγRIIc mRNA in these cells was tested after incubating neutrophils as well as PBMCs with the inflammatory activators LPS, GM-CSF, or TNFα. In particular, GM-CSF induced a strong up-regulation of FcγRIIc mRNA in neutrophils obtained from individuals with an FCGR2CORF/stop genotype, whereas this was found to a significantly lower extent in individuals with an FCGR2Cstop/stop genotype (P < .01; Figure 3D).
Redirected antibody-dependent cellular cytotoxicity
To assess whether FcγRIIc expression on innate immune cells was functionally active, we tested the capability of the NK cells to kill antibody-coated target cells. To selectively target either FcγRIIc or FcγRIIIa on NK cells, we used the rADCC test with the FcγR-bearing murine mastocytoma P815 cell line loaded with anti-FcγRII or anti-FcγRIII antibody.
We found that NK cells of an FCGR2CORF/stop genotype donor were able to kill both anti-FcγRII and anti-FcγRIII–loaded target cells (Figure 3E). In contrast, NK cells of an FCGR2Cstop/stop genotype donor were able to kill only anti-FcγRIII–loaded target cells. When the expression of FcγRIIc was reduced by culturing NK cells in the presence of activating IL-2 or IL-15 cytokines, the effect of FcγRIIc crosslinking in the rADCC assay showed even more pronounced killing capacity in IL-2– or IL-15–activated NK cells (Figure 3F).
Discussion
Genetic variation in immunologically relevant genes results in differential responsiveness to infection but might subsequently also predispose to autoimmune disease as a result of unbalanced immunity. We investigated the role of the FCGR gene cluster on the predisposition to ITP by using a novel MPLA assay to analyze the relevant SNPs as well as CNV in FCGR2A, FCGR2B, FCGR2C, FCGR3A, and FCGR3B.
By using MLPA, we were able to genotype more accurately than has been possible before, because now CNV and SNP analysis can be interpreted in one test system. FCGR2A and FCGR2B did not show gene copy number variation, in contrast to the other genes studied. We demonstrate a variation in the copy number for the FCGR3A gene, which, to our knowledge, has never been reported before. The relevance of CNV for the activating FCGR2C allele was indicated by the fact that in 91% of the white control population we found an SNP in exon 3 that results in a stop codon determining the presence of a pseudogene. However, the remaining 9% of the population expresses a functionally activating FcγRIIc on their immune cells. Although the presence of a pseudogene or ORF is determined by an SNP, this variation behaves like a copy number polymorphism with respect to the activating FCGR2C gene. Regarding the existence of an unbalanced immunity in autoinflammation, we found a significant overrepresentation of the FCGR2C-ORF allele in the ITP population (19%).
The relevance of this observation on the FCGR2C-ORF allele for ITP was subsequently shown by in vitro studies. We found expression of FcγRIIc mRNA in neutrophils, monocytes, and NK cells, but not in T and B cells. Neutrophils and monocytes also express other isoforms of FcγRII33,43 and, although they have a key role as phagocytes in ITP, they are not ideal candidates to study the expression and function of FcγRIIc. NK cells express FcγRIIc but no FcγRIIa or FcγRIIb, and these cells were therefore used as a functional read-out for FcγRIIc, however, no direct link with the pathogenesis of ITP can be made. Only individuals carrying an FCGR2C-ORF allele were found to express FcγRIIc protein on NK cells (Figure 3B). We furthermore demonstrated that NK cells bearing FcγRIIc can mediate antibody-dependent cellular cytotoxicity (ADCC) to antibody-coated targets, demonstrating that FcγRIIc is indeed an activating IgG receptor molecule on immune cells. Although expressed to a lower extent than FcγRIIIa, FcγRIIc seems to mediate more killing than FcγRIIIa (Figure 3E). This might be due to the fact that FcγRIIc contains its own signaling motif, whereas FcγRIIIa is dependent on the γ-ζ heterodimer for signaling.
When investigating the role of FcγRIIc under inflammatory conditions, we found that IL-15 (but not IL-2) modulated FcγRIIc expression on NK cells, and GM-CSF strongly up-regulated FcγRIIc mRNA in phagocytes. Again, this was only observed in individuals carrying an FCGR2C-ORF allele. FcγRIIc might play a vital role during inflammatory conditions, mediating enhanced uptake of immune complexes or killing of antibody-coated target cells. This may lead to increased platelet destruction in ITP, in which these cytokines are present in sera of patients.44,45
In conclusion, we found that FCGR2C-ORF predisposes to ITP. We have shown that FcγRIIc is expressed by phagocytes and NK cells and that this enhances effector function toward antibody-coated targets. Furthermore, we have shown that under inflammatory conditions (GM-CSF or IL-15), FcγRIIc function is even further enhanced. Thus, taken together, these results suggest that FCGR2C is a variably expressed gene highly relevant for immunity that may contribute to susceptibility and severity of infections and autoimmune disease. We believe that this novel FCGR-specific MLPA will be applicable to study other autoimmune/inflammatory diseases more extensively than has been possible before.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank J. Schouten for his assistance in the development of the FCGR-specific MLPA, and J.H.M. Verhagen for his help with the design of the gene-specific long-range PCR for determining the promoter polymorphisms in FCGR2B and FCGR2C.
This work was supported by grants from The Netherlands Genomics Initiative (NROG, 050-71-315). W.B.B. was supported by ZonMw (920-03-391). E.v. M. was supported by the Van Loghem Foundation (RvB-0503-0051B-EB).
Authorship
Contribution: W.B.B. and E.v.M. designed and performed research, analyzed data, and wrote the paper. M.B., M.P., H.R.K., and T.W.K. provided patient material. J.G. performed research, M.de B. provided vital analytical tools, and D.R. designed parts of the research. M.B., M.de H., and H.R.K. provided intellectual input, and T.W.K. designed the overall study, supervised the research, and helped write the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: T.W. Kuijpers, Emma Children's Hospital, Academic Medical Center, Rm G8-205, Meibergdreef 9, 1105 AZ, Amsterdam, The Netherlands; e-mail: t.w.kuijpers@amc.uva.nl.
References
Author notes
W.B.B. and E.v.M. contributed equally to this paper.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal