Abstract
D-cyclins are regulators of cell division that act in a complex with cyclin-dependent kinases to commit cells to a program of DNA replication. D-cyclins are overexpressed in many tumors, including multiple myeloma and leukemia, and contribute to disease progression and chemoresistance. To better understand the role and impact of D-cyclins in hematologic malignancies, we conducted a high throughput screen for inhibitors of the cyclin D2 promoter and identified the drug cyproheptadine. In myeloma and leukemia cells, cyproheptadine decreased expression of cyclins D1, D2, and D3 and arrested these cells in the G0/G1 phase. After D-cyclin suppression, cyproheptadine induced apoptosis in myeloma and leukemia cell lines and primary patient samples preferentially over normal hematopoietic cells. In mouse models of myeloma and leukemia, cyproheptadine inhibited tumor growth without significant toxicity. Cyproheptadine-induced apoptosis was preceded by activation of the mitochondrial pathway of caspase activation and was independent of the drug's known activity as an H1 histamine and serotonin receptor antagonist. Thus, cyproheptadine represents a lead for a novel therapeutic agent for the treatment of malignancy. Because the drug is well tolerated and already approved in multiple countries for clinical use as an antihistamine and appetite stimulant, it could be moved directly into clinical trials for cancer.
Introduction
D-type cyclins are critical regulators of the cell cycle that act in a complex with cyclin-dependent kinases (CDKs) to promote the phosphorylation of Rb and initiate cellular transition from G1 to S phase.1 Overexpression of D-cyclins occurs in many tumors and leads to increased cell proliferation2-4 and chemoresistance.5 In contrast, reducing D-cyclins directly or indirectly can decrease cellular proliferation and induce apoptosis.6-8
Aberrant expression of one or more D-cyclins is virtually universal in multiple myeloma (MM), with 54% of myeloma tumors overexpressing cyclin D1, 48% overexpressing cyclin D2, 3% overexpressing cyclin D3, and 8% overexpressing both cyclin D1 and D2.9 The overexpression of D-cyclins contributes to the pathogenesis and chemoresistance of this disease.3,4,9 The t(11;14) involving cyclin D1 is found in 15% to 20% of myeloma patients and is associated with improved survival.10 However, levels of cyclin D1 protein often do not correlate as well with prognosis.11 In contrast, patients with myeloma and increased cyclin D2 expression, especially in cooperation with the presence of other oncogenes, such as c-Maf, MafB, and FGFR3/MMSET, have an inferior event-free survival.12 Likewise, D-cyclins are increased in a subset of patients with acute myeloid leukemia and are associated with poor outcome.13
To better understand the regulation of D-cyclins and the effects of targeting their expression in myeloma and leukemia cells, we conducted a chemical screen for inhibitors of the human cyclin D2 promoter using NIH3T3 cells engineered to overexpress the cyclin D2 promoter driving firefly luciferase. From this screen, we identified glucocorticoids and demonstrated that these drugs inhibit cyclin D2 transactivation by increasing the proteasomal degradation of c-maf via increased ubiquitin expression.14
Here we identify that cyproheptadine inhibits the transactivation of cyclin D2 through mechanisms independent of c-maf. Cyproheptadine is a known inhibitor of the H1 histamine and 5-HT serotonin receptors. Previously, cyproheptadine has been evaluated in patients for the treatment of migraines,15 anorexia,16 and atopic dermatitis.17 In clinical trials, the drug was well tolerated without hematologic toxicity. The ability of cyproheptadine to inhibit D-cyclin expression has not been reported previously. In this report, we evaluated the effects of cyproheptadine on myeloma and leukemia cell lines as well as primary patient samples. Cyproheptadine decreased D-cyclin levels and induced cell death in myeloma and leukemia cell lines and patient samples preferentially over normal hematopoietic cells. Finally, cyproheptadine demonstrated activity in mouse models of leukemia and myeloma. Thus, cyproheptadine represents a lead for a novel therapeutic agent for the treatment of malignancy.
Methods
Chemicals and reagents
Cyproheptadine hydrochloride, amitriptyline, loratadine, thymidine, histamine, serotonin, and propidium iodine (PI) were purchased from Sigma-Aldrich (St Louis, MO). DiIC1(5) (1,1′3,3,3′,3′ hexamethylindodicarbocyanine) was purchased from Invitrogen (Carlsbad, CA).
Cell lines
All human MM cell lines were maintained in Iscove Modified Dulbecco Medium (IMDM). Human and murine leukemia cell lines were maintained in RPMI 1640 medium. Mouse fibroblast NIH3T3 cells were maintained in Dulbeco modified Eagle medium. All media were supplemented with 10% fetal calf serum (FCS), 100 μg/mL of penicillin, and 100 units/mL of streptomycin (all from Hyclone, Logan, UT).
Primary cells
Primary human MM and acute myeloid leukemia (AML) samples were isolated from fresh bone marrow and peripheral blood samples, respectively, and obtained from patients who consented to the donation of a research sample. Primary normal hematopoietic cells were obtained from healthy volunteers donating their peripheral blood mononuclear stem cells (PBSCs) for allotransplantation. Mononuclear cells were isolated from the samples by Ficoll density centrifugation. Primary cells were cultured at 37°C in IMDM supplemented with 10% FCS, 1 mM of l-glutamine, and appropriate antibiotics. The collection and use of human tissue for this study were approved by the University Health Network institutional review board. Informed consent was obtained in accordance with the Declaration of Helsinki.
Cell viability, apoptosis, proliferation, and clonogenic growth assays
Cell viability was assessed by the MTS assay (Promega, Madison, WI) according to the manufacturer's instructions and as previously described.18
In primary cells, cell viability and apoptosis were measured by annexin V–fluorescein isothiocyanate (FITC; Biovision Research Products, Mountain View, CA) staining and flow cytometry according to the manufacturer's instructions and as previously described.19 Mononuclear cells from patients with MM were costained with phycoerythrin-labeled anti-CD138 and FITC-labeled annexin V. The percentage of CD138+/annexin V− cells was quantified as a marker of cell viability as previously described.20
Cellular proliferation was assessed by measuring uptake of 5-bromodeoxyuridine (BrdU; Sigma-Aldrich) by flow cytometry as previously described.21 To assess clonogenic growth, primary AML cells or granulocyte colony-stimulating factor (G-CSF) mobilized PBSCs (6.25 × 105/mL) were treated with cyproheptadine or buffer control for 24 hours. After treatment, cells were washed and equal volumes were plated in triplicate in MethoCult GF H4434 medium (StemCell Technologies, Vancouver, BC) containing 1% methycellulose in IMDM, 30% FCS, 1% bovine serum albumin, 3 U/mL of recombinant human erythropoietin, 10−4 M of 2-mercaptoethanol, 2 mM of l-glutamine, 50 ng/mL of recombinant human stem cell factor, 10 ng/mL of GM-CSF, and 10 ng/mL of rh IL-3. Seven days (AML samples) or 14 days (normal PBCS) after plating, the number of colonies containing 20 or more cells was counted as previously described.19,22
Luciferase assay for cyclin-D2 transactivation
Full-length c-maf cDNA was subcloned into an IRES-GFP-MIEV retroviral vector. NIH3T3 cells were infected with this construct, and stable cells expressing GFP and c-maf were selected by flow cytometry and immunoblotting, respectively. The promoter of cyclin D2 (−894 to −4), containing c-maf responsive element sequence (MARE), was cloned from HeLa cell genomic DNA and subcloned into the pGL2 luciferase reporter vector (Promega). This construct was cotransfected with pcDNA3.1 containing a neomycin resistance gene into NIH3T3 wild-type cells and NIH3T3 cells stably overexpressing c-maf-IRES-GFP. Cells stably expressing c-maf, GFP, and luciferase were selected for further application.
Luciferase activity was measured according to the manufacturer's instructions (Promega) and as previously described.14 Briefly, the cell culture medium was removed using an EMBLA plate washer (Molecular Devices, Sunnyvale, CA), and 1X Glo Lysis buffer (Promega) was added by the robotic liquid handler. After 10 minutes of incubation, an equal volume of Bright-Glo Luciferase substrate (Promega) was added, and the luminescence signal was detected with a 96-well Luminoskan luminescence plate reader (Thermo Electron, Waltham, MA) with a 5-second integration.
Immunoblotting
Whole cell lysates were prepared from myeloma and leukemia cells as described previously.18 Briefly, cells were washed with phosphate-buffered saline (PBS, pH 7.4) and resuspended in lysis buffer (10 mM Tris [pH 7.4], 150 mM NaCl, 0.1% Triton ×-100, 0.5% sodium deoxycholate, and 5 mM ethylenediaminetetraacetic acid) containing protease inhibitors (Complete tablets; Roche Diagnostics, Indianapolis, IN). Protein concentrations were determined by the Bradford assay. Equal amounts of protein were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels followed by transfer to polyvinylidene difluoride membranes. Membranes were probed with antibodies including monoclonal anti–human cyclin D1 (1:200 vol/vol; Cell Signaling Technology, Danvers, MA), polyclonal anti–human cyclin D2, rabbit anti-C/EBPA, monoclonal anti-APA2A (1:2000 vol/vol), p21 (1:400 vol/vol; Santa Cruz Biotechnology, Santa Cruz, CA), monoclonal anti–human cyclin D3 (1:200 vol/vol; BD PharMingen, San Jose, CA), polyclonal anti–human caspase-3 (1:5000 vol/vol; Cell Signaling Technology), monoclonal anti–human caspase-9 (1:3000 vol/vol; R&D Systems, Minneapolis, MN), monoclonal anti–human caspase-8 (1:3000 vol/vol; BD PharMingen), monoclonal anti-HA (1:400 vol/vol) and p53 (1:400 vol/vol; Cell Signaling Technology), rabbit anti–human SP1 (1:1000 vol/vol; Upstate Biotechnology, Charlottesville, VA), or monoclonal anti-β actin (1:10 000 vol/vol; Sigma-Aldrich) followed by secondary horseradish peroxidase–conjugated goat antimouse (1:10 000 vol/vol) or anti–rabbit IgG (1:5000 vol/vol; GE Healthcare, Little Chalfont, United Kingdom). Detection was performed by the enhanced chemical luminescence method (Pierce, Rockford, IL). Protein levels were semiquantified by densitometry (Quantity One; Bio-Rad, Hercules, CA). Density was expressed as (Protein/Actin) and normalized to the control cells.
Reverse-transcriptase real-time PCR
First-strand cDNA was synthesized from 1 μg DNase-treated total cellular RNA using random primers and SuperScript II reverse transcriptase (Invitrogen) according to the manufacturer's protocols. Real-time PCR assays were performed in triplicate with 100 ng of RNA equivalent cDNA, SYBR Green PCR Master mix (Applied Biosystems, Foster City, CA), and 400 nM of gene-specific primers. Reactions were processed and analyzed on an ABI 7700 Sequence Detection System (Applied Biosystems). Forward/reverse PCR primer pairs for human cDNAs were as follows: cyclin D2 (5′-TGCAGAAGGACATCCAACC-3′/5′-AGGAACATGCAGACAGCACC-3′); cyclin D3 (5′-AGTATGGAGCTGCTGTGT TG C-3′ /5′-AAGACTTCCTCCTCACAGCG-3′); 18S (5′-GGACATCTAAGGG-CATCACA-3′/5′-AGGAATTGACGGAAGGGCAC-3′). Relative mRNA expression was determined using the ΔΔCT method as described.18
Cell-cycle analysis
Cell-cycle analysis was performed as previously described.6 Briefly, cells were harvested, washed with cold PBS, resuspended in 70% cold ethanol, and incubated overnight at −20°C. Cells were then treated with 100 ng/mL of DNase-free RNase (Invitrogen) at 37°C for 30 minutes, washed with cold PBS, and resuspended in PBS with 50 μg/mL of PI. DNA content was analyzed by flow cytometry (FACSCalibur; BD Biosciences, San Jose, CA). The percentage of cells in each phase of the cell cycle was calculated with ModFit software (Verity Software House, Topsham, ME).
Mitochondrial membrane potential (ΔΨM) assessment
Changes in mitochondrial membrane potential (ΔΨM) were detected by staining cells with 40 nM of DiIC1(5) (1,1′3,3,3′,3′hexamethylindodicarbocyanine) (Invitrogen) and 5 μg/mL of PI as previously described.23 Cells stained with DiIC1(5) were incubated at 37°C for 30 minutes and then analyzed by flow cytometry (Coulter Epics Elite; Beckman Coulter, Fullerton, CA) by exciting at 633 nm and measuring through a 675 plus or minus 20 nm bandpass filter.
Assessment of cyproheptadine's antileukemic and antimyeloma activity in vivo
MDAY-D2 (MDAY) murine leukemia cells (5 × 105) were injected intraperitoneally into DBA2 mice (The Jackson Laboratory, Bar Harbor, ME). Mice were then treated with cyproheptadine at 50 mg/kg per day in PBS with 2% ethanol or vehicle control intraperitoneally for 5 days. Ten days after injection of cells, mice were killed, and the volume and cell count in the malignant ascites was measured.
LP-1 MM cells (10 × 106) were injected subcutaneously into the flank of sublethally irradiated (3.5 Gy) NOD/SCID mice (Ontario Cancer Institute, Toronto, ON). When tumors where palpable, mice were treated with 36 mg/kg of cyproheptadine daily in PBS with 2% dimethyl sulfoxide or vehicle control intraperitoneally daily for 7 weeks. Tumor volume (tumor length × width2 × 0.5236)24 was measured weekly using calipers. Mouse body weight and blood counts were also monitored weekly.
To analyze the effects of cyproheptadine on levels D-cyclins in xenografted tumor tissues, LP1 or MDAY-D2 cells were injected subcutaneously into sublethally irradiated NOD/SCID mice. When tumors are palpable, mice were treated intraperitoneally with cyproheptadine (10 mg/kg per day) in Tween 80:0.9% NaCl (1:95) or vehicle control. Five (MDAY-D2) or 10 days (LP1) after cyproheptadine administration, mice were killed, and the tumors were excised and weighed; 20 mg of tumor sample was lysed in radioimmunoprecipitation assay buffer with a homogenizer. After clarification by high-speed centrifugation, equal protein amounts of the clear supernatant were analyzed for cyclin D2 and cyclin D3 expression by immunoblotting.
All animal studies were carried out according to the regulations of the Canadian Council on Animal Care and with the approval of the local ethics review board.
Statistical analysis
Results were expressed as mean plus or minus SD. Treatment effects were compared using Student t test, and differences between means were considered to be significant when P was less than .05. All in vitro experiments were repeated at least 3 times.
Results
Cyproheptadine reduces D-cyclin expression in myeloma and leukemia cell lines
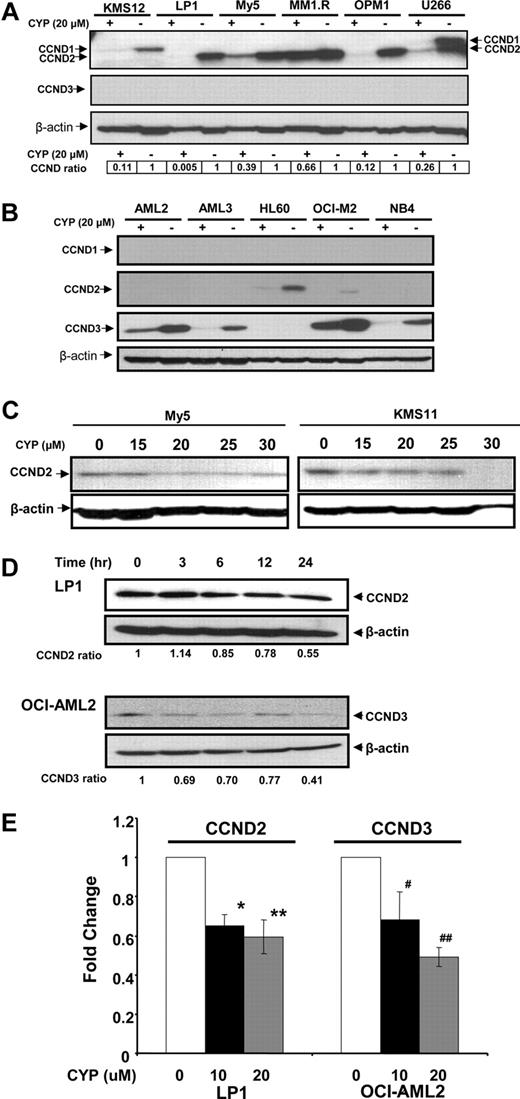
Previously, we used a chemical biology approach to identify c-maf-dependent and -independent inhibitors of transactivation of the cyclin D2 promoter.14 From this screen, we identified cyproheptadine as a c-maf-independent inhibitor of cyclin D2 transactivation14 (Figure S1, available on the Blood website; see the Supplemental Materials link at the top of the online article). To further examine cyproheptadine, we evaluated its effects on D-cyclin expression in myeloma and leukemia cells. Myeloma and leukemia cell lines were treated with cyproheptadine for 24 hours. After incubation, levels of cyclin D1, D2, and D3 protein were measured by immunoblotting (Figure 1). D-cyclin expression varied among the cell lines, but each cell line expressed at least one D-cyclin. By immunoblotting, cyproheptadine suppressed expression of all of the D-cyclins expressed in the tumor cells. Decreased levels of cyclin D2 and cyclin D3 mRNA were also observed in LP-1 and OCI-AML2 cells, respectively, consistent with our identification of cyproheptadine through a screen for inhibitors of D-cyclin transactivation (Figure 1E). Decreased D-cyclin levels occurred across a range of events that dysregulated D-cyclins, including cyclin D1 translocation (U266),25 c-maf overexpression (OCI-My5, LP-1, and KMS11),14,26 and FGFR3 translocation (KMS11).27
Cyproheptadine reduces expression of cyclin D2 in multiple myeloma lines. (A) KMS12, LP-1, OCI-MY5, MM1.R, OPM1, and U266 multiple myeloma cells and (B) OCI-AML2, OCI-AML3, HL60, OCI-M2, and NB4 leukemia cells (106) were treated with 20 μM of cyproheptadine (CYP). Twenty-four hours after incubation, cell lysates were prepared, normalized for total protein, and analyzed by SDS-PAGE/immunoblotting using antibodies specific for cyclin D1, D2, D3, and anti–β-actin. CCND ratio represents (density of the D-cyclin)/(density of β-actin) relative to control cells. (C) OCI-MY5 and KMS11 myeloma cells (106) were treated with increasing concentrations of cyproheptadine. Twenty-four hours after incubation, cell lysates were prepared, normalized for total protein, and analyzed by SDS-PAGE/immunoblotting using antibodies specific for cyclin D2 and anti–β-actin. (D) LP1 myeloma and OCI-AML2 leukemia cells (106) were treated with 15 μM of cyproheptadine. At increasing times after incubation, cell lysates were prepared, normalized for total protein, and analyzed by SDS-PAGE/immunoblotting using antibodies specific for cyclin D2 and cyclin D3. CCND ratio represents (density of the D-cyclin)/(density of β-actin) relative to control cells. (E) LP1 myeloma and OCI-AML2 leukemia cells (106) were treated with 10 and 20 μM of cyproheptadine. Twenty-four hours after incubation, total cellular RNA was isolated. Cyclin D2 (CCND2) and Cyclin D3 (CCND3) mRNA expression were measured relative to 18S RNA by real-time RT-PCR. Data represent the mean plus or minus SEM fold change of CCND2/18S or CCND3/18S expression relative to untreated controls (ΔΔCT normalization) from 4 independent experiments. *P = .017, **P = .004, #P = .015, ##P = .006, all by Student t test.
Cyproheptadine reduces expression of cyclin D2 in multiple myeloma lines. (A) KMS12, LP-1, OCI-MY5, MM1.R, OPM1, and U266 multiple myeloma cells and (B) OCI-AML2, OCI-AML3, HL60, OCI-M2, and NB4 leukemia cells (106) were treated with 20 μM of cyproheptadine (CYP). Twenty-four hours after incubation, cell lysates were prepared, normalized for total protein, and analyzed by SDS-PAGE/immunoblotting using antibodies specific for cyclin D1, D2, D3, and anti–β-actin. CCND ratio represents (density of the D-cyclin)/(density of β-actin) relative to control cells. (C) OCI-MY5 and KMS11 myeloma cells (106) were treated with increasing concentrations of cyproheptadine. Twenty-four hours after incubation, cell lysates were prepared, normalized for total protein, and analyzed by SDS-PAGE/immunoblotting using antibodies specific for cyclin D2 and anti–β-actin. (D) LP1 myeloma and OCI-AML2 leukemia cells (106) were treated with 15 μM of cyproheptadine. At increasing times after incubation, cell lysates were prepared, normalized for total protein, and analyzed by SDS-PAGE/immunoblotting using antibodies specific for cyclin D2 and cyclin D3. CCND ratio represents (density of the D-cyclin)/(density of β-actin) relative to control cells. (E) LP1 myeloma and OCI-AML2 leukemia cells (106) were treated with 10 and 20 μM of cyproheptadine. Twenty-four hours after incubation, total cellular RNA was isolated. Cyclin D2 (CCND2) and Cyclin D3 (CCND3) mRNA expression were measured relative to 18S RNA by real-time RT-PCR. Data represent the mean plus or minus SEM fold change of CCND2/18S or CCND3/18S expression relative to untreated controls (ΔΔCT normalization) from 4 independent experiments. *P = .017, **P = .004, #P = .015, ##P = .006, all by Student t test.
Cyproheptadine arrests cells in the G1 phase of the cell cycle
D-cyclins are required for cell commitment to proliferation and entry to the S phase of the cell cycle. Decreasing D-cyclin expression is associated with G1 arrest.28-30 Because cyproheptadine decreased levels of D-cyclins, we tested the effects of cyproheptadine on cell-cycle progression. Myeloma and leukemia cells were treated with increasing concentrations of cyproheptadine, and the percentage of cells in the different phases of the cell cycle was measured by PI staining and analysis by flow cytometry. Consistent with its effect on D-cyclin gene and protein expression, cyproheptadine arrested myeloma and leukemia cells in the G0/G1 phase (Figure 2). Cyproheptadine induced cell-cycle arrest in a dose-dependent manner and at concentrations that were similar to those required to decrease the expression of D-cyclins. Cell-cycle arrest was first detected at 12 hours after treatment with cyproheptadine, a time point later than the time required to reduce D-cyclin expression.
Cyproheptadine arrests cell lines in the G1 phase of the cell cycle. LP1 cells (A) and OCI-AML2 cells (B) (2 × 106) were treated with 5 and 20 μM of cyproheptadine. Twenty-four hours after treatment, the percentage of cells in each phase of the cell cycle was determined by PI staining and flow cytometry. A representative histogram is shown. (C) LP1 cells (2 × 106) were treated with 20 μM of cyproheptadine. At increasing times of incubation, the percentage of cells in the G0/G1 phase of the cell cycle was determined by PI staining and flow cytometry. Data represent the mean plus or minus SD percentage of cells of in the G0/G1 phase. (D) OCI-AML2 cells (1.5 × 106) were pretreated with 10 and 20 μM of cyproheptadine overnight, followed by incubation with 20 μM of BrdU for 1 hour. Labeled cells were analyzed on a flow cytometer as described in “Cell-cycle analysis.” A representative histogram is shown. (E) MM1.S and MM1.R myeloma cells (2 × 106) were treated with increasing concentrations of cyproheptadine. Twenty-four hours after treatment, cell lysates were prepared, normalized for total protein, and analyzed by SDS-PAGE/immunoblotting using antibodies specific for p53, p21, and anti–β-actin. (F) KMS11 and LP1 myeloma cells and OCI-AML2 and Jurkat leukemia cells (2 × 106) were treated with increasing concentrations of cyproheptadine. Twenty-four hours after treatment, cell lysates were prepared, normalized for total protein, and analyzed by SDS-PAGE/immunoblotting using antibodies specific for SP1, APA2, C/EBPA, cyclin D2 (CCND2), cyclin D3 (CCND3), and anti–β-actin. APA2, CCND2, and CCND3 ratios represent (density of the protein)/(density of β-actin) relative to control cells. (G) LP1 myeloma and OCI-AML2 leukemia (2 × 106) were treated with 20 μM of cyproheptadine. At increasing times after incubation, cell lysates were prepared, normalized for total protein, and analyzed by SDS-PAGE/immunoblotting using antibodies specific for APA2 and anti–β-actin.
Cyproheptadine arrests cell lines in the G1 phase of the cell cycle. LP1 cells (A) and OCI-AML2 cells (B) (2 × 106) were treated with 5 and 20 μM of cyproheptadine. Twenty-four hours after treatment, the percentage of cells in each phase of the cell cycle was determined by PI staining and flow cytometry. A representative histogram is shown. (C) LP1 cells (2 × 106) were treated with 20 μM of cyproheptadine. At increasing times of incubation, the percentage of cells in the G0/G1 phase of the cell cycle was determined by PI staining and flow cytometry. Data represent the mean plus or minus SD percentage of cells of in the G0/G1 phase. (D) OCI-AML2 cells (1.5 × 106) were pretreated with 10 and 20 μM of cyproheptadine overnight, followed by incubation with 20 μM of BrdU for 1 hour. Labeled cells were analyzed on a flow cytometer as described in “Cell-cycle analysis.” A representative histogram is shown. (E) MM1.S and MM1.R myeloma cells (2 × 106) were treated with increasing concentrations of cyproheptadine. Twenty-four hours after treatment, cell lysates were prepared, normalized for total protein, and analyzed by SDS-PAGE/immunoblotting using antibodies specific for p53, p21, and anti–β-actin. (F) KMS11 and LP1 myeloma cells and OCI-AML2 and Jurkat leukemia cells (2 × 106) were treated with increasing concentrations of cyproheptadine. Twenty-four hours after treatment, cell lysates were prepared, normalized for total protein, and analyzed by SDS-PAGE/immunoblotting using antibodies specific for SP1, APA2, C/EBPA, cyclin D2 (CCND2), cyclin D3 (CCND3), and anti–β-actin. APA2, CCND2, and CCND3 ratios represent (density of the protein)/(density of β-actin) relative to control cells. (G) LP1 myeloma and OCI-AML2 leukemia (2 × 106) were treated with 20 μM of cyproheptadine. At increasing times after incubation, cell lysates were prepared, normalized for total protein, and analyzed by SDS-PAGE/immunoblotting using antibodies specific for APA2 and anti–β-actin.
Consistent with an arrest in the G0/G1 phase, cyproheptadine also decreased cellular proliferation as measured by BrdU uptake (Figure 2D). We also measured changes in other cell-cycle regulatory proteins and detected an increase in p21 expression, but no significant change in levels of p53 (Figure 2E).
To assess upstream regulators of D-cyclin that may be affected by cyproheptadine, we searched for putative transcription factor binding sites that were present to the 1640-bp fragment of the cyclin D2 promoter used in the initial high throughput screen14 as well as the cyclin D1 and D3 promoters. From this search, we identified 7 transcription factors, of which APA2, C/EBPA, and Sp-1 had the greatest number of putative binding sites. To determine whether cyproheptadine altered the expression of these proteins, KMS11, LP1, Jurkat, and OCI-AML2 cells were treated with increasing concentrations of cyproheptadine; and 24 hours after treatment, levels of APA2, C/EBPA, and Sp-1 were measured by immunoblotting. Except for Jurkat cells, cyproheptadine decreased expression of APA2 at concentrations and times that preceded reductions in D-cyclin expression. Reductions in C/EBPA and Sp-1 were also seen, but these changes were not observed in all cell lines and occurred at times and concentrations after reductions in D-cyclins were detected (Figure 2F,G).
Cyproheptadine induces apoptosis in MM cell lines and primary patient samples
Reductions in D-cyclins and G0/G1 arrest can induce apoptosis in malignant cells.31,32 Therefore, we tested the effects of cyproheptadine on the viability of myeloma and leukemia cells. Myeloma and leukemia cell lines were treated with increasing concentrations of cyproheptadine, and cell viability was measured by the MTS assay after 24, 48, and 72 hours of incubation. Cyproheptadine reduced the number of viable myeloma and leukemia cell lines, such as LP-1, KMS11, OCI-AML1, OCI-AML2, and OCI-AML4 cells at 72 hours with an IC50 in the low micromolar range. In contrast, it was less toxic to nonleukemic and nonmyeloma cells, such as HeLa or NIH3T3 cells with an IC10 more than 30 μM (data not shown). The decreased number of viable cells by MTS assay was associated with a concentration- and time-dependent increase in apoptosis as measured by annexin V–FITC staining (Figures 3C, S2). The concentration of cyproheptadine required to induce cell death matched the concentrations of cyproheptadine required to decrease D-cyclins and arrest cells in the G0/G1 phase. In cell lines such as LP1, 20 μM of cyproheptadine reduced cell number by approximately 60%. In contrast, in MM1.R cells, only slight reductions in cell viability and D-cyclin expression were observed at 20 μM of cyproheptadine. However, in this cell line, cell number was reduced at 30 μM, and reductions in D-cyclin expression were also detected (Figures 1, 3; and data not shown). Moreover, reductions in D-cyclins occurred at times that preceded cell-cycle arrest, suggesting that the G0/G1 arrest is secondary to the decrease in D-cyclins.
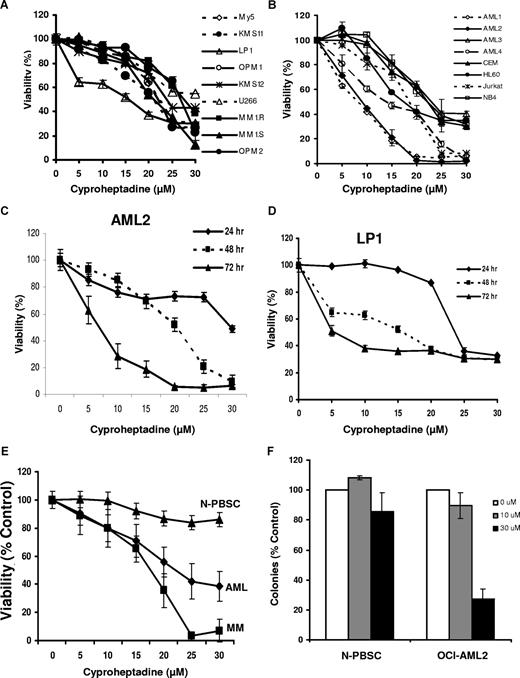
Cyproheptadine reduces the viability of myeloma and leukemia cell lines and primary patient samples. (A) Multiple myeloma cell lines (1.5 × 104/well) were seeded in 96-well plates and then treated with increasing concentrations of cyproheptadine. Cell viability was measured at 72 hours by MTS assay. Cell viability is expressed as a mean percentage plus or minus SD (n = 3) relative to untreated cells. (B) Leukemia cell lines were treated with increasing concentrations of cyproheptadine. Cell viability was measured at 72 hours by MTS assay. Cell viability is expressed as a mean percentage plus or minus SD (n = 3) relative to untreated cells. (C) OCI-AML2 (AML2) leukemia and (D) LP-1 myeloma cells were treated with increasing concentrations of cyproheptadine. At increasing incubation time, cell viability was measured by MTS assay. Cell viability is expressed as a mean percentage plus or minus SD (n = 3) relative to untreated cells at that time point. (E) Mononuclear cells from the peripheral blood of patients with AML (n = 9), the marrow of patients with myeloma (n = 8), or from the peripheral blood of volunteers donating G-CSF–mobilized hematopoietic stem cells for allotransplantation (N-PBSC; n = 5) were treated with increasing concentrations of cyproheptadine for 48 hours. After treatment, apoptosis was measured by staining with annexin V-FITC. Mononuclear cells from the marrow of patients with multiple myeloma were costained with phycoerythrin-labeled anti-CD138 to identify the plasma cells and the percentage of CD138+/annexin V− cells was quantified as a marker of myeloma cell viability. Data represent the mean percentage plus or minus SEM of viable cells relative to untreated control cells. (F) Mononuclear cells from patients with AML (n = 3) or from volunteers donating PBSC (n = 3) were treated with increasing concentrations of cyproheptadine for 24 hours. After treatment, cells were washed, plated in MethoCult, and counted as described in “Cell viability, apoptosis, proliferation, and clonogenic growth assays.” Data represent the mean percentage plus or minus SD of colonies relative to untreated control cells.
Cyproheptadine reduces the viability of myeloma and leukemia cell lines and primary patient samples. (A) Multiple myeloma cell lines (1.5 × 104/well) were seeded in 96-well plates and then treated with increasing concentrations of cyproheptadine. Cell viability was measured at 72 hours by MTS assay. Cell viability is expressed as a mean percentage plus or minus SD (n = 3) relative to untreated cells. (B) Leukemia cell lines were treated with increasing concentrations of cyproheptadine. Cell viability was measured at 72 hours by MTS assay. Cell viability is expressed as a mean percentage plus or minus SD (n = 3) relative to untreated cells. (C) OCI-AML2 (AML2) leukemia and (D) LP-1 myeloma cells were treated with increasing concentrations of cyproheptadine. At increasing incubation time, cell viability was measured by MTS assay. Cell viability is expressed as a mean percentage plus or minus SD (n = 3) relative to untreated cells at that time point. (E) Mononuclear cells from the peripheral blood of patients with AML (n = 9), the marrow of patients with myeloma (n = 8), or from the peripheral blood of volunteers donating G-CSF–mobilized hematopoietic stem cells for allotransplantation (N-PBSC; n = 5) were treated with increasing concentrations of cyproheptadine for 48 hours. After treatment, apoptosis was measured by staining with annexin V-FITC. Mononuclear cells from the marrow of patients with multiple myeloma were costained with phycoerythrin-labeled anti-CD138 to identify the plasma cells and the percentage of CD138+/annexin V− cells was quantified as a marker of myeloma cell viability. Data represent the mean percentage plus or minus SEM of viable cells relative to untreated control cells. (F) Mononuclear cells from patients with AML (n = 3) or from volunteers donating PBSC (n = 3) were treated with increasing concentrations of cyproheptadine for 24 hours. After treatment, cells were washed, plated in MethoCult, and counted as described in “Cell viability, apoptosis, proliferation, and clonogenic growth assays.” Data represent the mean percentage plus or minus SD of colonies relative to untreated control cells.
Given the effects of cyproheptadine on myeloma and leukemia cell lines, we extended our studies to evaluate the effects of cyproheptadine on primary myeloma and AML samples. Bone marrow and peripheral blood samples were obtained from consenting patients with myeloma (n = 8) and AML (n = 9), respectively. As controls, normal hematopoietic stem cells (n = 5) were obtained from peripheral blood samples from G-CSF–treated volunteers donating peripheral blood stem cells for allotransplantation. Mononuclear cells were isolated from these samples and treated with increasing concentrations of cyproheptadine. Forty-eight hours after treatment, cell viability was measured by annexin V staining. Cyproheptadine reduced the viability of primary myeloma and AML samples but was less toxic to normal hematopoietic cells (Figure 3E).
In addition to the short-term cytotoxicity assays, we evaluated the effects of cyproheptadine on the clonogenic growth of primary AML and normal hematopoietic cells. Cyproheptadine inhibited the clonogenic survival of the AML samples (n = 3) but not the normal hematopoietic cells (n = 3; Figure 3F).
Cyproheptadine abolishes malignant ascites formation and inhibits tumor growth in xenograft models
Given the selectivity of cyproheptadine cytotoxicity toward malignant cell lines and primary patient samples, we explored its efficacy in vivo using 2 separate mouse models. DBA2 mice were injected intraperitoneally with MDAY-D2 mouse leukemia cells and treated with cyproheptadine (40 mg/kg per day over 5 days) or vehicle control. Significantly, treatment with cyproheptadine completely abolished the formation of malignant ascites in this mouse model (Figure 4A,B) without decreasing body weight (data not shown).
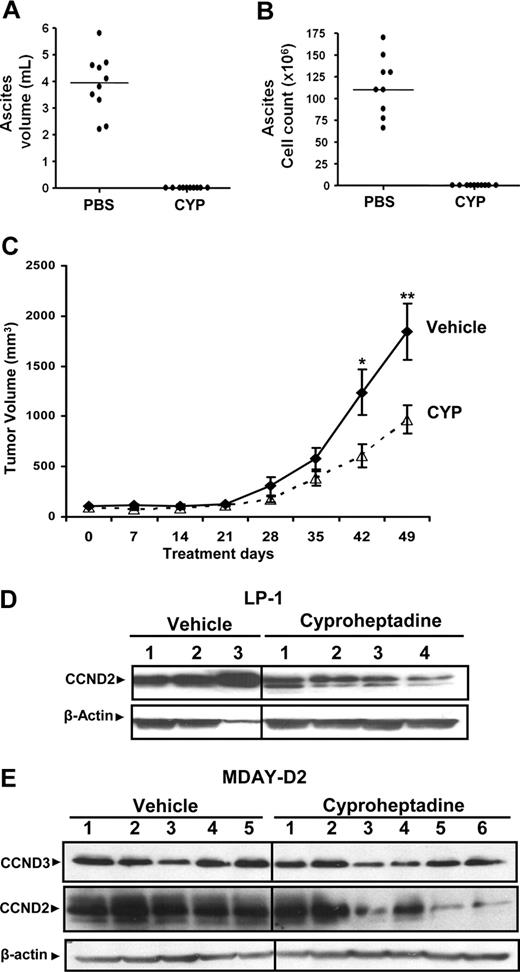
Cyproheptadine inhibits growth of leukemia and myeloma cells in mouse models. MDAY-D2 cells (5 × 105) were injected intraperitoneally into DBA2 mice. Mice were then treated with cyproheptadine (CYP; 40 mg/kg per day; n = 10) or vehicle control (n = 10) intraperitoneally for 1 week. At the end of treatment, the mice were killed. (A) The volume of malignant ascites in the peritoneal cavity was measured. (B) The total number of malignant MDAY-D2 cells in the malignant ascites was counted. The bar represents the median of the population. (C) LP-1 myeloma cells (10 × 106) were injected subcutaneously into the flank of sublethally irradiated NOD/SCID mice. Two weeks after injection, mice were treated with cyproheptadine 36 mg/kg (n = 10) or vehicle control (n = 10) as described in “Assessment of cyproheptadine's antileukemic and antimyeloma activity in vivo.” Tumor volume was measured weekly with a caliper. Data represent the mean plus or minus SEM. *P = .048, **P = .032, by the Student t test. (D) LP-1 myeloma cells (10 × 106) were injected subcutaneously into the flank of sublethally irradiated NOD/SCID mice. When tumors were palpable, mice were treated intraperitoneally with cyproheptadine (10 mg/kg per day) or vehicle control. Ten days after treatment, mice were killed and the tumor was excised. From the excised tumors, cell lysates were prepared, normalized for total protein, and analyzed by SDS-PAGE/immunoblotting using antibodies specific for cyclin D2 (CCND2) and anti–β-actin. Numbers represent tumors from individual mice. (E) MDAY-D2 murine leukemia cells (5 × 105) were injected subcutaneously into the flank of sublethally irradiated NOD/SCID mice. When tumors were palpable, mice were treated intraperitoneally with cyproheptadine (10 mg/kg per day) or vehicle control. Five days after treatment, mice were killed and the tumor was excised. From the excised tumors, cell lysates were prepared, normalized for total protein, and analyzed by SDS-PAGE/immunoblotting using antibodies specific for cyclin D2 (CCND2), cyclin D3 (CCND3), and anti–β-actin. Numbers represent tumors from individual mice.
Cyproheptadine inhibits growth of leukemia and myeloma cells in mouse models. MDAY-D2 cells (5 × 105) were injected intraperitoneally into DBA2 mice. Mice were then treated with cyproheptadine (CYP; 40 mg/kg per day; n = 10) or vehicle control (n = 10) intraperitoneally for 1 week. At the end of treatment, the mice were killed. (A) The volume of malignant ascites in the peritoneal cavity was measured. (B) The total number of malignant MDAY-D2 cells in the malignant ascites was counted. The bar represents the median of the population. (C) LP-1 myeloma cells (10 × 106) were injected subcutaneously into the flank of sublethally irradiated NOD/SCID mice. Two weeks after injection, mice were treated with cyproheptadine 36 mg/kg (n = 10) or vehicle control (n = 10) as described in “Assessment of cyproheptadine's antileukemic and antimyeloma activity in vivo.” Tumor volume was measured weekly with a caliper. Data represent the mean plus or minus SEM. *P = .048, **P = .032, by the Student t test. (D) LP-1 myeloma cells (10 × 106) were injected subcutaneously into the flank of sublethally irradiated NOD/SCID mice. When tumors were palpable, mice were treated intraperitoneally with cyproheptadine (10 mg/kg per day) or vehicle control. Ten days after treatment, mice were killed and the tumor was excised. From the excised tumors, cell lysates were prepared, normalized for total protein, and analyzed by SDS-PAGE/immunoblotting using antibodies specific for cyclin D2 (CCND2) and anti–β-actin. Numbers represent tumors from individual mice. (E) MDAY-D2 murine leukemia cells (5 × 105) were injected subcutaneously into the flank of sublethally irradiated NOD/SCID mice. When tumors were palpable, mice were treated intraperitoneally with cyproheptadine (10 mg/kg per day) or vehicle control. Five days after treatment, mice were killed and the tumor was excised. From the excised tumors, cell lysates were prepared, normalized for total protein, and analyzed by SDS-PAGE/immunoblotting using antibodies specific for cyclin D2 (CCND2), cyclin D3 (CCND3), and anti–β-actin. Numbers represent tumors from individual mice.
In a second experiment, we evaluated the in vivo efficacy of cyproheptadine against a xenograft model of myeloma. LP-1 myeloma cells were injected subcutaneously into the flanks of sublethally irradiated NOD/SCID mice. Two weeks later, once tumors were established, mice were treated with cyproheptadine or vehicle control. In this model, cyproheptadine delayed tumor growth with an approximately 2-fold reduction in tumor volume at the end of treatment (Figure 4C). Similar results were also obtained in xenograft animals bearing JJN3 myeloma cells (data not shown). Of note, cyproheptadine did not reduce the animals' body weight or complete blood counts (data not shown).
Next, we explored whether cyproheptadine decreased D-cyclin expression in xenografted tumors. Here, LP1 and MDAY-D2 cells were injected subcutaneously into NOD/SCID mice. When tumors are palpable, mice were treated intraperitoneally with cyproheptadine (10 mg/kg per day) or vehicle control. Five (MDAY-D2) or 10 days (LP1 cells) after cyproheptadine administration, mice were killed, the tumors were excised and lysed, and cyclin D2 and cyclin D3 were expressed by immunoblotting. Intraperitoneal treatment with cyproheptadine decreased expression of cyclin D2 and cyclin D3 in xenografted tumors, consistent with its effects on cells in culture (Figure 4D,E).
Cyproheptadine induces apoptosis via the mitochondrial pathway of caspase activation and independent of histamine H1 or serotonin receptor antagonism
To explore the mechanism of cyproheptadine's toxicity in malignant cells, we first evaluated the effects of the pan caspase inhibitor benzoyl-Val-Ala-Asp-fluoromethylketone (z-VAD-fmk; Enzyme Systems, Dublin, CA) on cyproheptadine-induced cell death. OCI-MY5 myeloma cells were treated with increasing concentrations of cyproheptadine with and without z-VAD-fmk. Cell viability was measured at 48 hours by MTS assay. z-VAD-fmk completely inhibited cyproheptadine-induced cell death, consistent with the induction of caspase-dependent apoptosis (Figure 5A).
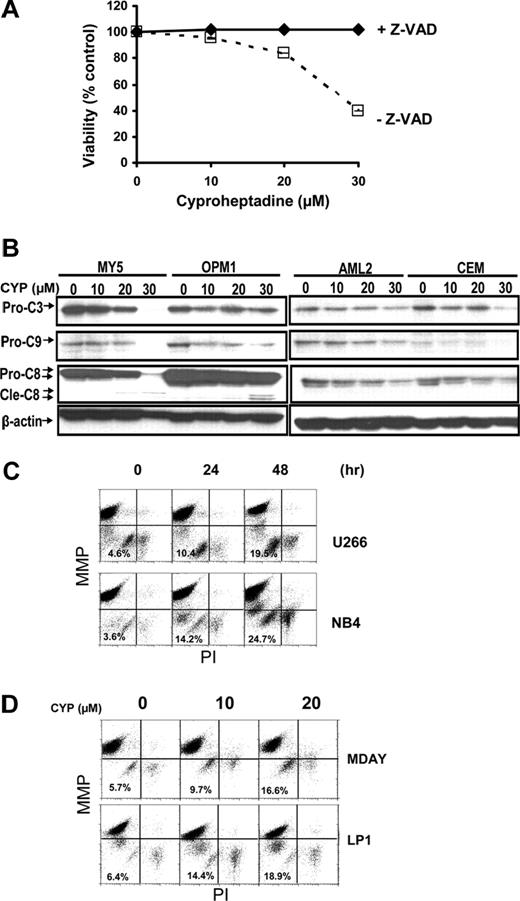
Cyproheptadine activates the mitochondrial pathway of caspase activation. (A) OCI-MY5 myeloma cells (2 × 104) were treated with increasing concentrations of cyproheptadine (CYP) with and without the pan-caspase inhibitor z-VAD-fmk (100 μM). Forty-eight hours after incubation, cell viability was measured by MTS assay. Data represent the mean percentage plus or minus SD of viable cells compared with control treated controls. (B) OCI-MY5 and OPM1 myeloma cells and OCI-AML2 and CEM leukemia cells were treated with increasing concentrations of cyproheptadine. Twenty-four hours after treatment, total cellular protein was isolated and analyzed by SDS-PAGE/immunoblotting using anti–caspase-3 (Pro-C3), anti–caspase-8 (Pro-C8), anti–caspase-9 (Pro-C9), and anti–β-actin. Cle-C8 indicates cleaved caspase-8. (C) U266 and NB4 cells (2 × 106) were treated with cyproheptadine (CYP, 20 μM) for increasing times. After incubation, cells were harvested and stained with DiIC1(5) and PI to measure mitochondrial membrane potential (ΔΨM) and plasma membrane integrity, respectively. Results were analyzed by flow cytometry. (D) MDAY-D2 and LP-1 cells (2 × 106 cells) were treated with increasing concentrations of cyproheptadine. Forty-eight hours after incubation, cells were harvested and stained with DiIC1(5) and PI to measure mitochondrial membrane potential (ΔΨM). Results were analyzed by flow cytometry.
Cyproheptadine activates the mitochondrial pathway of caspase activation. (A) OCI-MY5 myeloma cells (2 × 104) were treated with increasing concentrations of cyproheptadine (CYP) with and without the pan-caspase inhibitor z-VAD-fmk (100 μM). Forty-eight hours after incubation, cell viability was measured by MTS assay. Data represent the mean percentage plus or minus SD of viable cells compared with control treated controls. (B) OCI-MY5 and OPM1 myeloma cells and OCI-AML2 and CEM leukemia cells were treated with increasing concentrations of cyproheptadine. Twenty-four hours after treatment, total cellular protein was isolated and analyzed by SDS-PAGE/immunoblotting using anti–caspase-3 (Pro-C3), anti–caspase-8 (Pro-C8), anti–caspase-9 (Pro-C9), and anti–β-actin. Cle-C8 indicates cleaved caspase-8. (C) U266 and NB4 cells (2 × 106) were treated with cyproheptadine (CYP, 20 μM) for increasing times. After incubation, cells were harvested and stained with DiIC1(5) and PI to measure mitochondrial membrane potential (ΔΨM) and plasma membrane integrity, respectively. Results were analyzed by flow cytometry. (D) MDAY-D2 and LP-1 cells (2 × 106 cells) were treated with increasing concentrations of cyproheptadine. Forty-eight hours after incubation, cells were harvested and stained with DiIC1(5) and PI to measure mitochondrial membrane potential (ΔΨM). Results were analyzed by flow cytometry.
To better understand caspase activation by cyproheptadine, we measured levels of pro-caspases 3, 8, and 9 by immunoblotting after treatment with cyproheptadine. In both leukemia and myeloma cell lines, cyproheptadine caused reductions in all 3 caspases. However, reductions in pro-caspases 3 and 9 preceded reductions in pro-caspase-8 (Figure 5B). Activation of caspase 9 is associated with the mitochondrial pathway of caspase activation,33 so we examined the effects of cyproheptadine on the mitochondria. Changes in mitochondrial membrane potential were measured by flow cytometry after treatment with cyproheptadine. In a time- and dose-dependent manner, cyproheptadine caused loss of mitochondrial membrane potential before loss of plasma membrane integrity (Figure 5C,D; and data not shown). These results indicate activation of the mitochondrial caspase pathway by cyproheptadine.
Cyproheptadine is a histamine H1 and serotonin 5HT2 receptor antagonist.34,35 Therefore, we tested whether the addition of histamine or serotonin in excess could prevent cyproheptadine from inducing apoptosis. Preincubation of cells with high concentrations of histamine, serotonin, or a combination of histamine and serotonin did not abrogate cyproheptadine-induced cell death (Figure S3). Therefore, the proapoptotic activity of cyproheptadine is not the result of competitive inhibition of the H1 and/or serotonin receptors, suggesting that cyproheptadine has an additional molecular target that mediates its cytotoxic function.
To further verify that cyproheptadine does not exert its cell- cycle and cytotoxic effects via the histamine receptor, we compared its activity with a structurally unrelated antihistamine, triprolidine. Unlike cyproheptadine, triprolidine did not reduce the viability of leukemia or myeloma cells (Figure 6). Therefore, cyproheptadine's proapoptotic effects occur through a mechanism distinct from H1 receptor antagonism.
The structurally unrelated antihistamine triprolidine does not induce cell death. (A) The chemical structures of cyproheptadine and triprolidine are shown. (B) LP-1 and KMS11 myeloma and OCI-AML2 and Jurkat leukemia cells were treated with increasing concentrations of cyproheptadine (CYP) and triprolidine (TPL). Seventy-two hours after incubation, cell viability was measured by MTS staining. Data represent the mean percentage plus or minus SD of cell viability relative to untreated controls.
The structurally unrelated antihistamine triprolidine does not induce cell death. (A) The chemical structures of cyproheptadine and triprolidine are shown. (B) LP-1 and KMS11 myeloma and OCI-AML2 and Jurkat leukemia cells were treated with increasing concentrations of cyproheptadine (CYP) and triprolidine (TPL). Seventy-two hours after incubation, cell viability was measured by MTS staining. Data represent the mean percentage plus or minus SD of cell viability relative to untreated controls.
Discussion
In MM and acute leukemia, aberrant expression of D-cyclins stimulates progression of the cell cycle, thereby promoting cell growth and proliferation.4,36 Therefore, molecules that decrease D-cyclin expression would be useful chemical probes to better understand this biologic pathway and its relation to malignancy. In addition, some of these molecules could be leads for novel therapeutic agents. To identify such compounds, we conducted a chemical screen in NIH3T3 cells for compounds that inhibit transactivation of the cyclin D2 promoter.14 From this screen, we identified cyproheptadine that suppressed cyclin D2 transactivation even in the presence of increased levels of c-maf, an oncogenic transcription factor that directly binds the cyclin D2 promoter. In this report, we furthered our investigation of cyproheptadine and demonstrated that cyproheptadine decreased D-cyclin expression, arrested cells in the G0/G1 phase, and induced apoptosis in myeloma and leukemia cells.
Cyproheptadine preferentially induced cell death in malignant AML and myeloma cells over normal hematopoietic cells. Unfortunately, the D-cyclin expression profile was not available for these samples. In the future, it would be useful to determine whether the response to cyproheptadine in culture is related to levels of D-cyclin expression. Such data might be useful in developing biomarkers to predict patients most probable to respond to such drugs.
Given its selective cytotoxicity for malignant cells in culture and its ability to inhibit tumor growth in xenograft models, cyproheptadine represents a lead for a novel therapeutic agent for the treatment of myeloma and leukemia. Because the drug is well tolerated and already approved in multiple countries for clinical use as an antihistamine and appetite stimulant, it could be moved directly into clinical trials for cancer. It is unknown, however, whether antitumor concentrations of cyproheptadine can be obtained in humans. However, in healthy volunteers, a single dose of 5 mg produced a peak serum concentration of approximately 0.14 μM.37,38 However, patients have been routinely treated with cyproheptadine at doses up to 32 mg for the management of allergic conditions,39 and dose escalations to achieve antitumor concentrations have not been attempted. In addition, daily treatment with cyproheptadine could produce serum levels of the drug that are higher than those observed after a single dose because the elimination of cyproheptadine occurs slowly over 6 days.37 Finally, tissue concentrations of cyproheptadine exceed serum concentrations by up to 20-fold; and, in a patient who overdosed on cyproheptadine and ethanol, tissue concentrations of cyproheptadine were more than 4-fold higher than the concentration of cyproheptadine required to produce an antitumor effect in cultured cells.38 So, antitumor concentrations of cyproheptadine in malignant cells might be achievable after daily treatment with high doses of cyproheptadine. Thus, these factors and the ability to inhibit tumor growth in mice support the advancement of cyproheptadine as a therapeutic agent for the treatment of cancer. Finally, analogs of cyproheptadine could be developed that are more potent cytotoxins and more useful clinically for the treatment of malignancy.
Many mechanisms result in dysregulation of D-cyclins in malignancy, leading to increased cellular proliferation and chemoresistance. For example, in MM, overexpression of D-cyclins occurs through chromosomal translocation-dependent and -independent mechanisms. Translocations of the cyclin D1 and D3 genes with the IgH locus occur in 15% and 3% of patients with myeloma, respectively.36 Although translocations involving cyclin D2 and the IgH locus have not yet been reported in patients with myeloma, cyclin D2 is overexpressed in up to 80% of these patients.36 Overexpression of cyclin D2 can result from increased levels of the oncogene c-maf that directly promotes transcription of cyclin D2.14,26 Increased cyclin D2 expression can also result from enhanced signaling through the fibroblast growth factor receptor (FGFR) that promotes cyclin D expression via the MAPK phospho-signaling pathway.9,36,40 Targeting the transforming events that lead to increased cyclin D expression can decrease cellular proliferation and induce apoptosis. For example, inhibiting the FGFR signaling pathway leads to decreased levels of cyclin D41,42 and increased cell death in myeloma cells.43,44 Likewise, decreasing c-maf expression leads to decreased levels of cyclin D2.14 In our study, cyproheptadine decreased cyclin D expression and induced apoptosis in cells with a variety of transforming events, including cyclin D1 translocation (U266)25 , c-maf overexpression (JJN3),14,26 and FGFR3 translocation (KMS11).27 The ability of cyproheptadine to act across a broad spectrum of transforming events suggests that molecules such as this could be useful therapeutically in a wide spectrum of patients. These results also indicate that cyproheptadine reduces D-cyclin expression through a broad mechanism independent of specific transforming pathways.
In this study, cyproheptadine decreased expression of D-cyclin mRNA and protein. However, reductions in levels of D-cyclin proteins were greater than the reductions in D-cyclin mRNA. This difference may reflect the short half-life of D-cyclin proteins. However, we cannot exclude that cyproheptadine is also decreasing levels of D-cyclins through transcription-independent mechanisms.
Our data demonstrated that reductions in D-cyclin expression and G0/G1 arrest by cyproheptadine preceded its cytotoxicity. Moreover, the concentrations of cyproheptadine required to reduce D-cyclins and arrest cells matched the concentrations of drug required to induce apoptosis. These results suggest that the reductions in D-cyclin expression and cell-cycle arrest could be responsible for the ensuing cell death. In keeping with this hypothesis, knockdown of cyclin D2 with siRNA can induce cell death in myeloma cell lines.45 However, we cannot exclude the possibility that reductions in D-cyclins and apoptosis occur through separate mechanisms. As such, decreased D-cyclins may be a biomarker of an anticancer effect of cyproheptadine and, in some cell lines, not directly related to its ability to induce cell death. Therefore, additional studies will be required to understand the mechanism by which cyproheptadine exerts its effects on D-cyclins and the cell cycle and how these changes relate to its ability to induce apoptosis in malignant cells.
We demonstrated that cyproheptadine decreased the D-cyclin regulator APA2. However, additional studies will be required to determine whether the reductions in APA2 are functionally important for the effects of cyproheptadine on D-cyclins and for cyproheptadine's cytotoxicity. Potentially, cyproheptadine may also alter the binding of transcription factors to the D-cyclin promoter without altering the protein levels of these transcription factors.
Although cyproheptadine is a well-known antagonist of the H1 histamine and serotonin receptors, we demonstrated that its ability to decrease D-cyclins and induce apoptosis was independent of its known function to inhibit these receptors. As such, this work suggests that the molecule has additional binding targets responsible for its cytotoxic effects, and these proteins will be elucidated in future studies. By identifying these binding targets, second-generation compounds that bind with greater specificity and/or affinity could be rationally synthesized. These second compounds might be able to induce apoptosis at lower concentrations and avoid the side effects of cyproheptadine that are mediated by its inhibition of the H1 and serotonin receptors.
In conclusion, a chemical biology approach identified the off-patent drug cyproheptadine as an inhibitor of D-cyclin expression that induced cell death and delayed tumor growth in mouse models. Thus, we suggest that cyproheptadine could be a lead for a novel therapeutic agent for the treatment of these malignancies. Alternatively, because cyproheptadine has been previously approved for alternate indications, it could be repurposed and moved directly into clinical trial. Such trials would determine whether concentrations of cyproheptadine required to produce an antitumor effect could be safely achieved in patients.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Priscilla DeLuca for assistance with manuscript preparation.
This work was supported by grants from the Multiple Myeloma Research Foundation, the Canadian Institutes of Health Research, and the Leukemia and Lymphoma Society of Canada. A.D.S. is a CIHR Clinician Scientist and a Leukemia & Lymphoma Society Scholar in Clinical Research.
Authorship
Contribution: X.M. designed the research, analyzed data, performed research, and wrote the paper; S.-b.L., R.H., M.G., S.C., G.W.X., X.W., R.B.Z., A.D., C.-x.S., K.L., R.T., and Y.Z. performed research and analyzed data; N.J., H.M., and J.L.W. contributed vial new reagents; D.W.H., S.T., and A.K.S. designed research and analyzed data; and A.D.S. designed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Aaron D. Schimmer, Princess Margaret Hospital, Rm 9-516, 610 University Avenue, Toronto, ON, M5G 2M9, Canada; e-mail: aaron.schimmer@utoronto.ca.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal