Abstract
Acute myeloid leukemia is a heterogeneous disease from the molecular and biologic standpoints, and even patients with a specific gene expression profile may present clinical and molecular heterogeneity. We studied the epigenetic profiles of a cohort of patients who shared a common gene expression profile but differed in that only half of them harbored mutations of the CEBPA locus, whereas the rest presented with silencing of this gene and coexpression of certain T-cell markers. DNA methylation studies revealed that these 2 groups of patients could be readily segregated in an unsupervised fashion based on their DNA methylation profiles alone. Furthermore, CEBPA silencing was associated with the presence of an aberrant DNA hypermethylation signature, which was not present in the CEBPA mutant group. This aberrant hypermethylation occurred more frequently at sites within CpG islands. CEBPA-silenced leukemias also displayed marked hypermethylation compared with normal CD34+ hematopoietic cells, whereas CEBPA mutant cases showed only mild changes in DNA methylation compared with these normal progenitors. Biologically, CEBPA-silenced leukemias presented with a decreased response to myeloid growth factors in vitro.
Introduction
Aberrant transcriptional programming is a hallmark of leukemogenesis.1 Although it is well accepted that transcription factors play a crucial role in directing lineage decisions during normal hematopoiesis, it is becoming increasingly clear that epigenetic modifications also have a profound influence on regulation of gene transcription.2 It is probable that these levels of regulation are not mutually exclusive; for instance, it has been shown that certain lineage-specific transcription factors such as PU.1 can guide the locations in the genome that become methylated.3 Still, it remains largely unknown how DNA methylation patterns are determined
The basic leucine zipper transcription factor CCAAT/enhancer binding protein-α (C/EBPα) is a key regulator of hematopoietic stem cell homeostasis and normal myeloid development, and disruption of its normal function has been implicated in myeloid malignancies.4,5 The CEBPA gene is reported to be mutated in 5% to 15% of all cases of acute myeloid leukemia (AML).6-9 Furthermore, it can also be aberrantly repressed by oncogenes, such as AML1-ETO,10 or modified at the posttranslational level through the actions of FLT3 and Ras.11,12 Methylation of the CEBPA promoter has also been reported in AML as well as in other malignancies.13-16
In a genome-wide gene expression profiling study of 285 cases of de novo AML, it was found that AMLs with mutations in CEBPA (CEBPAmut) featured gene expression signatures that clearly discriminated them from other AML subgroups.17 However, a small subset of leukemias without CEBPA gene aberrations was found to express a similar gene expression signature. It was found that a frequent characteristic in this subset of leukemias was CpG hypermethylation of the CEBPA proximal promoter, which was associated with silencing of CEBPA expression (CEBPAsil). Subsequent analyses revealed an immature mixed myeloid/T-lymphoid immunophenotype and frequent NOTCH1 mutations as additional characteristics of these leukemias.16
Although these studies suggested a role for epigenetic silencing of CEBPA in the leukemic phenotype, they also raised several questions: (1) Is silencing of CEBPA through hypermethylation an isolated event or rather one aspect of broader epigenetic modifications? (2) Are particular biologic pathways specifically involved in the disease phenotype? (3) Are these leukemias more similar to T-cell acute lymphoblastic leukemia (T-ALL) than AML? To address these questions and further investigate the role of DNA methylation in this subgroup of leukemias, we analyzed DNA methylation on a genome-wide scale in CEBPAmut and CEBPAsil patients and also in T-ALL. We found that CEBPA methylation was associated with aberrant hypermethylation of many genes compared with CEBPAmut AMLs or to normal CD34+ hematopoietic progenitor cells. These hypermethylated genes included many factors involved in transcriptional regulation and mesenchymal cell differentiation and harbored several common genomic features. The epigenetic signature of this form of leukemia was distinct from T-ALL. Taken together, these data suggest the existence of a subtype of immature myeloid/T-lymphoid leukemia in which aberrant epigenetic programming results in a disease that is refractory to normal environmental signals and antileukemia therapy.
Methods
Patient samples
Twenty-five acute leukemia cases were studied. They included 14 of 15 leukemias from the original gene expression cluster 4 described by Valk et al,17 and represented 8 cases with CEBPA mutations and 6 cases with silenced CEBPA characteristics. Two additional cases of CEBPA-silenced leukemias, which had been identified in a second AML cohort and described in our previous work,16 were also studied. Nine adult T-ALLs were selected, representing various stages of maturation. Characteristics of these T-ALL cases are given in Table S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article). Normal CD34+ progenitor cells were purified from bone marrow specimens from 8 healthy donors: 4 acquired from the Translational Trials Development and Support Laboratory, Cincinnati Children's Hospital (Cincinnati, OH) and 4 purchased from Allcells (Emeryville, CA). This research was approved by the institutional review boards at Albert Einstein College of Medicine, Weill Cornell Medical College, and Erasmus University, and donor informed consent was obtained in accordance with the Declaration of Helsinki.
DNA methylation analysis by HELP
High-molecular-weight DNA was isolated from white cell fractions using a standard high salt procedure, and the HELP (HpaII tiny fragment enrichment by ligation-mediated PCR) assay was carried out as previously described.18,19 All samples for microarray hybridization were processed at the Roche-NimbleGen Service Laboratory. Samples were labeled using Cy-labeled random primers (9 mers) and then hybridized onto a human HG17 custom-designed oligonucleotide array (50-mers) covering 25 626 HpaII-amplifiable fragments located at gene promoters and imprinted regions. HpaII-amplifiable fragments are defined as genomic sequences contained between 2 flanking HpaII sites found within 200 to 2000 bp from each other. Each HpaII-amplifiable fragment on the array is represented by 15 individual probes distributed randomly across the microarray slide. Scanning was performed using a GenePix 4000B scanner (Molecular Devices, Sunnyvale, CA) as previously described.20 Quality control and data analysis of HELP microarrays were performed as described by Thompson et al.21 DNA methylation was measured as the log(HpaII/MspI) ratio, where HpaII reflects the hypomethylated fraction of the genome and MspI represents the whole genome reference. All microarray data have been submitted to the Gene Expression Omnibus (GEO) repository (accession number GSE14417).22
Gene expression microarrays
Detailed protocols are provided in Document S1.
Microarray data analysis
Unsupervised clustering of HELP and gene expression data by principal component analysis were performed using the statistical software R version 2.6.2 and the BioConductor package MADE4.23 Supervised analysis was carried out using a moderated t test with a significance level of P less than .001 (all P values were still significant at P < .05 after correcting for multiple testing using the Benjamini-Hochberg approach) and fold change more than 2 for gene expression. In the case of the methylation data, an absolute difference in methylation more than 2 between the means of the 2 populations was required to increase the likelihood of detection of biologically significant changes in methylation levels.
Quantitative DNA methylation analysis by MassARRAY EpiTyping
This analysis is described in Document S1.
CEBPA status classifier
The DNA methylation status for each HpaII-amplifiable fragment was categorized as follows: fragments having log 2 (HpaII/MspI) less than 0 were considered methylated, and those with log 2 ratios greater than 0 were considered hypomethylated. AML samples were categorized according to their CEBPA locus mutation status, and a Fisher exact test was performed at each locus to rank its predictive ability. Cutoffs for perfect, intermediate, and poor classifiers were set at P values of 1.55e−4 (which corresponds to Fisher exact P value based on perfect classification for this sample size), .2, and more than .2, respectively. The use of .20 as the threshold was set arbitrarily but represents an acceptable P value range for intermediate classification ability given the sample size.
Sequence analysis
Sequence retrieval, repeat element analysis, and motif analysis are described in Document S1.
In vitro cultures and thymidine incorporation assay
These are described in Document S1.
Results
CEBPA silencing is associated with an aberrant DNA methylation signature
To determine whether DNA methylation of the CEBPA promoter was an isolated event or part of widespread aberrant epigenetic gene silencing, we carried out a genome-wide DNA methylation study. For this, we used the HELP (HpaII tiny fragment enrichment by ligation-mediated PCR) assay,19 a method that accurately identifies the DNA methylation level of 25 626 HpaII-amplifiable fragments annotated to approximately 14 000 human gene promoters. All of the methylation profiles passed a rigorous quality control and normalization procedure as described by Thompson et al.21 The accuracy in detecting variance in DNA methylation was validated by performing single-locus quantitative EpiTyping on a panel of 10 genes (Figure S1).
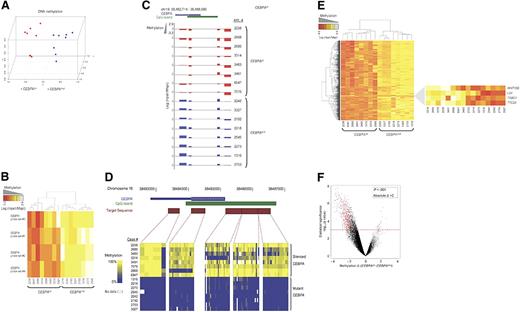
We first assessed the underlying variability in DNA methylation of 8 cases with CEBPAsil and 8 cases with CEBPAmut using principal component analysis (PCA). This analysis demonstrated separate clustering of the 2 groups, indicating that these groups of patients are epigenetically distinct (Figure 1A). The CEBPA locus itself is represented on the HELP microarray by 4 HpaII-amplifiable fragments. These fragments correspond to nucleotides +1334 to +1980, +857 to +1113, −1569 to −1819, and −2247 to −2863, relative to the transcription start site. Analysis of these fragments confirmed our previous observations: whereas the region was unmethylated in all CEBPAmut cases, it was highly methylated in all CEBPAsil cases (2-tailed t test: P < 1e−9; Figure 1B,C). DNA methylation of the CEBPA locus was confirmed using MassARRAY EpiTyping (Sequenom, San Diego, CA; Figure 1D).
A unique methylation profile distinguishes CEBPAsil from CEBPAmut AML. (A) Principal component analysis of DNA methylation data using the HELP assay on 8 CEBPAsil and 8 CEBPAmut AML cases revealed that the cases were readily segregated into 2 clusters, which matched exactly with CEBPA status. (B) Heatmap representation of the 4 probe sets annotated to the CEBPA locus on the HELP microarray; cases are clustered according to their methylation status. CEBPAsil cases cluster together (left node), and all show higher levels of methylation for at least 3 of the 4 probe sets. (C) Representation of the positioning of the 4 probe sets relative to the genomic localization of the CEBPA locus and its CpG island on chromosome 19. HELP methylation values for each leukemia case are represented in one row; the y-axis represents centered log2 (HpaII/MspI) ratios. Positive values correspond to hypomethylated fragments, whereas a negative deflection reflects a methylated fragment. The first 8 rows correspond to the CEBPAsil cases (in red), and the remaining rows to the CEBPAmut cases (in blue). (D) Heatmap representing the DNA methylation status at 5 different regions of the CEBPA locus. Percentage cytosine methylation was determined at these regions for all cases using MassARRAY EpiTyper. (E) Two-dimensional hierarchical clustering of genes differentially methylated between the 2 leukemia subgroups, illustrated by a heatmap. Supervised analysis identified 567 HpaII-amplifiable fragments (P < .001 and absolute difference in methylation > 2). Cases are represented in the columns; probe sets are represented in the rows. CEBPAsil cases are clustered in the left node and display high methylation levels for 563 HpaII-amplifiable fragments. (Right) Heatmap representation of the 4 probe sets that displayed the opposite behavior, ie, relative hypomethylation in CEBPAsil leukemia. (F) A plot of methylation difference between CEBPAsil and CEBPAmut cases (x-axis) versus statistical significance (y-axis) shows the marked asymmetry of the 2 branches, illustrating the overall tendency to higher methylation levels in the CEBPAsil cases. Red points represent probe sets that reached both criteria for differential methylation on our analysis (P < .001 and absolute methylation difference > 2).
A unique methylation profile distinguishes CEBPAsil from CEBPAmut AML. (A) Principal component analysis of DNA methylation data using the HELP assay on 8 CEBPAsil and 8 CEBPAmut AML cases revealed that the cases were readily segregated into 2 clusters, which matched exactly with CEBPA status. (B) Heatmap representation of the 4 probe sets annotated to the CEBPA locus on the HELP microarray; cases are clustered according to their methylation status. CEBPAsil cases cluster together (left node), and all show higher levels of methylation for at least 3 of the 4 probe sets. (C) Representation of the positioning of the 4 probe sets relative to the genomic localization of the CEBPA locus and its CpG island on chromosome 19. HELP methylation values for each leukemia case are represented in one row; the y-axis represents centered log2 (HpaII/MspI) ratios. Positive values correspond to hypomethylated fragments, whereas a negative deflection reflects a methylated fragment. The first 8 rows correspond to the CEBPAsil cases (in red), and the remaining rows to the CEBPAmut cases (in blue). (D) Heatmap representing the DNA methylation status at 5 different regions of the CEBPA locus. Percentage cytosine methylation was determined at these regions for all cases using MassARRAY EpiTyper. (E) Two-dimensional hierarchical clustering of genes differentially methylated between the 2 leukemia subgroups, illustrated by a heatmap. Supervised analysis identified 567 HpaII-amplifiable fragments (P < .001 and absolute difference in methylation > 2). Cases are represented in the columns; probe sets are represented in the rows. CEBPAsil cases are clustered in the left node and display high methylation levels for 563 HpaII-amplifiable fragments. (Right) Heatmap representation of the 4 probe sets that displayed the opposite behavior, ie, relative hypomethylation in CEBPAsil leukemia. (F) A plot of methylation difference between CEBPAsil and CEBPAmut cases (x-axis) versus statistical significance (y-axis) shows the marked asymmetry of the 2 branches, illustrating the overall tendency to higher methylation levels in the CEBPAsil cases. Red points represent probe sets that reached both criteria for differential methylation on our analysis (P < .001 and absolute methylation difference > 2).
To identify the genes other than CEBPA that were differentially methylated between the 2 groups, we performed a supervised analysis. This comparison identified 567 HpaII-amplifiable fragments (474 genes), which included CEBPA. Strikingly, almost all (n = 470) of these genes were hypermethylated in the CEBPAsil leukemias. Hypermethylation is thus a specific biologic feature of these leukemias that distinguishes them from CEBPAmut cases. Only 4 probe sets were hypomethylated in CEBPAsil leukemias, which included LCK and TRBC1, T-cell genes that we previously reported were overexpressed in this group of patients16 (Figure 1E,F; Table S2).
Using Ingenuity Pathway Analysis software (Redwood City, CA), we carried out a network analysis to investigate whether genes in this differentially methylated signature interrelated with each other. Interestingly, the 2 top-scoring networks were centered around CEBPA and NFκB and could almost exclusively be generated using genes that were differentially methylated (32 of 35 and 24 of 35 genes in the networks, respectively). The fact that the majority of genes in the network were affected by aberrant methylation is highly suggestive of the biologic importance of this observation. An additional network involved several bone morphogenetic protein (BMP) factors (in which 25 of 34 genes were involved), suggesting potential involvement of transforming growth factor-β signaling in these tumors as well. The genes involved in the 3 top-scoring networks are listed in Table 1 and Figure S2. Among the most represented gene ontology terms within the hypermethylated signature were regulation of transcription and mesenchymal cell development (Table 2). Despite the association of some CEBPAsil cases with Notch1 activating mutations, we did not find a significant overlap between this hypermethylated gene signature and the Notch1 signature reported by Palomero et al.24
Genes in the top 5 networks generated by the differentially methylated signature between CEBPAsil and CEBPAmut cases
| No. . | Molecules in network . | Focus molecules . |
|---|---|---|
| 1 | ADAMTS9, BARX1, CaMKII, CD180, CDH8, CHAT (includes EG:1103), DRD1, ELAVL3, FGF5, GNAS, GRIN2B, Gs-coupled receptor, HTR7, IGHM, IGKC, ISL1, KRT17, KRT19, LHX3, MAP6, MEOX2, MNX1, NEFL, NEFM, NEUROD1, NEUROG3, NFkB, NKX2–2, PAX1, PENK, PTPN13, RASGRF1, SATB2, TLR1, TLR8 | 32 |
| 2 | CCK, CDKN2B, CDX2, CEBPA, COL4A1, cyclin A, E2f, EID2, ESX1, FOXG1, HGF, HIST1H2BJ, histone h3, insulin, IRF8, JTV1, LRAT, Mapk, MARCKS (includes EG:4082), PGR, PITX2, POU2F3, POU3F2, PRLR, PTGFR, Rap1, Ras, RASA2, Rb, RELN, Rsk, Smad1/5/8, SMPDL3A, TFAP2C, VEGF | 24 |
| 3 | alkaline phosphatase, ALPL, APOB, ATOH1, BMP, BMP3, BMP6, BMP7, FCGR2B, FOXA2, FOXC2, GSC, IL1, LDL, LPL, LRP6, MHC class II, Mmp, MSX2, NELL1, ONECUT2, OSBPL6, P38 MAPK, PCDHGC3, PDGF BB, PLC-γ, PTHLH, SFRP1, SOX9, TGF-β, UCP2, Wnt, WNT2, WNT10B, ZAK | 24 |
| 4 | ACTN2, ADCY1, adenylate cyclase, AFAP1, α-actinin, BDNF, Cacna1, CACNA1A, CACNA1E, CACNA1I, CACNA2D1, CALML5, calmodulin, F actin, FOXL1, GAL, GALR1, GRIN3A, GRM1, Jnk, KCNN2, MARCKSL1, MBP, N-type calcium channel, NMDA receptor, OCLN, OPRK1, PDLIM3, Pkc(s), PP2A, SLC6A2, SLC6A5, SNAP25, TAC1, voltage gated calcium channel | 24 |
| 5 | ADAM12, Akt, BRCA2, cofilin, creatine kinase, Creb, CYR61, EFNA5, EPHA, EPHA2, EPHA3, EPHA5, EPHA7, EPHA/B, GABBR2, GDNF, GFRA1, GREM1, HABP4, HSPB8, HTR1B, IRS4, MAGI1, N-cor, NAE1, p70 S6k, Pdgf, PDGFC, PI3K, PIK3R1, PRKCZ, Rab5, RET, Rock, TOP2B | 23 |
| No. . | Molecules in network . | Focus molecules . |
|---|---|---|
| 1 | ADAMTS9, BARX1, CaMKII, CD180, CDH8, CHAT (includes EG:1103), DRD1, ELAVL3, FGF5, GNAS, GRIN2B, Gs-coupled receptor, HTR7, IGHM, IGKC, ISL1, KRT17, KRT19, LHX3, MAP6, MEOX2, MNX1, NEFL, NEFM, NEUROD1, NEUROG3, NFkB, NKX2–2, PAX1, PENK, PTPN13, RASGRF1, SATB2, TLR1, TLR8 | 32 |
| 2 | CCK, CDKN2B, CDX2, CEBPA, COL4A1, cyclin A, E2f, EID2, ESX1, FOXG1, HGF, HIST1H2BJ, histone h3, insulin, IRF8, JTV1, LRAT, Mapk, MARCKS (includes EG:4082), PGR, PITX2, POU2F3, POU3F2, PRLR, PTGFR, Rap1, Ras, RASA2, Rb, RELN, Rsk, Smad1/5/8, SMPDL3A, TFAP2C, VEGF | 24 |
| 3 | alkaline phosphatase, ALPL, APOB, ATOH1, BMP, BMP3, BMP6, BMP7, FCGR2B, FOXA2, FOXC2, GSC, IL1, LDL, LPL, LRP6, MHC class II, Mmp, MSX2, NELL1, ONECUT2, OSBPL6, P38 MAPK, PCDHGC3, PDGF BB, PLC-γ, PTHLH, SFRP1, SOX9, TGF-β, UCP2, Wnt, WNT2, WNT10B, ZAK | 24 |
| 4 | ACTN2, ADCY1, adenylate cyclase, AFAP1, α-actinin, BDNF, Cacna1, CACNA1A, CACNA1E, CACNA1I, CACNA2D1, CALML5, calmodulin, F actin, FOXL1, GAL, GALR1, GRIN3A, GRM1, Jnk, KCNN2, MARCKSL1, MBP, N-type calcium channel, NMDA receptor, OCLN, OPRK1, PDLIM3, Pkc(s), PP2A, SLC6A2, SLC6A5, SNAP25, TAC1, voltage gated calcium channel | 24 |
| 5 | ADAM12, Akt, BRCA2, cofilin, creatine kinase, Creb, CYR61, EFNA5, EPHA, EPHA2, EPHA3, EPHA5, EPHA7, EPHA/B, GABBR2, GDNF, GFRA1, GREM1, HABP4, HSPB8, HTR1B, IRS4, MAGI1, N-cor, NAE1, p70 S6k, Pdgf, PDGFC, PI3K, PIK3R1, PRKCZ, Rab5, RET, Rock, TOP2B | 23 |
GO terms highly represented among the differentially methylated genes between the 2 leukemia subgroups
| Go term . | P . | Benjamini . | FDR . |
|---|---|---|---|
| Mesenchymal cell development | 4.1e−5 | 1.9 e−2 | 0.1 |
| Neural crest cell differentiation | 3.5e−4 | 5.1e−2 | 0.6 |
| Regulation of transcription | 1.4e−4 | 4.6e−2 | 0.2 |
| Transcription DNA-dependent | 2.8e−4 | 4.7e−2 | 0.5 |
| RNA biosynthetic process | 2.9e−4 | 4.6e−2 | 0.5 |
| Go term . | P . | Benjamini . | FDR . |
|---|---|---|---|
| Mesenchymal cell development | 4.1e−5 | 1.9 e−2 | 0.1 |
| Neural crest cell differentiation | 3.5e−4 | 5.1e−2 | 0.6 |
| Regulation of transcription | 1.4e−4 | 4.6e−2 | 0.2 |
| Transcription DNA-dependent | 2.8e−4 | 4.7e−2 | 0.5 |
| RNA biosynthetic process | 2.9e−4 | 4.6e−2 | 0.5 |
CEBPAsil and CEBPAmut leukemia cells are epigenetically distinct from normal bone marrow CD34+ cells
Does hypermethylation in CEBPAsil leukemic blasts represent aberrant epigenetic programming compared with normal CD34+ cells? In a supervised comparison of CEBPAsil leukemia cells with CD34+ healthy donor bone marrow fractions (n = 8), a large number of differentially methylated loci (1035 HpaII-amplifiable fragments, 876 unique genes) was found. Similarly as in the previous analyses, most of the genes (841 of 876) were hypermethylated in the CEBPAsil leukemia subgroup (Figure 2A,B). Moreover, 281 of these 876 genes appeared to be hypermethylated both compared with normal CD34+ cells and to CEBPAmut leukemias, indicating that aberrant hypermethylation of these genes occurred exclusively in CEBPAsil cases. These included, among others, the tumor suppressors CDKN2B and IRF8, the cell cycle regulator MyoD1, and the WNT signaling antagonist SFRP1. Loss of all these genes has been previously implicated in tumorigenesis.25-29 Furthermore, hypermethylation of the promoter regions of CDKN2B, MyoD1, and SFRP1 has been described in association with several forms of leukemia.30-35 A complete list of the genes exclusively methylated in the CEBPAsil subgroup versus normal CD34+ cells is provided in Table S3. Both of the proximal HpaII-amplifiable fragments for the CEBPA locus reached statistical significance and showed hypermethylation in the CEBPAsil group versus the normal CD34+ cells, with an absolute methylation change slightly below our set threshold of 2 (change, −1.63 and 1.99; Figure S3). Only 45 of the 1035 HpaII-amplifiable fragments were hypomethylated in the CEBPAsil cases and hypermethylated in normal CD34+ cells. These loci included the T-cell genes CD3D and TRBC1.
Aberrant hypermethylation is a feature of CEBPAsil leukemia. (A) Two-dimensional hierarchical clustering of genes differentially methylated between the CEBPAsil cases and normal CD34+ hematopoietic progenitors, illustrated by a heatmap. Supervised analysis identified 1035 HpaII-amplifiable fragments (P < .001 and absolute difference in methylation > 2). Cases are represented in the columns; probe sets are represented in the rows. CEBPAsil cases are clustered in the left node and display marked hypermethylation compared with the normals, as illustrated by the predominance of probe sets with low log2 (HpaII/MspI) ratios. (B) Methylation difference between CEBPAsil and normal CD34+ cells (x-axis) versus statistical significance (y-axis) plot with marked asymmetry of the 2 branches, reflecting the tendency to higher methylation levels in this subgroup. Red points represent probe sets that reached both criteria for differential methylation in our analysis. (C) Two-dimensional hierarchical clustering of genes differentially methylated between the CEBPAmut AML and normal CD34+ hematopoietic progenitors, illustrated by a heatmap. Supervised analysis identified 322 probe sets (286 genes). CEBPAmut cases are clustered in the left node and display equal components of hypermethylation and hypomethylation compared with CD34+ normal cells. (D) A plot of methylation difference between mutant CEBPA and normal CD34+ cells (x-axis) versus statistical significance (y-axis) shows symmetric branches and less pronounced differences in methylation than in the case of the silenced CEBPA subgroup. Red points represent probe sets that reached both criteria for differential methylation on our analysis.
Aberrant hypermethylation is a feature of CEBPAsil leukemia. (A) Two-dimensional hierarchical clustering of genes differentially methylated between the CEBPAsil cases and normal CD34+ hematopoietic progenitors, illustrated by a heatmap. Supervised analysis identified 1035 HpaII-amplifiable fragments (P < .001 and absolute difference in methylation > 2). Cases are represented in the columns; probe sets are represented in the rows. CEBPAsil cases are clustered in the left node and display marked hypermethylation compared with the normals, as illustrated by the predominance of probe sets with low log2 (HpaII/MspI) ratios. (B) Methylation difference between CEBPAsil and normal CD34+ cells (x-axis) versus statistical significance (y-axis) plot with marked asymmetry of the 2 branches, reflecting the tendency to higher methylation levels in this subgroup. Red points represent probe sets that reached both criteria for differential methylation in our analysis. (C) Two-dimensional hierarchical clustering of genes differentially methylated between the CEBPAmut AML and normal CD34+ hematopoietic progenitors, illustrated by a heatmap. Supervised analysis identified 322 probe sets (286 genes). CEBPAmut cases are clustered in the left node and display equal components of hypermethylation and hypomethylation compared with CD34+ normal cells. (D) A plot of methylation difference between mutant CEBPA and normal CD34+ cells (x-axis) versus statistical significance (y-axis) shows symmetric branches and less pronounced differences in methylation than in the case of the silenced CEBPA subgroup. Red points represent probe sets that reached both criteria for differential methylation on our analysis.
In the CEBPAmut group, abnormal methylation was less abundant, with only 322 HpaII-amplifiable fragments (286 unique genes) detected as differentially methylated with respect to normal CD34+ cells. Moreover, the difference in DNA methylation was distributed evenly, where approximately half of these probe sets were hypermethylated and the other half hypomethylated (Figure 2C,D). This finding indicates that the markedly increased prevalence of abnormal promoter hypermethylation is highly specific to CEBPAsil cases among the CEBPA gene expression signature AML patients.
When we compared both subgroups of leukemias with normal CD34+ cells, 100 unique genes were found to be aberrantly methylated in the leukemia samples. These included CDKN2A, CCR7, MT2A, and NOS3 and were all linked together in a cancer/cell signaling network through Ingenuity Pathway Analysis (Table S4). This finding suggests that aberrant hypermethylation of these genes is a common event in both these leukemogenic processes and therefore deserves further study.
Hypermethylation in CEBPAsil leukemias is localized mainly to CpG islands and CG clusters
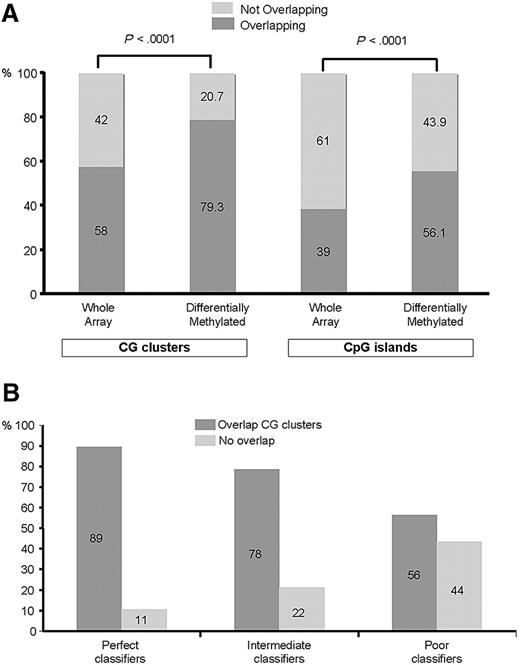
To obtain further insight into the differences in methylation profiles between CEBPAsil leukemias and CEBPAmut AMLs, we next asked whether the observed differences in DNA methylation were found at either CpG islands or CG clusters36 more frequently than expected based on the distribution of all fragments on the arrays. For this analysis, we categorized all HpaII-amplifiable fragments on the HELP array into 2 categories: those that overlapped with a known CpG island or CG cluster and those that did not. We found 15 043 of 25 626 (58%) fragments to be completely located within, or partly overlapping with, CG clusters. Furthermore, 10 006 of 25 626 (39%) fragments were located in or partly overlapped with CpG islands. Of the 567 fragments identified as differentially methylated between the 2 leukemia groups, 450 (79.3%) overlapped with at least one CG cluster, whereas 318 (56.1%) overlapped with a CpG island (Figure 3A). Thus, there was a significant enrichment in the signature for fragments overlapping with either CpG islands (56% vs 39%) as well as with CG clusters (79% vs 58%, proportion test; P < .001 for both), which is consistent with their proposed regulatory functions. On the other hand, these analyses show that a subset of the differentially methylated HpaII-amplifiable fragments did not overlap with CpG islands or CG clusters, underscoring the importance of extending DNA methylation studies beyond those areas of the genome containing CpG islands and CG clusters.
Aberrant hypermethylation colocalizes to CpG islands and CG clusters. (A) The genomic position of every HpaII-amplifiable fragment on the HELP array was compared with the location of known CpG islands and CG clusters, and the fragments on the array were divided into 2 categories: those overlapping with either one of these genomic elements and those not overlapping. To determine whether the differentially methylated genes between CEBPAsil and CEBPAmut leukemias were enriched for either one of these types of elements, a proportions test was used to compare the relative proportion of the 2 types of HpaII fragments in the signature (overlapping vs not overlapping) to the relative proportion on the array. Stacking bars are used to illustrate the finding of a significant enrichment for HpaII-amplifiable fragments overlapping with CpG islands (right) and CG clusters (left) in the hypermethylated signature of CEBPAsil leukemia, as it compares with the genomic localization of all HpaII-amplifiable fragments on the HELP array. (B) Each HpaII-amplifiable fragment represented on the HELP array was also categorized according to its ability to discriminate between CEBPAsil and CEBPAmut leukemias as perfect, intermediate, or poor classifiers, and the proportional amount of fragments overlapping with CG clusters was calculated for each group of classifiers. Better classifiers were more frequently associated with CG clusters.
Aberrant hypermethylation colocalizes to CpG islands and CG clusters. (A) The genomic position of every HpaII-amplifiable fragment on the HELP array was compared with the location of known CpG islands and CG clusters, and the fragments on the array were divided into 2 categories: those overlapping with either one of these genomic elements and those not overlapping. To determine whether the differentially methylated genes between CEBPAsil and CEBPAmut leukemias were enriched for either one of these types of elements, a proportions test was used to compare the relative proportion of the 2 types of HpaII fragments in the signature (overlapping vs not overlapping) to the relative proportion on the array. Stacking bars are used to illustrate the finding of a significant enrichment for HpaII-amplifiable fragments overlapping with CpG islands (right) and CG clusters (left) in the hypermethylated signature of CEBPAsil leukemia, as it compares with the genomic localization of all HpaII-amplifiable fragments on the HELP array. (B) Each HpaII-amplifiable fragment represented on the HELP array was also categorized according to its ability to discriminate between CEBPAsil and CEBPAmut leukemias as perfect, intermediate, or poor classifiers, and the proportional amount of fragments overlapping with CG clusters was calculated for each group of classifiers. Better classifiers were more frequently associated with CG clusters.
Next we categorized each HpaII-amplifiable fragment into methylated or hypomethylated and ranked them by their ability to discriminate between CEBPAmut and CEBPAsil cases (summarized by Fisher exact test P value). Fragments were then grouped into perfect, intermediate, or poor classifiers. A total of 66 fragments were categorized as perfect classifiers. Of these, 59 (89%) were located in CG clusters, 2 of which were also annotated as CpG islands). All 66 fragments exhibit consistent methylation in CEBPAsil cases and hypomethylation in CEBPAmut cases. Intermediate classifiers showed 78% of the fragments overlapping with CG clusters, and only 56% of the poor classifiers overlapped with a CG cluster (Figure 3B). Taken together, these data suggest that, among differentially methylated genes, those that contain CpG islands or clusters are most critical for defining the epigenetic signature of CEBPAsil leukemia cases.
Hypermethylated promoters in CEBPAsil display specific genomic features
We next examined whether genes susceptible to becoming methylated in CEBPAsil leukemias (compared with CEBPAmut) shared any common genomic features. The regions spanning the 2-kb sequence upstream of the transcription start site of the 327 RefSeq genes assigned to the differentially methylated 567 HpaII-amplifiable fragments were retrieved. Next, these sequences were compared with 2422 RefSeq gene promoter sequences (corresponding to 2672 HpaII-amplifiable fragments) that were randomly selected from the pool of HpaII-amplifiable fragments that did not show any methylation difference between CEBPAsil and CEBPAmut. Remarkably, hypermethylated genes were significantly less likely to contain an Alu sequence than the control sequences. (Genes containing Alu sequences: 60% [1453 of 2422] in the control sequences vs 26% [85 of 327] in the differentially methylated sequences; Fisher exact test, P < 2.2e−16; Table 3.) This was the only repeat element that showed a statistically significant distribution difference between the CEBPAsil signature and the randomly selected control group. All other repeat sequences (eg, SINE/MIRs, LINEs, or MERs) were present in approximately equal proportions. We also looked at the promoter regions from 780 RefSeq genes annotated to the 1035 HpaII-amplifiable fragments differentially methylated between CEBPAsil leukemias versus normal CD34+ bone marrow cells. We compared these with 3086 randomly selected RefSeq promoters (corresponding to 4470 control HpaII-amplifiable fragments). Once again, we found that differentially methylated sequences were less likely to contain Alu elements (60% in control sequences [1865 of 3086] vs 35% in the differentially methylated sequences [277 of 780]; Fisher exact test, P < 2.2e−16).
Alu elements are depleted from RefSeq promoters that become hypermethylated in CEBPAsil leukemia
| . | Silenced RefSeq (n = 327) . | Random RefSeq controls (n = 2422) . | P (Fisher exact test) . |
|---|---|---|---|
| Alu | 85 (26%) | 1453 (60%) | < 2.2e−16 |
| No Alu | 242 (74%) | 969 (40%) |
| . | Silenced RefSeq (n = 327) . | Random RefSeq controls (n = 2422) . | P (Fisher exact test) . |
|---|---|---|---|
| Alu | 85 (26%) | 1453 (60%) | < 2.2e−16 |
| No Alu | 242 (74%) | 969 (40%) |
To determine whether a specific DNA sequence was associated with the differentially methylated genes, we used the Finding Informative Regulatory Elements (FIRE) algorithm37 to look at a region containing 2 kb of sequence upstream of the reported transcription start site. This analysis identified a nondegenerate 5′-CGCGCTC-3′ motif, which was significantly enriched in the differentially methylated sequences compared with the control sequences. The nondegenerate motif could be detected in 45% of the differentially methylated sequences (147 of 327), whereas it was found in less than 24% of the control sequences (574 of 2422; Fisher exact test, P < 3.3e−13). An independent FIRE analysis was carried out to study the promoter sequences of the RefSeq genes annotated to the probe sets in the CEBPAsil versus normal CD34+ cell signature. Although this analysis discovered the same nondegenerate motif as before, it was slightly less successful at discriminating between the 2 groups (present in 32% of the differentially methylated sequences [251 of 780] vs 20% of the control sequences [628 of 3086]; Fisher exact test, P < 8.7e−12). Although this motif does not correspond to a known transcription factor binding site, FIRE annotated sequences containing this motif as being associated with the Gene Ontology term GO:0043565, involved in sequence-specific DNA binding (P < 1e−7). The enrichment of this element within the hypermethylated signature is suggestive of the existence of a putative DNA binding protein that might participate in establishing this epigenetic signature.
Methylated genes in CEBPAsil leukemias tend to be expressed at low levels
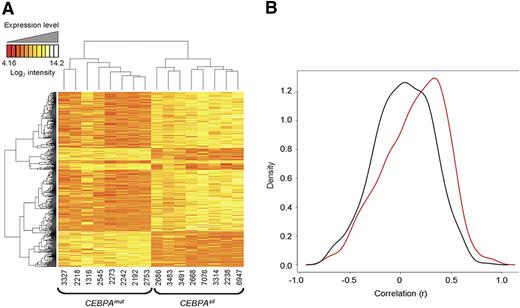
To determine the correlation of DNA methylation with gene expression, we examined genome-wide mRNA transcript abundance of the same CEBPAsil and CEBPAmut cases using Affymetrix HGU133 Plus 2.0 microarrays (Santa Clara, CA). An unsupervised analysis using PCA indicated that, in contrast to DNA methylation profiles, expression data did not result in segregation along the major axes (Figure S4). Nevertheless, a supervised analysis identified 587 probe sets, corresponding to 415 unique genes, as differentially expressed between the 2 groups of leukemias, which is in line with our previous findings16 (Figure 4A). Overlap between the differentially methylated and differentially expressed genes was only minimal (12 unique genes, including CEBPA itself). This is consistent with our previous report showing that gene expression and methylation profiling tend to capture biologic variance in different sets of genes.18 Interestingly, signal intensities of 62.5% of the genes detected as differentially hypermethylated were measured at low abundance (intensity < log2[100]) in both the CEBPAsil and the CEBPAmut groups. This may imply that these genes are epigenetically silenced by DNA methylation in one group and repressed through a different mechanism in the other. Along these lines, expression levels for most of these genes were detected at similarly low levels in an independent cohort of 400 AML cases of other subtypes (data not shown). Methylation and gene expression were validated by MassARRAY on 15 genes for 3 CEBPAsil and 4 CEBPAmut randomly selected cases and by quantitative reverse-transcribed polymerase chain reaction on 10 of these genes on 2 to 4 randomly selected cases from each group (Figure S5).
DNA methylation and gene expression capture complementary information. (A) Two-dimensional hierarchical clustering of genes differentially expressed between the 2 leukemia subgroups, illustrated by a heatmap. Supervised analysis identified 587 probe sets (415 genes) at a P < .001 and fold change more than 2. Cases are represented in the columns; probe sets are represented in the rows. CEBPAsil cases are clustered in the right node, and CEBPAmut cases are clustered in the left node. (B) Density (y-axis) plot for the gene-by-gene correlations (x-axis) between gene expression log intensity and log(HpaII/MspI) values. A positive correlation between these 2 measures translates into a negative biologic correlation, that is, hypermethylation in combination with lower expression levels, or hypomethylation in combination with higher expression levels. In black, density plot for the correlation between expression and methylation for a set of 600 randomly selected probe sets. In red, density plot for the correlation between expression and methylation measured by the 567 probe sets in the genes differentially methylated between the 2 subgroups. The shifting of the density plot to the right reflects a tendency to a stronger correlation of DNA methylation with gene expression levels in this subset of genes. The figure is representative of 5 analyses, each time using a different set of 600 randomly selected HpaII-amplifiable fragments for the calculation of correlations.
DNA methylation and gene expression capture complementary information. (A) Two-dimensional hierarchical clustering of genes differentially expressed between the 2 leukemia subgroups, illustrated by a heatmap. Supervised analysis identified 587 probe sets (415 genes) at a P < .001 and fold change more than 2. Cases are represented in the columns; probe sets are represented in the rows. CEBPAsil cases are clustered in the right node, and CEBPAmut cases are clustered in the left node. (B) Density (y-axis) plot for the gene-by-gene correlations (x-axis) between gene expression log intensity and log(HpaII/MspI) values. A positive correlation between these 2 measures translates into a negative biologic correlation, that is, hypermethylation in combination with lower expression levels, or hypomethylation in combination with higher expression levels. In black, density plot for the correlation between expression and methylation for a set of 600 randomly selected probe sets. In red, density plot for the correlation between expression and methylation measured by the 567 probe sets in the genes differentially methylated between the 2 subgroups. The shifting of the density plot to the right reflects a tendency to a stronger correlation of DNA methylation with gene expression levels in this subset of genes. The figure is representative of 5 analyses, each time using a different set of 600 randomly selected HpaII-amplifiable fragments for the calculation of correlations.
We next examined whether differential methylation in the genes contained within the CEBPAsil versus CEBPAmut subtypes was more correlated to expression levels than methylation occurring in randomly selected subsets of genes. For this we determined the correlation between transcript abundance and DNA methylation status for the 474 genes differentially methylated between the 2 leukemia subgroups and compared it with that of genes annotated to 600 randomly selected probe sets from the HELP microarray. This comparison was repeated for 5 different randomly selected groups of control probe sets. In every instance, the differentially methylated genes showed a slightly stronger correlation with gene expression levels (corresponding to a negative biologic correlation between DNA methylation and gene expression) than the randomly selected genes (Figure 4B).
CEBPAsil leukemias are epigenetically distinct from T-ALL
We hypothesized that CEBPAsil leukemias, although expressing certain T-cell genes, are biologically and epigenetically not only different from other AMLs, but also from T-ALL. To test this hypothesis, we generated HELP profiles from a selection of 9 T-ALL cases from various maturation stages and compared those with the methylation data of the CEBPAsil leukemias. Unsupervised analysis of HELP using PCA resulted in separate clustering of the CEBPAsil leukemias and the T-ALL cases (Figure 5A). A supervised analysis identified 164 genes (199 HpaII-amplifiable fragments) differentially methylated between the CEBPAsil leukemias and T-ALLs (Figure 5B; Table S5). Specific analysis of the CEBPA locus HpaII-amplifiable fragments indicated a mixed pattern in the T-ALL samples. Whereas the majority of cases (n = 6) showed hypermethylation of the CEBPA proximal promoter, the remaining 3 did not (Figure S6). A similar separation of CEBPAsil leukemias and T-ALL samples was observed based on gene expression data (Figure 5C), indeed suggesting that CEBPAsil leukemias are biologically and epigenetically distinct from T-ALL.
CEBPAsil leukemias differ genetically and epigenetically from T-ALL. (A) Principal component analysis of DNA methylation data comparing 8 immature acute myeloid/T lymphoid CEBPAsil cases with a selection of 9 T-ALL cases representing a spectrum of maturation stages, showing separate clustering of the 2 groups of leukemias. (B) Two-dimensional hierarchical clustering of genes differentially methylated between the CEBPAsil leukemias and the T-ALL cases, illustrated by a heatmap. Cases are represented in the columns; probe sets are represented in the rows. Supervised analysis identified 213 differentially methylated probe sets (199 genes). CEBPAsil cases are clustered in the left node and display a predominance of hypomethylated probe sets. (C) Principal component analysis of gene expression data for the same cases also demonstrates separate clustering of the 2 groups of leukemias, indicating that these 2 groups display distinct expression profiles.
CEBPAsil leukemias differ genetically and epigenetically from T-ALL. (A) Principal component analysis of DNA methylation data comparing 8 immature acute myeloid/T lymphoid CEBPAsil cases with a selection of 9 T-ALL cases representing a spectrum of maturation stages, showing separate clustering of the 2 groups of leukemias. (B) Two-dimensional hierarchical clustering of genes differentially methylated between the CEBPAsil leukemias and the T-ALL cases, illustrated by a heatmap. Cases are represented in the columns; probe sets are represented in the rows. Supervised analysis identified 213 differentially methylated probe sets (199 genes). CEBPAsil cases are clustered in the left node and display a predominance of hypomethylated probe sets. (C) Principal component analysis of gene expression data for the same cases also demonstrates separate clustering of the 2 groups of leukemias, indicating that these 2 groups display distinct expression profiles.
CEBPAsil leukemia cells are resistant to myeloid growth factors
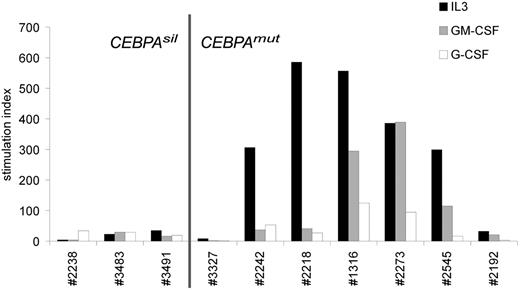
To study whether silencing by methylation of the myeloid specific transcription factor CEBPA corresponded with altered biologic response to natural growth stimuli, we compared growth factor stimulated 3H-TdR incorporation data obtained from pretreatment CEBPAsil and CEBPAmut AML samples. Remarkably, CEBPAsil leukemia cells showed little or no response to the myeloid growth stimuli interleukin-3 (IL-3), granulocyte-macrophage colony-stimulating factor (GM-CSF), or granulocyte colony-stimulating factor (G-CSF), whereas CEBPAmut samples were particularly sensitive to IL-3, but in certain cases also to GM-CSF (Figure 6; Table S6). Whereas 2 of the CEBPAmut cases (3327 and 2192) showed comparable results to the CEBPAsil cases, this did not appear to be explained by their methylation profiles, and we think that it is a reflection of the biologic heterogeneity of the group. In contrast, both CEBPAsil and CEBPAmut patient cells showed weak but significant response to factors that stimulate primitive cells, in particular thrombopoietin or stem cell factor (Table S6). These data are in agreement with previous data showing loss of the granulocyte macrophage progenitors fraction and enrichment of hematopoietic stem cell population in marrow of conditional Cebpa knockout animals.38 Interestingly, we did not observe promoter hypermethylation of the genes encoding the receptors for IL-3, GM-CSF, or G-CSF, suggesting other mechanisms of altered growth factor response. Several of the growth factor receptors appeared to be expressed as determined by gene expression profiling analysis, although we observed significantly lower expression of CSF2RB, the gene encoding the common beta chain of the receptors of IL-3 and GM-CSF (P = .02, Wilcoxon rank-sum test) in CEBPAsil patients compared with CEBPAmut AML samples. Together, these data suggest that CEBPAsil cases reflect a more primitive hematopoietic cell that does not enter cell cycle in response to myeloid growth factors, an event that could be associated with chemoresistance. In agreement, although the numbers of patients were too small to rule out the influence of covariables, the CEBPAsil patient outcomes (5-year overall survival 25%) were considerably worse than that of the CEBPAmut cases (5-year overall survival 88%; log-rank test P < .003).
CEBPAsil leukemia cells are resistant to myeloid growth. Tritiated thymidine incorporation experiment of 3 CEBPAsil and 7 CEBPAmut leukemia samples. Response of leukemia cells to IL-3, GM-CSF, and G-CSF is shown relative to irradiated nonstimulated cells (ie, stimulation index), based on the mean of 3 measurements.
CEBPAsil leukemia cells are resistant to myeloid growth. Tritiated thymidine incorporation experiment of 3 CEBPAsil and 7 CEBPAmut leukemia samples. Response of leukemia cells to IL-3, GM-CSF, and G-CSF is shown relative to irradiated nonstimulated cells (ie, stimulation index), based on the mean of 3 measurements.
Discussion
The subtype of immature myeloid/T-lymphoid leukemia reported in this study was first captured as belonging to a larger group of patients all sharing a specific gene expression signature (originally designated as cluster 4).17 A subset of these patients was subsequently observed to harbor hypermethylation and silencing of the CEBPA gene along with expression of T-cell genes.16 In the current study, we applied integrated epigenetic profiling to explore in greater depth the nature of this disease, and demonstrate that this is indeed a biologic entity that is distinct from both AML and T-ALL and has unique epigenetic, transcriptional, and biologic features.
Given the similarity in gene expression profiles of CEBPAsil and CEBPAmut AML found initially, we were primarily interested in determining whether DNA methylation profiles might more readily discriminate these 2 groups. Along these lines, we have previously shown that integrating information obtained from DNA methylation profiling and gene expression profiling in human leukemia specimens could yield more in-depth insight into biologic differences between patients.18 We find that PCA, an unbiased method to discriminate biologic differences, showed a marked separation between epigenetic profiles of patients with mutant or silenced CEBPA. This indicates that these are 2 biologically distinct entities, which is further supported by the differences in growth factor responses between the 2 leukemia subtypes. In contrast, gene expression was not as obviously distinct, demonstrating for the first time in a practical manner that these 2 methods of interpreting transcriptional programming are complementary and when combined can most robustly identify new disease subtypes.
One remarkable finding in the CEBPAsil cases was that their unique DNA methylation signature consisted almost entirely of hypermethylation of hundreds of loci compared with their CEBPAmut counterparts. By chance, one would expect a more or less balanced distribution between hypermethylation and hypomethylation, as observed in the comparison between CEBPAmut cases and normal CD34+ cells. The greater tendency toward hypermethylation of these leukemias with silenced CEBPA was further confirmed by the comparison of the CEBPAsil subgroup with normal CD34+ hematopoietic cells. Together, this suggests epigenetic deregulation to play a more critical role in leukemogenesis of those cases than of CEBPAmut cases. Such a hypothesis is supported by our observation of involvement of a majority of the members of gene regulatory networks centered around the CEBPA, NFκB, and BMP pathways, all implicated in the normal homeostasis of hematopoietic stem cells and the myeloid lineage.
When DNA methylation profiles and gene expression profiles were queried by supervised analysis, 474 and 415 genes were identified as significantly differentially methylated or differentially expressed, respectively. Yet there was only minimal overlap between these 2 gene lists. This is consistent with our previous observation that gene expression and DNA methylation array studies capture different sets of genes and thus are complementary to each other when collecting information on biologic variance.18 The fact that expression of more than 60% of the genes in the hypermethylated signature was not detected in both groups partly explains this finding because differences in low abundance transcripts may be hard to detect by expression arrays. Despite all these technical limitations, we still observed a greater tendency to overall inverse correlation between gene expression and DNA methylation status among those genes whose methylation status varied between the 2 groups. This result suggests that the biologic impact of epigenetic modifications may vary between gene subsets. Although this has been shown for specific histone modifications, such as the association between HOX cluster genes and methylation of lysine 27 on histone 3,39,40 a similar situation has not been previously shown for cytosine methylation beyond that of its distribution relative to CpG islands.
Although the existence of a hypermethylated phenotype in CEBPAsil immature acute myeloid/T-lymphoid leukemia could be related to a relatively more immature cell type involved in the leukemic transformation, an alternative explanation could lie in leukemia-specific mechanisms. The latter either could involve a global up-regulation of DNA methylation machinery, leading to more or less uncontrolled hypermethylation of multiple loci, or, alternatively, could be a specific effect of targeted methylation of selected genes. We took several approaches to investigate the various possibilities. Comparative analyses between the 2 leukemia subgroups and immature CD34+ control samples from healthy individuals indicated indeed a greater state of hypermethylation in the CEBPAsil leukemia blasts than in normal CD34+ cells. This is suggestive for the idea that hypermethylation is not primarily a read-out for cellular maturation status. In contrast, CEBPAmut AMLs had markedly fewer hypermethylated genes compared with CD34+, indicating that an increase in DNA methylation is not a general feature of all AMLs. Interestingly, mRNA expression levels of the de novo DNA methyltransferase DNMT3B were significantly increased in CEBPAsil versus CEBPAmut leukemias but showed similarly high levels in normal CD34+ cells (data not shown). Thus, although expression of methylating enzymes may be high in healthy immature cells as well, this does not necessarily lead to a similar hypermethylated signature as found in the leukemic subgroup. There were no differences in expression of other DNA methyltransferases (data not shown).
Specific targeting of methylation to certain genes by complexes involving DNA methyltransferases and transcription factors has been shown previously.41 Inversely, it could be hypothesized that loss of protective mechanisms, such as DNA-binding proteins and nucleosome positioning, could render certain promoters susceptible to the targeting by the DNA methylation machinery.42 A common characteristic of the leukemia subgroup studied here was silencing of the transcription factor C/EBPα. Using algorithms for motif analysis comparing significantly hypermethylated genes with control genes, we were unable to detect a significant enrichment for C/EBP binding sites among these genes, arguing against a direct role for the absence of C/EBPα. These analyses did, however, yield 2 potentially relevant findings. First, we identified enrichment of a novel sequence motif. This sequence could possibly play a role in either directing the binding certain protein complexes to DNA or in helping determine nucleosome positioning. Second, a significant depletion of Alu repeats was detected in the promoters of differentially methylated RefSeq genes. Although the causes behind such a finding can only be speculated on, similar observation have been made in the past, suggesting that this is probably not a random finding.43
CEBPAsil leukemias were previously found to have a mixed myeloid/T-lymphoid phenotype. We were therefore interested to determine how their DNA methylation profiles related to those from T-ALL samples. Comparisons of the DNA methylation and expression profiles of a representative collection of 9 T-ALL cases, ranging from very immature to mature, to those of the CEBPAsil leukemias revealed significant differences between the 2 types of leukemias. This is consistent with the idea that CEBPAsil leukemias represent an entity not entirely myeloid but also not entirely T-lymphoid and may represent a very immature myeloid/T-lymphoid subtype that is in current clinical practice sometimes diagnosed as AML and sometimes as T-ALL.
The discovery of a hypermethylation profile in a specific leukemia subgroup is particularly interesting in light of the recent developments of demethylating drugs. Notably, the CEBPAsil leukemia subgroup showed a markedly worse treatment response than CEBPAmut AML.16 Interestingly, this is in contrast with the previous report by Hackanson et al, who found that methylation of CEBPA was associated with favorable prognostic groups in AML.13 However, the cases reported by that group all displayed methylation restricted to an upstream region of the CEBPA locus, whereas the methylation in our samples was more extensive, covering both proximal and distal regions. Although the numbers are small and multivariable analysis with additional covariates such as cytogenetic and molecular subgroups is warranted, these results identify CEBPAsil leukemia cases as attractive candidates for investigative treatment with such demethylating agents. It is interesting to speculate that restoring the normal programming of CEBPA, NFκB, and BMP pathways could facilitate targeting of these tumors with either standard antileukemia regimens or with specific targeted therapy agents.
Taken together, we show that integrated epigenetic and gene expression analyses of leukemia can distinguish and illustrate phenotypes with biologic and potential clinical significance. Analysis of the genes involved in aberrant epigenetic programming allows for generation of hypotheses toward dissecting previously unrecognized mechanisms of leukemogenesis. A broader application of integrated gene expression and epigenetic profiling to acute leukemia might allow many other such entities to be resolved.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the colleagues of the bone marrow transplantation group and the molecular diagnostics laboratory of the Department of Hematology at Erasmus University Medical Center for storage of samples, molecular analysis, and in vitro culture of leukemia cells; Arjan van de Berg and Kirsten van Lom for morphologic and cytochemical analysis; Tom Cupedo for his assistance with flow cytometry; and Timurs Maculins for mutational analysis of NOTCH1 in T-ALL samples.
This work was supported by the National Institutes of Health (NIH; CA118316; R.D.), the Dutch Cancer Society Koningin Wilhelmina Fonds (EMCR 2006-3522; R.D., P.J.M.V., and B.L.), and ErasmusMC (MRace; R.D.). M.E.F. is supported by an American Society of Hematology Fellow Scholar Award. A.M. is supported by the National Cancer Institute (R01 CA104348), the Chemotherapy Foundation, the Sam Waxman Cancer Research Foundation, and the G&P Foundation and is a Leukemia & Lymphoma Society Scholar. J.M.G. is supported by NIH (R01 HD044078). J.G. is supported by NIH (MSTP training grant GM007288).
Authorship
Contribution: M.E.F., B.J.W., J.M.G., A.M., and R.D. conceived and designed the research; M.E.F., B.J.W., Y.L., and C.A.J.E.-V. performed the research; M.E.F., B.J.W., L.S., J.G., M.F., A.M., and R.D. analyzed the data; A.W.L., B.L., P.J.M.V., and R.D. contributed research material; B.L., J.M.G., and P.J.M.V. contributed to data interpretation; M.F. and J.M.G. contributed analytical tools; and M.E.F., B.J.W., L.S., J.G., A.M., and R.D. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ari Melnick, Weill Cornell Medical College, 525 E 68th St, WMC box 113 (C-620), New York, NY 10021; e-mail: amm2014@med.cornell.edu.
References
Author notes
*M.E.F. and B.J.W. contributed equally to this study.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal