Abstract
Compared with human platelets, rodent platelets are less responsive to peptides and peptidomimetics containing an arginine-glycine-aspartic acid (RGD) motif. Using chimeric human-rat αIIbβ3 molecules, we found that this difference in Arg-Gly-Asp-Ser (RGDS) sensitivity was the result of amino acid substitutions at residues 157, 159, and 162 in the W3:4-1 loop and an Asp-His replacement at residue 232 in the W4:4-1 loop of the αIIb β propeller. Introducing the entire rat W3:4-1 and W4:4-1 loops into human αIIbβ3 also decreased the inhibitory effect of the disintegrins, echistatin and eristostatin, and the αIIbβ3 antagonists, tirofiban and eptifibatide, on fibrinogen binding, whereas the specific point mutations did not. This suggests that RGDS interacts with αIIb in a different manner than with these small molecules. None of these species-based substitutions affected the ability of αIIbβ3 to interact with RGD-containing macromolecules. Thus, human von Willebrand factor contains an RGD motif and binds equally well to adenosine diphosphate-stimulated human and rodent platelets, implying that other motifs are responsible for maintaining ligand binding affinity. Many venoms contain RGD-based toxins. Our data suggest that these species amino acids differences in the αIIb β-propeller represent an evolutionary response by rodents to maintain hemostasis while concurrently protecting against RGD-containing toxins.
Introduction
Ligand binding to integrins initiates intracellular signals that regulate cellular growth and differentiation.1,2 Conversely, cells regulate the ability of integrins to recognize ligands. The prototypic example of integrin regulation is the platelet integrin αIIbβ3; on resting platelets, αIIbβ3 is inactive, but after platelet stimulation, it assumes an active conformation that enables it to bind macromolecular ligands, such as fibrinogen and von Willebrand factor (VWF).3 Many integrin ligands contain an arginine-glycine-aspartic acid (RGD) motif4-6 that participates in integrin binding. Conversely, RGD-containing peptides can act as competitive inhibitors of ligand binding.4-7 For example, the RGD motif located in the C1 domain of VWF8-10 appears to be necessary for VWF binding to αIIbβ3, and the RGD-mimetic small molecules tirofiban and eptifibatide are competitive inhibitors of fibrinogen binding to αIIbβ3.11,12 Nonetheless, the interaction of ligands with integrins, such as αIIbβ3, is substantially more complex than would be predicted from these experiments. For example, although RGD-containing macromolecular ligands, such as fibrinogen, readily bind to the αIIbβ3 of various mammalian species, RGD-based small molecules that are potent antagonists of fibrinogen binding to human αIIbβ35-7 have substantially less potent effects on αIIbβ3 from rabbit, mouse, and rat.13-15
The ligand-binding site on αIIbβ3 consists of specific regions located in the amino-terminal portions of its αIIb and β3 subunits. In the crystal structure of the αIIbβ3 headpiece, ligands bind to a “specificity determining” loop in the β3 βA domain and to a “cap” composed of 4 loops on the upper surface of the αIIb β-propeller domain.16 Analysis of αvβ3 crystals containing a cyclic RGD pentapeptide revealed that the Arg of the pentapeptide is inserted into a cleft between the third and fourth blades of the αvβ-propeller domain and forms salt bridges with D150 and D218 located in the loops connecting the second and third, and third and fourth, propeller blades, respectively.17,18 Although the RGD-peptidomimetic tirofiban11 and the cyclic Lys-Gly-Asp (KGD)-containing heptapeptide eptifibatide12 bind to the same regions in αIIbβ3 as Arg-Gly-Asp-Ser (RGDS), in the crystals of the αIIbβ3 headpiece, the positively charged residue of each antagonist was not in contact with residues homologous to αv D150 and D218 but interacted instead with D224 located deeper within the ligand-binding pocket.16
Previously, we reported that the molecular basis for the species-specific differences in RGD responsiveness is the result of sequence differences in the first 4 blades of the 7-bladed αIIb β-propeller domain.15 Here, we have identified the residues responsible for these differences, addressed whether the sequence differences affect the RGD-containing disintegrins, echistatin and eristostatin, and the RGD mimetics, tirofiban and eptifibatide, and asked whether they also affect RGD-mediated binding of VWF binding to αIIbβ3.
Methods
Construction of chimeric αIIb subunits
Full-length cDNAs for human, rat, and mouse αIIb, a full-length human β3, and a near-full-length rat β3 cDNA were used in these studies.15,19,20 The αIIb cDNAs were inserted into pcDNA3.1(+) Neo and the β3 cDNAs into pcDNA3.1(+) Zeo (Invitrogen, Carlsbad, CA). Single and multiple amino acid substitutions were introduced into the αIIb cDNAs using the QuickChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA). Oligonucleotides for the mutagenesis were 35 to 45 nucleotides in length and were prepared by Integrated DNA Technologies (Coralville, IA). All mutated sequences were confirmed by sequence analysis in at least one orientation as previously described.20
Stable expression of αIIbβ3 in CHO cells
Chinese hamster ovary (CHO) cells were cultured in Ham F-12 media (Invitrogen) supplemented with 10% fetal bovine serum (HyClone Laboratories, Logan, UT). Plasmids containing cDNAs for αIIb and β3 were introduced into the CHO cells using FuGENE 6 according to the manufacturer's instructions (Roche Molecular Biochemicals, Indianapolis, IN). Two days after transfection, cells were transferred to a selection medium containing 500 μg/mL G418 (Invitrogen) and 300 μg/mL Zeocin (Invitrogen). After 3 weeks of selection, αIIbβ3 expression was assessed by flow cytometry using P34, a monoclonal antibody (mAb) recognizing rat, mouse, and human αIIbβ3 (a gift from Dr H. Miyazaki, Kirin Brewery, Gunma, Japan),21 as well as A2A9, a mAb that recognizes the human αIIbβ3 heterodimer.22 Transfected cells were sorted by fluorescence-activated cell sorting to obtain cell lines expressing high levels of αIIbβ3.15
Fibrinogen binding to CHO cells expressing αIIbβ3
Purified human fibrinogen obtained from Enzyme Research Laboratories was labeled with fluorescein isothiocyanate (FITC) using a Calbiochem FITC Labeling Kit as described by the manufacturer (Calbiochem, San Diego, CA). Fibrinogen labeled in this manner was monomeric as assessed by gel-filtration chromatography, supported platelet aggregation as well as unlabeled fibrinogen, and was 95% clottable with thrombin.23 CHO cells (1.5 × 105), suspended in 100 μL 10 mM sodium phosphate buffer, pH 7.4, containing 137 mM NaCl, 1 mM CaCl2, and 1% bovine serum albumin, were incubated with 200 μg/mL FITC-fibrinogen and 5 mM dithiothreitol (DTT) for 30 minutes at 37°C.24,25 After one wash with suspension buffer, cells were fixed with 0.37% formalin, and the amount of bound FITC-fibrinogen was measured by flow cytometry as previously described.15 FITC-fibrinogen specifically bound to αIIbβ3 was calculated as the difference in fluorescence of transfected and untransfected cells incubated with FITC-fibrinogen in the presence of DTT minus the fluorescence of transfected cells incubated with FITC-fibrinogen in the absence of DTT. The ability of RGDS (Sigma-Aldrich, St Louis, MO), echistatin (Sigma-Aldrich), eristostatin (purified from the venom of Eristocophis macmahoni,26 tirofiban; Ben Venue Laboratories, Bradford, OH), or eptifibatide (Millennium Pharmaceuticals, Cambridge, MA) to inhibit fibrinogen binding was determined by adding increasing concentrations of each antagonist to the 30-minute incubations.
Characterization of transgenic murine platelets expressing human αIIb
The production of mice transgenic for the human αIIb gene locus has been previously described.27 All mice used in these studies had been backcrossed onto a C57BL/6J (Charles River Breeding Laboratories, Portage, MI) background for more than 10 generations. The genetic composition of mice homozygous for both the transgene and a knockout of murine Itga2b28 (mmHH mice) was confirmed by polymerase chain reaction. Absence of murine αIIb and the presence of human αIIb on the platelet surface were confirmed by flow cytometry.
Washed murine platelets were obtained from platelet-rich plasma as described previously.29 The presence of human αIIb was detected using MAB1990, an antihuman αIIb-specific mAb (Chemicon International, Temecula, CA), as a primary antibody and phycoerythrin-labeled goat anti–mouse IgG (Sigma-Aldrich) as a secondary antibody. Mouse αIIbβ3 was detected using an FITC-conjugated rat-antimurine αIIbβ3 mAb (BD Biosciences PharMingen, San Diego, CA). Human platelets, positive controls for these measurements, were collected from healthy volunteers after informed consent was obtained in accordance with the Declaration of Helsinki, under a protocol approved by the Institutional Review Board for Studies involving Human Subjects of the Children's Hospital of Philadelphia. Murine studies were approved by the same institution's Animal Care and Use Committee.
Measurements of platelet aggregation
Platelet-rich plasma was prepared from blood anticoagulated with a 0.1 volume of 0.13 M sodium citrate obtained from normal human volunteers by venipuncture and from anesthetized rats and mice by puncture of the exposed abdominal aorta. Platelets were isolated from the platelet-rich plasma by gel filtration on Sepharose 2B (GE Healthcare, Little Chalfont, United Kingdom) using an elution buffer containing 137 mM NaCl, 2.7 mM KCl, 1 mM MgCl2, 5.6 mM glucose, 0.35 mg/mL bovine serum albumin, 3.3 mM NaH2PO4, and 4 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid, pH 7.4, as previously described.30 Turbidometric measurements of adenosine diphosphate (ADP)–stimulated platelet aggregation were made in a Chrono-Log (Havertown, PA) Lumi Dual-Aggregometer. Platelet suspensions were supplemented with human fibrinogen (Sigma-Aldrich) at final concentrations of 200 μg/mL, 1 mM CaCl2, and various concentrations of RGDS (Sigma-Aldrich), echistatin, eristostatin, or tirofiban before adding ADP.
Measurement of fibrinogen and VWF binding to platelets
The affinity of fibrinogen and VWF for human, mouse, and murine mmHH platelets was measured using FITC-labeled fibrinogen and FITC-labeled VWF as described previously.15 Briefly, 0.5-mL aliquots of approximately 5 × 107 gel-filtered platelets were incubated with increasing concentrations of either ligand for 30 minutes at 37°C. The platelets were then gently washed once with 1% fetal bovine serum in phosphate-buffered saline and resuspended in a fixing buffer consisting of 10 mM sodium phosphate buffer, pH 7.4, with 137 mM NaCl, and 0.37% formalin. Specific mean fluorescence intensity values were calculated by subtracting the background fluorescence as described in “Fibrinogen binding to CHO cells expressing αIIbβ3” and expressed in percentages. The dissociation constants (Kd) of human and mouse αIIbβ3 for fibrinogen and VWF were obtained by Scatchard analysis of the binding data.
Results
The W3:4-1 and W4:4-1 loops of αIIb determine the response of human and rat αIIbβ3 to RGDS
We reported previously that the difference in the ability of the tetrapeptide RGDS to inhibit fibrinogen binding to human (H) and rat (R) αIIbβ3 was the result of species-specific differences in the third and fourth blades of αIIb β-propeller.15 Four loops on the upper surface of these blades constitute a “ligand-binding cap” on αIIb.31,32 We postulated that sequence differences in these loops account for the differential response of human and rat αIIbβ3 to RGDS. Two of the loops, W3:2-3 and W3:3-4, are highly conserved between human and rat, but loops W3:4-1 and W4:4-1 are only 50% identical.24
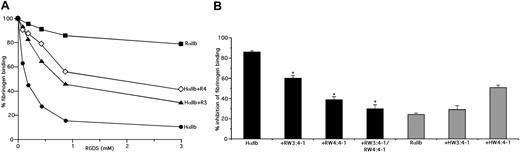
As shown in Figure 1, 1 mM RGDS inhibited DTT-induced fibrinogen binding to human αIIbβ3 expressed on CHO cells by 85.7% plus or minus 1.2% but only inhibited fibrinogen binding to rat αIIbβ3 by 23.8% plus or minus 2.0%, clearly differentiating human from rat αIIbβ3, similar to our prior publication.15 We then found that sequentially replacing the human W3:4-1 and W4:4-1 loops with the corresponding rat sequences progressively decreased the inhibitory effect of RGDS. Thus, the inhibitory effect of 1 mM RGDS on fibrinogen binding to human αIIbβ3 containing rat W3:4-1 was reduced to 60.2% plus or minus 2.3% (P < 10−13), the effect on human αIIbβ3 containing rat W4:4-1 was reduced to 38.9% plus or minus 2.4% (P < 10−22), and the effect on human αIIbβ3 containing both rat loops was reduced to 30.1% plus or minus 3.5%. The latter was not significantly different from the effect of 1 mM RGDS on wild-type (WT) rat αIIbβ3 (P = .24). Conversely, replacing the W3:4-1 and W4:4-1 loops in rat αIIb with corresponding human sequences increased the inhibitory effect of 1 mM RGDS to 29.9% plus or minus 4.4% and 50.8% plus or minus 4.8%, respectively (Figure 1B). These results indicate that most, if not all, of the difference in the effect of RGDS on human and rat αIIbβ3 can be accounted for by sequence differences in the W3:4-1 and W4:4-1 loops.
The W3:4-1 and W4:4-1 loops of αIIb determine the responsiveness of αIIbβ3 to RGDS. (A) Effect of increasing concentration of RGDS on FITC-labeled fibrinogen binding to CHO cells expressing recombinant αIIbβ3 containing WT HαIIb, HαIIb containing a RαIIb W3:4-1 loop (HαIIb + RW3:4-1), HαIIb containing a RαIIb W4:4-1 loop (HαIIb + RW4:4-1), or RαIIb. (B) Percent inhibition of FITC-fibrinogen binding to CHO cells expressing recombinant αIIbβ3 containing WT human or rat αIIb or the designated human/rat αIIb chimeras by 1 mM RGDS. The data shown are the mean plus or minus 1 SE of 5 or more separate experiments, each performed in triplicate. ■ represent WT HαIIb and HαIIb containing the designated RαIIb loops;  represent RαIIb and RαIIb containing the designated HαIIb loops. *P < 10−14 compared with WT HαIIb.
represent RαIIb and RαIIb containing the designated HαIIb loops. *P < 10−14 compared with WT HαIIb.
The W3:4-1 and W4:4-1 loops of αIIb determine the responsiveness of αIIbβ3 to RGDS. (A) Effect of increasing concentration of RGDS on FITC-labeled fibrinogen binding to CHO cells expressing recombinant αIIbβ3 containing WT HαIIb, HαIIb containing a RαIIb W3:4-1 loop (HαIIb + RW3:4-1), HαIIb containing a RαIIb W4:4-1 loop (HαIIb + RW4:4-1), or RαIIb. (B) Percent inhibition of FITC-fibrinogen binding to CHO cells expressing recombinant αIIbβ3 containing WT human or rat αIIb or the designated human/rat αIIb chimeras by 1 mM RGDS. The data shown are the mean plus or minus 1 SE of 5 or more separate experiments, each performed in triplicate. ■ represent WT HαIIb and HαIIb containing the designated RαIIb loops;  represent RαIIb and RαIIb containing the designated HαIIb loops. *P < 10−14 compared with WT HαIIb.
represent RαIIb and RαIIb containing the designated HαIIb loops. *P < 10−14 compared with WT HαIIb.
Identification of residues in the W3:4-1 and W4:4-1 loops that impair the effect of RGDS
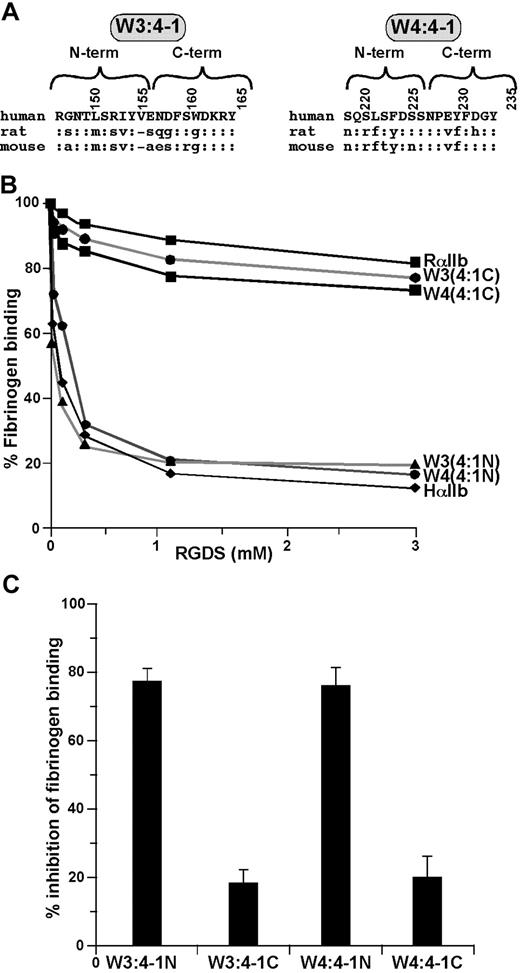
To screen the W3:4-1 and W4:4-1 loops for residues that alter the inhibitory effect of RGDS, we replaced the N-terminal and C-terminal halves of each loop of human αIIb with the corresponding rat sequences (Figure 2A). We found that replacing the N-terminal halves had no effect, whereas replacing the C-terminal halves reduced the inhibitory effect of RGDS on human αIIbβ3 to that on rat (Figure 2B,C).
Localization of the sites responsible for the responsiveness of αIIbβ3 to RGDS to the C-terminal halves of the αIIb W3:4-1 and W4:4-1 loops. (A) Amino acid sequences of the human, rat, and mouse αIIb W3:4-1 and W4:4-1 loops; “:” indicates sequence identity. Numbering is for human αIIb. (B) Effect of increasing concentration of RGDS on FITC-labeled fibrinogen binding to CHO cells expressing recombinant αIIbβ3 containing WT HαIIb, RαIIb, and HαIIb in which the N- or C-terminal halves of the human W3:4-1 and W4:4-1 loops have been replaced by the corresponding rat sequence. (C) Percent inhibition of fibrinogen binding to the human/rat chimeras shown in panel B by 1 mM RGDS. Data shown are the mean plus or minus 1 SE of 3 experiments.
Localization of the sites responsible for the responsiveness of αIIbβ3 to RGDS to the C-terminal halves of the αIIb W3:4-1 and W4:4-1 loops. (A) Amino acid sequences of the human, rat, and mouse αIIb W3:4-1 and W4:4-1 loops; “:” indicates sequence identity. Numbering is for human αIIb. (B) Effect of increasing concentration of RGDS on FITC-labeled fibrinogen binding to CHO cells expressing recombinant αIIbβ3 containing WT HαIIb, RαIIb, and HαIIb in which the N- or C-terminal halves of the human W3:4-1 and W4:4-1 loops have been replaced by the corresponding rat sequence. (C) Percent inhibition of fibrinogen binding to the human/rat chimeras shown in panel B by 1 mM RGDS. Data shown are the mean plus or minus 1 SE of 3 experiments.
There are 3 amino acid differences between the C-terminal human halves of human and rat W3:4-1 (Figure 2A). When the amino acids present in rat W3:4-1 were introduced sequentially into the human sequence, there were small decreases in the ability of RGDS to inhibit fibrinogen binding to αIIbβ3. Thus, compared with WT human αIIb, replacing E157 with S, D159 with G, or W162 with G reduced inhibition by 1 mM RGDS to 60.0% plus or minus 2.5%, 45.9% plus or minus 1.0%, and 60.5% plus or minus 3.2%, respectively (Table 1). These results were similar to the effect of replacing the entire C-terminal half of human W3:4-1 with the rat sequence. Surprisingly, deleting of V156 in human αIIb had no effect (Table 1). This result was unexpected because a Val analogous to V156 is not present in 3 known RGD-resistant species (rat, mouse, and rabbit; Poncz and Newman,19 Thornton and Poncz,20 and NCBI database AAD51954).
Inhibition of fibrinogen binding to recombinant αIIbβ3* containing a human, rat, or chimeric human/rat α subunit by 1 mM RGDS
| . | % inhibition† . |
|---|---|
| WT α subunits | |
| Human | 85.7 ± 1.2 |
| Rat | 23.8 ± 12.7 |
| αIIb W3:4-1 substitutions (rat→human) | |
| ΔV156 | 83.7 ± 4.4 |
| E157S | 60.0 ± 3.6 |
| D159G | 45.9 ± 1.4 |
| W162G | 60.5 ± 4.6 |
| αIIb W4:4-1 substitutions (rat→human) | |
| E229V | 91.1 ± 4.7 |
| Y230F | 82.2 ± 6.1 |
| D232H | 31.1 ± 3.2 |
| D232N | 36.1 ± 3.3 |
| D232E | 76.2 ± 2.3 |
| D232A | NFB‡ |
| Combined αIIb substitutions (rat→human) | |
| V156D,D232H | 39.1 ± 11.4 |
| W3:4-1, D232H | 30.9 |
| . | % inhibition† . |
|---|---|
| WT α subunits | |
| Human | 85.7 ± 1.2 |
| Rat | 23.8 ± 12.7 |
| αIIb W3:4-1 substitutions (rat→human) | |
| ΔV156 | 83.7 ± 4.4 |
| E157S | 60.0 ± 3.6 |
| D159G | 45.9 ± 1.4 |
| W162G | 60.5 ± 4.6 |
| αIIb W4:4-1 substitutions (rat→human) | |
| E229V | 91.1 ± 4.7 |
| Y230F | 82.2 ± 6.1 |
| D232H | 31.1 ± 3.2 |
| D232N | 36.1 ± 3.3 |
| D232E | 76.2 ± 2.3 |
| D232A | NFB‡ |
| Combined αIIb substitutions (rat→human) | |
| V156D,D232H | 39.1 ± 11.4 |
| W3:4-1, D232H | 30.9 |
Recombinant αIIbβ3 was expressed in CHO cells and was activated using DTT as described in “Fibrinogen binding to CHO cells expressing αIIbβ3.”
The results are the mean and 1 SE of three or more experiments.
NFB indicates no fibrinogen binding.
There are also 3 amino acid differences between the C-terminal halves of human and rat W4:4-1 (Figure 2A). However, replacing E229 in human αIIb with V or Y230 with F had no significant effect on the inhibitory effect of RGDS. By contrast, when D232 was replaced by H, 1 mM RGDS decreased fibrinogen binding to αIIbβ3 by 31.5% plus or minus 3.2%, similar to its effect on WT rat αIIbβ3 (P = .19; Table 1). Replacing D232 with N had a comparable effect (36.1% ± 3.3% inhibition), whereas replacing D232 with Glu had no effect. Thus, these results indicate that, although residues in both the W3:4-1 and W4:4-1 loops affects the ability of RGDS to inhibit fibrinogen binding to αIIbβ3, the presence of an acidic residue at position 232 in W4:4-1 appears to be essential.
Effect of RGD-containing disintegrins on the function of human and rat αIIbβ3
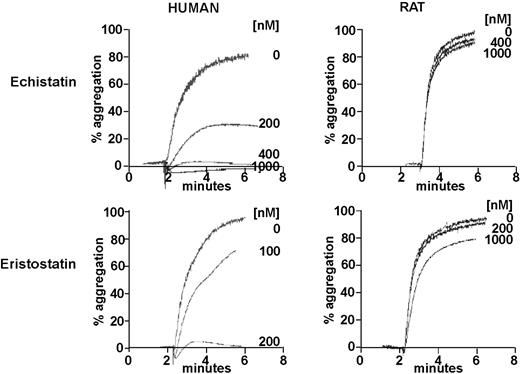
Disintegrins, a family of low molecular weight, cysteine-rich proteins present in venomous snakes, such as Viperidae and Crotalidae, block integrin-ligand interactions via specific tripeptide motifs.33 For disintegrins that interact with αIIbβ3, these motifs include RGD, KGD, and WGD.33 Thus, both of the RGD-containing disintegrins, echistatin and eristostatin, are potent inhibitors of human platelet aggregation, but neither inhibits the aggregation of rat platelets (Figure 3).
Inhibition of the ADP-stimulated aggregation of human and rat platelets by the disintegrins, echistatin and eristostatin. Turbidometric measurements of platelet aggregation were made using gel-filtered platelets stimulated by 20 μM ADP in the presence of the indicated concentrations of echistatin and eristostatin.
Inhibition of the ADP-stimulated aggregation of human and rat platelets by the disintegrins, echistatin and eristostatin. Turbidometric measurements of platelet aggregation were made using gel-filtered platelets stimulated by 20 μM ADP in the presence of the indicated concentrations of echistatin and eristostatin.
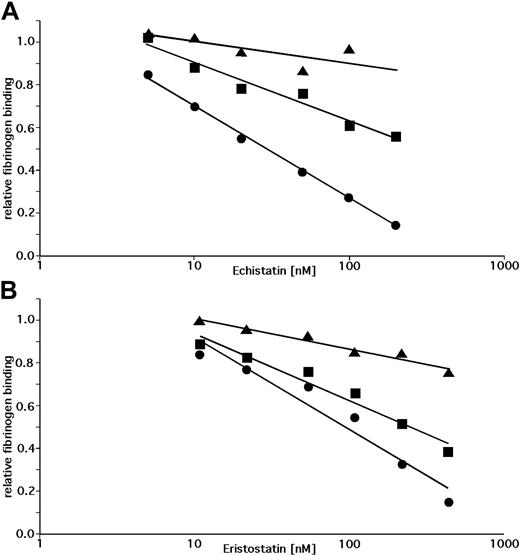
To determine whether the inability of echistatin and eristostatin to inhibit the aggregation of rat platelets is the result of differences in the W3:4-1 and W4:4-1 loops of human and rat αIIb, we measured the ability of these disintegrin to inhibit DTT-induced fibrinogen binding to WT αIIbβ3, to human αIIbβ3 containing the rat W3:4-1 and W4:4-1 loops, and to human αIIbβ3 containing the αIIb D232H polymorphism. The fibrinogen binding data were then plotted against the logarithm of the disintegrin concentration (Figure 4) to calculate IC50 (Table 2). We found that introducing rat W3:4-1 and W4:4-1 loops into human αIIb decreased the inhibitory effect of echistatin and eristostatin by approximately 4 and 2.5 orders of magnitude, respectively. By contrast, replacing D232 with H decreased the inhibitory effect 10.2- and 2.8-fold. These experiments indicate that the differential effects of echistatin and eristostatin on human and rat αIIbβ3 result almost entirely from sequence differences in the αIIb W3:4-1 and W4:4-1 loops. However, compared with RGDS, the inhibitory effects of these disintegrins on αIIbβ3 function are substantially less affected by the identity of the residue at position 232.
Inhibition of DTT-induced fibrinogen binding by echistatin or eristostatin. Inhibition of DTT-induced fibrinogen binding to CHO cells expressing wild human αIIbβ3 (•) or human αIIbβ3 containing either the RαIIb W3:4-1/W4:4-1 loops (▴) or an αIIb D232H mutation (■) by echistatin or eristostatin. Fibrinogen binding was normalized to binding in the absence of disintegrin and is plotted against the log of the disintegrin concentration. Trend lines were calculated using Microsoft Excel. Data shown are the mean of 3 (echistatin) and 4 (eristostatin) experiments.
Inhibition of DTT-induced fibrinogen binding by echistatin or eristostatin. Inhibition of DTT-induced fibrinogen binding to CHO cells expressing wild human αIIbβ3 (•) or human αIIbβ3 containing either the RαIIb W3:4-1/W4:4-1 loops (▴) or an αIIb D232H mutation (■) by echistatin or eristostatin. Fibrinogen binding was normalized to binding in the absence of disintegrin and is plotted against the log of the disintegrin concentration. Trend lines were calculated using Microsoft Excel. Data shown are the mean of 3 (echistatin) and 4 (eristostatin) experiments.
Inhibition of fibrinogen binding to human recombinant WT αIIbβ3 and to recombinant human αIIbβ3 containing a chimeric human/rat α subunit by echistatin and eristostatin
| . | WT human αIIbβ3 . | Human αIIbβ3 + αIIb D232H . | Human αIIbβ3 + rat αIIb W3:4-1/W4:4-1 . |
|---|---|---|---|
| IC50 for inhibiting fibrinogen binding to αIIbβ3, nM* | |||
| Echistatin | 29.2 | 298.9 | 1.09 × 106 |
| Erisostatin | 100.9 | 283.1 | 5.23 × 104 |
| Relative IC50 for inhibiting fibrinogen binding to αIIbβ3† | |||
| Echistatin | 1 | 10.2 | 3.7 × 104 |
| Erisostatin | 1 | 2.8 | 519 |
| . | WT human αIIbβ3 . | Human αIIbβ3 + αIIb D232H . | Human αIIbβ3 + rat αIIb W3:4-1/W4:4-1 . |
|---|---|---|---|
| IC50 for inhibiting fibrinogen binding to αIIbβ3, nM* | |||
| Echistatin | 29.2 | 298.9 | 1.09 × 106 |
| Erisostatin | 100.9 | 283.1 | 5.23 × 104 |
| Relative IC50 for inhibiting fibrinogen binding to αIIbβ3† | |||
| Echistatin | 1 | 10.2 | 3.7 × 104 |
| Erisostatin | 1 | 2.8 | 519 |
IC50 were calculated from the equations from the trend lines shown in Figure 4.
IC50 were normalized to fibrinogen binding to wild-type human αIIbβ3.
Effect of tirofiban and eptifibatide on fibrinogen binding to human and rat αIIbβ3
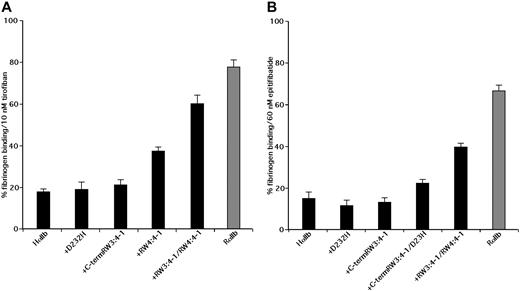
The RGD-based peptidomimetic analog of tyrosine, tirofiban, and the KGD-based cyclic heptapeptide eptifibatide inhibit human platelet aggregation with IC50 of approximately 10 nM and approximately 150 nM, respectively.34-36 Like RGDS, echistatin and eristostatin, tirofiban and eptifibatide have little effect on rat platelets. Thus, although 10 nM tirofiban reduced DTT-induced fibrinogen binding to human αIIbβ3 on CHO cells by 82.2% plus or minus 1.5%, it reduced fibrinogen binding to rat αIIb by only 22.2% plus or minus 2.4% (Figure 5A). Similarly, 60 nM eptifibatide reduced DTT-induced fibrinogen binding to human αIIbβ3 on CHO cells by 84.6% plus or minus 2.2% but inhibited fibrinogen binding to αIIbβ3 containing a rat α subunit by only 33.6% plus or minus 2.2% (Figure 5B).
Inhibition of DTT-induced fibrinogen binding by tirofiban and eptifibatide. Inhibition of DTT-induced fibrinogen binding to CHO cells expressing WT HαIIb with Hβ3 (HαIIb), WT rat αIIb with Rβ3, and the designated human/rat αIIb chimeras with Hβ3 by 10 nM tirofiban or 60 nM eptifibatide. Data shown are the mean plus or minus 1 SE of 3 or more separate experiments.
Inhibition of DTT-induced fibrinogen binding by tirofiban and eptifibatide. Inhibition of DTT-induced fibrinogen binding to CHO cells expressing WT HαIIb with Hβ3 (HαIIb), WT rat αIIb with Rβ3, and the designated human/rat αIIb chimeras with Hβ3 by 10 nM tirofiban or 60 nM eptifibatide. Data shown are the mean plus or minus 1 SE of 3 or more separate experiments.
To determine whether the sequence of the αIIb W3:4-1 and W4:4-1 loops influences the ability of tirofiban and eptifibatide to inhibit fibrinogen binding, we measured their effect on fibrinogen binding to chimeric human/rat αIIbβ3 molecules. Replacing the human W3:4-1 loop with the corresponding rat sequence had an insignificant effect on the ability of 10 nM tirofiban to inhibit fibrinogen binding, as did replacing D232 with His in W4:4-1 (Figure 5A). On the other hand, replacing the entire W4:4-1 loop reduced inhibition by tirofiban by 32% and replacing both W3:4-1 and W4:4-1 reduced inhibition by 70%. Similar results were seen when 60 nM eptifibatide was used as the αIIbβ3 antagonist (Figure 5B). Thus, like RGDS, echistatin and eristostatin, the differential effects of tirofiban and eptifibatide on human and rat αIIbβ3 result from sequence differences in the αIIb W3:4-1 and W4:4-1 loops. Unlike the other small molecules, however, the differential effects were unaffected by the D232H polymorphism in W4:4-1 and required the simultaneous presence of sequence differences in both the W3:4-1 and W4:4-1 loops.
Effect of RGDS and tirofiban on VWF binding to human and rodent αIIbβ3
VWF binding to αIIbβ3 requires the presence of an RGD motif.37 Thus, we expected its affinity for αIIbβ3 would reflect the intrinsic ability of αIIbβ3 to interact with RGDS. To test this assumption, we compared the binding of FITC-labeled human fibrinogen and VWF to ADP-stimulated human and rat platelets by flow cytometry. However, despite the presence of RGD in identical positions in human and rat VWF38 (and NCBI databases XP 342760 and ABC86573), human VWF did not bind to rat platelets. On the other hand, human WWF bound equally well to human and mouse platelets (Table 3; and data not shown). Thus, the presence of an RGD motif may be necessary, but is not sufficient, to mediate VWF binding to αIIbβ3.
ADP-stimulated binding of fibrinogen and VWF to human, murine, and mmHH platelets and inhibition by RGDS and tirofiban
| . | Human platelets . | Murine platelets . | mmHH platelets* . |
|---|---|---|---|
| Ligand binding (Kd, nM) | |||
| Fibrinogen | 66.5 ± 3.2 | 67.1 ± 2.8 | ND |
| VWF | 20.6 ± 2.8 | 25.6 ± 3.1 | ND |
| IC50 for fibrinogen binding | |||
| RGDS, μM | 3.5 ± 1.6 | 35.9 ± 7.1 | 2.3 ± 0.2 |
| Tirofiban, nM | 0.9 ± 0.2 | 1.6 ± 0.9 | 1.3 ± 0.3 |
| IC50 for VWF binding | |||
| RGDS, μM | 21.0 ± 1.2 | 68.6 ± 20.8 | 14.4 ± 10.3 |
| Tirofiban, nM | 1.2 ± 0.4 | 2.0 ± 1.0 | 1.5 ± 0.2 |
| . | Human platelets . | Murine platelets . | mmHH platelets* . |
|---|---|---|---|
| Ligand binding (Kd, nM) | |||
| Fibrinogen | 66.5 ± 3.2 | 67.1 ± 2.8 | ND |
| VWF | 20.6 ± 2.8 | 25.6 ± 3.1 | ND |
| IC50 for fibrinogen binding | |||
| RGDS, μM | 3.5 ± 1.6 | 35.9 ± 7.1 | 2.3 ± 0.2 |
| Tirofiban, nM | 0.9 ± 0.2 | 1.6 ± 0.9 | 1.3 ± 0.3 |
| IC50 for VWF binding | |||
| RGDS, μM | 21.0 ± 1.2 | 68.6 ± 20.8 | 14.4 ± 10.3 |
| Tirofiban, nM | 1.2 ± 0.4 | 2.0 ± 1.0 | 1.5 ± 0.2 |
Values are the mean and 1 SE of 3 experiments.
ND indicates not done.
Transgenic mice are homozygous for a disabled murine αIIb gene and express a human αIIb transgene at a level approaching that of human platelets.27
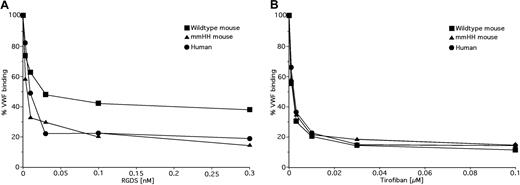
Next, we compared the ability of RGDS and tirofiban to inhibit the binding of human VWF and fibrinogen to human and mouse αIIbβ3. As shown in Table 3 and Figure 6A, we found that RGDS was a less potent inhibitor of VWF and fibrinogen binding to mouse αIIbβ3 than to human αIIbβ3 (P < .08 and P < .01). By contrast, there was no difference in the ability of tirofiban to inhibit VWF and fibrinogen binding to αIIbβ3 from either species (Table 3; Figure 6B). Taken in the context of the lack of response of rat αIIbβ3 to these inhibitors, these data are consistent with the notion that the ability of RGD-based small molecules to interact with αIIbβ3 is determined by the amino acid composition of the walls of the αIIb ligand binding cleft.
Effects of RGDS and tirofiban on VWF binding to αIIbβ3 on human, and murine WT and mmHH platelets. Binding of FITC-labeled human VWF to ADP-stimulated human platelets (•), murine platelets (■), and mmHH platelets (▴) was measured in the presence of increasing concentrations of RGDS (A) or tirofiban (B). The data shown are the mean of 3 experiments.
Effects of RGDS and tirofiban on VWF binding to αIIbβ3 on human, and murine WT and mmHH platelets. Binding of FITC-labeled human VWF to ADP-stimulated human platelets (•), murine platelets (■), and mmHH platelets (▴) was measured in the presence of increasing concentrations of RGDS (A) or tirofiban (B). The data shown are the mean of 3 experiments.
Discussion
To gain insight into the interaction of αIIbβ3 with small RGD-containing molecules, we took advantage of species-specific differences in the ability of these molecules to inhibit αIIbβ3 function. Thus, although such molecules are potent inhibitors of fibrinogen binding to αIIbβ3 on human platelets,39 they interact poorly, if at all, with αIIbβ3 on rat platelets. Previously, we reported that the differential ability of the tetrapeptide RGDS to inhibit fibrinogen binding to human and rat αIIbβ3 resulted from species-specific differences in the αIIb β-propeller domain.15 Here, we have identified these differences and extended their effect to the RGD-containing disintegrins, echistatin and erisostatin, to the peptidomimetics, tirofiban and eptifibatide, and to a second RGD-containing αIIbβ3 ligand, VWF.
We found that species-specific differences at residues 156, 157, 159, and 162 located in a loop connecting the second and third blades of the αIIb β propeller, as well as the D232H substitution in the loop connecting blades 3 and 4 affected the ability of RGDS to inhibit fibrinogen binding to αIIbβ3. However, none of the substitutions affected fibrinogen binding to αIIbβ3.31 These results are compatible with the observations that neither RGD motifs in the fibrinogen α chain is necessary for fibrinogen binding to αIIbβ3.40-42 By contrast, replacing D232 with alanine abrogated fibrinogen binding31 (Table 1), implying that the residue at position 232 is either part of an overlapping site for fibrinogen binding or its presence is necessary to maintain αIIb in a conformation competent to bind fibrinogen. F231A and Y234A mutations also substantially reduce fibrinogen binding to αIIbβ3. However, neither F231A, nor D232A, nor Y234A affects the binding of nonligand mimetic monoclonal antibodies to αIIbβ3,31 implying that they do not induce gross conformational changes in αIIb, and their effect may be to directly perturb the αIIb fibrinogen-binding site.
Although the D232H substitution significantly decreased the ability of RGDS to inhibit fibrinogen binding to αIIbβ3, but it had less of an effect on echistatin- and eristostatin-mediated inhibition and essentially no effect on tirofiban and eptifibatide. To provide a structural explanation for these unexpected observations, we took advantage of the available crystal structures of the αvβ3 and αIIbβ3 headpieces containing RGD-based small molecules.16,18 Analysis of a crystal structure of the cyclic pentapeptide RGD-[D-Phe]-[N-methyl-V-] bound to the extracellular domain of the αvβ3 revealed that electrostatic interactions involving its Arg and Asp residues represented the predominant contacts between it and the αv and β3 integrin chains.18 In particular, the Arg residue bound to both αv D150 and D218, which are homologous to αIIb D159 and D232. As we discovered, these 2 residues are also involved in RGD sensitivity of αIIbβ3: D159 is in the W3:4-1 loop and D232 in the W4:4-1 loop have a major effect on RGD sensitivity.
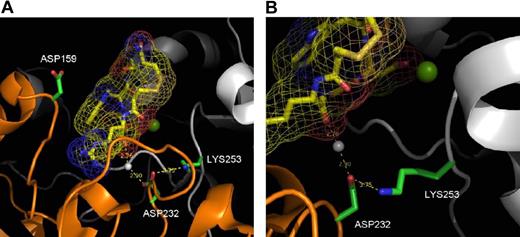
There is no available crystal structure of the αIIbβ3 headpiece complexed with RGDS or a related peptide. However, eptifibatide, a cyclic peptide containing a homo-RGD sequence, is a close homolog of RGDS in which homo-arginine has one extra methylene inserted into its side chain. The presence of homoarginine in eptifibatide is noteworthy because it provides extreme selectivity for αIIbβ3 over αvβ3.43 As noted in Table 1, mutation of D159 in the human αIIb W3:4-1 loop to glycine decreases the sensitivity of human αIIbβ3 to RGDS by approximately 44%. Interestingly, the side chain of D159 is highly solvent-exposed (Figure 7A) and does not directly contact the guanidino side chain of eptifibatide. Hence, D159G does not have a large affect on binding to eptifibatide, although it does have a large effect on binding to RGDS. Thus, RGDS appears to interact with the αIIb ligand binding cap in a different manner from eptifibatide; in particular, the Arg side chain of RGDS clearly interacts more favorably with D159 than the homo-arginine of eptifibatide, either via a long-range electrostatic interaction or via a more direct interaction.
X-ray crystal structure of eptifibatide bound to human αIIbβ3. Crystal structure is based on published data of eptifibatide docked into the ligand pocket of the extracellular head of human αIIbβ3.16 In both panels, the eptifibatide molecule is shown in yellow. The white sphere represents crystal water; green sphere, magnesium ion. (A) This image focuses on Asp159 and shows no direct interaction with the eptifibatide. (B) This image focuses on Asp232 and shows that its interaction with eptifibatide is through a water molecule.
X-ray crystal structure of eptifibatide bound to human αIIbβ3. Crystal structure is based on published data of eptifibatide docked into the ligand pocket of the extracellular head of human αIIbβ3.16 In both panels, the eptifibatide molecule is shown in yellow. The white sphere represents crystal water; green sphere, magnesium ion. (A) This image focuses on Asp159 and shows no direct interaction with the eptifibatide. (B) This image focuses on Asp232 and shows that its interaction with eptifibatide is through a water molecule.
Mutation of D232 in the W4:4-1 loop to a positively charged residue resulted in a 63% decrease in the sensitivity of human αIIbβ3 sensitivity to RGDS inhibition, but not to inhibition by eptifibatide. Although D232, like D159, is within the vicinity of eptifibatide, it does not directly interact with it (Figure 7B). D232 is located beneath the eptifibatide molecule, where it forms an intersubunit salt bridge with K253 in β3. In addition, a crystal water bridges a hydrogen bond between D232 and a carboxylate group from eptifibatide. Removing a negatively charged residue at this position (ie, D232H) leads to the unsatisfied buried charge that might change the structure of the protein, resulting in decreased RGDS binding. Nonetheless, such an alteration in structure must be subtle because the mutant retains the ability to interact with both fibrinogen and eptifibatide. These data indicate that RGDS interacts with αIIb in a manner different from eptifibatide; in particular, the Arg side chain of RGDS interacts more favorably with D159 and D232 than eptifibatide and other αIIbβ3-specific small molecule inhibitors. Based on the single published crystal structure of αIIbβ3, the latter molecules interact instead with D224 deeper in the ligand-binding cleft.16 On the other hand, the available crystal structure of αIIbβ3 is insufficient to explain the changes in RGD ligand affinity between various species.
Although RGD-containing molecules can be potent inhibitors of fibrinogen binding to αIIbβ3, the interaction of fibrinogen itself with αIIbβ3 is unaffected by deletion of both RGD motifs from the fibrinogen α chain and appears to be mediated primarily by sequences located at the carboxyl-terminus of the fibrinogen γ chain instead.44-46 An explanation for this apparent paradox is unclear. We have reported that the interaction of RGD and γ chain peptides with αIIbβ3 is mutually exclusive,15 suggesting that the peptides bind to either the same or allosterically linked sites. In favor of the latter explanation, we previously found that RGDS binding to αIIbβ3 caused the exposure of the epitope for a conformation-specific anti-β3 monoclonal antibody.15 The widespread distribution of mutations that impair fibrinogen binding over the upper surface of the αIIbβ3 headpiece is also consistent with this explanation.31
Unlike human fibrinogen, which binds equally well to human and rat αIIbβ3,15 human VWF does not bind to rat αIIbβ3. Accordingly, the observation that human VWF binds equally well to αIIbβ3 on human and mouse platelets was unexpected. Thus, because human, mouse, and rat VWF each contain an RGD motif,37 it is probable that other portions of the VWF molecule contribute to VWF binding to αIIbβ3. In fibronectin, an RGD sequence located in the 10th type III homology repeat and a region containing the pentapeptide sequence PHSRN located in the adjacent ninth type III repeat function synergistically to support fibronectin binding to α5β1.47,48 Thus, mutating RGD in fibronectin fusion proteins or the synergy site in the ninth repeat had similar effects on integrin binding to fibronectin.47 We found no difference in the affinity of VWF for human and mouse αIIbβ3 despite significant differences in the ability of human and mouse αIIbβ3 to interact with RGDS. Thus, by analogy with fibronectin binding to integrins, these observations argue that VWF factor contains a synergy site that acts in concert with RGD to determine the affinity of VWF binding to αIIbβ3.
In conclusion, we found that the differences in the ability of RGD-containing small molecules to interact with the integrin αIIbβ3 on human, rat, and mouse platelets are the result of amino acid substitutions located in the loops connecting the second, third, and fourth blades of the αIIb β propeller. In particular, the αIIbβ on rodent platelets is unresponsive to these molecules. On the other hand, the substitutions do not affect the ability of αIIbβ3 to interact with RGD-containing macromolecular ligands, implying that other sites on these ligands, acting in synergy with or independent of RGD, are responsible for maintaining the affinity of ligand binding. It is noteworthy that venoms often contain RGD-containing toxins. Thus, the decreased inhibitory effect of RGD on rodent αIIbβ3 may be an evolutionary response by rodents to maintain hemostasis while concurrently protecting themselves against toxic RGD-containing venom proteins.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported in part by the National Institutes of Health (grant HL40387; J.S.B., M.P.) and generous contributions from the Plummer family and Feldman family (Shulman Foundation; M.P.).
National Institutes of Health
Authorship
Contribution: R.B.B. carried out most of the vector and cell line constructions and a major part of studying RGD inhibition of fibrinogen binding; H.Z. helped complete the RGD, disintegrin, and agonist studies; M.A.T. established the mmHH mouse line and characterized them and carried out initial VWF binding studies; C.S.S. carried out the detailed modeling; W.F.D. designed and interpreted many of the molecular modeling analyses; M.A.K. assisted in the fibrinogen binding studies, the human and murine platelet studies, and in data analysis; and J.S.B. and M.P. contributed to study design, data interpretation, and manuscript preparation.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
R.B.B. was deceased at the time of manuscript preparation.
Correspondence: Mortimer Poncz, Division of Hematology, Children's Hospital of Philadelphia, 34th Street and Civic Center Boulevard, Philadelphia, PA 19104; e-mail: poncz@email.chop.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal