Abstract
Overexpression of methylguanine methyltransferase P140K (MGMTP140K) has been successfully used for in vivo selection and chemoprotection in mouse and large animal studies, and has promise for autologous and allogeneic gene therapy. We examined the long-term safety of MGMTP140K selection in a clinically relevant dog model. Based on the association of provirus integration and proto-oncogene activation leading to leukemia in the X-linked immunodeficiency trial, we focused our analysis on the distribution of retrovirus integration sites (RIS) relative to proto-oncogene transcription start sites (TSS). We analyzed RIS near proto-oncogene TSS before (n = 157) and after (n = 129) chemotherapy in dogs that received MGMTP140K gene-modified cells and identified no overall increase of RIS near proto-oncogene TSS after chemotherapy. We also wanted to determine whether in vivo selected cells retained fundamental characteristics of hematopoietic stem cells. To that end, we performed secondary transplantation of MGMTP140K gene-modified cells after in vivo selection in dog leukocyte antigen (DLA)–matched dogs. Gene-modified cells achieved multilineage repopulation, and we identified the same gene-modified clone in both dogs more than 800 and 900 days after transplantation. These data suggest that MGMTP140K selection is well tolerated and should allow clinically for selection of gene-corrected cells in genetic or infectious diseases or chemoprotection for treatment of malignancy.
Introduction
Hematopoietic stem cell (HSC) gene therapy has advanced to the point at which engraftment of gene-modified cells in large animal models has reached clinically therapeutic levels (> 10%-15%) for a variety of diseases (for review, see Neff et al1 ). In addition, several significant clinical gene therapy trials have clearly demonstrated the potential of retrovirus gene-modified cells as an effective cure for the genetic diseases of X-linked severe combined immunodeficiency (SCID-X1),2 adenosine deaminase-deficient severe combined immunodeficiency,3 and X-linked chronic granulomatous disease (X-CGD).4 These large animal and clinical studies are aided by either the use of ablative radiation or an inherent growth/survival advantage of the gene-modified cells after transplantation. For many other single-gene genetic diseases (ie, β-thalassemias and pyruvate kinase deficiency), acquired diseases (ie, glioblastoma and acute myeloid leukemia), or infectious diseases (ie, AIDS) in which retrovirus gene therapy is appropriate, a potential limiting curative factor is gene-marking levels below therapeutic thresholds due to the inability to use ablative conditioning and no substantial growth advantage/in vivo selection of gene-modified cells (for reviews, see Neff et al1 and Trobridge et al5 ). This requires alternate in vivo selection strategies after transplantation to increase the percentage of gene-modified cells to therapeutic levels. To realize the full clinical potential of retrovirus gene therapy, safe in vivo selection strategies using drug resistance gene therapy will be critical to propel these and other promising gene transfer applications.
The basic premise of drug resistance gene therapy is to stably deliver a transgene that confers resistance to a cytotoxic drug via several different mechanisms (for reviews, see Neff et al,1 Trobridge et al,5 and Sorrentino6 ). Several selectable markers have been extensively evaluated because of their ability to protect cells from cytotoxic agents, including dihydrofolate reductase (DHFR),7 multidrug resistance gene 1 (MDR1),8 cytidine deaminase (CDA),9 and methylguanine-DNA-methyltransferase (MGMT).10-13 The most promising in vivo selection strategy is methyl guanine methyltransferase mutant P140K (MGMTP140K). Several groups have demonstrated efficient, stable, multilineage in vivo selection in mice,14 dogs,10,11 and nonhuman primates (B.C.B., C.I., G.D.T., H.-P.K., manuscript in preparation) that received cells gene modified with MGMTP140K and received chemotherapy regimens consisting of either O6-benzylguanine (O6BG) and 1,3-bis-(2-chloroethyl)-1-nitrosourea (BCNU) or temozolomide. Stable multilineage increases in hematopoietic lineages after chemotherapy suggest that the selection occurs at the stem cell/early progenitor, and that selection will be critical for the applications mentioned above to avoid the need for chronic administration of drugs or pharmacologic agents. Although MGMTP140K-mediated in vivo selection and chemoprotection is an attractive approach, the power and consequence (benefit and detriment) of in vivo selection is clearly demonstrated by SCID-X1.
The potent intrinsic proliferative advantage of cells gene modified with the common γ-chain has led to the well-documented cure of SCID-X1 in 2 clinical trials,2 but, at the same time, led to several cases of gene therapy–induced leukemia.15,16 The success and subsequent leukemic transformation in multiple SCID-X1 patients were both intimately linked with the intrinsic in vivo selective advantage that the gene-modified cells possess. The clonal expansion that led to the adverse events in the SCID-X1 trial, the benign expansion in the X-CGD trial, and leukemia in large animal models have all had retroviral integration in or near known proto-oncogenes that at least partially led to the adverse events.4,15,17,18 Thus, we have focused our analysis on retrovirus integration sites (RIS) near transcription start sites (TSS) of proto-oncogenes.
Specifically, we directly compared the RIS profile around proto-oncogene TSS before and after multiple chemotherapy regimens in the dog. We also extended the studies to include a thorough examination of the engraftment, repopulation potential, and general fitness of MGMTP140K-selected HSCs. A true test of HSCs is elusive, but transplantation in the large animal (ie, dog or nonhuman primate) is the widely recognized “gold standard” of HSCs. Therefore, we evaluated engraftment and repopulation potential of MGMTP140K-selected cells in a secondary dog. The combination of these studies will test whether the approach using overexpression of MGMTP140K followed by multiple rounds of chemotherapy skews the RIS profile toward dangerous RIS by analysis of the genomic locus and the proximity of proto-oncogenes, and whether the selected cells still are able to function as multipotent HSCs.
Methods
Animals
Dogs were raised and housed at the Fred Hutchinson Cancer Research Center under conditions approved by the American Association for Accreditation of Laboratory Animal Care. Animal experiments were reviewed and approved by the Fred Hutchinson Cancer Research Institutional Animal Care and Use Committee.
Myeloablative transplantation with gammaretrovirus-transduced cells
Nonmyeloablative transplantation with lentivirus-transduced cells
Transplantation procedures and protocols have been previously described.19
Myeloablative secondary transplantation with lentivirus-transduced cells
Primary transplantation recipients received 5 consecutive days of recombinant canine granulocyte colony-stimulating factor (cG-CSF, 5 μg/kg body weight subcutaneously, twice daily) and recombinant canine stem cell factor (cSCF, 25 μg/kg body weight subcutaneously, once daily). On the sixth day before bone marrow harvest, a single dose of cG-CSF (5 μg/kg body weight subcutaneously) was administered. After bone marrow aspiration, the red blood cells were removed using standard ammonium chloride lysis. In preparation for infusion of the bone marrow product, the secondary recipient received oral cyclosporine beginning on day −3 (day 0 is the day of cell infusion) at 15 mg/kg body weight orally, twice daily. The secondary recipient received a myeloablative conditioning regimen of 920 cGy total body irradiation at 7 cGy/min delivered by a linear accelerator, and the cells were infused intravenously thereafter.
Retroviral vectors and transductions
Variable number of tandem repeats
In the allogeneic recipients, donor chimerism status was assessed using a polymerase chain reaction (PCR) analysis that measures the percentage of polymorphic (GAAA)n repeats. Briefly, the PCR reaction contained 10 pmol of the forward and reverse primer, 10% 10× PCR buffer with magnesium chloride, 200 μM deoxyribonucleotide mix, 0.1 units/μL of Platinum Taq DNA polymerase, and 60 ng of genomic sample DNA. The samples were amplified, and subsequently, Gene-Scan software (Applied Biosystems, Branchburg, NJ) was used to analyze donor- and host-specific peak areas and estimate the donor chimerism.
Flow cytometric analysis of gene-marked cells
Green fluorescent protein (GFP)–expressing white blood cells (WBCs) were quantitated by flow cytometric analysis of at least 20 000 events (propidium iodide [1 μg/mL] excluding, forward and right angle light scatter gated) on a FACSCalibur flow cytometer (BD Biosciences). Flow cytometric data were analyzed by CellQuest version 3.1f software (BD Biosciences) with gating to exclude fewer than 0.1% control cells in the relevant region.
Fluorescence-activated cell sorting of hematopoietic subsets
Hemolyzed blood was used as starting material for WBC analysis. WBCs were counted and labeled with antibodies specific to CD3 (obtained from Peter Moore, University of California, Davis, CA), CD14 (phycoerythrin [PE]–conjugated; DakoCytomation R0864, Carpinteria, CA), and DM5 (obtained from the Sandmaier laboratory, Fred Hutchinson Cancer Research Center, Seattle, WA) in separate tubes. The cells were washed, and cells labeled with CD3 and DM5 were stained with a PE-conjugated secondary antibody (polyclonal goat anti–mouse immunoglobulin PE-conjugated; DakoCytomation R0864). The cells were sorted on a FACSAria (BD Biosciences). The cells were gated using propidium iodide exclusion (as in “Flow cytometric analysis of gene-marked cells”); forward and right-angle light scatter was used to distinguish between bulk lymphocytes, bulk granulocytes, and bulk monocytes. The cell lineage gates were further separated by 2-color flow cytometry using PE-labeled antibodies to the surface markers for CD3+ lymphocytes, CD14+ monocytes, and DM5+ granulocytes. The sorted cells were reanalyzed on the FACSAria to check purity.
Quantitative PCR
Gene marking was also analyzed with the TaqMan 5′ nuclease quantitative real-time PCR assay. Sample DNA was analyzed in duplicate with a GFP-specific primer/probe combination (5′-CTG CAC CAC CGG CAA-3′; 5′-GTA GCG GCT GAA GCA CTG-3′; probe, CCA CCC TGA CCT ACG GCC TG-3′) or lentivirus-specific primer/probe combination (5′-TGA AAG CGA AAG GGA AAC CA-3′; 5′-CCG TGC GCG CTT CAG-3′; probe, 5′-6-carboxyfluorescein (FAM)-AGC TCT CTC GAC GCA GGA CTC GGC-5-(and 6)-carboxytetramethylrhodamine (TAMRA)–3′) (Synthegen, Houston, TX). In a separate reaction, a canine interleukin-3–specific primer/probe combination (5′-ATG AGC AGC TTC CCC ATC C-3′; 5′-GTC GAA AAA GGC CTC CCC-3′; probe, 5′-FAM-TCC TGC TTG GAT GCC AAG TCC CAC-TAMRA-3′) was used to adjust for equal loading of genomic DNA per reaction. Standards consisted of dilutions of DNA extracted from cell lines transduced with a single copy of a GFP-containing lentivirus vector and DNA from a normal dog. Reactions were run by the ABI master mix (Applied Biosystems) on the ABI Prism 7500 Sequence Detection System (Applied Biosystems) under the following thermal cycling conditions: 50°C for 2 minutes and 95°C for 10 minutes, then 40 cycles of 95°C for 15 seconds and 60°C for 1 minute.
Colony-forming unit assay
Bone marrow (∼ 10 mL) was obtained from dogs sedated by general anesthesia (ketamine, 10 mg/kg intravenously [IV]; diazepam, 0.5 mg/kg IV; butorphanol, 0.4 mg/kg IV; and rimadyl, 2.2 mg/kg subcutaneously). The cells were cultured in α minimal essential medium supplemented with FBS (HyClone, Logan UT), bovine serum albumin (Sigma-Aldrich, St Louis, MO), 0.5% (wt/vol) agar (Cambrex BioScience, Rockland, ME) overlaid on medium with 0.3% agar (wt/vol) containing 100 ng/mL cSCF, cG-CSF, canine granulocyte-macrophage colony-stimulating factor, and 4 U/mL erythropoietin. Cultures were incubated at 37°C in 5% CO2 and 95% air in a humidified incubator. Whole bone marrow cells were plated at a density of 105 cells per plate. Individual colonies were plucked after 14 days, and DNA from colony-forming units (CFUs) was extracted in a PCR tube by adding 10 μL of a picked colony into 90 μL of sterile H2O and 500 μg/mL proteinase K. After thorough vortexing, the sample was placed in a PCR machine running an extraction program (56°C, 120 minutes; 99°C, 10 minutes).
CFU retrovirus-specific PCR
To enumerate gene transfer DNA from CFUs and amplified to detect provirus sequences and general DNA, approximately 20 μL of the crude DNA extract was analyzed by PCR using retrovirus-specific forward and reverse primers (Lenti2F, 5′-AGA GAT GGG TGC GAG AGC GTC A-3′ and Lenti2R, 5′-TGC CTT GGT GGG TGC TAC TCC TAA-3′). In addition, another 20 μL of each colony extract is used to amplify the β-actin gene using specific forward and reverse primers (actin2 + 5′-CTA GAA GCA TTT GCG GTG GAC GAT-3′ and actin3 + 5′-GCT CCT CCC TGG AGA AGA GCT A-3′) for general DNA detection. PCR conditions include initial denaturation at 94°C for 1 minute, 40 cycles of 94°C for 1 minute, 65°C for 30 seconds, and 72°C for 1 minute, and a final extension at 72°C for 10 minutes. Negative controls are run without DNA (water) or with DNA extracted from normal canine cells. Percentage positive CFUs were calculated as the number of samples that were positive for both provirus and β-actin versus the number of samples positive for only β-actin.
Linear amplification-mediated PCR
Analysis of RIS
Integration site–specific PCR
Peripheral blood from different time points (G340 primary recipient: PBL 1101 days posttransplant (dpt), bulk granulocytes 1099 dpt, bulk lymphocytes 1098 dpt, DM5+ granulocytes 1100 dpt, CD14+ monocytes 1099 dpt, CD3+ lymphocytes 1101 dpt; G346 secondary recipient: PBL 644 dpt, bulk granulocytes 659 dpt, bulk lymphocytes 659 dpt, DM5+ granulocytes 660 dpt, CD14+ monocytes 651 dpt, CD3+ lymphocytes 663 dpt) was obtained, DNA was extracted with a QIAamp DNA blood kit following the manufacturer's directions, and 300 ng of DNA was used in subsequent PCR reactions. Primers used previously during LAM-PCR specific to the virus long terminal repeat (LTR) were used as the forward primer, and canine-specific reverse primers to designated integration sites were designed between 150 and 900 bp downstream from the identified LTR genome junction site. Standard PCR conditions were used to amplify the specific integration sites (94°C, 5 minutes; 40 cycles of 94°C for 60 seconds; 60°C for 60 seconds; 72°C for 90 seconds; 72°C for 10 minutes). PCR products were visualized on 2% agarose 1× Tris-acetic acid-EDTA (TAE) gels. Normal dog DNA samples were run under identical conditions. Only single bands of samples with negative controls were excised, TOPO TA (Invitrogen, Carlsbad, CA) cloned, and sequenced with M13 primers, as described.
Chemotherapy treatment with O6BG, BCNU, or temozolomide
Results
Sustained multilineage hematopoietic repopulation of MGMTP140K gene-marked cells
The crucial component of successful retrovirus-mediated in vivo selection is sustained multilineage selection without requirement of chronic drug treatment. In this study, we show long-term follow-up of several dogs (Figure 1; Figure S2 and Table S1, available on the Blood website; see the Supplemental Materials link at the top of the online article) after sustained in vivo selection with either O6BG and BCNU or temozolomide with a median follow-up of approximately 4.8 years (n = 11) after transplantation. In all dogs, peripheral blood counts remained in a normal range during this time period, apart from transient drops in counts related to chemotherapy treatment. Morphologic bone marrow examinations were performed at least twice in each dog. All samples showed trilineage maturation and a normal blast count with no dysplastic features and, in a recent report, normal peripheral blood maturation into all hematopoietic lineages was analyzed.17 For additional information regarding clinical analysis of 2 dogs that presented with nonhematopoietic complications, see below.
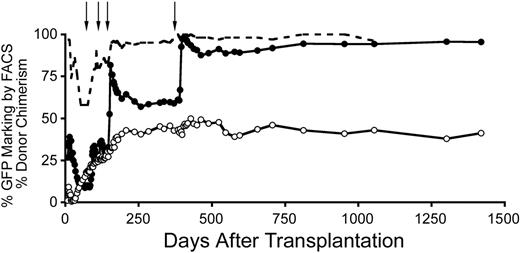
Stable increase in gene marking after in vivo selection of MGMTP140K gene-modified cells. Gene marking in granulocytes (●), lymphocytes (○), and donor chimerism (----) before and after in vivo selection of MGMTP140K-GFP gene-marked cells in a dog. Treatment with O6BG and BCNU denoted by ↓.
Stable increase in gene marking after in vivo selection of MGMTP140K gene-modified cells. Gene marking in granulocytes (●), lymphocytes (○), and donor chimerism (----) before and after in vivo selection of MGMTP140K-GFP gene-marked cells in a dog. Treatment with O6BG and BCNU denoted by ↓.
Durable MGMTP140K-mediated in vivo selection and chemoprotection
We have previously shown10,11 efficient in vivo selection and chemoprotection in the dog using MGMTP140K and wanted to extend these studies to test durability/longevity of this approach. Two dogs (G179 and G197) received autologous CD34+-selected cells transduced with RD114 pseudotype retroviral vector MIEG(P140K), whereas 2 dogs received dog leukocyte antigen (DLA)–identical allogeneic CD34+-selected cells transduced with the same vector. These results have been published previously. In all dogs, we were able to achieve successful in vivo selection and chemoprotection.10,11
The 2 recipients of autologous cells received a final treatment of O6BG and temozolomide after approximately 17 months between treatments. In each case, the dose was 100 mg/m2 higher than the previous dose. In G179, the level of gene marking in granulocytes increased from 83% to 99%. In G197, gene marking rose from 47% to 99% in granulocytes (Figure 2A). In the 2 autologous animals that received a further cycle of treatment, we once again observed chemoprotection mediated by the gene-marked cells. G179 received 800 mg/m2 temozolomide, but the absolute neutrophil count (ANC) did not drop below 2000/μL, and the platelets remained above 150 000/μL. In G197, which received a dose of 900 mg/m2, ANC did not drop below 1500/μL, and platelets remained above 100 000/μL (Figure 2B). As a control, we have administered 800 mg/m2 temozolomide to a dog with very low gene marking (below 0.5%). In this animal, ANC dropped to less than 100/μL and remained below 500/μL for 9 days. Platelets went down to 3000/μL and remained below 50 000/μL for 21 days (data not shown). This confirms that MGMTP140K-mediated in vivo selection and chemoprotection are durable even after prolonged durations between chemotherapy.
Durable in vivo selection and chemoprotection. (A) Gene marking in granulocytes (●) and lymphocytes (○) before and after in vivo selection of MGMTP140K-GFP gene-marked cells in a dog. Treatment with O6BG and temozolomide denoted by black arrows. (B) The corresponding platelet count of the same dog during the chemotherapy treatment cycles.
Durable in vivo selection and chemoprotection. (A) Gene marking in granulocytes (●) and lymphocytes (○) before and after in vivo selection of MGMTP140K-GFP gene-marked cells in a dog. Treatment with O6BG and temozolomide denoted by black arrows. (B) The corresponding platelet count of the same dog during the chemotherapy treatment cycles.
MGMTP140K-mediated in vivo selection does not select for RIS near proto-oncogene TSS
The major adverse consequence of HSC gene therapy has been dominance and subsequent leukemic outgrowth of gene-modified clones with provirus integration in or near proto-oncogenes (for reviews, see Baum et al25,26 ). Gene modification with MGMTP140K confers a profound selective advantage in the presence of chemotherapy agents (ie, BCNU and temozolomide), and we wanted to study whether the selection created a bias for clones with RIS near proto-oncogene TSS. The common thread between leukemic transformations in the SCID-X1 clinical trials has been provirus integration near the TSS of proto-oncogenes, so this is what we focused on for safety evaluation. To accomplish this, we carried out integration site analysis both before and after chemotherapy (Table 1). Initially, it was clear that multiple clones contributed to hematopoiesis from the polyclonal banding pattern of the LAM-PCR products (Figure 3A,B). Grouping the mapped retroviral integrants into populations before (n = 157) and after (n = 127) chemotherapy revealed that there was not a consistent significant increase of clones surrounding the TSS of proto-oncogenes (Figure 3C, and Tables S2, S3). The only significant increase of mapped RIS from the postchemotherapy group relative to the prechemotherapy group that was also significantly higher than random integration was in the window below 10 kb upstream of proto-oncogene TSS. Using the subset of RIS that were most frequently captured during sequencing as a surrogate assay for clonal contribution did not directly correlate with proximity from a proto-oncogene TSS (Tables S2, S3). The average distance from proto-oncogene TSS of the top 10% most frequently captured RIS was approximately 1 Mb before chemotherapy and approximately 1.7 Mb after chemotherapy. Furthermore, a frequently captured RIS before chemotherapy did not identify a RIS that would also dominate after chemotherapy because this was never identified (Table S4). There was also no distinct difference between the numbers of RIS isolated from dogs that received posttransplant BCNU or temozolomide (Table 1). Additional long-term dogs (Figure S2) not included in the genomic mapping detailed above displayed polyclonal banding patterns of RIS amplified by LAM-PCR from the most recent samples analyzed with no indication of oligo- or monoclonality. All dogs analyzed continue to display polyclonal repopulation even up to 6 years after transplantation and after numerous rounds of in vivo selection with either O6BG and BCNU or temozolomide.
RIS profile before and after chemotherapy
| Treatment . | No. of RIS . | Range of dpt . | No. of chemotherapy treatments . |
|---|---|---|---|
| Prechemotherapy | 157 | 61-112 | |
| Temozolomide* | 73 | 80-112 | |
| BCNU† | 84 | 61-104 | |
| No. of RIS <10 kb from proto-oncogene TSS | 3 | ||
| No. of RIS <100 kb from proto-oncogene TSS | 8 | ||
| No. of RIS <500 kb from proto-oncogene TSS | 56 | ||
| Average distance from proto-oncogene TSS | ∼1.81 Mb | ||
| Postchemotherapy | 127 | 701-1448 | |
| Temozolomide* | 64 | 722-1425 | 9 (G179) and 9 (G197)‡ |
| BCNU† | 63 | 701-1448 | 2 (G069) and 4 (G154) |
| BCNU§ | 36 | 740-1037 | 7 (G187) and 4 (G250) |
| No. of RIS <10 kb from proto-oncogene TSS | 2 | ||
| No. of RIS <100 kb from proto-oncogene TSS | 6 | ||
| No. of RIS <500 kb from proto-oncogene TSS | 37 | ||
| Average distance from proto-oncogene TSS | ∼2.19 Mb | ||
| Total sites | 284 | ||
| Shared sites | 19 |
| Treatment . | No. of RIS . | Range of dpt . | No. of chemotherapy treatments . |
|---|---|---|---|
| Prechemotherapy | 157 | 61-112 | |
| Temozolomide* | 73 | 80-112 | |
| BCNU† | 84 | 61-104 | |
| No. of RIS <10 kb from proto-oncogene TSS | 3 | ||
| No. of RIS <100 kb from proto-oncogene TSS | 8 | ||
| No. of RIS <500 kb from proto-oncogene TSS | 56 | ||
| Average distance from proto-oncogene TSS | ∼1.81 Mb | ||
| Postchemotherapy | 127 | 701-1448 | |
| Temozolomide* | 64 | 722-1425 | 9 (G179) and 9 (G197)‡ |
| BCNU† | 63 | 701-1448 | 2 (G069) and 4 (G154) |
| BCNU§ | 36 | 740-1037 | 7 (G187) and 4 (G250) |
| No. of RIS <10 kb from proto-oncogene TSS | 2 | ||
| No. of RIS <100 kb from proto-oncogene TSS | 6 | ||
| No. of RIS <500 kb from proto-oncogene TSS | 37 | ||
| Average distance from proto-oncogene TSS | ∼2.19 Mb | ||
| Total sites | 284 | ||
| Shared sites | 19 |
Sites identified in dogs that were treated with O6BG and temozolomide.
Sites identified in dogs that were treated with O6BG and BCNU.
Dogs G179 and G197 received a total of 10 chemotherapy treatments, but only 9 before retrovirus integration site analysis.
Sites identified in dogs (G187 and G250) that were treated with O6BG and BCNU but not included in the analysis of total sites or postchemotherapy.
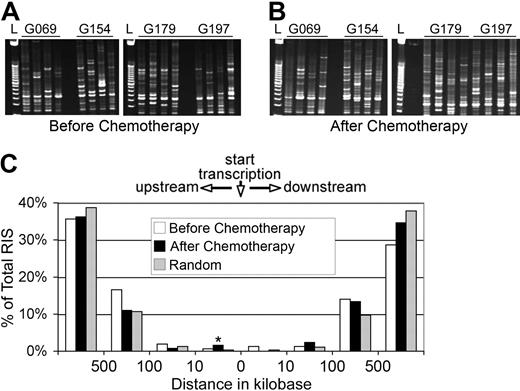
Similar distribution of retroviral integrants relative to proto-oncogenes before and after chemotherapy. Representative gel of RIS amplified by LAM-PCR before (A) and after (B) chemotherapy with either O6BG and BCNU (G069 and G154) or O6BG and temozolomide (G179 and G197). (C) The positions of RIS mapped relative to the RefSeq gene TSS of proto-oncogenes as defined in either the Sanger Cancer Gene Census or the Retroviral Tagged Cancer Gene Database. *P < .001 (SD).
Similar distribution of retroviral integrants relative to proto-oncogenes before and after chemotherapy. Representative gel of RIS amplified by LAM-PCR before (A) and after (B) chemotherapy with either O6BG and BCNU (G069 and G154) or O6BG and temozolomide (G179 and G197). (C) The positions of RIS mapped relative to the RefSeq gene TSS of proto-oncogenes as defined in either the Sanger Cancer Gene Census or the Retroviral Tagged Cancer Gene Database. *P < .001 (SD).
Clinical analysis of 2 dogs with health complications
As stated above, normal hematopoiesis was observed in all long-term dogs transplanted with MGMTP140K gene-modified cells. Recently, 2 dogs had to be euthanized due to a general decline in health. One dog, approximately 5.5 years (Table S1 G069) after receiving a DLA-identical allogeneic graft and posttransplant in vivo selection with BCNU (G069), presented with imbalance, bloody diarrhea, and declining body score, as well as a low blood count hematocrit (HCT) of 25%. Thorough examination by a board-certified veterinarian revealed no specific symptom responsible for the observed decline in health. After examination, this dog was observed having a seizure, for which diazepam was administered (0.1 mg/kg). Chemistry panels demonstrated mild elevation of liver enzymes, including alkaline phosphatase at 99 U/L (normal range 10-84 U/L), alanine aminotransferase at 74 U/L (normal range 5-65 U/L), aspartate aminotransferase at 73 U/L (normal range 16-60 U/L), and low albumin at 1.3 g/dL (normal range 2.3-4.0 g/dL); however, no values were considered critical. Blood cultures returned negative, and blood counts, aside from HCT, continued to be normal. Necropsy and subsequent pathologic analyses revealed only encephalitis of the temporal lobe with focal demyelinization. A second dog approximately 5.5 years (Table S1 G179), after receiving an autologous graft and posttransplant in vivo selection with temozolomide (G179), also presented with decreasing activity. Upon examination including ultrasound, the animal was found to have an abdominal mass. Based on these findings, the animal was euthanized. Autopsy revealed a metastatic hemangiosarcoma with no marrow involvement. LAM-PCR on marrow and tissues from the primary and metastatic tumors showed a polyclonal marking pattern, thus demonstrating that there was no monoclonal vector insertion contributing to the hemangiosarcoma (data not shown). This is a tumor that can develop spontaneously in dogs (generally after 8-10 years of age) and is somewhat common (for review, see Smith27 ). Although large datasets on specific breeds of dog and frequency of hemangiosarcoma development are limited, approximately 0.3% to 2% of all dog necropsies note hemangiosarcoma and 7% of all malignancies in dogs are hemangiosarcoma.28 In addition, dogs are more prone to tumors after total body irradiation.29 Thus, to the best of our knowledge and analysis, this tumor has no association with the genetic modification of the repopulating cells. Generally, the long-term data in this study support the safety of this approach, and the only adverse events appear to be unrelated to the MGMTP140K gene-marked cells. The remaining dogs described in this work are all healthy at the last examination with normal blood counts.
Gene-modified cells maintain multilineage hematopoietic repopulation potential after in vivo selection
The most convincing assay for HSC is multilineage hematopoietic repopulation in a large animal model. To address this, we used the DLA-identical dog model and carried out primary and secondary transplants, with the secondary transplants coming after multiple rounds of in vivo selection in the primary recipient (Figure 4A schematic representation). Dogs were only considered for secondary transplantation if their original donor or a DLA-identical dog from the colony was available, which reduced the number of dogs available. Primary recipients received multiple rounds of in vivo selection and gene marking stabilized in dogs G340 and G403 after the final round of in vivo selection at approximately 60% and 4%, respectively (Figure 4B,C top panels). After transplantation, the secondary recipients (Figure 4B bottom panel G346, and 4C bottom panel G404) recovered neutrophil and platelet counts within expected time frames compared with historical controls. Shortly after transplantation, the gene marking stabilized at approximately 80% and 1% in dogs G346 and G404, respectively (Figure 4). Interestingly, the magnitude of expansion/in vivo selection, most notably achieved in G340 (Figure 4B top panel), was maintained in the secondary recipient G346 (Figure 4B bottom panel). PCR of CFUs in this first pair of primary and secondary recipients (Figure 4B) confirmed that the gene marking was from similar numbers of gene-marked immature progenitors or stem cells (data not shown) and not simply selection for clones with multiple integration sites.
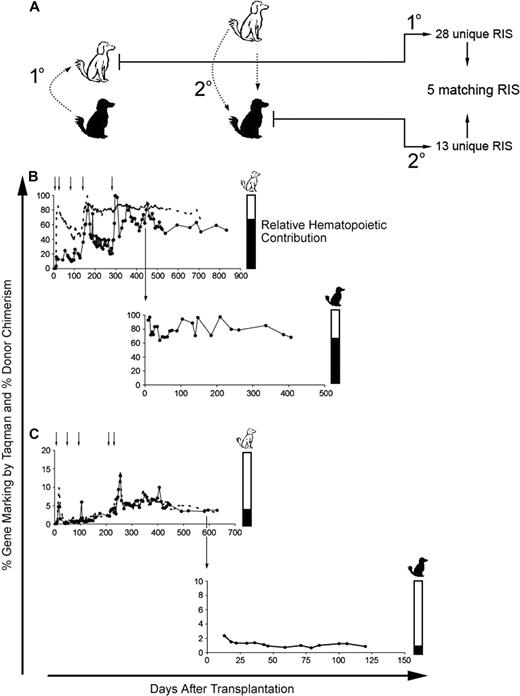
Engraftment and multilineage repopulation potential of MGMTP140K gene-modified cells in secondary recipients after in vivo selection. (A) Schematic representation of secondary transplantation in DLA-matched dogs. The white dog represents the initial recipient of the gene-marked hematopoietic cells from the black dog (1°). The 2° transfer of the MGMTP140K gene-modified cells back into the original donor (black dog). (B,C top panels) Percentage of donor-positive WBC (----) and donor gene-modified WBC (●) in primary recipients (white dogs) is shown as a function of time. Chemotherapy denoted by small black arrows above the graph. Large black arrow indicates the time point at which the bone marrow was transplanted back into the original donors (black dogs). (B,C bottom panels) Percentage of gene-modified WBC in the secondary recipients (black dogs). Bar graphs to the right of the gene-marking plots are an alternate representation of hematopoietic contribution from each dog (white and black).
Engraftment and multilineage repopulation potential of MGMTP140K gene-modified cells in secondary recipients after in vivo selection. (A) Schematic representation of secondary transplantation in DLA-matched dogs. The white dog represents the initial recipient of the gene-marked hematopoietic cells from the black dog (1°). The 2° transfer of the MGMTP140K gene-modified cells back into the original donor (black dog). (B,C top panels) Percentage of donor-positive WBC (----) and donor gene-modified WBC (●) in primary recipients (white dogs) is shown as a function of time. Chemotherapy denoted by small black arrows above the graph. Large black arrow indicates the time point at which the bone marrow was transplanted back into the original donors (black dogs). (B,C bottom panels) Percentage of gene-modified WBC in the secondary recipients (black dogs). Bar graphs to the right of the gene-marking plots are an alternate representation of hematopoietic contribution from each dog (white and black).
Hematopoietic subset staining, FACS sorting, and retrovirus-specific TaqMan PCR were used to confirm contribution of gene-modified cells in all hematopoietic lineages. After stable engraftment in secondary recipients, retrovirus-specific TaqMan PCR was carried out on FACS-sorted bulk lymphocytes, granulocytes, DM5+ granulocytes, CD3+ lymphocytes, and CD14+ monocytes (Figure S1) in primary and secondary recipients. TaqMan PCR confirmed that all hematopoietic lineages tested had contribution from gene-modified cells (data not shown).
Multiple gene-marked clones contribute to hematopoiesis in primary and secondary recipients
To determine whether the high level of gene marking in the secondary recipient G340 (Figure 4B bottom panel) was a result of MGMTP140K-mediated selection in the primary recipient and not outgrowth of a dominant clone, we carried out integration site analysis in both animals 106 days after the secondary transplantation along with initial integration analysis in the second pair of dogs (Figure 4A,B). Preliminary retrovirus integration analysis confirmed that multiple clones contributed to hematopoietic repopulation in the primary and secondary recipients (Figure 5A,B), and genomic mapping of sites isolated from the dogs in Figure 4B identified multiple unique RIS and 5 identical clones in primary and secondary recipients from LAM-PCR analysis of DNA prepared from peripheral blood cells (Figure 4A). In addition, using primers designed to amplify a specific integration site, we were able to identify the identical clone in bulk sorted lymphocytes, granulocytes, DM5+ granulocytes, CD3+ lymphocytes, and CD14+ monocytes in primary and secondary recipients (Figure 5C). Using the same specific clone tracking approach, we were also able to identify the same clone as early as 58 days after transplantation in the primary recipient until the most recent follow-up 1386 days after primary transplantation and 938 days after secondary transplantation (data not shown). This clone was also identified in the secondary recipient at multiple time points ranging from 69 to 938 days after transplantation (data not shown).
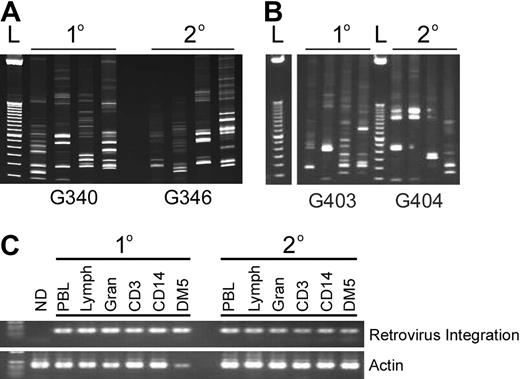
Multiple clones and common clones contribute to hematopoiesis in primary and secondary recipients. (A,B) Representative gel of RIS amplified by LAM-PCR in primary (1°) and secondary (2°) recipients. (C) Detection of a common integration site in different hematopoietic lineages using LTR-specific (forward) and dog genomic (reverse) primers of the primary and secondary dogs pictured in panel A. “ND” is a different dog that received lentivirus-transduced cells.
Multiple clones and common clones contribute to hematopoiesis in primary and secondary recipients. (A,B) Representative gel of RIS amplified by LAM-PCR in primary (1°) and secondary (2°) recipients. (C) Detection of a common integration site in different hematopoietic lineages using LTR-specific (forward) and dog genomic (reverse) primers of the primary and secondary dogs pictured in panel A. “ND” is a different dog that received lentivirus-transduced cells.
Chemoprotection in secondary recipients of MGMTP140K gene-modified cells
To determine whether the MGMTP140K gene-modified cells maintained their ability to facilitate chemoprotection and in vivo selection in secondary recipients, we treated one of the secondary dogs (Figure 4B bottom panel) with a dose of O6BG and BCNU that from previous studies in our laboratory11 would cause substantial neutropenia and thrombocytopenia in a dog with low to no gene marking. The dog was treated with 5 mg/kg O6BG and 0.4 mg/kg BCNU, as previously described,11 and total blood cell counts, including ANC and platelets, were monitored for 3 weeks after treatment. There was no pronounced cytopenia (Figure S3), with ANC never dropping below 3500/μL and platelets never dropping below 240 000/μL. In addition, gene marking levels, determined by TaqMan PCR, increased after treatment with O6BG/BCNU (Figure S4).
Discussion
The development of leukemia in clinical trials involving patients with SCID-X1 after HSC gene therapy and the demonstration that HoxB4 gene-modified cells lead to leukemia in large animals emphasize the importance of a thorough examination of strategies that elicit positive selection to gene-modified cells.15,17 These findings raise the question whether all ex vivo and in vivo selection strategies carry an increased risk of leukemic transformation or whether this is a result of specific genetic backgrounds and/or the transgenes involved. To this end, we wanted to examine the MGMTP140K-based in vivo selection strategy that is currently the most robust selection system based on mouse and large animal data.10,11 In a clinically relevant dog model of HSC gene therapy, we were able to show that multiple rounds of chemotherapy did not substantially alter the RIS profile with respect to the proximity to proto-oncogene TSS. In addition, after multiple rounds of chemotherapy, MGMTP140K gene-modified cells were able to engraft a second dog, contribute to polyclonal multilineage hematopoiesis, and maintain chemoprotective functions.
Early attempts at drug resistance gene therapy, resulting in sustained in vivo selection at the stem cell level, defined as stable multilineage increases in gene-marked cells after treatment with a chemical or biologic agent, were not successful mainly due to the type of drug (mechanism of action) and the associated transgene that confers resistance or growth advantage (for review, see Neff et al1 and Sorrentino6 ). A variety of systems has been evaluated for the ability to confer drug resistance after retrovirus delivery/overexpression to target cells and includes, but is not limited to, DHFR mutants,30 MDR1,8,31 CDA,9,32 aldehyde dehydrogenase (ALDH),33 glutathione S-transferase (GST),34 and MGMT mutants.10,11,14,35 Early studies with DHFR, MDR1, and more recently CDA clearly demonstrated a selective advantage of circulating gene-modified cells in murine models, but in all cases this selection was transient, suggesting the selection was at the committed progenitor and/or terminally differentiated cell. In addition, preliminary studies with ALDH and GST have shown that retrovirus delivery/overexpression to target cells does lead to chemoprotection in vitro, but in vivo studies of these 2 drug resistance genes have not been reported. These combined results point to the crux of the dilemma: what constitutes a potent stem cell selection system? All of the studies mentioned above showed efficient gene delivery and expression of the functional transgene. Therefore, it is apparent that the lack of sustained selection is not due solely to the gene transfer approach, but is also critically influenced by the method of action of the associated drug(s) and the magnitude of positive selection of the gene-modified cells and negative selection of the wild-type cells after drug treatment.14 Specifically, chemoprotection of gene-modified cells conferred by DHFR mutants to methotrexate, MDR1 to etoposide, daunomycin, taxol, doxorubicin, and vinblastine, and CDA to cytarabine and gemcitabine works efficiently, but there is no or minimal HSC toxicity of the associated drug(s). This suggests that, whereas these drug resistance gene therapy approaches are attractive for a variety of disease models, using them for HSC selection is unlikely to be successful. Alternatively, ALDH and GST elicit chemoprotection from drugs, cyclophosphamide and busulfan/melphalan, respectively, which have been shown to act at least in part through HSC toxicity. This suggests ALDH and GST overexpression in HSCs may be promising, but translation of these systems to in vivo models is as yet untested. Although these systems have their utility for alternative approaches, HSC selection requires a transgene that confers substantial chemoprotection from a defined stem cell toxin.
Drugs such as BCNU and, more recently, temozolomide are well-established stem cell and early progenitor toxins.36-38 A seminal observation was that retrovirus delivery/overexpression of bacterial O6-alklguanine-DNA alkyltransferase39 and subsequently by human alkyltransferase40,41 protected a variety of cells, including hematopoietic progenitors, from BCNU. This was the first indication that MGMT might be a useful strategy for in vivo selection and chemoprotection. Subsequently, specific active site inhibitors of wild-type MGMT were developed, namely O6BG, which sensitized cells in a synergistic fashion when used in conjunction with BCNU.42,43 The development of specific mutants of MGMT that were resistant to O6BG44-49 further improved the positive selection of gene-modified cells relative to cells expressing the wild-type form of MGMT, and also reduced the mutagenesis rate.49 These mutants of MGMT have led to the current strategy of MGMT-mediated drug resistance gene therapy using retrovirus-delivered mutant versions of MGMT (ie, P140K and G156A) to confer resistance of HSCs to chemotherapy regimens of O6BG and BCNU or temozolomide14,35,50 and allow for in vivo selection and chemoprotection. We first described the translation of this work to the large animal model,10,11 and in this study demonstrate sustained and durable in vivo selection and chemoprotection. These are convincing data that MGMTP140K-mediated in vivo selection and chemoprotection occur at the level of HSCs and can be achieved with either a gammaretovirus- or lentivirus-based vector. In addition, it is important to note that we have demonstrated efficient engraftment and in vivo selection with lentivirus vectors using either the elongation factor 1α or spleen focus-forming virus (SFFV) promoters to express MGMTP140K. The differences that we report in this study, and previously in the macaque model,51 to murine studies52 with regard to engraftment of cells expressing MGMTP140K from the SFFV promoter could be due to different expression levels between the animal models. This is not the only possibility and warrants further investigation. But, because of the clinical experience with SCID-X1 in which potent in vivo selection led to leukemic transformation, we wanted to extend these studies of MGMTP140K in the clinically relevant dog model to test the stability of the gene-modified graft. It was important to assess whether the repeated drug treatments had compromised or substantially altered the gene-modified pool, potentially leading to leukemic transformation.
All of the instances of leukemic transformation in clinical gene therapy studies and large animal models have involved provirus integration in or near proto-oncogenes.15-18 With the current integrating vector systems commonly used for gene therapy, integration in the proximity of proto-oncogenes is inevitable regardless of the type of integrating vector system (ie, gammaretrovirus, lentivirus, or foamy virus23 ). In the absence of a survival advantage for gene-modified cells, leukemic transformation in large animal models has been very rare,18 even though integration analysis has demonstrated that there are numerous provirus integration events in and near numerous proto-oncogenes.23,53,54 Previous work in a murine model of MGMTP140K demonstrated that clones survived serial transplantation and multiple rounds of chemotherapy, but the RIS profile was not studied due to a relatively limited number of identified integration sites.55 Therefore, we wanted to determine whether in vivo selection using MGMTP140K caused a disproportionate bias for RIS in or near proto-oncogene TSS as an indication of a preleukemic state. Although numerous RIS were identified less than 500 K from proto-oncogene TSS, including genes such as MDS1/Evi1 and LMO2, there was no overall selection for these sites after multiple rounds of chemotherapy, indicating that selection is dependent upon the protection elicited by the overexpression of MGMTP140K during chemotherapy and not due to outgrowth of potentially dangerous clones with an intrinsic growth advantage. This, in turn, suggests that MGMTP140K-mediated in vivo selection/chemoprotection is a relatively safe approach. Another potential safety risk associated with drug resistance gene therapy is the overexpression of the transgene, affording protection of gene-modified cells from what would otherwise be toxic levels of chemotherapy. By using such a system instead of biologic means for selection (ie, growth factors), there is the potential to compromise the selected cells. Several studies have documented deleterious effects of cytotoxic therapy to HSCs.56,57 So, to address the fundamental question whether multiple rounds of chemotherapy had compromised the basic “stemness” of the HSCs, we conducted secondary transplantations in the dog model after in vivo selection in primary recipients.
Large animal transplantation is the most thorough test of the multilineage engraftment and repopulation potential of HSC and gene-modified HSCs. For instance, relative to murine transplantation (even serial transplantation), the proliferation requirement of a HSC in the large animal is orders of magnitude higher.58 Therefore, secondary transplantation in the large animal model is a comprehensive test of the self-renewal and multilineage proliferation capacities of gene-modified cells and an excellent assay to test whether in vivo selection in primary recipients compromised the gene-modified cells. If multiple chemotherapy treatments compromised the “stemness” of MGMTP140K gene-modified HSCs, potential outcomes could be a failure to engraft or, over time, an exhaustion of gene-modified cells leading to graft failure in secondary recipients. On the contrary, however, we observed quite the opposite of the MGMTP140K gene-modified cells, with efficient engraftment in primary recipients, survival, and in vivo selection after multiple rounds of chemotherapy with O6BG and temozolomide or BCNU. Maintenance of engraftment and multilineage repopulation potential in secondary recipients and follow-up of the dogs are ongoing. These findings demonstrate that the engraftment and repopulation in secondary recipients are derived from gene-modified HSCs with multilineage repopulation potential.
Using safe and effective in vivo selection and chemoprotection strategies will be essential to translate the most promising gene therapy applications for treatment of a variety of diseases. In the wake of severe adverse events in a variety of gene therapy studies in which the common thread was improved survival/proliferation of gene-modified cells, the safety of any future approach is critically important before attempting clinical trials. We have previously demonstrated potent in vivo selection and chemoprotection of MGMTP140K in the dog model, which suggests this method should translate well to humans. The proof of principle has now been well established for MGMTP140K, but a thorough examination of the safety of this approach was required. Generally, the long-term data reported in this study are promising, and the only adverse events appear to be unrelated to the MGMTP140K gene-marked cells. Furthermore, the RIS profiling around proto-oncogene TSS and large animal model secondary transplantation is convincing evidence that MGMTP140K-mediated in vivo selection is relatively safe, but continued improvement in vector design, targeted integration, regulated expression, and a thorough understanding of what effect underlying genetic background may have on in vivo selection are required to continue to alleviate safety concerns. Progress in all of these areas is required to translate promising in vivo selection strategies, such as MGMT, to clinical applications.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Bonnie Larson and Helen Crawford for help in preparing the manuscript. We thank Michele Spector and the Fred Hutchinson Cancer Research Center Dog Lab staff for the care of the dogs.
This work was supported in part by National Institutes of Health (Bethesda, MD) grants HL36444, HL74162, DK56465, HL092554, and DK47754.
National Institutes of Health
Authorship
Contibution: B.C.B. designed and performed research, performed statistical analysis, and wrote the manuscript; R.S., K.A.K., and S.G. performed research; C.I. collected data; T.N. and G.D.T. designed and performed research; and H.-P.K. designed research and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Dr Hans-Peter Kiem, Fred Hutchinson Cancer Research Center, Clinical Research Division, D1-100, 1100 Fairview Ave N, PO Box 19024, Seattle, WA 98109-1024; e-mail: hkiem@fhcrc.org.