Abstract
The treatment of healthy donors with granulocyte colony-stimulating factor (G-CSF) and dexamethasone results in sufficient numbers of circulating granulocytes to prepare granulocyte concentrates for clinical purposes. Granulocytes obtained in this way demonstrate relatively normal functional behavior combined with a prolonged life span. To study the influence of mobilizing agents on granulocytes, we used oligonucleotide microarrays to identify genes that are differentially expressed in mobilized granulocytes compared with control granulocytes. More than 1000 genes displayed a differential expression pattern, with at least a 3-fold difference. Among these, a large number of genes was induced that encode proteins involved in inflammation and the immune response, such as C-type lectins and leukocyte immunoglobulin-like receptors. Because mobilized granulocytes have a prolonged life span, we focused on genes involved in the regulation of apoptosis. One of the most prominent among these was CAST, the gene encoding calpastatin. Calpastatins are the endogenous inhibitors of calpains, a family of calcium-dependent cysteine proteases recently shown to be involved in neutrophil apoptosis. Transcriptional activity of the CAST gene was induced by G-CSF/dexamethasone treatment both in vivo and in vitro, whereas the protein expression of CAST was stabilized during culture. These studies provide new insight in the genotypic changes as well as in the regulation of the immunologic functions and viability of mobilized granulocytes used for clinical transfusion purposes.
Introduction
Granulocyte concentrates constitute a promising adjuvant tool in the treatment of neutropenic and immunocompromised patients experiencing life-threatening infections in which the exclusive use of modern antimicrobial drugs and additional growth factors is ineffective.1-3 Donor stimulation with a combination of granulocyte colony-stimulating factor (G-CSF) and dexamethasone has become the standard procedure to increase the number of neutrophils in the circulation of the donors and, thus, to collect a sufficient amount of cells for the preparation of granulocyte concentrates.4
In addition to its important function in granulopoiesis, G-CSF has been reported to modulate several granulocyte functions in vitro. For instance, G-CSF increases the chemotactic ability, enhances cell adhesion to vascular endothelium, promotes phagocytosis, and primes the NADPH-oxidase activity (for a review, see Eyles et al5 ). However, glucocorticosteroids such as dexamethasone have been suggested to suppress some granulocyte functions, including mobility, adhesion, and microbial killing.6-8
Despite all the aforementioned studies, we and others have found that donor granulocytes, when mobilized for transfusion purposes, show virtually normal functional characteristics in vitro in the presence of some minor phenotypic changes, whereas their life span is consistently prolonged.9,10
Both G-CSF and dexamethasone are well-established prosurvival factors for neutrophilic granulocytes,11-13 and this effect involves various survival signaling pathways. G-CSF has been shown to increase in neutrophils the mRNA expression of A1/Bfl-1, an antiapoptotic member of the Bcl-2 family of proteins.14 The prosurvival effect of dexamethasone in neutrophils has been connected to the stabilization of Mcl-1, another antiapoptotic Bcl-2 family member, during neutrophil culture.15 Furthermore, G-CSF inhibits the mitochondria-dependent activation of caspase-3 in neutrophils via control of the calpain-dependent degradation of the X-linked inhibitor of apoptosis (XIAP).16,17 In these studies, the antiapoptotic effect of G-CSF was shown to depend on de novo protein synthesis.
Calpains form a family of calcium-dependent cysteine proteases of which calpain-1 (μ-calpain, calpain I), calpain-2 (m-calpain, calpain II), and the natural inhibitor of calpains, calpastatin, are ubiquitously expressed. Several studies have implicated calpain activity in spontaneous neutrophil apoptosis,18,19 and there is a growing number of indications that calpains play an important role in the early phase of programmed cell death.17,20
Although some of the major functional characteristics relevant for host defense seem well preserved in the donor granulocytes, the extent to which other relevant properties of these cells are altered by the in vivo preactivation is as yet unclear. To understand how G-CSF and dexamethasone may induce the granulocytes to obtain their increased survival capacity, we performed a comparative study with the use of Agilent microarrays (Agilent Technologies Netherlands, Amstelveen, The Netherlands) coupled with real-time reverse transcription–polymerase chain reaction (RT-PCR), flow cytometry, and immunoblotting. In the present study, we demonstrate that mobilization of granulocytes with G-CSF and dexamethasone has a considerable impact on gene expression of these cells, with more than 1000 genes being strongly affected. These changes could be partially mimicked by in vitro culture of neutrophils in the presence of G-CSF and dexamethasone. However, more than 75% of changes in gene expression were unique for in vivo mobilization. The affected genes encoded proteins involved in cellular transcriptional activity and protein synthesis, immune response and inflammation, as well as cell survival and apoptosis. We show that among the genes involved in control of cell apoptosis, treatment with G-CSF and dexamethasone induced an increased expression of calpastatin, the endogenous inhibitor of calpains. This increased expression also could be accomplished by culturing neutrophils with G-CSF/dexamethasone in vitro. Furthermore, calpains are demonstrated to contribute importantly to neutrophil apoptosis, whereas the increase in calpastatin mRNA and protein levels directly corresponds to the prolonged lifespan of neutrophils when treated with G-CSF/dexamethasone in vivo or in vitro.
Methods
Experimental design
Granulocytes from 3 different healthy donors (2 males and 1 female) were studied before and after treatment with G-CSF and dexamethasone. Donors received G-CSF (5 μg/kg subcutaneously) and dexamethasone (8 mg orally). The study was approved by the ethical medical committee at Sanquin Research and Landsteiner Laboratory and was conducted with informed consent given in accordance with the Declaration of Helsinki. Blood samples were taken just before the donor treatment (control sample, fresh) and 16 to 20 hours after G-CSF and dexamethasone administration (in vivo treatment).
Neutrophils from the control sample were isolated and directly prepared for RNA isolation (see the section “RNA isolation, amplification, labeling, and hybridization”) or cultured overnight in the absence (control sample, apoptosis) or presence of G-CSF (Amgen Europe, Breda, The Netherlands) and dexamethasone (Sigma-Aldrich, St Louis, MO; in vitro treatment). Thus, 4 pools of RNA were obtained for comparison (Figure S1, available on the Blood website; see the Supplemental Materials link at the top of the online article).
Granulocyte isolation and culture
Heparinized venous blood was collected from the donors, and the granulocytes were isolated as described.21,22 In short, the granulocytes and mononuclear cells were separated over isotonic Percoll with a specific density of 1.076 g/mL. Erythrocytes in the pellet were lysed in ice-cold medium containing 155 mmol/L NH4Cl, 10 mol/L KHCO3, and 0.1 mmol/L ethylene diamine tetraacetic acid (EDTA), pH 7.4. Granulocytes were washed and resuspended in Hepes-buffered saline solution (HBSS, containing 132 mmol/L NaCl, 6.0 mmol/L KCl, 1.0 mmol/L CaCl2, 1.0 mmol/L MgSO4, 1.2 mmol/L potassium phosphate, 20 mmol/L Hepes, 5.5 mmol/L glucose, and 0.5% (wt/vol) human serum albumin, pH 7.4). The purity of granulocytes isolated with this method was more than 95%. Overnight culture was performed in HBSS with or without addition of 100 ng/mL G-CSF and 1 μmol/L dexamethasone.
RNA isolation, amplification, labeling, and hybridization
Total cellular RNA was extracted from a minimum of 20 × 106 cells with TRIzol reagent (Invitrogen, Breda, The Netherlands) according to the protocol provided by the manufacturer, with the following minor modifications. An additional phenol-chloroform extraction was performed and the isopropanol precipitation at −20°C was facilitated by the addition of 20 μg/mL glycogen (Roche Applied Science, Almere, The Netherlands). Purity and integrity of the RNA samples were confirmed on the Agilent 2100 bioanalyzer (Agilent Technologies Netherlands) using the RNA 6000 Nano LabChip kit. Finally, mRNA was amplified with the MessageAmp II Kit (Applied Biosystems/Ambion, Foster City, CA). Labeling, hybridization, and data extraction were performed at ServiceXS (Leiden, The Netherlands), as has been described elsewhere.23
Microarray imaging and data analysis
The microarray slides were scanned with the Agilent dual-laser DNA microarray scanner. Default settings of Agilent Feature Extraction preprocessing protocols were used to obtain normalized expression values from the raw scans. Exact protocol and parameter settings are described in the Agilent Feature Extraction Software User Manual 8.5 (http://chem.agilent.com/scripts/LiteraturePDF.asp?iWHID = 37 629). The default Agilent normalization procedure, called Linear & Lowess, was applied. Rosetta Resolver (Rosetta Biosoftware, Seattle, WA) was used for analysis of the data. Genes were defined as differentially transcribed if the average expression level changed at least 3-fold compared with unstimulated cells (0 hours, control sample) in all 3 donors. The microarray data have been deposited in National Center for Biotechnology Information's Gene Expression Omnibus24 and are accessible through GEO Series accession number GSE12841 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE12841).
Quantitative RT-PCR validation of gene expression
PCR amplification was performed on a LightCycler instrument (Roche Applied Science), and analyzed with LightCycler Software version 3.5 (Roche Molecular Biochemicals, Mannheim, Germany). The reaction was performed with Lightcycler FastStart DNA MasterPLUS SYBR Green I (Roche Diagnostics, Indianapolis, IN). The annealing temperature used for all primers was 65°C. The reaction mixture consisted of 4 μL of cDNA, 1 μL of relevant primer combination, and 4 μL of SYBR Green I mix in a total volume of 20 μL. All amplified cDNA was compared with the standard within the same run, and in every run the same standard was used, although there was very little variation in the standard between runs.
For amplification, the following LightCycler protocol was used. The chemical cleft of the Taq polymerase was removed by preincubation for 10 minutes at 95°C; the template was amplified for 40 cycles, with annealing of the primers at 65°C. The fluorescence was measured at the end of each cycle at 72°C. At the end of 40 cycles, a melting curve was generated to determine the unique features of the DNA amplified. The specific size of the product was determined on a 1% (wt/vol) agarose gel. Subsequently, the obtained band was purified using the GFX PCR DNA and Gel Band purification kit (Amersham Biosciences, a division of GE Healthcare, Little Chalfont, United Kingdom) according to the manufacturer's instructions to remove excess dNTPs and primers. The product was sequenced by Big-dye Terminator Sequencing and ABI Prism software (Applied Biosystems, Foster City, CA). The sequence was verified with BLAST (http://www.ncbi.nlm.nih.gov/BLAST/) to determine specificity. All products obtained were unique and had no overlap with other isoforms.
Standard curves and relative quantitation
As a source of cDNA for standard curves to which all samples were normalized, neutrophils were isolated from an apheresis buffy coat obtained from the Sanquin Blood Bank North-West Region (Amsterdam, The Netherlands). Serial 10-fold dilutions from the cDNA obtained were made to which each sample was quantified with the method described in Technical Note No. LC 13/2001 (Roche Applied Science), as has been described elsewhere.25
Immunostaining and FACS analysis
Cell surface expression of various receptors on granulocytes was assayed in total leukocyte samples by flow cytometry (ie, fluorescence-activated cell-sorting [FACS]), with saturating concentrations of commercially available monoclonal antibodies (MoAbs), either directly labeled with fluorescein isothiocyanate (FITC), phycoerythrin (PE), or indirectly labeled with allophycocyanin (APC). CD52-FITC, CD55-FITC, CD59-FITC, and CD69-PE were from Sanquin Reagents (Amsterdam, The Netherlands); EMR3-FITC and CD177-unconjugated were from AbD Serotec (Oxford, United Kingdom); and goat F(ab′)2 anti–mouse-IgG-APC was from Southern Biotech (Birmingham, AL).
Samples were analyzed on an LSRII flow cytometer equipped with FACSDiva software (BD, Franklin Lakes, NJ). Cells were gated based on their forward and side scatter, and 10 000 gated events were collected per sample (100% positive staining for CD16 and negative for CD36 [monocytes] or CD56 [NK cells] confirmed purity of the analyzed population).
Annexin V binding
To detect apoptosis, cells were labeled for 10 minutes on ice with FITC-labeled annexin V (Bender Med Systems, Vienna, Austria), diluted 1:500 in HBSS, and supplemented with 2.5 mM CaCl2. Annexin V labeling was followed by a single wash step with the same medium, whereupon the cells were resuspended in HBSS 2.5 mM CaCl2 containing 1 μg/mL propidium iodide (PI; Sigma-Aldrich). After an additional 5 minutes on ice, the samples were analyzed on a FACScan flow cytometer (BD). Surviving cells were defined as the cells in the lower left quadrant that stained negative for both annexin V and PI. A total of 10 000 events were collected for each sample, and data were analyzed with the use of CellQuest Pro software (BD).
Western blot analysis
Total cell lysates were prepared by treating the cells with a lysis buffer (250 mmol/L sucrose, 70 mmol/L KCl, 0.5% Triton X-100 (vol/vol), 0.5% β-octylglucoside (vol/vol), 2 mmol/L NaVO4, 1 mmol/L NaF, 1 mM EDTA, supplemented with a complete protease inhibitor cocktail mix (PIM; Roche Diagnostic, Almere, The Netherlands) and 2 mmol/L diisopropylfluorophosphate (DFP; Fluka Chemica, Steinheim, Switzerland) in phosphate-buffered saline [PBS]) for 30 minutes on ice. Afterward, samples were mixed with 4× Laemli sample buffer (LSB; 50 mmol/L Tris-HCl, pH 6.8, 10% glycerol [vol/vol], 5 mmol/L DTT [DL-dithiothreitol, Sigma] 1% β-mercaptoethanol, 1% sodium dodecylsulfate [SDS; m/v], 10 μg/mL bromophenol blue) and boiled for 15 minutes at 95°C. All samples were stored at −20°C before subjection to SDS–polyacrylamide gel electrophoresis (SDS-PAGE). Samples were run on 10%, 1.5-mm polyacrylamide gels in a protean-3 mini system (Bio-Rad Laboratories, Veenendaal, The Netherlands). The equivalent of 1.5 × 106 cells was loaded in each lane.
After electrophoresis, proteins were transferred to polyvinyl difluoride membranes (PVDF, Bio-Rad), which were subsequently blocked for 60 minutes with blocking buffer (5% nonfat dry milk [m/v, Elk; Campina, Zaltbommel, The Netherlands] in Tris-buffered saline, 0.1% Tween-20 [vol/vol]). The membranes were immune-labeled with specific antibodies in blocking buffer containing 2 mmol/L NaN3 overnight at 4°C. After washing, the membranes were labeled with fluorescently labeled secondary antibodies (either IRDye 800CW or IRDye 700 [Li-COR Bioscience, Lincoln, NE]), and the protein bands were visualized with the Odyssey Infrared Imaging System (Li-COR), and analyzed with the accompanying software (version 2.1).
Statistics
Significantly regulated genes were selected with Rosetta Resolver (Rosetta Biosoftware, Seattle, WA). Genes with a fold change greater than or equal to 3, together with a P value cutoff of .01 (by 1-way analysis of variance [ANOVA] test with the Benjamini-Hochberg false-discovery rate correction), were considered significantly different across the different cell populations. Genes differentially expressed in mobilized granulocytes were categorized by reported or putative functions by the use of the OntoExpress program (http://vortex.cs.wayne.edu/projects.htm#Onto-Express). Graphs were drawn and statistical analysis was performed with GraphPad Prism version 5.00 for Windows (GraphPad Software, San Diego, CA). Data were evaluated by paired, one-tailed Student t test where indicated. The results are presented as the mean plus or minus SEM, as indicated.
Results
G-CSF/dexamethasone mobilization of neutrophils induces global changes in gene expression
Treatment of healthy donors with a combination of a single dose of G-CSF and dexamethasone has been described to result in an increase in the number of circulating neutrophils within 2 to 4 hours after administration, which peaks within 12 to 16 hours (Figure 1A) and returns back to normal levels within 48 to 72 hours.26 Granulocytes for transfusion usually are collected during the peak phase of mobilization (ie, after overnight treatment).
Effect of G-CSF and dexamethasone on granulocytes. (A) Comparison of the granulocyte concentration per milliliter of blood and the EMR3 expression level in time, after donor treatment with G-CSF and dexamethasone (representative graph). PMNs indicate polymorphonuclear leukocytes; ○, EMR3 expression; ■, PMNs per milliliter. (B) Changes in granulocyte gene expression after stimulation with G-CSF and dexamethasone. At 18 to 20 hours after stimulation of the donors or culture of granulocytes with G-CSF and dexamethasone in vitro, neutrophil gene expression was determined by the use of Agilent Whole Humane Genome microarrays. Numbers refer to differentially expressed genes in neutrophils isolated 18 to 20 hours after the administration of G-CSF and dexamethasone in vivo and/or those cultured overnight in medium supplemented with G-CSF and dexamethasone.
Effect of G-CSF and dexamethasone on granulocytes. (A) Comparison of the granulocyte concentration per milliliter of blood and the EMR3 expression level in time, after donor treatment with G-CSF and dexamethasone (representative graph). PMNs indicate polymorphonuclear leukocytes; ○, EMR3 expression; ■, PMNs per milliliter. (B) Changes in granulocyte gene expression after stimulation with G-CSF and dexamethasone. At 18 to 20 hours after stimulation of the donors or culture of granulocytes with G-CSF and dexamethasone in vitro, neutrophil gene expression was determined by the use of Agilent Whole Humane Genome microarrays. Numbers refer to differentially expressed genes in neutrophils isolated 18 to 20 hours after the administration of G-CSF and dexamethasone in vivo and/or those cultured overnight in medium supplemented with G-CSF and dexamethasone.
We used a microarray approach to analyze gene expression patterns in mobilized cells in comparison with the cells obtained from the same healthy control donors before G-CSF/dexamethasone administration. The administration of G-CSF/dexamethasone results in both mobilization of immature neutrophils from the bone marrow, as indicated by staining with EMR3, a myeloid-specific member of the epidermal growth factor-7-transmembrane (EGF-TM7) family of adhesion class TM7 receptors, which has been recently described as a marker for mature granulocytes (Figure 1A),27 as well as direct effects on the already circulating cells. To estimate the contribution of the latter, we also performed in vitro G-CSF/dexamethasone incubations and analyses on neutrophils taken before the in vivo treatment (see also the scheme for experimental design in Figure S1). This step also allows the evaluation of changes in gene expression associated with the G-CSF/dexamethasone–induced delay in apoptosis.
By the use of Agilent Whole Human Genome microarrays, we screened approximately 32 000 gene transcripts. This analysis revealed that approximately 1000 genes were differentially expressed in the cells after the administration in vivo of G-CSF/dexamethasone compared with those isolated before mobilization (Figure 1B and Document S1). Even more genes were differentially expressed after treatment in vitro of neutrophils with G-CSF/dexamethasone (Figure 1B and Document S1).
Approximately 150 genes were similarly regulated in both experimental systems (in vivo vs in vitro, Table 1), but still more than 800 genes demonstrated a unique expression pattern for the in vivo stimulation.
Genes similarly regulated by the treatment with G-CSF/dexamemethasone in vivo and in vitro
| Sequence ID . | Primary sequence name . | Sequence code . | Sequence description . | Fold change . | |
|---|---|---|---|---|---|
| In vivo . | In vitro . | ||||
| 1507161 | CD163 | NM_004244 | CD163 molecule | 54.27 | 11.88 |
| 1984958 | CD177 | CR592446 | CD177 molecule | 24.38 | 8.50 |
| 1519170 | GADD45A | NM_001924 | Growth arrest and DNA-damage-inducible, alpha | 20.49 | 10.75 |
| 1515523 | SLC1A3 | NM_004172 | Solute carrier family 1 (glial high affinity glutamate transporter), | 16.98 | 11.45 |
| 1521404 | IL18R1 | NM_003855 | Interleukin 18 receptor 1 | 16.54 | 8.19 |
| 1523267 | FKBP5 | NM_004117 | FK506 binding protein 5 | 13.12 | 7.36 |
| 1528485 | SORT1 | NM_002959 | Sortilin 1 | 12.11 | 20.42 |
| 1534791 | SAMSN1 | NM_022136 | SAM domain, SH3 domain and nuclear localization signals, 1 | 11.87 | 8.34 |
| 1506996 | ADORA3 | NM_020683 | Adenosine A3 receptor | 10.90 | 10.49 |
| 1525991 | TRPS1 | NM_014112 | Trichorhinophalangeal syndrome I | 9.13 | 4.45 |
| 1538316 | ACN9 | NM_020186 | ACN9 homolog (Saccharomyces cerevisiae) | 9.13 | 6.87 |
| 1508652 | KLF9 | NM_001206 | Kruppel-like factor 9 | 8.65 | 7.46 |
| 1524454 | XBP1 | NM_005080 | X-box binding protein 1 | 7.48 | 6.51 |
| 1510813 | IRAK3 | NM_007199 | Interleukin-1 receptor-associated kinase 3 | 7.28 | 10.56 |
| 1531325 | ORF1-FL49 | NM_032412 | Putative nuclear protein ORF1-FL49 | 7.04 | 4.25 |
| 1516682 | CAST | NM_001750 | Calpastatin | 6.97 | 10.64 |
| 1506968 | ST6GALNAC3 | NM_152996 | ST6 | 6.56 | 4.94 |
| 1513738 | SPFH1 | NM_006459 | SPFH domain family, member 1 | 6.39 | 6.20 |
| 1991986 | TRPM2 | NM_001001 188 | Transient receptor potential cation channel, subfamily M, member 2 | 6.38 | 13.63 |
| 1989099 | GRINA | NM_000837 | Poly (ADP-ribose) polymerase family, member 10 | 5.60 | 11.34 |
| 1522658 | VNN1 | NM_004666 | Vanin 1 | 5.35 | 4.47 |
| 1533591 | SIRT5 | NM_012241 | Sirtuin | 4.60 | 4.50 |
| 1521022 | HIPK2 | AK074291 | Homeodomain interacting protein kinase 2 | 4.28 | 8.24 |
| 1505831 | CIAS1 | NM_004895 | Cold autoinflammatory syndrome 1 | 4.10 | 19.21 |
| 1527074 | PEX11G | NM_080662 | Peroxisomal biogenesis factor 11 gamma | 4.06 | 4.21 |
| 1507368 | CCND3 | NM_001760 | Cyclin D3 | 3.69 | 5.58 |
| 1519571 | CDC42EP3 | NM_006449 | CDC42 effector protein (Rho GTPase binding) 3 | 3.58 | 7.45 |
| 1531872 | TMEM140 | AK056910 | Transmembrane protein 140 | 3.48 | 6.02 |
| 1534101 | IL1R1 | NM_000877 | Interleukin 1 receptor, type I | 3.35 | 6.15 |
| 1511860 | ZDHHC2 | NM_016353 | Zinc finger, DHHC-type containing 2 | 3.24 | 4.21 |
| 1530609 | QSCN6 | NM_002826 | Quiescin Q6 | 3.08 | 4.22 |
| 1518979 | CPD | NM_001304 | Carboxypeptidase D | 3.03 | 4.63 |
| 1506387 | MAPK6 | NM_002748 | Mitogen-activated protein kinase 6 | 3.03 | 5.03 |
| 1515049 | ZNF278 | NM_032051 | Zinc finger protein 278 | −3.05 | −4.11 |
| 1983183 | NDUFB6 | NM_182739 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 6, 17 kDa | −3.10 | −4.04 |
| 1507683 | FANCA | NM_000135 | Fanconi anemia, complementation group A | −3.19 | −4.79 |
| 1985202 | IGKC | BC067092 | Immunoglobulin kappa constant | −3.21 | −5.50 |
| 1522438 | GLIPR1 | U16307 | GLI pathogenesis-related 1 (glioma) | −3.25 | −12.71 |
| 1525398 | RFX5 | NM_000449 | Selenium binding protein 1 | −3.25 | −5.02 |
| 1522187 | TOPORS | NM_005802 | Topoisomerase I binding, arginine/serine-rich | −3.26 | −4.96 |
| 1536170 | TST | NM_003312 | Thiosulfate sulfurtransferase (rhodanese) | −3.28 | −6.47 |
| 1530334 | CST3 | NM_000099 | Cystatin C | −3.29 | −5.23 |
| 1534756 | PSMB10 | NM_002801 | Proteasome (prosome, macropain) subunit, beta type, 10 | −3.30 | −5.60 |
| 1519302 | STAT1 | NM_139266 | Signal transducer and activator of transcription 1, 91 kDa | −3.39 | −10.76 |
| 1522898 | MVP | AK097472 | Major vault protein | −3.51 | −4.58 |
| 1524812 | CEP63 | NM_025180 | Centrosomal protein 63kDa | −3.54 | −4.22 |
| 1538148 | RAB37 | NM_175738 | RAB37, member RAS oncogene family | −3.55 | −5.17 |
| 1986068 | RNF43 | CR620892 | Ring finger protein 43 | −3.60 | −7.06 |
| 1984701 | CARD9 | NM_052813 | Caspase recruitment domain family, member 9 | −3.61 | −27.75 |
| 1523761 | CBR1 | NM_001757 | Carbonyl reductase 1 | −3.61 | −4.75 |
| 1511505 | RABGAP1L | AB007940 | RAB GTPase activating protein 1-like | −3.63 | −10.78 |
| 1511424 | EMR3 | NM_032571 | Egf-like module containing, mucin-like, hormone receptor-like 3 | −3.71 | −8.61 |
| 1513189 | TRERF1 | NM_033502 | Transcriptional regulating factor 1 | −3.76 | −16.11 |
| 1990225 | IGF2BP2 | AA451676 | Insulin-like growth factor 2 mRNA binding protein 2 | −3.79 | −6.35 |
| 1513264 | CST5 | NM_001900 | Cystatin D | −3.82 | −5.63 |
| 1990830 | FAM111A | BU684362 | Family with sequence similarity 111, member A | −3.83 | −8.95 |
| 1534340 | CD46 | NM_002389 | CD46 molecule, complement regulatory protein | −3.83 | −6.11 |
| 1517555 | CYP27A1 | NM_000784 | Cytochrome P450, family 27, subfamily A, polypeptide 1 | −3.88 | −7.43 |
| 1516190 | SAMHD1 | NM_015474 | SAM domain and HD domain 1 | −3.89 | −8.86 |
| 1528817 | MME | NM_007289 | Membrane metalloendopeptidase (neutral endopeptidase) | −3.92 | −8.11 |
| 1536250 | CCT2 | NM_006431 | Chaperonin containing TCP1, subunit 2 (beta) | −4.04 | −5.43 |
| 1515232 | POLB | NM_002690 | Polymerase (DNA directed), beta | −4.07 | −5.28 |
| 1527068 | PSME1 | NM_006263 | Proteasome (prosome, macropain) activator subunit 1(PA28 alpha) | −4.15 | −4.84 |
| 1986072 | COL27A1 | AI382322 | Collagen, type XXVII, alpha 1 | −4.21 | −4.54 |
| 1532664 | PDLIM1 | NM_020992 | PDZ and LIM domain 1 (elfin) | −4.25 | −5.30 |
| 1524915 | OR51E1 | NM_152430 | Olfactory receptor, family 51, subfamily E, member 1 | −4.27 | −4.44 |
| 1507227 | P2RY14 | NM_014879 | Purinergic receptor P2Y, G-protein coupled, 14 | −4.28 | −12.99 |
| 1508056 | EGR3 | NM_004430 | Early growth response 3 | −4.41 | −4.50 |
| 1515615 | PRRG4 | NM_024081 | Proline rich Gla (G-carboxyglutamic acid) 4 (transmembrane) | −4.42 | −4.38 |
| 1517506 | EGR2 | NM_000399 | Early growth response 2 (Krox-20 homolog, Drosophila) | −4.67 | −14.32 |
| 1513793 | CATSPER2 | NM_172097 | Cation channel, sperm associated 2 | −4.74 | −4.19 |
| 1533707 | RG9MTD2 | NM_152292 | RNA (guanine-9-) methyltransferase domain containing 2 | −4.80 | −7.17 |
| 1507738 | FBP1 | NM_000507 | Fructose-1.6-bisphosphatase 1 | −4.82 | −7.32 |
| 1535598 | NCOA4 | NM_005437 | Nuclear receptor coactivator 4 | −4.87 | −21.40 |
| 1507851 | CTGLF1 | NM_133446 | Centaurin, gamma-like family, member 1 | −4.88 | −5.47 |
| 1506502 | S100A4 | NM_002961 | S100 calcium binding protein A4 | −4.91 | −16.73 |
| 1522210 | TIGD7 | NM_033208 | Tigger transposable element derived 7 | −5.02 | −10.17 |
| 1522120 | MATN1 | AK057725 | Matrilin 1, cartilage matrix protein | −5.06 | −4.48 |
| 1519593 | IFIT5 | NM_012420 | Interferon-induced protein with tetratricopeptide repeats 5 | −5.07 | −11.79 |
| 1533259 | RBP7 | NM_052960 | Ubiquitination factor E4B (UFD2 homolog, yeast) | −5.16 | −12.38 |
| 1532326 | DDX58 | NM_014314 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 | −5.17 | −14.04 |
| 1510042 | NHS | NM_198270 | Nance-Horan syndrome | −5.40 | −12.90 |
| 1983408 | ZFP36L1 | CD619445 | Zinc finger protein 36, C3H type-like 1 | −5.41 | −4.09 |
| 1518324 | FAM111A | NM_022074 | Family with sequence similarity 111, member A | −5.59 | −5.82 |
| 1990540 | CLU | NM_203339 | Clusterin | −5.66 | −5.71 |
| 1520999 | DGKG | NM_001346 | Diacylglycerol kinase, gamma 90 kDa | −5.71 | −5.82 |
| 1983869 | ADAM28 | NM_014265 | ADAM metallopeptidase domain 28 | −5.76 | −7.39 |
| 1518750 | PARP16 | NM_017851 | Poly (ADP-ribose) polymerase family, member 16 | −5.84 | −11.81 |
| 1505761 | TNFAIP2 | NM_006291 | Beta-amylase | −5.90 | -9.81 |
| 1534912 | CMTM5 | NM_181618 | Chemokine-like factor superfamily 5 | −5.91 | −4.63 |
| 1508430 | SLC25A22 | NM_024698 | Solute carrier family 25 (mitochondrial carrier: glutamate), | −5.94 | −13.23 |
| 1525314 | S100A10 | NM_002966 | S100 calcium binding protein A10 | −5.94 | −6.72 |
| 1521056 | GP1BB | NM_000407 | Septin 5 | −5.97 | −4.76 |
| 1539206 | SPECC1 | NM_152904 | Spectrin domain with coiled-coils 1 | −6.24 | −4.94 |
| 1527296 | HERC5 | NM_016323 | Hect domain and RLD 5 | −6.27 | −7.29 |
| 1506913 | MBIP | NM_016586 | MAP3K12 binding inhibitory protein 1 | −6.28 | −21.40 |
| 1515002 | CYSLTR1 | NM_006639 | Cysteinyl leukotriene receptor 1 | −6.45 | −9.25 |
| 1512448 | ISOC1 | NM_016048 | Isochorismatase domain containing 1 | −6.51 | −7.66 |
| 1513897 | GIMAP5 | NM_018384 | GTPase, IMAP family member 5 | −6.56 | −4.68 |
| 1520165 | AF242507 | AF242507 | Homosapiens m-tumor necrosis factor-alpha induced protein mRNA, | −6.58 | −5.74 |
| 1534914 | MPEG1 | AK074166 | macrophage expressed gene 1 | −6.60 | −5.82 |
| 1988054 | RAD50 | AK128882 | RAD50 homolog (S cerevisiae) | −6.71 | −6.23 |
| 1513813 | G0S2 | NM_015714 | G0/G1 switch 2 | −6.74 | −10.55 |
| 1530332 | MX1 | NM_002462 | Myxovirus (influenza virus) resistance 1, | −6.76 | −12.23 |
| 1522509 | OR2B2 | NM_033057 | Olfactory receptor, family 2, subfamily B, member 2 | −6.88 | −4.91 |
| 1991493 | PARP14 | NM_017554 | Poly (ADP-ribose) polymerase family, member 14 | −7.03 | −4.29 |
| 1527378 | CSF1R | NM_005211 | Colony stimulating factor 1 receptor, | −7.08 | −8.69 |
| 1514500 | CEP57 | BC039711 | Centrosomal protein 57 kDa | −7.08 | −7.41 |
| 1522188 | PTGS2 | NM_000963 | Prostaglandin-endoperoxide synthase 2 | −7.22 | −90.22 |
| 1537251 | OR52K3P | AF143328 | Olfactory receptor, family 52, subfamily K, member 3 pseudogene | −7.25 | −5.96 |
| 1514502 | CCDC37 | NM_182628 | Coiled-coil domain containing 37 | −7.27 | −5.51 |
| 1528770 | RPGRIP1 | NM_020366 | Retinitis pigmentosa GTPase regulator interacting protein 1 | −7.32 | −6.15 |
| 1987032 | TREX1 | NM_016381 | Three prime repair exonuclease 1 | −7.45 | −5.26 |
| 1510272 | RNASE3 | NM_002935 | Ribonuclease, RNase A family, 3 (eosinophil cationic protein) | −7.61 | −11.42 |
| 1991208 | TUSC1 | NM_001004125 | Tumor suppressor candidate 1 | −7.62 | −5.54 |
| 1539129 | SDPR | NM_004657 | Serum deprivation response (phosphatidylserine binding protein) | −7.80 | −5.25 |
| 1538745 | SGK | NM_005627 | Serum/glucocorticoid regulated kinase | −7.90 | −5.90 |
| 1524314 | IFI44 | NM_006417 | Interferon-induced protein 44 | −7.91 | −10.63 |
| 1989096 | SSBP3 | NM_001009955 | Single stranded DNA binding protein 3 | −8.76 | −4.70 |
| 1520076 | MFI2 | BC032042 | Antigen p97 | −8.93 | −6.15 |
| 1518480 | DDHD1 | AK091528 | DDHD domain containing 1 | −9.02 | −8.19 |
| 1512017 | RNASE2 | NM_002934 | Ribonuclease, RNase A family, 2 (liver, eosinophil-derived neurotoxin) | −9.05 | −8.41 |
| 1536355 | LPAL2 | AK125910 | Homo sapiens cDNA FLJ43922 fis | −9.08 | −13.45 |
| 1533135 | CXCL10 | NM_001565 | Chemokine (C-X-C motif) ligand 11 | −9.10 | −8.41 |
| 1522417 | TRIO | AY358688 | Triple functional domain (PTPRF interacting) | −9.21 | −10.68 |
| 1518425 | PSME2 | NM_002818 | Proteasome (prosome, macropain) activator subunit 2 (PA28 beta) | −9.29 | −12.60 |
| 1512119 | KNTC2 | NM_006101 | Kinetochore associated 2 | −9.33 | −4.25 |
| 1506804 | PLD4 | NM_138790 | Phospholipase D family, member 4 | −9.53 | −4.93 |
| 1520503 | NKX3–1 | NM_006167 | NK3 transcription factor related, locus 1 | −9.56 | −7.38 |
| 1533140 | NME5 | NM_003551 | nucleoside-diphosphate kinase | −9.99 | −8.02 |
| 1536919 | HLA-DMB | NM_002118 | Major histocompatibility complex, class II, DM beta | −10.73 | −4.66 |
| 1538875 | AF15Q14 | NM_020380 | AF15q14 protein | −10.83 | −7.68 |
| 1519782 | LAP3 | NM_015907 | Leucine aminopeptidase 3 | −11.23 | −24.38 |
| 1529525 | LYSMD2 | NM_153374 | LysM, putative peptidoglycan-binding, domain containing 2 | −11.50 | −5.09 |
| 1516830 | P2RY10 | NM_014499 | Purinergic receptor P2Y, G-protein coupled, 10 | −11.67 | −4.14 |
| 1519975 | PRSS33 | NM_152891 | Protease, serine, 33 | −11.84 | −5.57 |
| 1507811 | PMAIP1 | NM_021127 | Phorbol-12-myristate-13-acetate-induced protein 1 | −12.76 | −6.44 |
| 1506299 | MMP12 | ENST000003 26227 | Macrophage metalloelastase precursor | −12.98 | −7.44 |
| 1536513 | IFIT3 | NM_001549 | Interferon-induced protein with tetratricopeptide repeats 3 | −13.08 | −9.40 |
| 1513158 | HRH4 | NM_021624 | Histamine receptor H4 | −13.66 | −6.92 |
| 1537338 | FGL2 | NM_006682 | Fibrinogen-like 2 | −13.76 | −31.09 |
| 1509838 | GBP1 | NM_002053 | Guanylate binding protein 1, interferon-inducible, 67kDa | −13.91 | −11.20 |
| 1512112 | EPSTI1 | NM_033255 | Epithelial stromal interaction 1 (breast) | −14.02 | −9.27 |
| 1522895 | GOLPH4 | NM_014498 | Golgi phosphoprotein 4 | −14.30 | −12.18 |
| 1508419 | CD52 | NM_001803 | CD52 molecule | −14.95 | −4.38 |
| 1506912 | RARRES1 | NM_002888 | Retinoic acid receptor responder (tazarotene induced) 1 | −15.02 | −9.44 |
| 1509737 | IFIT2 | NM_001547 | Interferon-induced protein with tetratricopeptide repeats 2 | −15.06 | −31.11 |
| 1505505 | CCL23 | NM_005064 | Chemokine (C-C motif) ligand 23 | −15.06 | −4.22 |
| 1518780 | AF131788 | AF131788 | Clone 24875 mRNA sequence | −16.78 | −12.00 |
| 1522258 | ISG15 | NM_005101 | Interferon, alpha-inducible protein (clone IFI-15K) | −18.16 | −4.81 |
| 1519399 | GBP5 | NM_052942 | Guanylate binding protein 5 | −18.22 | −10.65 |
| 1531078 | RSAD2 | NM_080657 | Radical S-adenosyl methionine domain containing 2 | −18.54 | −8.17 |
| 1515740 | SLC45A3 | NM_033102 | Solute carrier family 45, member 3 | −26.27 | −28.60 |
| 1517997 | IFIT1 | NM_001548 | Interferon-induced protein with tetratricopeptide repeats 1 | −34.47 | −35.07 |
| 1533457 | FCER1A | NM_002001 | Fc fragment of IgE, high affinity I, receptor for; alpha polypeptide | −61.82 | −31.20 |
| 1532327 | CLC | NM_001828 | Charcot-Leyden crystal protein | −69.08 | −16.00 |
| Sequence ID . | Primary sequence name . | Sequence code . | Sequence description . | Fold change . | |
|---|---|---|---|---|---|
| In vivo . | In vitro . | ||||
| 1507161 | CD163 | NM_004244 | CD163 molecule | 54.27 | 11.88 |
| 1984958 | CD177 | CR592446 | CD177 molecule | 24.38 | 8.50 |
| 1519170 | GADD45A | NM_001924 | Growth arrest and DNA-damage-inducible, alpha | 20.49 | 10.75 |
| 1515523 | SLC1A3 | NM_004172 | Solute carrier family 1 (glial high affinity glutamate transporter), | 16.98 | 11.45 |
| 1521404 | IL18R1 | NM_003855 | Interleukin 18 receptor 1 | 16.54 | 8.19 |
| 1523267 | FKBP5 | NM_004117 | FK506 binding protein 5 | 13.12 | 7.36 |
| 1528485 | SORT1 | NM_002959 | Sortilin 1 | 12.11 | 20.42 |
| 1534791 | SAMSN1 | NM_022136 | SAM domain, SH3 domain and nuclear localization signals, 1 | 11.87 | 8.34 |
| 1506996 | ADORA3 | NM_020683 | Adenosine A3 receptor | 10.90 | 10.49 |
| 1525991 | TRPS1 | NM_014112 | Trichorhinophalangeal syndrome I | 9.13 | 4.45 |
| 1538316 | ACN9 | NM_020186 | ACN9 homolog (Saccharomyces cerevisiae) | 9.13 | 6.87 |
| 1508652 | KLF9 | NM_001206 | Kruppel-like factor 9 | 8.65 | 7.46 |
| 1524454 | XBP1 | NM_005080 | X-box binding protein 1 | 7.48 | 6.51 |
| 1510813 | IRAK3 | NM_007199 | Interleukin-1 receptor-associated kinase 3 | 7.28 | 10.56 |
| 1531325 | ORF1-FL49 | NM_032412 | Putative nuclear protein ORF1-FL49 | 7.04 | 4.25 |
| 1516682 | CAST | NM_001750 | Calpastatin | 6.97 | 10.64 |
| 1506968 | ST6GALNAC3 | NM_152996 | ST6 | 6.56 | 4.94 |
| 1513738 | SPFH1 | NM_006459 | SPFH domain family, member 1 | 6.39 | 6.20 |
| 1991986 | TRPM2 | NM_001001 188 | Transient receptor potential cation channel, subfamily M, member 2 | 6.38 | 13.63 |
| 1989099 | GRINA | NM_000837 | Poly (ADP-ribose) polymerase family, member 10 | 5.60 | 11.34 |
| 1522658 | VNN1 | NM_004666 | Vanin 1 | 5.35 | 4.47 |
| 1533591 | SIRT5 | NM_012241 | Sirtuin | 4.60 | 4.50 |
| 1521022 | HIPK2 | AK074291 | Homeodomain interacting protein kinase 2 | 4.28 | 8.24 |
| 1505831 | CIAS1 | NM_004895 | Cold autoinflammatory syndrome 1 | 4.10 | 19.21 |
| 1527074 | PEX11G | NM_080662 | Peroxisomal biogenesis factor 11 gamma | 4.06 | 4.21 |
| 1507368 | CCND3 | NM_001760 | Cyclin D3 | 3.69 | 5.58 |
| 1519571 | CDC42EP3 | NM_006449 | CDC42 effector protein (Rho GTPase binding) 3 | 3.58 | 7.45 |
| 1531872 | TMEM140 | AK056910 | Transmembrane protein 140 | 3.48 | 6.02 |
| 1534101 | IL1R1 | NM_000877 | Interleukin 1 receptor, type I | 3.35 | 6.15 |
| 1511860 | ZDHHC2 | NM_016353 | Zinc finger, DHHC-type containing 2 | 3.24 | 4.21 |
| 1530609 | QSCN6 | NM_002826 | Quiescin Q6 | 3.08 | 4.22 |
| 1518979 | CPD | NM_001304 | Carboxypeptidase D | 3.03 | 4.63 |
| 1506387 | MAPK6 | NM_002748 | Mitogen-activated protein kinase 6 | 3.03 | 5.03 |
| 1515049 | ZNF278 | NM_032051 | Zinc finger protein 278 | −3.05 | −4.11 |
| 1983183 | NDUFB6 | NM_182739 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 6, 17 kDa | −3.10 | −4.04 |
| 1507683 | FANCA | NM_000135 | Fanconi anemia, complementation group A | −3.19 | −4.79 |
| 1985202 | IGKC | BC067092 | Immunoglobulin kappa constant | −3.21 | −5.50 |
| 1522438 | GLIPR1 | U16307 | GLI pathogenesis-related 1 (glioma) | −3.25 | −12.71 |
| 1525398 | RFX5 | NM_000449 | Selenium binding protein 1 | −3.25 | −5.02 |
| 1522187 | TOPORS | NM_005802 | Topoisomerase I binding, arginine/serine-rich | −3.26 | −4.96 |
| 1536170 | TST | NM_003312 | Thiosulfate sulfurtransferase (rhodanese) | −3.28 | −6.47 |
| 1530334 | CST3 | NM_000099 | Cystatin C | −3.29 | −5.23 |
| 1534756 | PSMB10 | NM_002801 | Proteasome (prosome, macropain) subunit, beta type, 10 | −3.30 | −5.60 |
| 1519302 | STAT1 | NM_139266 | Signal transducer and activator of transcription 1, 91 kDa | −3.39 | −10.76 |
| 1522898 | MVP | AK097472 | Major vault protein | −3.51 | −4.58 |
| 1524812 | CEP63 | NM_025180 | Centrosomal protein 63kDa | −3.54 | −4.22 |
| 1538148 | RAB37 | NM_175738 | RAB37, member RAS oncogene family | −3.55 | −5.17 |
| 1986068 | RNF43 | CR620892 | Ring finger protein 43 | −3.60 | −7.06 |
| 1984701 | CARD9 | NM_052813 | Caspase recruitment domain family, member 9 | −3.61 | −27.75 |
| 1523761 | CBR1 | NM_001757 | Carbonyl reductase 1 | −3.61 | −4.75 |
| 1511505 | RABGAP1L | AB007940 | RAB GTPase activating protein 1-like | −3.63 | −10.78 |
| 1511424 | EMR3 | NM_032571 | Egf-like module containing, mucin-like, hormone receptor-like 3 | −3.71 | −8.61 |
| 1513189 | TRERF1 | NM_033502 | Transcriptional regulating factor 1 | −3.76 | −16.11 |
| 1990225 | IGF2BP2 | AA451676 | Insulin-like growth factor 2 mRNA binding protein 2 | −3.79 | −6.35 |
| 1513264 | CST5 | NM_001900 | Cystatin D | −3.82 | −5.63 |
| 1990830 | FAM111A | BU684362 | Family with sequence similarity 111, member A | −3.83 | −8.95 |
| 1534340 | CD46 | NM_002389 | CD46 molecule, complement regulatory protein | −3.83 | −6.11 |
| 1517555 | CYP27A1 | NM_000784 | Cytochrome P450, family 27, subfamily A, polypeptide 1 | −3.88 | −7.43 |
| 1516190 | SAMHD1 | NM_015474 | SAM domain and HD domain 1 | −3.89 | −8.86 |
| 1528817 | MME | NM_007289 | Membrane metalloendopeptidase (neutral endopeptidase) | −3.92 | −8.11 |
| 1536250 | CCT2 | NM_006431 | Chaperonin containing TCP1, subunit 2 (beta) | −4.04 | −5.43 |
| 1515232 | POLB | NM_002690 | Polymerase (DNA directed), beta | −4.07 | −5.28 |
| 1527068 | PSME1 | NM_006263 | Proteasome (prosome, macropain) activator subunit 1(PA28 alpha) | −4.15 | −4.84 |
| 1986072 | COL27A1 | AI382322 | Collagen, type XXVII, alpha 1 | −4.21 | −4.54 |
| 1532664 | PDLIM1 | NM_020992 | PDZ and LIM domain 1 (elfin) | −4.25 | −5.30 |
| 1524915 | OR51E1 | NM_152430 | Olfactory receptor, family 51, subfamily E, member 1 | −4.27 | −4.44 |
| 1507227 | P2RY14 | NM_014879 | Purinergic receptor P2Y, G-protein coupled, 14 | −4.28 | −12.99 |
| 1508056 | EGR3 | NM_004430 | Early growth response 3 | −4.41 | −4.50 |
| 1515615 | PRRG4 | NM_024081 | Proline rich Gla (G-carboxyglutamic acid) 4 (transmembrane) | −4.42 | −4.38 |
| 1517506 | EGR2 | NM_000399 | Early growth response 2 (Krox-20 homolog, Drosophila) | −4.67 | −14.32 |
| 1513793 | CATSPER2 | NM_172097 | Cation channel, sperm associated 2 | −4.74 | −4.19 |
| 1533707 | RG9MTD2 | NM_152292 | RNA (guanine-9-) methyltransferase domain containing 2 | −4.80 | −7.17 |
| 1507738 | FBP1 | NM_000507 | Fructose-1.6-bisphosphatase 1 | −4.82 | −7.32 |
| 1535598 | NCOA4 | NM_005437 | Nuclear receptor coactivator 4 | −4.87 | −21.40 |
| 1507851 | CTGLF1 | NM_133446 | Centaurin, gamma-like family, member 1 | −4.88 | −5.47 |
| 1506502 | S100A4 | NM_002961 | S100 calcium binding protein A4 | −4.91 | −16.73 |
| 1522210 | TIGD7 | NM_033208 | Tigger transposable element derived 7 | −5.02 | −10.17 |
| 1522120 | MATN1 | AK057725 | Matrilin 1, cartilage matrix protein | −5.06 | −4.48 |
| 1519593 | IFIT5 | NM_012420 | Interferon-induced protein with tetratricopeptide repeats 5 | −5.07 | −11.79 |
| 1533259 | RBP7 | NM_052960 | Ubiquitination factor E4B (UFD2 homolog, yeast) | −5.16 | −12.38 |
| 1532326 | DDX58 | NM_014314 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 | −5.17 | −14.04 |
| 1510042 | NHS | NM_198270 | Nance-Horan syndrome | −5.40 | −12.90 |
| 1983408 | ZFP36L1 | CD619445 | Zinc finger protein 36, C3H type-like 1 | −5.41 | −4.09 |
| 1518324 | FAM111A | NM_022074 | Family with sequence similarity 111, member A | −5.59 | −5.82 |
| 1990540 | CLU | NM_203339 | Clusterin | −5.66 | −5.71 |
| 1520999 | DGKG | NM_001346 | Diacylglycerol kinase, gamma 90 kDa | −5.71 | −5.82 |
| 1983869 | ADAM28 | NM_014265 | ADAM metallopeptidase domain 28 | −5.76 | −7.39 |
| 1518750 | PARP16 | NM_017851 | Poly (ADP-ribose) polymerase family, member 16 | −5.84 | −11.81 |
| 1505761 | TNFAIP2 | NM_006291 | Beta-amylase | −5.90 | -9.81 |
| 1534912 | CMTM5 | NM_181618 | Chemokine-like factor superfamily 5 | −5.91 | −4.63 |
| 1508430 | SLC25A22 | NM_024698 | Solute carrier family 25 (mitochondrial carrier: glutamate), | −5.94 | −13.23 |
| 1525314 | S100A10 | NM_002966 | S100 calcium binding protein A10 | −5.94 | −6.72 |
| 1521056 | GP1BB | NM_000407 | Septin 5 | −5.97 | −4.76 |
| 1539206 | SPECC1 | NM_152904 | Spectrin domain with coiled-coils 1 | −6.24 | −4.94 |
| 1527296 | HERC5 | NM_016323 | Hect domain and RLD 5 | −6.27 | −7.29 |
| 1506913 | MBIP | NM_016586 | MAP3K12 binding inhibitory protein 1 | −6.28 | −21.40 |
| 1515002 | CYSLTR1 | NM_006639 | Cysteinyl leukotriene receptor 1 | −6.45 | −9.25 |
| 1512448 | ISOC1 | NM_016048 | Isochorismatase domain containing 1 | −6.51 | −7.66 |
| 1513897 | GIMAP5 | NM_018384 | GTPase, IMAP family member 5 | −6.56 | −4.68 |
| 1520165 | AF242507 | AF242507 | Homosapiens m-tumor necrosis factor-alpha induced protein mRNA, | −6.58 | −5.74 |
| 1534914 | MPEG1 | AK074166 | macrophage expressed gene 1 | −6.60 | −5.82 |
| 1988054 | RAD50 | AK128882 | RAD50 homolog (S cerevisiae) | −6.71 | −6.23 |
| 1513813 | G0S2 | NM_015714 | G0/G1 switch 2 | −6.74 | −10.55 |
| 1530332 | MX1 | NM_002462 | Myxovirus (influenza virus) resistance 1, | −6.76 | −12.23 |
| 1522509 | OR2B2 | NM_033057 | Olfactory receptor, family 2, subfamily B, member 2 | −6.88 | −4.91 |
| 1991493 | PARP14 | NM_017554 | Poly (ADP-ribose) polymerase family, member 14 | −7.03 | −4.29 |
| 1527378 | CSF1R | NM_005211 | Colony stimulating factor 1 receptor, | −7.08 | −8.69 |
| 1514500 | CEP57 | BC039711 | Centrosomal protein 57 kDa | −7.08 | −7.41 |
| 1522188 | PTGS2 | NM_000963 | Prostaglandin-endoperoxide synthase 2 | −7.22 | −90.22 |
| 1537251 | OR52K3P | AF143328 | Olfactory receptor, family 52, subfamily K, member 3 pseudogene | −7.25 | −5.96 |
| 1514502 | CCDC37 | NM_182628 | Coiled-coil domain containing 37 | −7.27 | −5.51 |
| 1528770 | RPGRIP1 | NM_020366 | Retinitis pigmentosa GTPase regulator interacting protein 1 | −7.32 | −6.15 |
| 1987032 | TREX1 | NM_016381 | Three prime repair exonuclease 1 | −7.45 | −5.26 |
| 1510272 | RNASE3 | NM_002935 | Ribonuclease, RNase A family, 3 (eosinophil cationic protein) | −7.61 | −11.42 |
| 1991208 | TUSC1 | NM_001004125 | Tumor suppressor candidate 1 | −7.62 | −5.54 |
| 1539129 | SDPR | NM_004657 | Serum deprivation response (phosphatidylserine binding protein) | −7.80 | −5.25 |
| 1538745 | SGK | NM_005627 | Serum/glucocorticoid regulated kinase | −7.90 | −5.90 |
| 1524314 | IFI44 | NM_006417 | Interferon-induced protein 44 | −7.91 | −10.63 |
| 1989096 | SSBP3 | NM_001009955 | Single stranded DNA binding protein 3 | −8.76 | −4.70 |
| 1520076 | MFI2 | BC032042 | Antigen p97 | −8.93 | −6.15 |
| 1518480 | DDHD1 | AK091528 | DDHD domain containing 1 | −9.02 | −8.19 |
| 1512017 | RNASE2 | NM_002934 | Ribonuclease, RNase A family, 2 (liver, eosinophil-derived neurotoxin) | −9.05 | −8.41 |
| 1536355 | LPAL2 | AK125910 | Homo sapiens cDNA FLJ43922 fis | −9.08 | −13.45 |
| 1533135 | CXCL10 | NM_001565 | Chemokine (C-X-C motif) ligand 11 | −9.10 | −8.41 |
| 1522417 | TRIO | AY358688 | Triple functional domain (PTPRF interacting) | −9.21 | −10.68 |
| 1518425 | PSME2 | NM_002818 | Proteasome (prosome, macropain) activator subunit 2 (PA28 beta) | −9.29 | −12.60 |
| 1512119 | KNTC2 | NM_006101 | Kinetochore associated 2 | −9.33 | −4.25 |
| 1506804 | PLD4 | NM_138790 | Phospholipase D family, member 4 | −9.53 | −4.93 |
| 1520503 | NKX3–1 | NM_006167 | NK3 transcription factor related, locus 1 | −9.56 | −7.38 |
| 1533140 | NME5 | NM_003551 | nucleoside-diphosphate kinase | −9.99 | −8.02 |
| 1536919 | HLA-DMB | NM_002118 | Major histocompatibility complex, class II, DM beta | −10.73 | −4.66 |
| 1538875 | AF15Q14 | NM_020380 | AF15q14 protein | −10.83 | −7.68 |
| 1519782 | LAP3 | NM_015907 | Leucine aminopeptidase 3 | −11.23 | −24.38 |
| 1529525 | LYSMD2 | NM_153374 | LysM, putative peptidoglycan-binding, domain containing 2 | −11.50 | −5.09 |
| 1516830 | P2RY10 | NM_014499 | Purinergic receptor P2Y, G-protein coupled, 10 | −11.67 | −4.14 |
| 1519975 | PRSS33 | NM_152891 | Protease, serine, 33 | −11.84 | −5.57 |
| 1507811 | PMAIP1 | NM_021127 | Phorbol-12-myristate-13-acetate-induced protein 1 | −12.76 | −6.44 |
| 1506299 | MMP12 | ENST000003 26227 | Macrophage metalloelastase precursor | −12.98 | −7.44 |
| 1536513 | IFIT3 | NM_001549 | Interferon-induced protein with tetratricopeptide repeats 3 | −13.08 | −9.40 |
| 1513158 | HRH4 | NM_021624 | Histamine receptor H4 | −13.66 | −6.92 |
| 1537338 | FGL2 | NM_006682 | Fibrinogen-like 2 | −13.76 | −31.09 |
| 1509838 | GBP1 | NM_002053 | Guanylate binding protein 1, interferon-inducible, 67kDa | −13.91 | −11.20 |
| 1512112 | EPSTI1 | NM_033255 | Epithelial stromal interaction 1 (breast) | −14.02 | −9.27 |
| 1522895 | GOLPH4 | NM_014498 | Golgi phosphoprotein 4 | −14.30 | −12.18 |
| 1508419 | CD52 | NM_001803 | CD52 molecule | −14.95 | −4.38 |
| 1506912 | RARRES1 | NM_002888 | Retinoic acid receptor responder (tazarotene induced) 1 | −15.02 | −9.44 |
| 1509737 | IFIT2 | NM_001547 | Interferon-induced protein with tetratricopeptide repeats 2 | −15.06 | −31.11 |
| 1505505 | CCL23 | NM_005064 | Chemokine (C-C motif) ligand 23 | −15.06 | −4.22 |
| 1518780 | AF131788 | AF131788 | Clone 24875 mRNA sequence | −16.78 | −12.00 |
| 1522258 | ISG15 | NM_005101 | Interferon, alpha-inducible protein (clone IFI-15K) | −18.16 | −4.81 |
| 1519399 | GBP5 | NM_052942 | Guanylate binding protein 5 | −18.22 | −10.65 |
| 1531078 | RSAD2 | NM_080657 | Radical S-adenosyl methionine domain containing 2 | −18.54 | −8.17 |
| 1515740 | SLC45A3 | NM_033102 | Solute carrier family 45, member 3 | −26.27 | −28.60 |
| 1517997 | IFIT1 | NM_001548 | Interferon-induced protein with tetratricopeptide repeats 1 | −34.47 | −35.07 |
| 1533457 | FCER1A | NM_002001 | Fc fragment of IgE, high affinity I, receptor for; alpha polypeptide | −61.82 | −31.20 |
| 1532327 | CLC | NM_001828 | Charcot-Leyden crystal protein | −69.08 | −16.00 |
Confirmation of microarray data by RT-PCR and flow cytometry
We used LightCycler RT-PCR to verify the changes in gene expression as identified by the microarray analysis. We selected 6 representative genes, encoding well-characterized cell surface receptors, from the microarray dataset for subsequent confirmation by RT-PCR and flow cytometry (Figure 2A). For these genes, there was a strong correlation (r = 0.94) between mRNA levels determined by either microarray or RT-PCR analysis. Mobilization of granulocytes resulted in increased expression levels of complement regulatory factors (CD55 and CD59) and a strong increase in the level of CD177 mRNA, as well as the simultaneous down-regulation of CLEC2A (CD69), CD52, and EMR3 mRNA levels.
G-CSF/dexamethasone mobilization changes the phenotype of granulocytes. (A) Light-cycler confirmation of representative genes. Genes (n = 6 identified as differentially expressed by Agilent Whole Humane Genome microarrays (□) were selected from the overall dataset for confirmation by light cycler real-time PCR ( ) after mobilization with G-CSF/dexamethasone. Data represent the mean ± SD of fold changes in gene expression from the 3 donors used in microarray experiments. Microarray data are presented as the mean fold change of the 3 donors. (B) Flow cytometric analysis of different neutrophil surface receptors. Neutrophils isolated from control donors (□) and those treated with G-CSF/dexamethasone (■) were analyzed for the expression of various surface receptors. Cells were stained with directly labeled monoclonal antibodies and measured by flow cytometry. Results represent the data from 3 independent experiments (mean ± SEM). *P < .05 (significant difference).
) after mobilization with G-CSF/dexamethasone. Data represent the mean ± SD of fold changes in gene expression from the 3 donors used in microarray experiments. Microarray data are presented as the mean fold change of the 3 donors. (B) Flow cytometric analysis of different neutrophil surface receptors. Neutrophils isolated from control donors (□) and those treated with G-CSF/dexamethasone (■) were analyzed for the expression of various surface receptors. Cells were stained with directly labeled monoclonal antibodies and measured by flow cytometry. Results represent the data from 3 independent experiments (mean ± SEM). *P < .05 (significant difference).
G-CSF/dexamethasone mobilization changes the phenotype of granulocytes. (A) Light-cycler confirmation of representative genes. Genes (n = 6 identified as differentially expressed by Agilent Whole Humane Genome microarrays (□) were selected from the overall dataset for confirmation by light cycler real-time PCR ( ) after mobilization with G-CSF/dexamethasone. Data represent the mean ± SD of fold changes in gene expression from the 3 donors used in microarray experiments. Microarray data are presented as the mean fold change of the 3 donors. (B) Flow cytometric analysis of different neutrophil surface receptors. Neutrophils isolated from control donors (□) and those treated with G-CSF/dexamethasone (■) were analyzed for the expression of various surface receptors. Cells were stained with directly labeled monoclonal antibodies and measured by flow cytometry. Results represent the data from 3 independent experiments (mean ± SEM). *P < .05 (significant difference).
) after mobilization with G-CSF/dexamethasone. Data represent the mean ± SD of fold changes in gene expression from the 3 donors used in microarray experiments. Microarray data are presented as the mean fold change of the 3 donors. (B) Flow cytometric analysis of different neutrophil surface receptors. Neutrophils isolated from control donors (□) and those treated with G-CSF/dexamethasone (■) were analyzed for the expression of various surface receptors. Cells were stained with directly labeled monoclonal antibodies and measured by flow cytometry. Results represent the data from 3 independent experiments (mean ± SEM). *P < .05 (significant difference).
Because all transcripts selected for the validation of the microarray data by quantitative RT-PCR represented surface receptors expressed on the plasma membrane, we determined the protein expression levels by flow cytometry (Figure 2B). Both complement regulatory factors CD55 (also known as decay-accelerating factor [DAF]) and CD59 were found to be present on fresh neutrophils. Their expression increased significantly after treatment with G-CSF/dexamethasone in vivo. Granulocyte mobilization also resulted in the increased expression levels of CD177 (also known as the human neutrophil antigen [HNA]-2a or NB1). Increased expression of CD177 on neutrophils has previously been reported in response to G-CSF treatment for hematopoietic stem cell mobilization.28 Control neutrophils clearly expressed the GPI-anchored proteins CD52 and, to a lesser extent, CD69, whereas the surface expression was either diminished (CD52) or had become completely null (CD69) on donor neutrophils mobilized in vivo, confirming the microarray data.
The expression of EMR3 also was found to be lower on the surface of in vivo mobilized cells. This may correspond with the lower maturation state of the granulocytes “purged” from the bone marrow by the combined treatment with G-CSF and dexamethasone, as also indicated by the concomitant neutrophilic left shift (data not shown).
Regulation of different biologic processes by G-CSF/dexamethasone mobilization
Significant numbers of genes with different expression patterns between the control granulocytes and those isolated after G-CSF/dexamethasone administration fell into 6 main functional gene ontology (GO) categories: (1) transcription and regulation of transcription, (2) signal transduction, (3) immune responses, (4) cell survival and apoptosis, (5) cell adhesion and motility, and (6) cell metabolism (Table 2). Similar analysis has been applied to the genes presented in Table 1 to compare these categories of gene patterns (Table S1).
G-CSF/dexamethasone mobilization of granulocytes modulates changes in expression profile of genes involved in different biological processes
| Sequence ID . | Primary sequence name . | Sequence code . | Sequence description . | Fold change . |
|---|---|---|---|---|
| Transcription and regulation of transcription | ||||
| 1514893 | CCNA1 | NM_003914 | Cyclin A1 | 21.01 |
| 1983249 | SMARCA3 | BF513730 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 3 | 10.43 |
| 1513827 | MLXIP | AB020674 | MLX interacting protein | 8.66 |
| 1519529 | MSL3L1 | NM_078629 | Male-specific lethal 3-like 1 (Drosophila) | 6.75 |
| 1521872 | MEF2A | NM_005587 | MADS box transcription enhancer factor 2, polypeptide A (myocyte enhancer factor 2A) | 6.47 |
| 1985292 | ETS2 | BX433326 | V-ets erythroblastosis virus E26 oncogene homolog 2 | 5.59 |
| 1525013 | BATF | NM_006399 | Basic leucine zipper transcription factor, ATF-like | 5.58 |
| 1537803 | ENO1 | NM_001428 | Enolase 1, (alpha) | 5.50 |
| 1533218 | HIPK2 | BC041926 | CDNA clone IMAGE:5300349 | 5.07 |
| 1522326 | PHTF1 | NM_006608 | Putative homeodomain transcription factor 1 | 4.80 |
| 1533591 | SIRT5 | NM_012241 | Sirtuin (silent mating type information regulation 2 homolog) 5 (Saccharomyces cerevisiae) | 4.60 |
| 1512793 | ZNF167 | NM_018651 | Zinc finger protein 167 | 4.30 |
| 1511846 | PLAG1 | NM_002655 | Pleomorphic adenoma gene 1 | 4.26 |
| 1512354 | ZNF438 | AK095436 | hypothetical protein LOC220929 | 4.20 |
| 1507417 | DACH1 | NM_080759 | Dachshund homolog 1 (Drosophila) | 4.04 |
| 1526663 | ZNF438 | NM_182755 | Zinc finger protein 438 | 4.02 |
| 1537859 | ZBTB20 | NM_015642 | Zinc finger and BTB domain containing 20 | 3.77 |
| 1522362 | ZNF658 | NM_033160 | Zinc finger protein 658 | 3.75 |
| 1534837 | ZNF254 | NM_004876 | Zinc finger protein 539 | 3.73 |
| 1507368 | CCND3 | NM_001760 | Cyclin D3 | 3.69 |
| 1509049 | VENTX | NM_014468 | VENT homeobox homolog (Xenopus laevis) | 3.61 |
| 1985753 | CRTC3 | AK090443 | CREB regulated transcription coactivator 3 | 3.58 |
| 1529278 | XRN2 | NM_012255 | 5′-3′ exoribonuclease 2 | 3.43 |
| 1522102 | E2F2 | NM_004091 | E2F transcription factor 2 | 3.36 |
| 1517472 | CCNC | NM_005190 | Cyclin C | 3.07 |
| 1535521 | KLF6 | U51869 | Kruppel-like factor 6 | 3.03 |
| 1537902 | TSC22D1 | NM_183422 | TSC22 domain family, member 1 | −3.01 |
| 1983175 | MCM7 | NM_182776 | MCM7 minichromosome maintenance deficient | −3.02 |
| 1529350 | SSRP1 | NM_003146 | Structure specific recognition protein 1 | −3.03 |
| 1505187 | RELB | NM_006509 | V-rel reticuloendotheliosis viral oncogene homolog B, nuclear factor of kappa light polypeptide gene enhancer in B cells 3 (avian) | −3.04 |
| 1537144 | DEXI | AK055529 | dexamethasone-induced transcript | −3.07 |
| 1516882 | MEF2C | NM_002397 | MADS box transcription enhancer factor 2, polypeptide C (myocyte enhancer factor 2C) | −3.09 |
| 1527019 | CHAF1A | NM_005483 | Chromatin assembly factor 1, subunit A (p150) | −3.25 |
| 1535620 | NCOR2 | NM_006312 | Nuclear receptor co-repressor 2 | −3.34 |
| 1523857 | ZNF395 | NM_018660 | Zinc finger protein 395 | −3.34 |
| 1512878 | ZMYND11 | NM_006624 | Zinc finger, MYND domain containing 11 | −3.35 |
| 1991240 | HMGA1 | NM_145904 | High mobility group AT-hook 1 | −3.37 |
| 1531990 | HEY1 | NM_012258 | Hairy/enhancer-of-split related with YRPW motif 1 | −3.39 |
| 1533586 | ELK1 | NM_005229 | ELK1, member of ETS oncogene family | −3.50 |
| 1515979 | NR4A3 | NM_173199 | Nuclear receptor subfamily 4, group A, member 3 | −3.58 |
| 1510333 | THRA | NM_003250 | Thyroid hormone receptor, alpha (erythroblastic leukemia viral (v-erb-a) oncogene homolog, avian) | −3.68 |
| 1537025 | BTG2 | NM_006763 | BTG family, member 2 | −3.73 |
| 1513189 | TRERF1 | NM_033502 | Transcriptional regulating factor 1 | −3.76 |
| 1511905 | JARID2 | NM_004973 | Jumonji, AT-rich interactive domain 2 | −3.81 |
| 1526826 | ATF3 | NM_004024 | Activating transcription factor 3 | −3.82 |
| 1523890 | RASSF7 | NM_003475 | Ras association (RalGDS/AF−6) domain family 7 | −3.84 |
| 1533807 | MAFF | NM_012323 | V-maf musculoaponeurotic fibrosarcoma oncogene homolog F (avian) | −3.89 |
| 1525862 | JUN | NM_002228 | V-jun sarcoma virus 17 oncogene homolog (avian) | −3.94 |
| 1507971 | NRL | NM_006177 | Neural retina leucine zipper | −4.09 |
| 1533916 | LEF1 | NM_016269 | Lymphoid enhancer-binding factor 1 | −4.22 |
| 1506607 | ASF1B | NM_018154 | ASF1 antisilencing function 1 homolog B (S cerevisiae) | −4.36 |
| 1509496 | AES | NM_198969 | Amino-terminal enhancer of split | −4.39 |
| 1533535 | TAF4 | NM_003185 | TAF4 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 135kDa | −4.40 |
| 1508056 | EGR3 | NM_004430 | Early growth response 3 | −4.41 |
| 1533262 | BRF1 | BC016743 | BRF1 homolog, subunit of RNA polymerase III transcription initiation factor IIIB (S cerevisiae) | −4.48 |
| 1533571 | PHF6 | NM_032458 | PHD finger protein 6 | −4.53 |
| 1517506 | EGR2 | NM_000399 | Early growth response 2 (Krox−20 homolog, Drosophila) | −4.66 |
| 1989375 | SMAD3 | NM_005902 | SMAD, mothers against DPP homolog 3 (Drosophila) | −4.97 |
| 1507856 | MYB | NM_005375 | V-myb myeloblastosis viral oncogene homolog (avian) | −5.06 |
| 1528623 | OLIG2 | NM_005806 | Oligodendrocyte lineage transcription factor 2 | −6.11 |
| 1524585 | GATA1 | NM_002049 | GATA binding protein 1 (globin transcription factor 1) | −6.64 |
| 1991493 | PARP14 | NM_017554 | Poly (ADP-ribose) polymerase family, member 14 | −7.03 |
| 1510055 | PCGF5 | BC007377 | Polycomb group ring finger 5 | −7.57 |
| 1510369 | ASCL2 | NM_005170 | Achaete-scute complex-like 2 (Drosophila) | −7.72 |
| 1522208 | ZNF584 | NM_173548 | Zinc finger protein 584 | −7.93 |
| 1523937 | RPS6KA5 | NM_004755 | Ribosomal protein S6 kinase, 90 kDa, polypeptide 5 | −8.07 |
| 1529851 | ETV7 | NM_016135 | Ets variant gene 7 (TEL2 oncogene) | −8.56 |
| 1506298 | GATA6 | NM_005257 | GATA binding protein 6 | −8.65 |
| 1989096 | SSBP3 | NM_001009955 | Single stranded DNA binding protein 3 | −8.76 |
| 1520503 | NKX31 | NM_006167 | NK3 transcription factor related, locus 1 (Drosophila) | −9.56 |
| 1522991 | ZNF649 | NM_023074 | Zinc finger protein 649 | −11.07 |
| 1514700 | NR4A1 | NM_002135 | Nuclear receptor subfamily 4, group A, member 1 | −92.91 |
| Signal transduction | ||||
| 1513543 | GPR84 | NM_020370 | G protein-coupled receptor 84 | 25.51 |
| 1505483 | EDG1 | NM_001400 | Endothelial differentiation, sphingolipid G-protein-coupled receptor, 1 | 18.05 |
| 1511321 | PTGER2 | NM_000956 | Prostaglandin E receptor 2 (subtype EP2), 53 kDa | 8.26 |
| 1511570 | ARHGAP24 | AK130576 | Rho GTPase activating protein 24 | 7.28 |
| 1523886 | PDE4D | NM_006203 | Phosphodiesterase 4D, cAMP-specific (phosphodiesterase E3 dunce homolog, Drosophila) | 6.64 |
| 1527053 | MYO10 | BC041694 | Myosin X | 6.48 |
| 1512860 | MS4A4A | NM_024021 | Membrane-spanning 4-domains, subfamily A, member 4 | 6.12 |
| 1539087 | DLC1 | NM_182643 | Deleted in liver cancer 1 | 5.96 |
| 1528643 | MS4A6A | NM_022349 | Membrane-spanning 4-domains, subfamily A, member 6A | 5.87 |
| 1524040 | GPR141 | NM_181791 | G protein-coupled receptor 141 | 5.28 |
| 1989101 | EDARADD | NM_080738 | EDAR-associated death domain | 4.64 |
| 1508615 | EXT1 | NM_000127 | Exostoses (multiple) 1 | 4.30 |
| 1521881 | STK3 | NM_006281 | Serine/threonine kinase 3 (STE20 homolog, yeast) | 3.79 |
| 1510158 | ITPKC | NM_025194 | Inositol 1.4,5-trisphosphate 3-kinase C | 3.63 |
| 1519571 | CDC42EP3 | NM_006449 | CDC42 effector protein (Rho GTPase binding) 3 | 3.58 |
| 1534557 | DPYSL3 | NM_001387 | Dihydropyrimidinase-like 3 | 3.54 |
| 1536781 | RPS6KA3 | NM_004586 | Ribosomal protein S6 kinase, 90 kDa, polypeptide 3 | 3.53 |
| 1533999 | HHIP | NM_022475 | Hedgehog interacting protein | 3.50 |
| 1512578 | GPR97 | NM_170776 | G protein-coupled receptor 97 | 3.42 |
| 1514738 | SLC26A6 | NM_022911 | Inositol hexaphosphate kinase 2 | 3.28 |
| 1517434 | IL4R | NM_000418 | Interleukin 4 receptor | 3.21 |
| 1539209 | GABBR1 | NM_001470 | Gamma-aminobutyric acid (GABA) B receptor, 1 | 3.19 |
| 1511435 | STK16 | NM_003691 | Serine/threonine kinase 16 | 3.17 |
| 1520958 | PRKAR2A | BC002763 | Inositol hexaphosphate kinase 2 | 3.12 |
| 1506387 | MAPK6 | NM_002748 | Mitogen-activated protein kinase 6 | 3.03 |
| 1523162 | ITPK1 | NM_014216 | Inositol 1.3,4-triphosphate 5/6 kinase | −3.05 |
| 1507956 | GNGT2 | NM_031498 | Guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2 | −3.09 |
| 1514326 | TAGAP | NM_138810 | T-cell activation GTPase activating protein | −3.09 |
| 1530414 | GPR171 | NM_013308 | G protein-coupled receptor 171 | −3.16 |
| 1527339 | P2RY5 | NM_005767 | Purinergic receptor P2Y, G-protein coupled, 5 | −3.17 |
| 1517355 | ARHGAP25 | BC039591 | Rho GTPase activating protein 25 | −3.23 |
| 1520830 | KIT | NM_000222 | V-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog | −3.28 |
| 1517660 | KLRK1 | NM_007360 | Killer cell lectin-like receptor subfamily C, member 4 | −3.37 |
| 1514077 | KLRB1 | NM_002258 | Killer cell lectin-like receptor subfamily B, member 1 | −3.43 |
| 1518328 | GRP | NM_002091 | Gastrin-releasing peptide | −3.64 |
| 1516849 | EPHB1 | NM_004441 | EPH receptor B1 | −3.80 |
| 1514753 | PRKAG2 | NM_016203 | Protein kinase, AMP-activated, gamma 2 non-catalytic subunit | −4.09 |
| 1512127 | SOCS1 | NM_003745 | Suppressor of cytokine signaling 1 | −4.13 |
| 1518864 | FLT3LG | NM_001459 | Fms-related tyrosine kinase 3 ligand | −4.26 |
| 1524915 | OR51E1 | NM_152430 | Olfactory receptor, family 51, subfamily E, member 1 | −4.27 |
| 1507227 | P2RY14 | NM_014879 | Purinergic receptor P2Y, G-protein coupled, 14 | −4.28 |
| 1523084 | GUCA1A | NM_000409 | Guanylate cyclase activator 1A (retina) | −4.37 |
| 1525046 | PILRA | NM_178273 | Paired immunoglobulin-like type 2 receptor alpha | −4.48 |
| 1534223 | PPP1R12B | NM_032105 | Protein phosphatase 1, regulatory (inhibitor) subunit 12B | −4.66 |
| 1518048 | GPR34 | NM_005300 | G-protein–coupled receptor 34 | −4.78 |
| 1534320 | ARRB1 | NM_004041 | Arrestin, beta 1 | −5.02 |
| 1534974 | KLRD1 | NM_002262 | Killer cell lectin-like receptor subfamily D, member 1 | −5.16 |
| 1525314 | S100A10 | NM_002966 | S100 calcium binding protein A10 | −5.94 |
| 1530940 | PRKAR2B | NM_002736 | Protein kinase, cAMP-dependent, regulatory, type II, beta | −5.97 |
| 1522410 | IL2RB | NM_000878 | Interleukin 2 receptor, beta | −6.06 |
| 1518652 | CD74 | NM_004355 | CD74 molecule, major histocompatibility complex, class II invariant chain | −6.23 |
| 1515002 | CYSLTR1 | NM_006639 | Cysteinyl leukotriene receptor 1 | −6.45 |
| 1530332 | MX1 | NM_002462 | Myxovirus (influenza virus) resistance 1, interferon-inducible protein p78 (mouse) | −6.76 |
| 1522509 | OR2B2 | NM_033057 | Olfactory receptor, family 2, subfamily B, member 2 | −6.88 |
| 1984459 | PDE4B | AB209081 | Phosphodiesterase 4B, cAMP-specific | −6.89 |
| 1527378 | CSF1R | NM_005211 | Colony stimulating factor 1 receptor, formerly McDonough feline sarcoma viral (v-fms) oncogene homolog | −7.08 |
| 1527858 | NRGN | NM_006176 | Neurogranin (protein kinase C substrate, RC3) | −7.49 |
| 1524392 | ANGPT1 | NM_001146 | Angiopoietin 1 | −8.84 |
| 1516837 | MS4A3 | NM_006138 | Membrane-spanning 4-domains, subfamily A, member 3 | −9.24 |
| 1989999 | IL5RA | NM_175725 | Interleukin 5 receptor, alpha | −9.66 |
| 1524637 | GPR162 | NM_019858 | Triosephosphate isomerase 1 | −10.10 |
| 1519217 | P2RY2 | NM_176072 | Purinergic receptor P2Y, G-protein coupled, 2 | −11.66 |
| 1516830 | P2RY10 | NM_014499 | Purinergic receptor P2Y, G-protein coupled, 10 | −11.67 |
| 1518123 | CD83 | NM_004233 | CD83 molecule | −11.67 |
| 1517216 | KLRF1 | NM_016523 | Killer cell lectin-like receptor subfamily F, member 1 | −12.68 |
| 1512228 | EBI2 | NM_004951 | Epstein-Barr virus–induced gene 2 (lymphocyte-specific G protein-coupled receptor) | −13.11 |
| 1513158 | HRH4 | NM_021624 | Histamine receptor H4 | −13.66 |
| 1537338 | FGL2 | NM_006682 | Fibrinogen-like 2 | −13.76 |
| 1515714 | GNG11 | NM_004126 | Guanine nucleotide binding protein (G protein), gamma 11 | −16.57 |
| 1527111 | GPR44 | NM_004778 | G protein-coupled receptor 44 | −23.51 |
| 1528909 | RGS1 | NM_002922 | Regulator of G-protein signaling 1 | −32.65 |
| Immune responses | ||||
| 1507161 | CD163 | NM_004244 | CD163 molecule | 54.27 |
| 1531927 | CYP1B1 | NM_000104 | Cytochrome P450, family 1, subfamily B, polypeptide 1 | 46.19 |
| 1984958 | CD177 | CR592446 | CD177 molecule | 24.38 |
| 1521404 | IL18R1 | NM_003855 | Interleukin 18 receptor 1 | 16.54 |
| 1983056 | MERTK | U08023 | C-mer proto-oncogene tyrosine kinase | 16.00 |
| 1532739 | CLEC5A | NM_013252 | C-type lectin domain family 5, member A | 13.44 |
| 1511835 | CLEC4D | NM_080387 | C-type lectin domain family 4, member D | 12.02 |
| 1535646 | CST7 | NM_003650 | Cystatin F (leukocystatin) | 11.43 |
| 1506996 | ADORA3 | NM_020683 | Adenosine A3 receptor | 10.89 |
| 1530098 | IL18RAP | NM_003853 | Interleukin 18 receptor accessory protein | 10.79 |
| 1532148 | LAIR1 | NM_021706 | Leukocyte-associated immunoglobulin-like receptor 1 | 10.45 |
| 1506562 | HP | NM_005143 | Haptoglobin | 10.31 |
| 1505718 | IL1R2 | NM_004633 | Interleukin 1 receptor, type II | 9.95 |
| 1507483 | LILRA5 | NM_181879 | Leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 5 | 7.56 |
| 1524454 | XBP1 | NM_005080 | X-box binding protein 1 | 7.48 |
| 1510813 | IRAK3 | NM_007199 | Interleukin−1 receptor-associated kinase 3 | 7.28 |
| 1538778 | TLR5 | NM_003268 | Toll-like receptor 5 | 5.97 |
| 1521047 | CR1 | NM_000651 | Complement component (3b/4b) receptor 1 (Knops blood group) | 5.87 |
| 1520792 | MAPK14 | NM_139013 | Mitogen-activated protein kinase 14 | 5.75 |
| 1535716 | CD59 | NM_203330 | CD59 molecule, complement regulatory protein | 5.67 |
| 1513648 | FCER1G | CB529629 | Fc fragment of IgE, high affinity I, receptor for; gamma polypeptide | 5.26 |
| 1521687 | TLR4 | BF670973 | Toll-like receptor 4 | 5.15 |
| 1530114 | MAP2K6 | NM_002758 | Mitogen-activated protein kinase kinase 6 | 4.87 |
| 1533789 | SERPINB1 | NM_030666 | Serpin peptidase inhibitor, clade B (ovalbumin), member 1 | 4.84 |
| 1522193 | FCAR | NM_002000 | Fc fragment of IgA, receptor for | 4.52 |
| 1527121 | PROK2 | NM_021935 | Prokineticin 2 | 4.33 |
| 1505831 | CIAS1 | NM_004895 | Cold autoinflammatory syndrome 1 | 4.09 |
| 1524842 | IL10 | NM_000572 | Interleukin 10 | 3.89 |
| 1538909 | IL1RAP | NM_002182 | Interleukin 1 receptor accessory protein | 3.80 |
| 1506853 | CHIT1 | NM_003465 | Chitinase 1 (chitotriosidase) | 3.48 |
| 1531091 | CD55 | NM_000574 | CD55 molecule, decay accelerating factor for complement (Cromer blood group) | 3.39 |
| 1534101 | IL1R1 | NM_000877 | Interleukin 1 receptor, type I | 3.34 |
| 1515424 | CYBB | S67289 | Cytochrome b-245, beta polypeptide (chronic granulomatous disease) | 3.29 |
| 1985998 | ALOX5 | BQ548663 | Arachidonate 5-lipoxygenase | 3.16 |
| 1510372 | IRAK4 | NM_016123 | Interleukin-1 receptor-associated kinase 4 | 3.10 |
| 1514172 | CD48 | NM_001778 | CD48 molecule | 3.09 |
| 1527340 | TLR8 | NM_016610 | Toll-like receptor 8 | 3.07 |
| 1511452 | SERPING1 | NM_000062 | Serpin peptidase inhibitor, clade G (C1 inhibitor), member 1, (angioedema, hereditary) | −3.00 |
| 1530131 | IRF2BP2 | BC020516 | Interferon regulatory factor 2 binding protein 2 | −3.03 |
| 1512742 | CLEC7A | NM_197954 | C-type lectin domain family 7, member A | −3.06 |
| 1528251 | NCR3 | NM_147130 | Natural cytotoxicity triggering receptor 3 | −3.13 |
| 1513772 | HLA-DRB3 | NM_022555 | Major histocompatibility complex, class II, DR beta 1 | −3.17 |
| 1518778 | ALOX15 | NM_001140 | Arachidonate 15-lipoxygenase | −3.24 |
| 1524640 | OAS3 | NM_006187 | 2'−5'-oligoadenylate synthetase 3, 100 kDa | −3.31 |
| 1985491 | HLA-DRB1 | X12544 | Major histocompatibility complex, class II, DR beta 1 | −3.31 |
| 1519302 | STAT1 | NM_139266 | Signal transducer and activator of transcription 1, 91 kDa | −3.39 |
| 1509654 | CTSW | NM_001335 | Cathepsin W (lymphopain) | −3.45 |
| 1992017 | PLAUR | NM_001005377 | Plasminogen activator, urokinase receptor | −3.50 |
| 1526588 | LY75 | NM_002349 | Lymphocyte antigen 75 | −3.51 |
| 1536508 | LST1 | NM_007161 | Leukocyte specific transcript 1 | −3.64 |
| 1515890 | FAIM3 | NM_005449 | Interleukin 24 | −3.65 |
| 1511424 | EMR3 | NM_032571 | Egf-like module containing, mucin-like, hormone receptor-like 3 | −3.71 |
| 1534340 | CD46 | NM_002389 | CD46 molecule, complement regulatory protein | −3.83 |
| 1537056 | GBP3 | NM_018284 | Guanylate binding protein 1, interferon-inducible, 67 kDa | −3.86 |
| 1517555 | CYP27A1 | NM_000784 | Cytochrome P450, family 27, subfamily A, polypeptide 1 | −3.88 |
| 1516190 | SAMHD1 | NM_015474 | SAM domain and HD domain 1 | −3.89 |
| 1519488 | HLA-DQB2 | NM_182549 | Major histocompatibility complex, class II, DQ beta 2 | −3.94 |
| 1983904 | IL32 | NM_001012631 | Interleukin 32 | −3.99 |
| 1505541 | GZMB | NM_004131 | Granzyme B | −4.04 |
| 1536691 | GZMK | NM_002104 | Granzyme K (granzyme 3; tryptase II) | −4.67 |
| 1522713 | GZMA | NM_006144 | Granzyme A | −4.80 |
| 1510975 | HLA-DRB5 | NM_002125 | Major histocompatibility complex, class II, DR beta 1 | −5.13 |
| 1527466 | HLA-DPA1 | NM_033554 | Major histocompatibility complex, class II, DP alpha 1 | −5.16 |
| 1516918 | B2M | AK022379 | Beta−2-microglobulin | −5.43 |
| 1538818 | HLA-DQB1 | NM_002123 | Major histocompatibility complex, class II, DQ beta 1 | −5.87 |
| 1521122 | SPON2 | NM_012445 | Spondin 2, extracellular matrix protein | −5.95 |
| 1517271 | PF4V1 | NM_002620 | Platelet factor 4 variant 1 | −6.48 |
| 1508682 | HLA-DPB1 | NM_002121 | Major histocompatibility complex, class II, DP beta 1 | −6.67 |
| 1529151 | OASL | NM_003733 | 2′−5′-oligoadenylate synthetase-like | −7.10 |
| 1987518 | CYB561 | NM_001017916 | Cytochrome b-561 | −7.39 |
| 1524314 | IFI44 | NM_006417 | Interferon-induced protein 44 | −7.91 |
| 1533656 | DEFA4 | NM_001925 | Defensin, alpha 4, corticostatin | −9.04 |
| 1526794 | HLA-DRA | NM_019111 | Major histocompatibility complex, class II, DR alpha | −10.12 |
| 1536919 | HLA-DMB | NM_002118 | Major histocompatibility complex, class II, DM beta | −10.73 |
| 1525510 | PF4 | NM_002619 | Platelet factor 4 (chemokine (C-X-C motif) ligand 4) | −11.03 |
| 1514544 | DEFA3 | NM_005217 | Defensin, alpha 1 | −11.85 |
| 1520461 | CD69 | NM_001781 | CD69 molecule | −28.65 |
| 1528045 | IL8 | NM_000584 | Interleukin 8 | −35.22 |
| 1533457 | FCER1A | NM_002001 | Fc fragment of IgE, high affinity I, receptor for; alpha polypeptide | −61.82 |
| Cell survival and apoptosis | ||||
| 1519170 | GADD45A | NM_001924 | Growth arrest and DNA-damage-inducible, alpha | 20.49 |
| 1531073 | CARD12 | NM_021209 | Caspase recruitment domain family, member 12 | 11.37 |
| 1513896 | BIRC1 | NM_004536 | Baculoviral IAP repeat-containing 1 | 10.55 |
| 1525148 | ANXA1 | NM_000700 | Annexin A1 | 7.39 |
| 1533289 | ANXA3 | NM_005139 | Annexin A3 | 7.33 |
| 1516682 | CAST | NM_001750 | Calpastatin | 6.97 |
| 1532823 | PECR | NM_018441 | Peroxisomal trans−2-enoyl-CoA reductase | 5.32 |
| 1539226 | CARD6 | NM_032587 | Caspase recruitment domain family, member 6 | 5.15 |
| 1531614 | ELMO2 | NM_022086 | Engulfment and cell motility 2 | 4.67 |
| 1512780 | LGALS8 | NM_006499 | Lectin, galactoside-binding, soluble, 8 (galectin 8) | 4.24 |
| 1505831 | CIAS1 | NM_004895 | Cold autoinflammatory syndrome 1 | 4.09 |
| 1521881 | STK3 | NM_006281 | Serine/threonine kinase 3 (STE20 homolog, yeast) | 3.79 |
| 1526256 | DDAH2 | NM_013974 | Dimethylarginine dimethylaminohydrolase 2 | 3.75 |
| 1522394 | TNFRSF10A | NM_003844 | Tumor necrosis factor receptor superfamily, member 10a | 3.69 |
| 1988846 | AIM1 | NM_001624 | Absent in melanoma 1 | 3.52 |
| 1509305 | LTB4R | NM_181657 | Leukotriene B4 receptor | 3.20 |
| 1984749 | HIP1 | BC082970 | Huntingtin interacting protein 1 | 3.19 |
| 1534293 | LGALS3 | NM_002306 | Lectin, galactoside-binding, soluble, 3 (galectin 3) | 3.03 |
| 1535837 | SQSTM1 | NM_003900 | Sequestosome 1 | −3.01 |
| 1537407 | NFKBIE | NM_004556 | Nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, epsilon | −3.12 |
| 1987065 | TNFRSF25 | NM_148968 | Homo sapiens tumor necrosis factor receptor superfamily | −3.36 |
| 1516125 | PRDX2 | NM_005809 | Peroxiredoxin 2 | −3.40 |
| 1531960 | GSTP1 | NM_000852 | Glutathione S-transferase pi | −3.42 |
| 1984701 | CARD9 | NM_052813 | Caspase recruitment domain family, member 9 | −3.61 |
| 1532501 | PIM2 | NM_006875 | Pim−2 oncogene | −3.76 |
| 1528041 | IL31RA | NM_139017 | Interleukin 31 receptor A | −3.91 |
| 1533374 | PHLDA2 | NM_003311 | Pleckstrin homology-like domain, family A, member 2 | −3.97 |
| 1526625 | CARD4 | NM_006092 | Caspase recruitment domain family, member 4 | −3.99 |
| 1520783 | DYRK2 | NM_006482 | Dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2 | −4.29 |
| 1516378 | TNFSF12 | NM_153012 | Tumor necrosis factor (ligand) superfamily, member 12 | −4.29 |
| 1530737 | LGALS2 | NM_006498 | Lectin, galactoside-binding, soluble, 2 (galectin 2) | −4.47 |
| 1510435 | LGALS12 | NM_033101 | Lectin, galactoside-binding, soluble, 12 (galectin 12) | −4.47 |
| 1523074 | HSPA9B | NM_004134 | Heat shock 70-kDa protein 9B (mortalin-2) | −5.16 |
| 1532326 | DDX58 | NM_014314 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 | −5.17 |
| 1508150 | AOC3 | NM_003734 | Amine oxidase, copper containing 3 (vascular adhesion protein 1) | −5.52 |
| 1538822 | IFI6 | NM_022873 | Interferon, alpha-inducible protein (clone IFI-6–16) | −5.56 |
| 1990540 | CLU | NM_203339 | Clusterin | −5.66 |
| 1517391 | BIRC3 | NM_001165 | Baculoviral IAP repeat-containing 3 | −6.55 |
| 1538745 | SGK | NM_005627 | Serum/glucocorticoid regulated kinase | −7.90 |
| 1523311 | BIRC4BP | NM_017523 | XIAP associated factor-1 | −9.09 |
| 1510278 | NGFRAP1 | NM_014380 | Nerve growth factor receptor (TNFRSF16) associated protein 1 | −9.35 |
| 1533140 | NME5 | NM_003551 | Non-metastatic cells 5, protein expressed in (nucleoside-diphosphate kinase) | −9.98 |
| 1533249 | HIG2 | NM_013332 | hypoxia-inducible protein 2 | −15.17 |
| Cell adhesion and motility | ||||
| 1536096 | MMP8 | NM_002424 | Matrix metallopeptidase 8 (neutrophil collagenase) | 97.77 |
| 1519153 | OLFM4 | NM_006418 | Olfactomedin 4 | 23.81 |
| 1514453 | CD44 | NM_000610 | CD44 molecule (Indian blood group) | 10.05 |
| 1532879 | EMILIN2 | NM_032048 | Elastin microfibril interfacer 2 | 9.85 |
| 1521082 | CEACAM1 | NM_001712 | Carcinoembryonic antigen-related cell adhesion molecule 1 (biliary glycoprotein) | 5.97 |
| 1509526 | MMP9 | NM_004994 | Matrix metallopeptidase 9 | 5.64 |
| 1522658 | VNN1 | NM_004666 | Vanin 1 | 5.35 |
| 1984856 | ADAM15 | NM_207191 | ADAM metallopeptidase domain 15 (metargidin) | 3.79 |
| 1507356 | F5 | NM_000130 | Coagulation factor V (proaccelerin, labile factor) | 3.78 |
| 1516175 | JAK2 | NM_004972 | Janus kinase 2 (a protein tyrosine kinase) | 3.78 |
| 1508488 | SPP1 | NM_000582 | Secreted phosphoprotein 1 (osteopontin, bone sialoprotein I, early T-lymphocyte activation 1) | 3.72 |
| 1523771 | ITGAM | NM_000632 | Integrin, alpha M (complement component 3 receptor 3 subunit) | 3.52 |
| 1508621 | PVRL2 | NM_002856 | Poliovirus receptor-related 2 (herpesvirus entry mediator B) | 3.42 |
| 1531024 | SIGLEC9 | NM_014441 | Sialic acid binding Ig-like lectin 9 | 3.34 |
| 1505504 | VIM | NM_003380 | Vimentin | 3.34 |
| 1986801 | LAMB3 | NM_001017402 | Laminin, beta 3 | 3.11 |
| 1518582 | CEACAM6 | BC005008 | Carcinoembryonic antigen-related cell adhesion molecule 6 (non-specific cross reacting antigen) | −3.07 |
| 1506966 | SIGLEC8 | NM_014442 | Sialic acid binding immunoglobulin-like lectin 8 | −3.16 |
| 1534776 | CXCL1 | NM_00–1511 | Chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha) | −3.20 |
| 1520341 | AZU1 | NM_001700 | Azurocidin 1 (cationic antimicrobial protein 37) | −3.23 |
| 1520433 | ITGB7 | NM_000889 | Integrin, beta 7 | −3.59 |
| 1505769 | TUBB1 | NM_030773 | Tubulin, beta 1 | −4.35 |
| 1528734 | ICAM2 | NM_000873 | Intercellular adhesion molecule 2 | −5.24 |
| 1991393 | CXCR4 | NM_001008540 | Chemokine (C-X-C motif) receptor 4 | −5.90 |
| 1989016 | CCL3L3 | NM_001001437 | Chemokine (C-C motif) ligand 3-like 3 | −5.98 |
| 1510688 | CCL3 | NM_002983 | Chemokine (C-C motif) ligand 3 | −6.35 |
| 1517696 | CCL5 | NM_002985 | Chemokine (C-C motif) ligand 5 | −7.71 |
| 1533135 | CXCL10 | NM_001565 | Chemokine (C-X-C motif) ligand 11 | −9.10 |
| 1505505 | CCL23 | NM_005064 | Chemokine (C-C motif) ligand 23 | −15.06 |
| 1526404 | CCL4 | NM_002984 | Chemokine (C-C motif) ligand 4 | −15.45 |
| 1533611 | PPBP | NM_002704 | Pro-platelet basic protein (chemokine (C-X-C motif) ligand 7) | −16.30 |
| 1515169 | ITGA4 | NM_000885 | Integrin, alpha 4 (antigen CD49D), alpha 4 subunit of VLA-4 receptor | −16.74 |
| 1537942 | CCR3 | NM_001837 | Chemokine (C-C motif) receptor 3 | −19.63 |
| Cell metabolism | ||||
| 1538977 | HPGD | NM_000860 | Hydroxyprostaglandin dehydrogenase 15-(NAD) | 36.27 |
| 1989284 | PFKFB2 | NM_001018053 | 6-phosphofructo-2-kinase/fructose-2.6-biphosphatase 2 | 31.40 |
| 1986330 | ATP9A | NM_006045 | ATPase, Class II, type 9A | 25.82 |
| 1511217 | LDHA | NM_005566 | Lactate dehydrogenase A | 10.11 |
| 1527335 | PCSK9 | NM_174936 | Proprotein convertase subtilisin/kexin type 9 | 7.95 |
| 1991955 | PLB1 | CR600701 | Phospholipase B1 | 7.59 |
| 1983476 | ATP13A3 | AJ306929 | ATPase type 13A3 | 7.56 |
| 1513367 | DPH5 | NM_0–15958 | DPH5 homolog (S cerevisiae) | 6.38 |
| 1526202 | NMNAT2 | NM_0–15039 | Nicotinamide nucleotide adenylyltransferase 2 | 6.31 |
| 1536086 | DPYD | NM_000110 | Dihydropyrimidine dehydrogenase | 6.11 |
| 1990338 | ATP11B | NM_014616 | ATPase, Class VI, type 11B | 6.09 |
| 1515968 | ALPL | NM_000478 | Alkaline phosphatase, liver/bone/kidney | 5.59 |
| 1506037 | MGAM | NM_004668 | Maltase-glucoamylase (alpha-glucosidase) | 5.41 |
| 1532823 | PECR | NM_018441 | Peroxisomal trans-2-enoyl-CoA reductase | 5.32 |
| 1986414 | ATP2B4 | NM_001001396 | ATPase, Ca2+ transporting, plasma membrane 4 | 4.99 |
| 1506589 | PFKFB3 | NM_004566 | 6-phosphofructo-2-kinase/fructose-2.6-biphosphatase 3 | 4.75 |
| 1512399 | AGPAT6 | NM_178819 | 1-acylglycerol-3-phosphate O-acyltransferase 6 (lysophosphatidic acid acyltransferase, zeta) | 4.74 |
| 1523383 | PGD | NM_002631 | Phosphogluconate dehydrogenase | 4.70 |
| 1519383 | ATP6V1C1 | NM_001695 | ATPase, H+ transporting, lysosomal 42 kDa, V1 subunit C1 | 4.11 |
| 1520655 | SMPDL3A | NM_006714 | Sphingomyelin phosphodiesterase, acid-like 3A | 4.09 |
| 1989848 | ACSL4 | NM_004458 | Acyl-CoA synthetase long-chain family member 4 | 4.02 |
| 1522656 | LDLR | NM_000527 | Low-density lipoprotein receptor | 3.63 |
| 1538541 | DHRS9 | NM_005771 | Dehydrogenase/reductase (SDR family) member 9 | 3.55 |
| 1514810 | ATP1A3 | NM_152296 | ATPase, Na+/K+ transporting, alpha 3 polypeptide | 3.53 |
| 1524666 | FABP5 | NM_001444 | Fatty acid binding protein 5 (psoriasis-associated) | 3.38 |
| 1538372 | ATP1A1 | NM_000701 | ATPase, Na+/K+ transporting, alpha 1 polypeptide | 3.38 |
| 1524650 | GNS | NM_002076 | Glucosamine (N-acetyl)-6- | 3.37 |
| 1523432 | ACSL1 | NM_001995 | Acyl-CoA synthetase long-chain family member 1 | 3.35 |
| 1529572 | SGSH | NM_000199 | N-sulfoglucosamine sulfohydrolase (sulfamidase) | 3.29 |
| 1538734 | ACSL3 | NM_004457 | Acyl-CoA synthetase long-chain family member 3 | 3.27 |
| 1985998 | ALOX5 | BQ548663 | Arachidonate 5-lipoxygenase | 3.16 |
| 1527407 | GAPDH | NM_002046 | Glyceraldehyde3-phosphate dehydrogenase | 3.16 |
| 1534240 | SULT1B1 | D89479 | Sulfotransferase family, cytosolic, 1B, member 1 | 3.10 |
| 1983183 | NDUFB6 | NM_182739 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 6 | −3.10 |
| 1525237 | ALDH6A1 | NM_005589 | Aldehyde dehydrogenase 6 family, member A1 | −3.22 |
| 1518778 | ALOX15 | NM_001140 | Arachidonate 15-lipoxygenase | −3.24 |
| 1523761 | CBR1 | NM_001757 | Carbonyl reductase 1 | −3.61 |
| 1510672 | MCCC1 | NM_020166 | Methylcrotonoyl-Coenzyme A carboxylase 1 (alpha) | −4.01 |
| 1990453 | NDUFV2 | BI520375 | NADH dehydrogenase (ubiquinone) flavoprotein 2, 24 kDa | −4.31 |
| 1521732 | ATP12A | NM_001676 | ATPase, H+/K+ transporting, nongastric, alpha polypeptide | −4.74 |
| 1526184 | LDHB | NM_002300 | Lactate dehydrogenase B | −6.08 |
| 1512448 | ISOC1 | NM_016048 | Isochorismatase domain containing 1 | −6.51 |
| 1989369 | METTL7A | BC004492 | Methyltransferase like 7A | −6.99 |
| 1518627 | FA2H | NM_024306 | Fatty acid 2-hydroxylase | −7.75 |
| 1506804 | PLD4 | NM_138790 | Phospholipase D family, member 4 | −9.53 |
| 1521949 | SORD | NM_003104 | Sorbitol dehydrogenase | −10.20 |
| 1512822 | SULF2 | NM_018837 | Sulfatase 2 | −21.22 |
| Sequence ID . | Primary sequence name . | Sequence code . | Sequence description . | Fold change . |
|---|---|---|---|---|
| Transcription and regulation of transcription | ||||
| 1514893 | CCNA1 | NM_003914 | Cyclin A1 | 21.01 |
| 1983249 | SMARCA3 | BF513730 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 3 | 10.43 |
| 1513827 | MLXIP | AB020674 | MLX interacting protein | 8.66 |
| 1519529 | MSL3L1 | NM_078629 | Male-specific lethal 3-like 1 (Drosophila) | 6.75 |
| 1521872 | MEF2A | NM_005587 | MADS box transcription enhancer factor 2, polypeptide A (myocyte enhancer factor 2A) | 6.47 |
| 1985292 | ETS2 | BX433326 | V-ets erythroblastosis virus E26 oncogene homolog 2 | 5.59 |
| 1525013 | BATF | NM_006399 | Basic leucine zipper transcription factor, ATF-like | 5.58 |
| 1537803 | ENO1 | NM_001428 | Enolase 1, (alpha) | 5.50 |
| 1533218 | HIPK2 | BC041926 | CDNA clone IMAGE:5300349 | 5.07 |
| 1522326 | PHTF1 | NM_006608 | Putative homeodomain transcription factor 1 | 4.80 |
| 1533591 | SIRT5 | NM_012241 | Sirtuin (silent mating type information regulation 2 homolog) 5 (Saccharomyces cerevisiae) | 4.60 |
| 1512793 | ZNF167 | NM_018651 | Zinc finger protein 167 | 4.30 |
| 1511846 | PLAG1 | NM_002655 | Pleomorphic adenoma gene 1 | 4.26 |
| 1512354 | ZNF438 | AK095436 | hypothetical protein LOC220929 | 4.20 |
| 1507417 | DACH1 | NM_080759 | Dachshund homolog 1 (Drosophila) | 4.04 |
| 1526663 | ZNF438 | NM_182755 | Zinc finger protein 438 | 4.02 |
| 1537859 | ZBTB20 | NM_015642 | Zinc finger and BTB domain containing 20 | 3.77 |
| 1522362 | ZNF658 | NM_033160 | Zinc finger protein 658 | 3.75 |
| 1534837 | ZNF254 | NM_004876 | Zinc finger protein 539 | 3.73 |
| 1507368 | CCND3 | NM_001760 | Cyclin D3 | 3.69 |
| 1509049 | VENTX | NM_014468 | VENT homeobox homolog (Xenopus laevis) | 3.61 |
| 1985753 | CRTC3 | AK090443 | CREB regulated transcription coactivator 3 | 3.58 |
| 1529278 | XRN2 | NM_012255 | 5′-3′ exoribonuclease 2 | 3.43 |
| 1522102 | E2F2 | NM_004091 | E2F transcription factor 2 | 3.36 |
| 1517472 | CCNC | NM_005190 | Cyclin C | 3.07 |
| 1535521 | KLF6 | U51869 | Kruppel-like factor 6 | 3.03 |
| 1537902 | TSC22D1 | NM_183422 | TSC22 domain family, member 1 | −3.01 |
| 1983175 | MCM7 | NM_182776 | MCM7 minichromosome maintenance deficient | −3.02 |
| 1529350 | SSRP1 | NM_003146 | Structure specific recognition protein 1 | −3.03 |
| 1505187 | RELB | NM_006509 | V-rel reticuloendotheliosis viral oncogene homolog B, nuclear factor of kappa light polypeptide gene enhancer in B cells 3 (avian) | −3.04 |
| 1537144 | DEXI | AK055529 | dexamethasone-induced transcript | −3.07 |
| 1516882 | MEF2C | NM_002397 | MADS box transcription enhancer factor 2, polypeptide C (myocyte enhancer factor 2C) | −3.09 |
| 1527019 | CHAF1A | NM_005483 | Chromatin assembly factor 1, subunit A (p150) | −3.25 |
| 1535620 | NCOR2 | NM_006312 | Nuclear receptor co-repressor 2 | −3.34 |
| 1523857 | ZNF395 | NM_018660 | Zinc finger protein 395 | −3.34 |
| 1512878 | ZMYND11 | NM_006624 | Zinc finger, MYND domain containing 11 | −3.35 |
| 1991240 | HMGA1 | NM_145904 | High mobility group AT-hook 1 | −3.37 |
| 1531990 | HEY1 | NM_012258 | Hairy/enhancer-of-split related with YRPW motif 1 | −3.39 |
| 1533586 | ELK1 | NM_005229 | ELK1, member of ETS oncogene family | −3.50 |
| 1515979 | NR4A3 | NM_173199 | Nuclear receptor subfamily 4, group A, member 3 | −3.58 |
| 1510333 | THRA | NM_003250 | Thyroid hormone receptor, alpha (erythroblastic leukemia viral (v-erb-a) oncogene homolog, avian) | −3.68 |
| 1537025 | BTG2 | NM_006763 | BTG family, member 2 | −3.73 |
| 1513189 | TRERF1 | NM_033502 | Transcriptional regulating factor 1 | −3.76 |
| 1511905 | JARID2 | NM_004973 | Jumonji, AT-rich interactive domain 2 | −3.81 |
| 1526826 | ATF3 | NM_004024 | Activating transcription factor 3 | −3.82 |
| 1523890 | RASSF7 | NM_003475 | Ras association (RalGDS/AF−6) domain family 7 | −3.84 |
| 1533807 | MAFF | NM_012323 | V-maf musculoaponeurotic fibrosarcoma oncogene homolog F (avian) | −3.89 |
| 1525862 | JUN | NM_002228 | V-jun sarcoma virus 17 oncogene homolog (avian) | −3.94 |
| 1507971 | NRL | NM_006177 | Neural retina leucine zipper | −4.09 |
| 1533916 | LEF1 | NM_016269 | Lymphoid enhancer-binding factor 1 | −4.22 |
| 1506607 | ASF1B | NM_018154 | ASF1 antisilencing function 1 homolog B (S cerevisiae) | −4.36 |
| 1509496 | AES | NM_198969 | Amino-terminal enhancer of split | −4.39 |
| 1533535 | TAF4 | NM_003185 | TAF4 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 135kDa | −4.40 |
| 1508056 | EGR3 | NM_004430 | Early growth response 3 | −4.41 |
| 1533262 | BRF1 | BC016743 | BRF1 homolog, subunit of RNA polymerase III transcription initiation factor IIIB (S cerevisiae) | −4.48 |
| 1533571 | PHF6 | NM_032458 | PHD finger protein 6 | −4.53 |
| 1517506 | EGR2 | NM_000399 | Early growth response 2 (Krox−20 homolog, Drosophila) | −4.66 |
| 1989375 | SMAD3 | NM_005902 | SMAD, mothers against DPP homolog 3 (Drosophila) | −4.97 |
| 1507856 | MYB | NM_005375 | V-myb myeloblastosis viral oncogene homolog (avian) | −5.06 |
| 1528623 | OLIG2 | NM_005806 | Oligodendrocyte lineage transcription factor 2 | −6.11 |
| 1524585 | GATA1 | NM_002049 | GATA binding protein 1 (globin transcription factor 1) | −6.64 |
| 1991493 | PARP14 | NM_017554 | Poly (ADP-ribose) polymerase family, member 14 | −7.03 |
| 1510055 | PCGF5 | BC007377 | Polycomb group ring finger 5 | −7.57 |
| 1510369 | ASCL2 | NM_005170 | Achaete-scute complex-like 2 (Drosophila) | −7.72 |
| 1522208 | ZNF584 | NM_173548 | Zinc finger protein 584 | −7.93 |
| 1523937 | RPS6KA5 | NM_004755 | Ribosomal protein S6 kinase, 90 kDa, polypeptide 5 | −8.07 |
| 1529851 | ETV7 | NM_016135 | Ets variant gene 7 (TEL2 oncogene) | −8.56 |
| 1506298 | GATA6 | NM_005257 | GATA binding protein 6 | −8.65 |
| 1989096 | SSBP3 | NM_001009955 | Single stranded DNA binding protein 3 | −8.76 |
| 1520503 | NKX31 | NM_006167 | NK3 transcription factor related, locus 1 (Drosophila) | −9.56 |
| 1522991 | ZNF649 | NM_023074 | Zinc finger protein 649 | −11.07 |
| 1514700 | NR4A1 | NM_002135 | Nuclear receptor subfamily 4, group A, member 1 | −92.91 |
| Signal transduction | ||||
| 1513543 | GPR84 | NM_020370 | G protein-coupled receptor 84 | 25.51 |
| 1505483 | EDG1 | NM_001400 | Endothelial differentiation, sphingolipid G-protein-coupled receptor, 1 | 18.05 |
| 1511321 | PTGER2 | NM_000956 | Prostaglandin E receptor 2 (subtype EP2), 53 kDa | 8.26 |
| 1511570 | ARHGAP24 | AK130576 | Rho GTPase activating protein 24 | 7.28 |
| 1523886 | PDE4D | NM_006203 | Phosphodiesterase 4D, cAMP-specific (phosphodiesterase E3 dunce homolog, Drosophila) | 6.64 |
| 1527053 | MYO10 | BC041694 | Myosin X | 6.48 |
| 1512860 | MS4A4A | NM_024021 | Membrane-spanning 4-domains, subfamily A, member 4 | 6.12 |
| 1539087 | DLC1 | NM_182643 | Deleted in liver cancer 1 | 5.96 |
| 1528643 | MS4A6A | NM_022349 | Membrane-spanning 4-domains, subfamily A, member 6A | 5.87 |
| 1524040 | GPR141 | NM_181791 | G protein-coupled receptor 141 | 5.28 |
| 1989101 | EDARADD | NM_080738 | EDAR-associated death domain | 4.64 |
| 1508615 | EXT1 | NM_000127 | Exostoses (multiple) 1 | 4.30 |
| 1521881 | STK3 | NM_006281 | Serine/threonine kinase 3 (STE20 homolog, yeast) | 3.79 |
| 1510158 | ITPKC | NM_025194 | Inositol 1.4,5-trisphosphate 3-kinase C | 3.63 |
| 1519571 | CDC42EP3 | NM_006449 | CDC42 effector protein (Rho GTPase binding) 3 | 3.58 |
| 1534557 | DPYSL3 | NM_001387 | Dihydropyrimidinase-like 3 | 3.54 |
| 1536781 | RPS6KA3 | NM_004586 | Ribosomal protein S6 kinase, 90 kDa, polypeptide 3 | 3.53 |
| 1533999 | HHIP | NM_022475 | Hedgehog interacting protein | 3.50 |
| 1512578 | GPR97 | NM_170776 | G protein-coupled receptor 97 | 3.42 |
| 1514738 | SLC26A6 | NM_022911 | Inositol hexaphosphate kinase 2 | 3.28 |
| 1517434 | IL4R | NM_000418 | Interleukin 4 receptor | 3.21 |
| 1539209 | GABBR1 | NM_001470 | Gamma-aminobutyric acid (GABA) B receptor, 1 | 3.19 |
| 1511435 | STK16 | NM_003691 | Serine/threonine kinase 16 | 3.17 |
| 1520958 | PRKAR2A | BC002763 | Inositol hexaphosphate kinase 2 | 3.12 |
| 1506387 | MAPK6 | NM_002748 | Mitogen-activated protein kinase 6 | 3.03 |
| 1523162 | ITPK1 | NM_014216 | Inositol 1.3,4-triphosphate 5/6 kinase | −3.05 |
| 1507956 | GNGT2 | NM_031498 | Guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2 | −3.09 |
| 1514326 | TAGAP | NM_138810 | T-cell activation GTPase activating protein | −3.09 |
| 1530414 | GPR171 | NM_013308 | G protein-coupled receptor 171 | −3.16 |
| 1527339 | P2RY5 | NM_005767 | Purinergic receptor P2Y, G-protein coupled, 5 | −3.17 |
| 1517355 | ARHGAP25 | BC039591 | Rho GTPase activating protein 25 | −3.23 |
| 1520830 | KIT | NM_000222 | V-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog | −3.28 |
| 1517660 | KLRK1 | NM_007360 | Killer cell lectin-like receptor subfamily C, member 4 | −3.37 |
| 1514077 | KLRB1 | NM_002258 | Killer cell lectin-like receptor subfamily B, member 1 | −3.43 |
| 1518328 | GRP | NM_002091 | Gastrin-releasing peptide | −3.64 |
| 1516849 | EPHB1 | NM_004441 | EPH receptor B1 | −3.80 |
| 1514753 | PRKAG2 | NM_016203 | Protein kinase, AMP-activated, gamma 2 non-catalytic subunit | −4.09 |
| 1512127 | SOCS1 | NM_003745 | Suppressor of cytokine signaling 1 | −4.13 |
| 1518864 | FLT3LG | NM_001459 | Fms-related tyrosine kinase 3 ligand | −4.26 |
| 1524915 | OR51E1 | NM_152430 | Olfactory receptor, family 51, subfamily E, member 1 | −4.27 |
| 1507227 | P2RY14 | NM_014879 | Purinergic receptor P2Y, G-protein coupled, 14 | −4.28 |
| 1523084 | GUCA1A | NM_000409 | Guanylate cyclase activator 1A (retina) | −4.37 |
| 1525046 | PILRA | NM_178273 | Paired immunoglobulin-like type 2 receptor alpha | −4.48 |
| 1534223 | PPP1R12B | NM_032105 | Protein phosphatase 1, regulatory (inhibitor) subunit 12B | −4.66 |
| 1518048 | GPR34 | NM_005300 | G-protein–coupled receptor 34 | −4.78 |
| 1534320 | ARRB1 | NM_004041 | Arrestin, beta 1 | −5.02 |
| 1534974 | KLRD1 | NM_002262 | Killer cell lectin-like receptor subfamily D, member 1 | −5.16 |
| 1525314 | S100A10 | NM_002966 | S100 calcium binding protein A10 | −5.94 |
| 1530940 | PRKAR2B | NM_002736 | Protein kinase, cAMP-dependent, regulatory, type II, beta | −5.97 |
| 1522410 | IL2RB | NM_000878 | Interleukin 2 receptor, beta | −6.06 |
| 1518652 | CD74 | NM_004355 | CD74 molecule, major histocompatibility complex, class II invariant chain | −6.23 |
| 1515002 | CYSLTR1 | NM_006639 | Cysteinyl leukotriene receptor 1 | −6.45 |
| 1530332 | MX1 | NM_002462 | Myxovirus (influenza virus) resistance 1, interferon-inducible protein p78 (mouse) | −6.76 |
| 1522509 | OR2B2 | NM_033057 | Olfactory receptor, family 2, subfamily B, member 2 | −6.88 |
| 1984459 | PDE4B | AB209081 | Phosphodiesterase 4B, cAMP-specific | −6.89 |
| 1527378 | CSF1R | NM_005211 | Colony stimulating factor 1 receptor, formerly McDonough feline sarcoma viral (v-fms) oncogene homolog | −7.08 |
| 1527858 | NRGN | NM_006176 | Neurogranin (protein kinase C substrate, RC3) | −7.49 |
| 1524392 | ANGPT1 | NM_001146 | Angiopoietin 1 | −8.84 |
| 1516837 | MS4A3 | NM_006138 | Membrane-spanning 4-domains, subfamily A, member 3 | −9.24 |
| 1989999 | IL5RA | NM_175725 | Interleukin 5 receptor, alpha | −9.66 |
| 1524637 | GPR162 | NM_019858 | Triosephosphate isomerase 1 | −10.10 |
| 1519217 | P2RY2 | NM_176072 | Purinergic receptor P2Y, G-protein coupled, 2 | −11.66 |
| 1516830 | P2RY10 | NM_014499 | Purinergic receptor P2Y, G-protein coupled, 10 | −11.67 |
| 1518123 | CD83 | NM_004233 | CD83 molecule | −11.67 |
| 1517216 | KLRF1 | NM_016523 | Killer cell lectin-like receptor subfamily F, member 1 | −12.68 |
| 1512228 | EBI2 | NM_004951 | Epstein-Barr virus–induced gene 2 (lymphocyte-specific G protein-coupled receptor) | −13.11 |
| 1513158 | HRH4 | NM_021624 | Histamine receptor H4 | −13.66 |
| 1537338 | FGL2 | NM_006682 | Fibrinogen-like 2 | −13.76 |
| 1515714 | GNG11 | NM_004126 | Guanine nucleotide binding protein (G protein), gamma 11 | −16.57 |
| 1527111 | GPR44 | NM_004778 | G protein-coupled receptor 44 | −23.51 |
| 1528909 | RGS1 | NM_002922 | Regulator of G-protein signaling 1 | −32.65 |
| Immune responses | ||||
| 1507161 | CD163 | NM_004244 | CD163 molecule | 54.27 |
| 1531927 | CYP1B1 | NM_000104 | Cytochrome P450, family 1, subfamily B, polypeptide 1 | 46.19 |
| 1984958 | CD177 | CR592446 | CD177 molecule | 24.38 |
| 1521404 | IL18R1 | NM_003855 | Interleukin 18 receptor 1 | 16.54 |
| 1983056 | MERTK | U08023 | C-mer proto-oncogene tyrosine kinase | 16.00 |
| 1532739 | CLEC5A | NM_013252 | C-type lectin domain family 5, member A | 13.44 |
| 1511835 | CLEC4D | NM_080387 | C-type lectin domain family 4, member D | 12.02 |
| 1535646 | CST7 | NM_003650 | Cystatin F (leukocystatin) | 11.43 |
| 1506996 | ADORA3 | NM_020683 | Adenosine A3 receptor | 10.89 |
| 1530098 | IL18RAP | NM_003853 | Interleukin 18 receptor accessory protein | 10.79 |
| 1532148 | LAIR1 | NM_021706 | Leukocyte-associated immunoglobulin-like receptor 1 | 10.45 |
| 1506562 | HP | NM_005143 | Haptoglobin | 10.31 |
| 1505718 | IL1R2 | NM_004633 | Interleukin 1 receptor, type II | 9.95 |
| 1507483 | LILRA5 | NM_181879 | Leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 5 | 7.56 |
| 1524454 | XBP1 | NM_005080 | X-box binding protein 1 | 7.48 |
| 1510813 | IRAK3 | NM_007199 | Interleukin−1 receptor-associated kinase 3 | 7.28 |
| 1538778 | TLR5 | NM_003268 | Toll-like receptor 5 | 5.97 |
| 1521047 | CR1 | NM_000651 | Complement component (3b/4b) receptor 1 (Knops blood group) | 5.87 |
| 1520792 | MAPK14 | NM_139013 | Mitogen-activated protein kinase 14 | 5.75 |
| 1535716 | CD59 | NM_203330 | CD59 molecule, complement regulatory protein | 5.67 |
| 1513648 | FCER1G | CB529629 | Fc fragment of IgE, high affinity I, receptor for; gamma polypeptide | 5.26 |
| 1521687 | TLR4 | BF670973 | Toll-like receptor 4 | 5.15 |
| 1530114 | MAP2K6 | NM_002758 | Mitogen-activated protein kinase kinase 6 | 4.87 |
| 1533789 | SERPINB1 | NM_030666 | Serpin peptidase inhibitor, clade B (ovalbumin), member 1 | 4.84 |
| 1522193 | FCAR | NM_002000 | Fc fragment of IgA, receptor for | 4.52 |
| 1527121 | PROK2 | NM_021935 | Prokineticin 2 | 4.33 |
| 1505831 | CIAS1 | NM_004895 | Cold autoinflammatory syndrome 1 | 4.09 |
| 1524842 | IL10 | NM_000572 | Interleukin 10 | 3.89 |
| 1538909 | IL1RAP | NM_002182 | Interleukin 1 receptor accessory protein | 3.80 |
| 1506853 | CHIT1 | NM_003465 | Chitinase 1 (chitotriosidase) | 3.48 |
| 1531091 | CD55 | NM_000574 | CD55 molecule, decay accelerating factor for complement (Cromer blood group) | 3.39 |
| 1534101 | IL1R1 | NM_000877 | Interleukin 1 receptor, type I | 3.34 |
| 1515424 | CYBB | S67289 | Cytochrome b-245, beta polypeptide (chronic granulomatous disease) | 3.29 |
| 1985998 | ALOX5 | BQ548663 | Arachidonate 5-lipoxygenase | 3.16 |
| 1510372 | IRAK4 | NM_016123 | Interleukin-1 receptor-associated kinase 4 | 3.10 |
| 1514172 | CD48 | NM_001778 | CD48 molecule | 3.09 |
| 1527340 | TLR8 | NM_016610 | Toll-like receptor 8 | 3.07 |
| 1511452 | SERPING1 | NM_000062 | Serpin peptidase inhibitor, clade G (C1 inhibitor), member 1, (angioedema, hereditary) | −3.00 |
| 1530131 | IRF2BP2 | BC020516 | Interferon regulatory factor 2 binding protein 2 | −3.03 |
| 1512742 | CLEC7A | NM_197954 | C-type lectin domain family 7, member A | −3.06 |
| 1528251 | NCR3 | NM_147130 | Natural cytotoxicity triggering receptor 3 | −3.13 |
| 1513772 | HLA-DRB3 | NM_022555 | Major histocompatibility complex, class II, DR beta 1 | −3.17 |
| 1518778 | ALOX15 | NM_001140 | Arachidonate 15-lipoxygenase | −3.24 |
| 1524640 | OAS3 | NM_006187 | 2'−5'-oligoadenylate synthetase 3, 100 kDa | −3.31 |
| 1985491 | HLA-DRB1 | X12544 | Major histocompatibility complex, class II, DR beta 1 | −3.31 |
| 1519302 | STAT1 | NM_139266 | Signal transducer and activator of transcription 1, 91 kDa | −3.39 |
| 1509654 | CTSW | NM_001335 | Cathepsin W (lymphopain) | −3.45 |
| 1992017 | PLAUR | NM_001005377 | Plasminogen activator, urokinase receptor | −3.50 |
| 1526588 | LY75 | NM_002349 | Lymphocyte antigen 75 | −3.51 |
| 1536508 | LST1 | NM_007161 | Leukocyte specific transcript 1 | −3.64 |
| 1515890 | FAIM3 | NM_005449 | Interleukin 24 | −3.65 |
| 1511424 | EMR3 | NM_032571 | Egf-like module containing, mucin-like, hormone receptor-like 3 | −3.71 |
| 1534340 | CD46 | NM_002389 | CD46 molecule, complement regulatory protein | −3.83 |
| 1537056 | GBP3 | NM_018284 | Guanylate binding protein 1, interferon-inducible, 67 kDa | −3.86 |
| 1517555 | CYP27A1 | NM_000784 | Cytochrome P450, family 27, subfamily A, polypeptide 1 | −3.88 |
| 1516190 | SAMHD1 | NM_015474 | SAM domain and HD domain 1 | −3.89 |
| 1519488 | HLA-DQB2 | NM_182549 | Major histocompatibility complex, class II, DQ beta 2 | −3.94 |
| 1983904 | IL32 | NM_001012631 | Interleukin 32 | −3.99 |
| 1505541 | GZMB | NM_004131 | Granzyme B | −4.04 |
| 1536691 | GZMK | NM_002104 | Granzyme K (granzyme 3; tryptase II) | −4.67 |
| 1522713 | GZMA | NM_006144 | Granzyme A | −4.80 |
| 1510975 | HLA-DRB5 | NM_002125 | Major histocompatibility complex, class II, DR beta 1 | −5.13 |
| 1527466 | HLA-DPA1 | NM_033554 | Major histocompatibility complex, class II, DP alpha 1 | −5.16 |
| 1516918 | B2M | AK022379 | Beta−2-microglobulin | −5.43 |
| 1538818 | HLA-DQB1 | NM_002123 | Major histocompatibility complex, class II, DQ beta 1 | −5.87 |
| 1521122 | SPON2 | NM_012445 | Spondin 2, extracellular matrix protein | −5.95 |
| 1517271 | PF4V1 | NM_002620 | Platelet factor 4 variant 1 | −6.48 |
| 1508682 | HLA-DPB1 | NM_002121 | Major histocompatibility complex, class II, DP beta 1 | −6.67 |
| 1529151 | OASL | NM_003733 | 2′−5′-oligoadenylate synthetase-like | −7.10 |
| 1987518 | CYB561 | NM_001017916 | Cytochrome b-561 | −7.39 |
| 1524314 | IFI44 | NM_006417 | Interferon-induced protein 44 | −7.91 |
| 1533656 | DEFA4 | NM_001925 | Defensin, alpha 4, corticostatin | −9.04 |
| 1526794 | HLA-DRA | NM_019111 | Major histocompatibility complex, class II, DR alpha | −10.12 |
| 1536919 | HLA-DMB | NM_002118 | Major histocompatibility complex, class II, DM beta | −10.73 |
| 1525510 | PF4 | NM_002619 | Platelet factor 4 (chemokine (C-X-C motif) ligand 4) | −11.03 |
| 1514544 | DEFA3 | NM_005217 | Defensin, alpha 1 | −11.85 |
| 1520461 | CD69 | NM_001781 | CD69 molecule | −28.65 |
| 1528045 | IL8 | NM_000584 | Interleukin 8 | −35.22 |
| 1533457 | FCER1A | NM_002001 | Fc fragment of IgE, high affinity I, receptor for; alpha polypeptide | −61.82 |
| Cell survival and apoptosis | ||||
| 1519170 | GADD45A | NM_001924 | Growth arrest and DNA-damage-inducible, alpha | 20.49 |
| 1531073 | CARD12 | NM_021209 | Caspase recruitment domain family, member 12 | 11.37 |
| 1513896 | BIRC1 | NM_004536 | Baculoviral IAP repeat-containing 1 | 10.55 |
| 1525148 | ANXA1 | NM_000700 | Annexin A1 | 7.39 |
| 1533289 | ANXA3 | NM_005139 | Annexin A3 | 7.33 |
| 1516682 | CAST | NM_001750 | Calpastatin | 6.97 |
| 1532823 | PECR | NM_018441 | Peroxisomal trans−2-enoyl-CoA reductase | 5.32 |
| 1539226 | CARD6 | NM_032587 | Caspase recruitment domain family, member 6 | 5.15 |
| 1531614 | ELMO2 | NM_022086 | Engulfment and cell motility 2 | 4.67 |
| 1512780 | LGALS8 | NM_006499 | Lectin, galactoside-binding, soluble, 8 (galectin 8) | 4.24 |
| 1505831 | CIAS1 | NM_004895 | Cold autoinflammatory syndrome 1 | 4.09 |
| 1521881 | STK3 | NM_006281 | Serine/threonine kinase 3 (STE20 homolog, yeast) | 3.79 |
| 1526256 | DDAH2 | NM_013974 | Dimethylarginine dimethylaminohydrolase 2 | 3.75 |
| 1522394 | TNFRSF10A | NM_003844 | Tumor necrosis factor receptor superfamily, member 10a | 3.69 |
| 1988846 | AIM1 | NM_001624 | Absent in melanoma 1 | 3.52 |
| 1509305 | LTB4R | NM_181657 | Leukotriene B4 receptor | 3.20 |
| 1984749 | HIP1 | BC082970 | Huntingtin interacting protein 1 | 3.19 |
| 1534293 | LGALS3 | NM_002306 | Lectin, galactoside-binding, soluble, 3 (galectin 3) | 3.03 |
| 1535837 | SQSTM1 | NM_003900 | Sequestosome 1 | −3.01 |
| 1537407 | NFKBIE | NM_004556 | Nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, epsilon | −3.12 |
| 1987065 | TNFRSF25 | NM_148968 | Homo sapiens tumor necrosis factor receptor superfamily | −3.36 |
| 1516125 | PRDX2 | NM_005809 | Peroxiredoxin 2 | −3.40 |
| 1531960 | GSTP1 | NM_000852 | Glutathione S-transferase pi | −3.42 |
| 1984701 | CARD9 | NM_052813 | Caspase recruitment domain family, member 9 | −3.61 |
| 1532501 | PIM2 | NM_006875 | Pim−2 oncogene | −3.76 |
| 1528041 | IL31RA | NM_139017 | Interleukin 31 receptor A | −3.91 |
| 1533374 | PHLDA2 | NM_003311 | Pleckstrin homology-like domain, family A, member 2 | −3.97 |
| 1526625 | CARD4 | NM_006092 | Caspase recruitment domain family, member 4 | −3.99 |
| 1520783 | DYRK2 | NM_006482 | Dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2 | −4.29 |
| 1516378 | TNFSF12 | NM_153012 | Tumor necrosis factor (ligand) superfamily, member 12 | −4.29 |
| 1530737 | LGALS2 | NM_006498 | Lectin, galactoside-binding, soluble, 2 (galectin 2) | −4.47 |
| 1510435 | LGALS12 | NM_033101 | Lectin, galactoside-binding, soluble, 12 (galectin 12) | −4.47 |
| 1523074 | HSPA9B | NM_004134 | Heat shock 70-kDa protein 9B (mortalin-2) | −5.16 |
| 1532326 | DDX58 | NM_014314 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 | −5.17 |
| 1508150 | AOC3 | NM_003734 | Amine oxidase, copper containing 3 (vascular adhesion protein 1) | −5.52 |
| 1538822 | IFI6 | NM_022873 | Interferon, alpha-inducible protein (clone IFI-6–16) | −5.56 |
| 1990540 | CLU | NM_203339 | Clusterin | −5.66 |
| 1517391 | BIRC3 | NM_001165 | Baculoviral IAP repeat-containing 3 | −6.55 |
| 1538745 | SGK | NM_005627 | Serum/glucocorticoid regulated kinase | −7.90 |
| 1523311 | BIRC4BP | NM_017523 | XIAP associated factor-1 | −9.09 |
| 1510278 | NGFRAP1 | NM_014380 | Nerve growth factor receptor (TNFRSF16) associated protein 1 | −9.35 |
| 1533140 | NME5 | NM_003551 | Non-metastatic cells 5, protein expressed in (nucleoside-diphosphate kinase) | −9.98 |
| 1533249 | HIG2 | NM_013332 | hypoxia-inducible protein 2 | −15.17 |
| Cell adhesion and motility | ||||
| 1536096 | MMP8 | NM_002424 | Matrix metallopeptidase 8 (neutrophil collagenase) | 97.77 |
| 1519153 | OLFM4 | NM_006418 | Olfactomedin 4 | 23.81 |
| 1514453 | CD44 | NM_000610 | CD44 molecule (Indian blood group) | 10.05 |
| 1532879 | EMILIN2 | NM_032048 | Elastin microfibril interfacer 2 | 9.85 |
| 1521082 | CEACAM1 | NM_001712 | Carcinoembryonic antigen-related cell adhesion molecule 1 (biliary glycoprotein) | 5.97 |
| 1509526 | MMP9 | NM_004994 | Matrix metallopeptidase 9 | 5.64 |
| 1522658 | VNN1 | NM_004666 | Vanin 1 | 5.35 |
| 1984856 | ADAM15 | NM_207191 | ADAM metallopeptidase domain 15 (metargidin) | 3.79 |
| 1507356 | F5 | NM_000130 | Coagulation factor V (proaccelerin, labile factor) | 3.78 |
| 1516175 | JAK2 | NM_004972 | Janus kinase 2 (a protein tyrosine kinase) | 3.78 |
| 1508488 | SPP1 | NM_000582 | Secreted phosphoprotein 1 (osteopontin, bone sialoprotein I, early T-lymphocyte activation 1) | 3.72 |
| 1523771 | ITGAM | NM_000632 | Integrin, alpha M (complement component 3 receptor 3 subunit) | 3.52 |
| 1508621 | PVRL2 | NM_002856 | Poliovirus receptor-related 2 (herpesvirus entry mediator B) | 3.42 |
| 1531024 | SIGLEC9 | NM_014441 | Sialic acid binding Ig-like lectin 9 | 3.34 |
| 1505504 | VIM | NM_003380 | Vimentin | 3.34 |
| 1986801 | LAMB3 | NM_001017402 | Laminin, beta 3 | 3.11 |
| 1518582 | CEACAM6 | BC005008 | Carcinoembryonic antigen-related cell adhesion molecule 6 (non-specific cross reacting antigen) | −3.07 |
| 1506966 | SIGLEC8 | NM_014442 | Sialic acid binding immunoglobulin-like lectin 8 | −3.16 |
| 1534776 | CXCL1 | NM_00–1511 | Chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha) | −3.20 |
| 1520341 | AZU1 | NM_001700 | Azurocidin 1 (cationic antimicrobial protein 37) | −3.23 |
| 1520433 | ITGB7 | NM_000889 | Integrin, beta 7 | −3.59 |
| 1505769 | TUBB1 | NM_030773 | Tubulin, beta 1 | −4.35 |
| 1528734 | ICAM2 | NM_000873 | Intercellular adhesion molecule 2 | −5.24 |
| 1991393 | CXCR4 | NM_001008540 | Chemokine (C-X-C motif) receptor 4 | −5.90 |
| 1989016 | CCL3L3 | NM_001001437 | Chemokine (C-C motif) ligand 3-like 3 | −5.98 |
| 1510688 | CCL3 | NM_002983 | Chemokine (C-C motif) ligand 3 | −6.35 |
| 1517696 | CCL5 | NM_002985 | Chemokine (C-C motif) ligand 5 | −7.71 |
| 1533135 | CXCL10 | NM_001565 | Chemokine (C-X-C motif) ligand 11 | −9.10 |
| 1505505 | CCL23 | NM_005064 | Chemokine (C-C motif) ligand 23 | −15.06 |
| 1526404 | CCL4 | NM_002984 | Chemokine (C-C motif) ligand 4 | −15.45 |
| 1533611 | PPBP | NM_002704 | Pro-platelet basic protein (chemokine (C-X-C motif) ligand 7) | −16.30 |
| 1515169 | ITGA4 | NM_000885 | Integrin, alpha 4 (antigen CD49D), alpha 4 subunit of VLA-4 receptor | −16.74 |
| 1537942 | CCR3 | NM_001837 | Chemokine (C-C motif) receptor 3 | −19.63 |
| Cell metabolism | ||||
| 1538977 | HPGD | NM_000860 | Hydroxyprostaglandin dehydrogenase 15-(NAD) | 36.27 |
| 1989284 | PFKFB2 | NM_001018053 | 6-phosphofructo-2-kinase/fructose-2.6-biphosphatase 2 | 31.40 |
| 1986330 | ATP9A | NM_006045 | ATPase, Class II, type 9A | 25.82 |
| 1511217 | LDHA | NM_005566 | Lactate dehydrogenase A | 10.11 |
| 1527335 | PCSK9 | NM_174936 | Proprotein convertase subtilisin/kexin type 9 | 7.95 |
| 1991955 | PLB1 | CR600701 | Phospholipase B1 | 7.59 |
| 1983476 | ATP13A3 | AJ306929 | ATPase type 13A3 | 7.56 |
| 1513367 | DPH5 | NM_0–15958 | DPH5 homolog (S cerevisiae) | 6.38 |
| 1526202 | NMNAT2 | NM_0–15039 | Nicotinamide nucleotide adenylyltransferase 2 | 6.31 |
| 1536086 | DPYD | NM_000110 | Dihydropyrimidine dehydrogenase | 6.11 |
| 1990338 | ATP11B | NM_014616 | ATPase, Class VI, type 11B | 6.09 |
| 1515968 | ALPL | NM_000478 | Alkaline phosphatase, liver/bone/kidney | 5.59 |
| 1506037 | MGAM | NM_004668 | Maltase-glucoamylase (alpha-glucosidase) | 5.41 |
| 1532823 | PECR | NM_018441 | Peroxisomal trans-2-enoyl-CoA reductase | 5.32 |
| 1986414 | ATP2B4 | NM_001001396 | ATPase, Ca2+ transporting, plasma membrane 4 | 4.99 |
| 1506589 | PFKFB3 | NM_004566 | 6-phosphofructo-2-kinase/fructose-2.6-biphosphatase 3 | 4.75 |
| 1512399 | AGPAT6 | NM_178819 | 1-acylglycerol-3-phosphate O-acyltransferase 6 (lysophosphatidic acid acyltransferase, zeta) | 4.74 |
| 1523383 | PGD | NM_002631 | Phosphogluconate dehydrogenase | 4.70 |
| 1519383 | ATP6V1C1 | NM_001695 | ATPase, H+ transporting, lysosomal 42 kDa, V1 subunit C1 | 4.11 |
| 1520655 | SMPDL3A | NM_006714 | Sphingomyelin phosphodiesterase, acid-like 3A | 4.09 |
| 1989848 | ACSL4 | NM_004458 | Acyl-CoA synthetase long-chain family member 4 | 4.02 |
| 1522656 | LDLR | NM_000527 | Low-density lipoprotein receptor | 3.63 |
| 1538541 | DHRS9 | NM_005771 | Dehydrogenase/reductase (SDR family) member 9 | 3.55 |
| 1514810 | ATP1A3 | NM_152296 | ATPase, Na+/K+ transporting, alpha 3 polypeptide | 3.53 |
| 1524666 | FABP5 | NM_001444 | Fatty acid binding protein 5 (psoriasis-associated) | 3.38 |
| 1538372 | ATP1A1 | NM_000701 | ATPase, Na+/K+ transporting, alpha 1 polypeptide | 3.38 |
| 1524650 | GNS | NM_002076 | Glucosamine (N-acetyl)-6- | 3.37 |
| 1523432 | ACSL1 | NM_001995 | Acyl-CoA synthetase long-chain family member 1 | 3.35 |
| 1529572 | SGSH | NM_000199 | N-sulfoglucosamine sulfohydrolase (sulfamidase) | 3.29 |
| 1538734 | ACSL3 | NM_004457 | Acyl-CoA synthetase long-chain family member 3 | 3.27 |
| 1985998 | ALOX5 | BQ548663 | Arachidonate 5-lipoxygenase | 3.16 |
| 1527407 | GAPDH | NM_002046 | Glyceraldehyde3-phosphate dehydrogenase | 3.16 |
| 1534240 | SULT1B1 | D89479 | Sulfotransferase family, cytosolic, 1B, member 1 | 3.10 |
| 1983183 | NDUFB6 | NM_182739 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 6 | −3.10 |
| 1525237 | ALDH6A1 | NM_005589 | Aldehyde dehydrogenase 6 family, member A1 | −3.22 |
| 1518778 | ALOX15 | NM_001140 | Arachidonate 15-lipoxygenase | −3.24 |
| 1523761 | CBR1 | NM_001757 | Carbonyl reductase 1 | −3.61 |
| 1510672 | MCCC1 | NM_020166 | Methylcrotonoyl-Coenzyme A carboxylase 1 (alpha) | −4.01 |
| 1990453 | NDUFV2 | BI520375 | NADH dehydrogenase (ubiquinone) flavoprotein 2, 24 kDa | −4.31 |
| 1521732 | ATP12A | NM_001676 | ATPase, H+/K+ transporting, nongastric, alpha polypeptide | −4.74 |
| 1526184 | LDHB | NM_002300 | Lactate dehydrogenase B | −6.08 |
| 1512448 | ISOC1 | NM_016048 | Isochorismatase domain containing 1 | −6.51 |
| 1989369 | METTL7A | BC004492 | Methyltransferase like 7A | −6.99 |
| 1518627 | FA2H | NM_024306 | Fatty acid 2-hydroxylase | −7.75 |
| 1506804 | PLD4 | NM_138790 | Phospholipase D family, member 4 | −9.53 |
| 1521949 | SORD | NM_003104 | Sorbitol dehydrogenase | −10.20 |
| 1512822 | SULF2 | NM_018837 | Sulfatase 2 | −21.22 |
Genes that were found to be differentially expressed after in vivo treatment with G-CSF-dexamethasone were analyzed with use of the OntoExpress program (http://vortex.cs.wayne.edu/projects.htm#Onto-Express), and categorized into functional profiles. The majority of analyzed genes are categorized into 6 main functional categories: transcription, signal transduction, immune response, cell faith & apoptosis, cell adhesion and mobility, and metabolism. Results are presented as the mean fold-increase or -decrease of 3 separate donors (comparison is with freshly isolated neutrophils t = 0).
Twenty-six genes involved in the regulation of transcription or the process of transcription itself (eg, CCNA1, SMARCA3, and MEF2A) were up-regulated after administration of G-CSF and dexamethasone. However, the expression of more than 40 of such genes was reduced, with NR4A1, NR4A3, and ZNF649 being most strongly affected. This finding suggests that granulocytes obtained from G-CSF/dexamethasone-treated donors express a transcriptional program that diverges from normal, circulating neutrophils. These data are indicative of coordinated changes in transcription and suggest the existence of both positive and negative regulatory mechanisms in the G-CSF/dexamethasone–induced transcriptional response.
Granulocyte mobilization has an effect on genes involved in inflammation
The expression of dozens of genes encoding molecules involved in the process of inflammation and the immune response was shown to be modulated by the in vivo administration of G-CSF/dexamethasone (Table 2). For example, the transcription of the gene CYBB was strongly induced. This gene encodes gp91phox, the enzymatic subunit of the NADPH oxidase enzyme complex that is responsible for superoxide production and contributes to microbial killing by neutrophils (Table 2).
The expression of several receptors responsible for ligand or pathogen sensing was induced as well, as indicated, for example, by the increase in transcripts for several C-type lectins such as CLEC5A, also known as myeloid DAP12-associated protein (MDL-1) and involved in the activation of cells,29 or CLEC4D (CLECSF8), which is also expressed on myeloid cells and has been suggested to take part in the process of endocytosis.30 Expression of another set of immune receptors that is associated with the activation of cells, eg CD48, LAIR1 (leukocyte-associated Ig-like receptor 1) and LILRA5 (LIR9, a member of the immunoglobulin-like receptor family), also was induced after mobilization.
However, we observed a significant reduction in the expression of genes traditionally associated with the adaptive immune response (eg, HLA-DQB2, HLA-DRB5, HLA-DPA1, HLA-DRA, HLA-DMB, and granzymes; Table 2).
In addition, the transcriptome of mobilized cells was characterized by the modulation of a cluster of genes coding for cytokines/chemokines and/or their receptors. Within the chemokine system, we identified CCL3, CCL4, CCL5, CXCL1, CXCL10, as well as the stroma-derived factor-1 receptor CXCR4, and CCR3, as being repressed by G-CSF and dexamethasone treatment at the transcriptional level. Moreover, the transcripts for Annexin-1 and -3 were highly induced by the G-CSF/dexamethasone treatment. Unexpectedly, we also found a strong reduction in IL8 transcripts after mobilization, whereas to our knowledge, no defects in the release of IL-8 from mobilized granulocytes have been reported to date.
Modulation of genes involved in the apoptotic machinery
Because mobilized granulocytes show delayed apoptosis upon in vitro culture,9,10 we carefully examined genes involved in the regulation of cell fate or apoptosis (Table 2). One group of molecules that is known to be regulated at the transcriptional level and is involved in the apoptotic process is the group of galectins. Galectins form a family of lectins that are defined by their ability to recognize β-galactose and the presence of consensus amino-acid sequences.31 Different members of this family have been shown to modulate various steps of the inflammatory response, such as cell-matrix interactions, cell trafficking, cell survival, cell-growth regulation, chemotaxis, and proinflammatory cytokine secretion.32,33 We observed a strong induction of galectin-8 (LGAL8) and moderate induction of galectin-3 (LGALS3), as well as a reduction of galectin-2 and -12 (LGALS2, LGALS12).
Most striking was the strong induction observed for the transcript of GADD45A, a member of the Gadd45 family of genes that have been implicated in stress signaling in response to physiologic or environmental stressors, resulting in either cell-cycle arrest, DNA repair, cell survival, and senescence or apoptosis. Hematopoietic cells from Gadd45a-deficient mice are more susceptible to apoptosis induced by ultraviolet (UV) radiation or genotoxic stress.34,35
Expression of the CAST gene encoding calpastatin was also strongly induced in mobilized cells. Whereas calpain activity has previously been shown to be involved in neutrophil cell death,17,20 calpastatin is a well-documented and specific endogenous inhibitor of calpains.36 It is encoded by a single gene that produces several isoforms via differential splicing or use of promoters. Calpastatins are known to be associated with an antiapoptotic effect in neutrophils,20 and we have recently observed that inhibition of calpain activity by G-CSF strongly affects neutrophil survival.17 Because mobilized cells have been reported to have a better survival capacity, calpastatin was investigated in greater detail.
Induction of calpastatin correlates with a delay in apoptosis
G-CSF causes rapid neutrophilia but is also known to act as a survival factor on mature, circulating neutrophils both in vitro16,17,37 and in vivo.9 Dexamethasone, although shown to induce apoptosis in various cell types, including lymphocytes, monocytes,38 and eosinophils,15,39 also acts as a prosurvival factor for neutrophils.13,40 This finding is in line with our observation that, upon in vitro culture, granulocytes obtained from donors treated with a combination of the 2 drugs showed a prolonged life span after overnight culture compared with neutrophils isolated from the blood of the same donors before treatment (Figure 3). The same extent of survival was achieved by the in vitro addition of G-CSF and dexamethasone to the culture medium during overnight culture (data not shown).
Prolonged survival of granulocytes isolated from donors mobilized with G-CSF/dexamethasone. Granulocytes were isolated from healthy, control donors (□) and from the same donors treated with G-CSF and dexamethasone (■), and cultured for 20 hours in HBSS. Afterward, the cell viability was assessed by the use of annexin V/PI staining. Results represents data from 6 independent experiments (mean ± SD). *P < .05 (significant difference).
Prolonged survival of granulocytes isolated from donors mobilized with G-CSF/dexamethasone. Granulocytes were isolated from healthy, control donors (□) and from the same donors treated with G-CSF and dexamethasone (■), and cultured for 20 hours in HBSS. Afterward, the cell viability was assessed by the use of annexin V/PI staining. Results represents data from 6 independent experiments (mean ± SD). *P < .05 (significant difference).
The calpastatin transcripts in neutrophils were significantly induced by treatment with G-CSF/dexamethasone, either when applied in vivo or in vitro (Table 1). First, we used RT-PCR to confirm the increase in transcription of the CAST gene that was observed in the microarray analysis. Human calpastatin is expressed as 12 different isoforms of various lengths. The major variation between the isoforms is found in the first 11 exons. The probe set in the microarray only recognizes the main high molecular weight isoforms. We have designed various primer pairs to detect all different isoforms separately in neutrophils and found that all the known transcripts described thus far were expressed (not shown). However, evidence from the literature indicates that all isoforms are able to inhibit calpain activity.41 Therefore, we decided to use a primer set that recognizes all different isoforms in our experiments to confirm the microarray data.
Neutrophils from individuals treated with a combination of G-CSF and dexamethasone, as well as neutrophils cultured with this combination in vitro, both displayed an increase in CAST gene expression (Figure 4A). In freshly purified neutrophils, several isoforms of calpastatin are expressed. In agreement with previously published work, also in other cells, the molecular sizes of these isoforms were found at 110, 108, 90, and 41 kDa.17,42 The most abundant isoforms expressed in neutrophils were the high molecular weight (HMW) forms found as 110- and 108-kDa bands. Neutrophils isolated from granulocyte transfusion donors expressed increased levels of HMW calpastatins compared with the control cells (Figure 4B).
Modulation of calpain/calpastatin pathway by treatment with G-CSF/dexamethasone. (A) Changes in the expression of the CAST gene after treatment with G-CSF and dexamethasone in vivo ( ) or in vitro (■) were measured by the light cycler RT-PCR. Results are presented as the mean ± SEM from 3 independent donors. (B) Different expression of calpastatin after administration of G-CSF and dexamethasone. Samples of neutrophils isolated from control donors (□) and donors stimulated with G-CSF and dexamethasone (■) were subjected to SDS-PAGE and analyzed on Western blots, stained with antibodies against calpastatin. Equal amounts of cells (1.5 × 106) were dissolved in sample buffer, and loaded in each lane. After staining with fluorescently labeled secondary antibody, the blots were scanned with Odyssey and analyzed with Licor Odyssey 2.1 software. Signals were normalized for p38 expression. The intensity of the 2 highest bands recognized by the antibody, which represent 2 major forms of calpastatin in neutrophils, was analyzed. Results represent data from 5 different donors (mean ± SEM). *P < .05 (significant difference). A representative blot comparing calpastatin levels in control cells and mobilized neutrophils is presented. (C) Samples of neutrophils incubated in the presence or absence of G-CSF and dexamethasone were taken at the indicated times and analyzed by Western blot. When indicated, CHX (10 μg/mL) was added to prevent new protein synthesis. Results represent data from 5 independent experiments (mean ± SEM). *P < .05 (significant difference). (D) Samples of neutrophils incubated in the presence or absence of calpain inhibitor 3 (CI3, 20 μmol/L) or caspase inhibitor zVAD (20 μmol/L) were taken at the indicated time points and analyzed by Western Blot. Results represent data from 5 independent experiments (mean ± SEM). *P < .05 (significant difference). (E) Inhibition of Calpains prolongs neutrophil viability. Neutrophils were isolated from healthy donors and cultured overnight alone or with the addition of G-CSF (10 ng/mL), CI3 (20 μmol/L), or zVAD (20 μM). When indicated, CHX (10 μg/mL) was added to prevent new protein synthesis. Cells negative for annexin V staining were considered to be alive. Results represent data from 6 independent experiments (mean ± SEM). *P < .05 (significant difference).
) or in vitro (■) were measured by the light cycler RT-PCR. Results are presented as the mean ± SEM from 3 independent donors. (B) Different expression of calpastatin after administration of G-CSF and dexamethasone. Samples of neutrophils isolated from control donors (□) and donors stimulated with G-CSF and dexamethasone (■) were subjected to SDS-PAGE and analyzed on Western blots, stained with antibodies against calpastatin. Equal amounts of cells (1.5 × 106) were dissolved in sample buffer, and loaded in each lane. After staining with fluorescently labeled secondary antibody, the blots were scanned with Odyssey and analyzed with Licor Odyssey 2.1 software. Signals were normalized for p38 expression. The intensity of the 2 highest bands recognized by the antibody, which represent 2 major forms of calpastatin in neutrophils, was analyzed. Results represent data from 5 different donors (mean ± SEM). *P < .05 (significant difference). A representative blot comparing calpastatin levels in control cells and mobilized neutrophils is presented. (C) Samples of neutrophils incubated in the presence or absence of G-CSF and dexamethasone were taken at the indicated times and analyzed by Western blot. When indicated, CHX (10 μg/mL) was added to prevent new protein synthesis. Results represent data from 5 independent experiments (mean ± SEM). *P < .05 (significant difference). (D) Samples of neutrophils incubated in the presence or absence of calpain inhibitor 3 (CI3, 20 μmol/L) or caspase inhibitor zVAD (20 μmol/L) were taken at the indicated time points and analyzed by Western Blot. Results represent data from 5 independent experiments (mean ± SEM). *P < .05 (significant difference). (E) Inhibition of Calpains prolongs neutrophil viability. Neutrophils were isolated from healthy donors and cultured overnight alone or with the addition of G-CSF (10 ng/mL), CI3 (20 μmol/L), or zVAD (20 μM). When indicated, CHX (10 μg/mL) was added to prevent new protein synthesis. Cells negative for annexin V staining were considered to be alive. Results represent data from 6 independent experiments (mean ± SEM). *P < .05 (significant difference).
Modulation of calpain/calpastatin pathway by treatment with G-CSF/dexamethasone. (A) Changes in the expression of the CAST gene after treatment with G-CSF and dexamethasone in vivo ( ) or in vitro (■) were measured by the light cycler RT-PCR. Results are presented as the mean ± SEM from 3 independent donors. (B) Different expression of calpastatin after administration of G-CSF and dexamethasone. Samples of neutrophils isolated from control donors (□) and donors stimulated with G-CSF and dexamethasone (■) were subjected to SDS-PAGE and analyzed on Western blots, stained with antibodies against calpastatin. Equal amounts of cells (1.5 × 106) were dissolved in sample buffer, and loaded in each lane. After staining with fluorescently labeled secondary antibody, the blots were scanned with Odyssey and analyzed with Licor Odyssey 2.1 software. Signals were normalized for p38 expression. The intensity of the 2 highest bands recognized by the antibody, which represent 2 major forms of calpastatin in neutrophils, was analyzed. Results represent data from 5 different donors (mean ± SEM). *P < .05 (significant difference). A representative blot comparing calpastatin levels in control cells and mobilized neutrophils is presented. (C) Samples of neutrophils incubated in the presence or absence of G-CSF and dexamethasone were taken at the indicated times and analyzed by Western blot. When indicated, CHX (10 μg/mL) was added to prevent new protein synthesis. Results represent data from 5 independent experiments (mean ± SEM). *P < .05 (significant difference). (D) Samples of neutrophils incubated in the presence or absence of calpain inhibitor 3 (CI3, 20 μmol/L) or caspase inhibitor zVAD (20 μmol/L) were taken at the indicated time points and analyzed by Western Blot. Results represent data from 5 independent experiments (mean ± SEM). *P < .05 (significant difference). (E) Inhibition of Calpains prolongs neutrophil viability. Neutrophils were isolated from healthy donors and cultured overnight alone or with the addition of G-CSF (10 ng/mL), CI3 (20 μmol/L), or zVAD (20 μM). When indicated, CHX (10 μg/mL) was added to prevent new protein synthesis. Cells negative for annexin V staining were considered to be alive. Results represent data from 6 independent experiments (mean ± SEM). *P < .05 (significant difference).
) or in vitro (■) were measured by the light cycler RT-PCR. Results are presented as the mean ± SEM from 3 independent donors. (B) Different expression of calpastatin after administration of G-CSF and dexamethasone. Samples of neutrophils isolated from control donors (□) and donors stimulated with G-CSF and dexamethasone (■) were subjected to SDS-PAGE and analyzed on Western blots, stained with antibodies against calpastatin. Equal amounts of cells (1.5 × 106) were dissolved in sample buffer, and loaded in each lane. After staining with fluorescently labeled secondary antibody, the blots were scanned with Odyssey and analyzed with Licor Odyssey 2.1 software. Signals were normalized for p38 expression. The intensity of the 2 highest bands recognized by the antibody, which represent 2 major forms of calpastatin in neutrophils, was analyzed. Results represent data from 5 different donors (mean ± SEM). *P < .05 (significant difference). A representative blot comparing calpastatin levels in control cells and mobilized neutrophils is presented. (C) Samples of neutrophils incubated in the presence or absence of G-CSF and dexamethasone were taken at the indicated times and analyzed by Western blot. When indicated, CHX (10 μg/mL) was added to prevent new protein synthesis. Results represent data from 5 independent experiments (mean ± SEM). *P < .05 (significant difference). (D) Samples of neutrophils incubated in the presence or absence of calpain inhibitor 3 (CI3, 20 μmol/L) or caspase inhibitor zVAD (20 μmol/L) were taken at the indicated time points and analyzed by Western Blot. Results represent data from 5 independent experiments (mean ± SEM). *P < .05 (significant difference). (E) Inhibition of Calpains prolongs neutrophil viability. Neutrophils were isolated from healthy donors and cultured overnight alone or with the addition of G-CSF (10 ng/mL), CI3 (20 μmol/L), or zVAD (20 μM). When indicated, CHX (10 μg/mL) was added to prevent new protein synthesis. Cells negative for annexin V staining were considered to be alive. Results represent data from 6 independent experiments (mean ± SEM). *P < .05 (significant difference).
Next, the calpastatin protein levels were monitored during neutrophil spontaneous apoptosis in the presence or absence of G-CSF and dexamethasone. Culturing of the cells resulted in reduced levels of all isoforms, but especially the HMW forms of calpastatin were diminished (Figure 4C and Figure S2), suggesting proteolysis of the protein during apoptosis. The addition of G-CSF and dexamethasone to culture media resulted in an increase in the protein expression after 8 hours of culture and prevented the absolute loss of the protein after 24 hours in culture. Inhibition of new protein synthesis by the addition of cycloheximide (CHX, 10 μg/mL) prevented the increase and led to the total loss of protein expression after prolonged culture, similar to the control cells (Figure 4C and Figure S2A). The reduction in CAST protein level was apparently caused by degradation by either the calpains that have overcome the inhibitory effect of the calpastatin43 or by caspases activated via a different, calpain-independent route,42 because the addition of either the cell-permeable calpain inhibitor III (CI3) or the cell-permeable pan-caspase inhibitor zVAD partially prevented the loss of calpastatin during overnight culture (Figure 4D and Figure S2B). Moreover, the combined addition of CI3 or zVAD to the cells cultured with G-CSF/dexamethasone induced an additive effect, with zVAD having a more pronounced effect (data not shown).
Inhibition of calpains prevents neutrophil apoptosis
To confirm the involvement of calpain activity in neutrophil apoptosis, cells were incubated overnight in the presence of the combination of G-CSF/dexamethasone, the cell-permeable calpain inhibitor III (CI3), or the cell-permeable pan-caspase inhibitor zVAD as a positive control, whereas cell viability was measured by annexin V staining and verified by cellular morphology (data not shown). As reported before,11,15 the addition of G-CSF/dexamethasone to the culture medium decreased the number of apoptotic cells after overnight culture. This prosurvival effect was abolished by the addition of CHX. In addition, CI3 delayed neutrophil apoptosis to a similar extent (Figure 4E). Together, these data suggest a direct link between the calpain activity, endogenous calpastatin levels, and apoptosis.
Discussion
Mobilization of granulocytes by the administration of G-CSF and corticosteroids is a well-established procedure to achieve the number of granulocytes required for transfusion to neutropenic patients with severe, nonresponsive infections.2,4 We studied the effects of G-CSF and dexamethasone administration on the gene expression pattern of granulocytes to be used for transfusion. We found that a high number of genes was differentially regulated after in vivo mobilization of granulocytes. Approximately 20% of those genes changed similarly upon in vitro culture of granulocytes with the combination of G-CSF and dexamethasone. Still, the expression of more than 700 genes was changed significantly only after in vivo administration, which is perhaps not surprising. It seems reasonable to assume that, within the time frame of the experiment (18-20 hours), secondary donor factors released upon administration of the mobilizing agents, from either granulocytes themselves or from other cell types, could have contributed to the observed changes in granulocyte gene expression. In this context, it is important to realize that the G-CSF receptor is not only present on precursor and mature neutrophilic granulocytes, but also on monocytes and platelets, as well as on endothelial cells and on adult neuronal stem cells.44-46 In contrast, the glucocorticoid receptors are ubiquitously expressed and known to induce or modulate gene transcription in various cell types.47
An additional or, in some cases alternative, explanation may be that the granulocytes recently mobilized from the bone marrow represent a less mature neutrophilic phenotype, as indicated by the lower expression of EMR3, a late marker of granulocytic differentiation,27 and the distinct left shift of the mobilized neutrophils. Another supportive feature of the induced neutrophil egress from the bone marrow was found in the increased transcriptional activity of the genes MMP9 and MMP8, both encoding matrix metalloproteases. The products of these MMP genes are important for creating a localized and highly enriched proteolytic environment in the bone marrow, thus facilitating the release of granulocytes from the stromal microenvironment.48
Finally, there may be combinatorial effects of G-CSF and dexamethasone and other environmental factors specific for the in vivo or in vitro situation. In any case, it seems reasonable to assume that most, if not all, of the gene regulatory effects observed upon the in vitro treatment with G-CSF and dexamethasone represent direct effects of these factors on granulocytes, but there are multiple other explanations possible for the effects found only in the in vivo or only in the in vitro condition, which may relate to differences in cell composition and/or the combinatorial effects of specific (experimental) conditions.
The basic effector functions of granulocytes collected for transfusion after administration of G-CSF and dexamethasone, including interaction with endothelial cells, migration, respiratory burst or killing capacity, seem to be unaffected.4,10,49,50 However, the life span of those cells is prolonged upon in vitro culture as well as in vivo. In addition, supplementation of the culture medium of previously untreated control cells with G-CSF/dexamethasone also prolongs cell survival. This apparently direct effect depends on gene expression and new protein synthesis because it is abolished by the addition of cycloheximide.
From the groups of genes differentially regulated in mobilized granulocytes, we focused on the regulatory genes of apoptosis. The most impressive change was observed for calpastatin, the endogenous inhibitor of calpains. Inhibition of calpains with pharmacologic inhibitors results in a significant delay of neutrophil apoptosis.19 We have recently found that G-CSF is implicated in the regulation of calpain activity by influencing the intracellular levels of Ca2+ upon in vitro culture of neutrophils,17 an effect depending on new protein synthesis. It has been proposed that, upon mild stimulation, calpastatin binds to calpains, preventing the proteases from degrading their substrates. However, the intramolecular, autolytic activation of calpains is not prevented by calpastatin binding,43,51 and if the levels of intracellular Ca2+ increase even higher, so stimulation becomes stronger, the protease will degrade its inhibitor and thereafter proceed to cleave other substrates. Calpastatin degradation has been shown to occur during spontaneous neutrophil apoptosis by activated calpains or proapoptotic caspases (eg, caspase-3 and -7).20,42,52 Calpastatin expression also has been shown to be preferentially induced in neutrophils isolated from patients with cystic fibrosis simultaneously with decreased levels of pro-calpain-1, coinciding with a delay in neutrophil cell death.20
Increased levels of mRNA for calpastatin were detected in mobilized neutrophils, isolated from granulocyte donors pretreated for 18 to 20 hours with G-CSF/dexamethasone as well as in neutrophils cultured in vitro in the presence of G-CSF/dexamethasone. A similar increase in calpastatin levels was observed at the protein level when mobilized neutrophils were compared with the control cells.
Immunoblot analysis of neutrophil lysates showed a loss of calpastatin expression upon in vitro culture. The addition of G-CSF and dexamethasone to the culture media increased the level of calpastatin after 8 hours of incubation, which was prevented by cycloheximide. However, after 24 hours in culture, the cells treated with prosurvival factors such as G-CSF and dexamethasone also displayed strongly reduced levels of calpastatin, albeit still significantly greater than cultured control cells. It appears that the stronger and prolonged activation of calpains allows these proteases to overcome the inhibitory effect of cellular calpastatin, which eventually results in its cleavage. Pharmacologic inhibition of calpains partially preserved calpastatin expression during 24 hours in culture, albeit to a lesser extent than with G-CSF and dexamethasone. Most likely, the activated caspases are responsible for this effect because neutrophils incubated in the presence of a pan-caspase inhibitor also sustained their level of calpastatin. Moreover, the combination of G-CSF/dexamethasone with zVAD led to an additive increase in calpastatin levels and survival of neutrophils.
Taken together, our data suggest that the calpain/calpastatin system plays an important role in neutrophil apoptosis. Inhibition of calpain activation by increasing the levels of its endogenous inhibitor may be one of the prosurvival mechanisms by which G-CSF and dexamethasone delay the apoptosis in neutrophils used for transfusion purposes. Finally, we showed that intracellular levels of calpastatin correlate with the viability of neutrophils in vitro and that pharmacologic inhibition of calpains decreases the rate of apoptosis during 24 hours of incubation. A role for calpastatin in apoptosis in neutrophils is supported by data demonstrating that a reduction in its expression levels by antisense nucleotides accelerates apoptosis.53
In summary, we have shown that mobilization of granulocytes for transfusion by the combination of G-CSF and dexamethasone strongly alters the gene expression pattern in circulating neutrophils compared with untreated donor neutrophils. The characteristics of cells used for transfusion can now be studied based on the changed transcriptional program. Using the changed transcript levels, we can now recognize that mobilized neutrophils display changes in subtle functional aspects that previously remained unidentified. These changes include genes involved in immune reactivity, motility, signal transduction, and gene transcription, as well as cell viability.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We gratefully acknowledge Prof D. Roos for critical reading and discussion of the manuscript and Dr Perry Moerland for excellent assistance with the bioinformatics and statistics of the microarray analysis.
This work was supported by a grant from the Sanquin Foundation for Cellular Blood Product Development (PPO-C-03-011-2003).
Authorship
Contribution: A.D. designed and performed research, analyzed data, and wrote the paper; B.J.v.R., J.G., and A.T.J.T helped perform parts of the research; O.R.F.M. helped analyze data; and F.B., T.K.v.d.B., and T.W.K. supervised the project and reviewed the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Agata Drewniak, Department of Blood Cell Research, Phagocyte Laboratory, Sanquin Research and Landsteiner Laboratory, Plesmanlaan 125, 1066CX Amsterdam, The Netherlands; e-mail: a.drewniak@sanquin.nl.
References
Supplemental data
Genes, which expression was similarly affected after in vivo and in vitro treatment with G-CSF- dexamethasone, were analyzed by the OntoExpress program (http://vortex.cs.wayne.edu/projects.htm#Onto-Express), and categorized into proper functional profiles (e.g. transcription, signal transduction, immune response, cell faith & apoptosis, cell adhesion & mobility, metabolism). Only selection of genes falls into those categories. Results are presented as the mean fold-increase or –decrease of three separate donors (comparison is with freshly isolated neutrophils t=0).

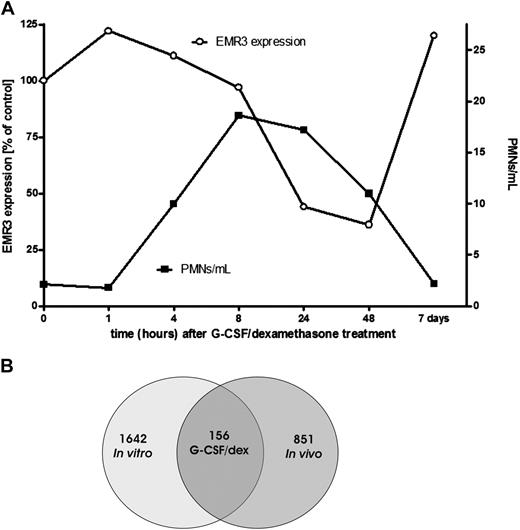
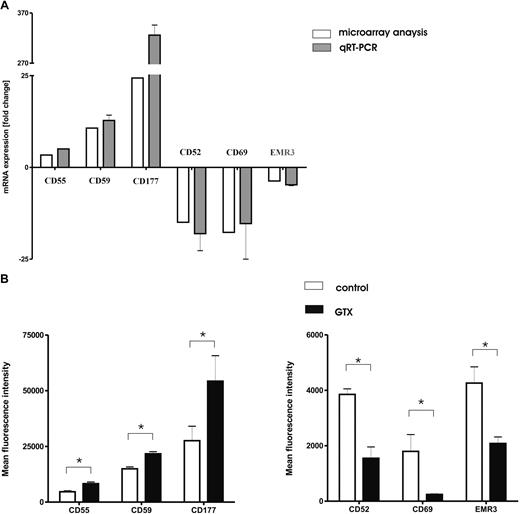
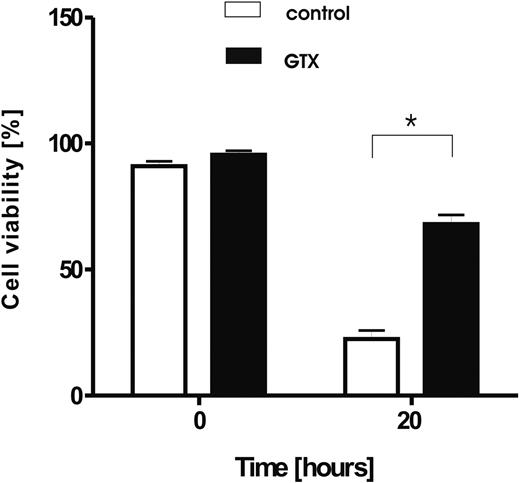
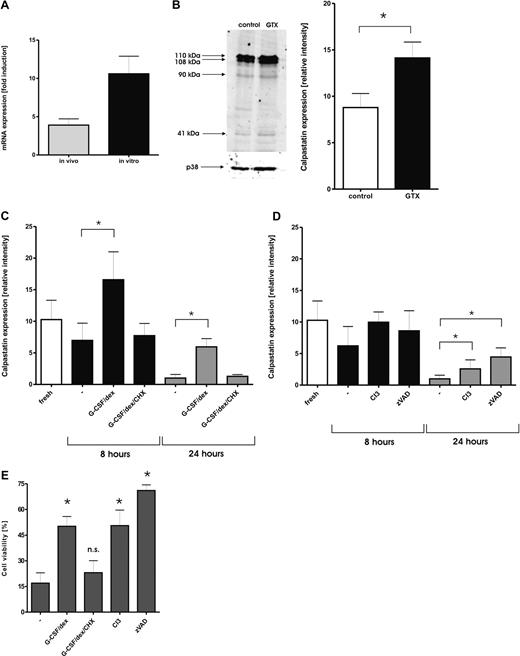
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal