Abstract
NF1 inactivation occurs in specific human cancers, including juvenile myelomonocytic leukemia, an aggressive myeloproliferative disorder of childhood. However, evidence suggests that Nf1 loss alone does not cause leukemia. We therefore hypothesized that inactivation of the Nf1 tumor suppressor gene requires cooperating mutations to cause acute leukemia. To search for candidate genes that cooperate with Nf1 deficiency in leukemogenesis, we performed a forward genetic screen using retroviral insertion mutagenesis in Nf1 mutant mice. We identified 43 common proviral insertion sites that contain candidate genes involved in leukemogenesis. One of these genes, Bcl11a, confers a growth advantage in cultured Nf1 mutant hematopoietic cells and causes early onset of leukemia of either myeloid or lymphoid lineage in mice when expressed in Nf1-deficient bone marrow. Bcl11a-expressing cells display compromised p21Cip1 induction, suggesting that Bcl11a's oncogenic effects are mediated, in part, through suppression of p21Cip1. Importantly, Bcl11a is expressed in human chronic myelomonocytic leukemia and juvenile myelomonocytic leukemia samples. A subset of AML patients, who had poor outcomes, of 16 clusters, displayed high levels of BCL11A in leukemic cells. These findings suggest that deregulated Bcl11a cooperates with Nf1 in leukemogenesis, and a therapeutic strategy targeting the BCL11A pathway may prove beneficial in the treatment of leukemia.
Introduction
Neurofibromatosis type 1 syndrome (NF1) is an inherited disease caused by germline mutations of the NF1 gene.1 NF1 encodes neurofibromin, a GTPase-activating protein that negatively regulates N-, H-, K-, and R-RAS signaling.2 Loss of NF1 function results in elevated Ras-GTP levels in neoplastic cells of patients with NF1.3,4 Thus, NF1 deficiency has been proposed to be functionally equivalent to activation of a RAS oncogene. Neurofibromin may have additional roles in growth control.5-8 Thus, further investigations into pathways altered in NF1-associated malignancies may shed light on these mechanisms.
Children with NF1 syndrome show a markedly increased incidence of myeloid malignancies, particularly juvenile myelomonocytic leukemia (JMML), which can further progress into acute myeloid leukemia (AML).9 Similarly, mice with a germline Nf1 mutation manifest some phenotypes of NF1 disease and thus provide a tractable model for investigating NF1-associated complications.10 Notably, evolution to AML is not observed in mice bearing mutant Nf1 alone. Instead, murine data from competitive repopulation experiments11 and the long latency of myeloid disease observed in recipients transplanted with Nf1−/− fetal liver cells12 support the possibility that a Nf1−/− leukemia-initiating cell must acquire one or more additional mutations to cause leukemia. However, little is known about the nature of these genetic abnormalities. We therefore set out to identify genetic events cooperating with inactivation of Nf1 in leukemogenesis.
The B-cell leukemia 11A gene (BCL11A/Evi9/CTIP1) is essential for normal lymphoid development13 and has been associated with hematologic malignancies.14,15 The BCL11A gene was initially identified from aberrant chromosomal translocations involving the immunoglobulin heavy chain locus detected in B-cell non-Hodgkin lymphomas.14 It encodes a Krüppel-like zinc-finger protein containing 3 C2H2 zinc finger motifs, a proline-rich region, and an acidic domain. BCL11A functions as a transcriptional regulator via directly binding to a guanine cytosine (GC)–rich DNA motif. BCL11A is expressed predominantly in brain, spleen, and testis and is down-regulated in a leukemia cell line under induction of myeloid differentiation.16
BXH-2 mice have proven to be a powerful model system to identify genetic lesions causally associated with leukemia.17 Approximately 15% of BXH-2 myeloid leukemias have proviral insertions at ecotropic viral integration site 2 (Evi2), all of which are located within a single large intron of the Nf1 gene.18 Consequently, Nf1 gene function is disrupted, and no wild-type gene product could be detected in Evi2-bearing leukemias.19 These studies support the idea that BXH-2 AML is an excellent system to explore the mechanisms of Nf1-associated leukemia.
We performed a genetic screen in the BXH-2 AML model using retroviral insertion mutagenesis to search for cooperating genetic events in Nf1-associated myeloid leukemia. Here we report isolation of 43 common insertion sites (CISs). One such CIS, Evi9, is associated with Nf1-deficient BXH-2 AML and targets Bcl11a. We found that overexpression of Bcl11a causes leukemia in mice under the condition of Nf1 deficiency. Bcl11a also promotes cell growth and suppresses p21Cip1 induction. High expression levels of Bcl11a are seen in certain subsets of AML patients.
Methods
Cell culture and mice
The Nf1-deficient and Nf1-proficient mouse hematopoietic cells immortalized with RED-Myb were maintained in the ASM media.12,20,21 The 293T were purchased from the ATCC (Manassas, VA) and maintained in Dulbecco modified Eagle medium (DMEM), supplemented with heat-inactivated 10% fetal bovine serum and 1% penicillin/streptomycin. Cell counts were determined manually using a hemocytometer (Reichert, Buffalo, NY). For all the studies described herein, cells growing in the logarithmic phase of growth were used. The Nf1(flox/+) and Nf1(Fcr/+) mouse lines have been previously described.22,23 The C57BL/6J and C57BL/6J.BoyJ strain of mouse was purchased from the National Cancer Institute (Bethesda, MD). Mice were housed, bred, and manipulated under specific pathogen-free conditions, following the University of Minnesota's regulations on institutional animal care for research use.
Patient samples
The JMML and chronic myelomonocytic leukemia patient samples used in this study were obtained following protocols approved by the University of Minnesota Internal Review Board, with informed consent obtained in accordance with the Declaration of Helsinki.
Isolation of proviral integration sites
A total of 2 μg genomic DNA was digested overnight with the combination of restriction enzymes TaqI, BspLU11I, and BclI (Roche Diagnostics, Indianapolis, IN), and subjected to the STA–polymerase chain reaction (PCR) and SplinkBlunt-PCR protocol, as previously reported.24 After electrophoresis on 1.2% of agarose gel, individual secondary PCR bands were cloned into TOPO TA vector (Invitrogen, Carlsbad, CA) for sequencing.
Sequence analysis
Proviral insertion sites (PISs) were mapped by searching against Ensembl and Celera Mouse Genome Database using the BLASTN algorithm.25
RT-PCR
Total RNA was extracted from tumor samples using TRIzol (Invitrogen), followed by treatment with DNase I, according to the manufacturer's instructions. Reverse-transcription PCR (RT-PCR) was conducted using the SuperScript II First Strand cDNA Synthesis system (Invitrogen), followed by PCR using gene-specific primers, as reported previously.20 Sequences of primers used in the RT-PCR for amplification of genes examined in this study are available on request.
Southern blotting analysis
A total of 10 μg genomic DNA was digested with the indicated restriction enzymes and resolved on a 1.0% agarose gel, followed by standard blotting, hybridization, and radioautography procedure.20 The pMIGR-Bcl11a construct-specific probe and the cDNA probe specific for the Ing1 gene were labeled with α-32P–dCTP using the random priming labeling approach.20
Construction of pMIGR-Bcl11a, retrovirus production, and establishment of stable transductants
Bcl11a cDNA (a gift from Dr Takuro Nakamura, The Cancer Institute, Japanese Foundation for Cancer Research, Tokyo, Japan) was fused with His tag by PCR and subcloned into the BglII-XhoI site of a retroviral expression vector, pMIGR (a gift from Dr Juli Miller, The University of Pennsylvania, Philadelphia, PA), to create pMIGR-Bcl11a, which expresses both Bcl11a and enhanced green fluorescence protein (EGFP).26 Constructs were verified by sequencing. pMIGR-Bcl11a and empty vector were used to transfect 293T cells for production of vesicular stomatitis virus G (VSV-G) pseudotyped retrovirus, as previously described.26 The retrovirus was used to infect cultured cells in the presence of 4 μg/mL polybrene. The EGFP-positive cells were sorted from the population of transduced cells by flow cytometry (BD Biosciences, San Jose, CA).
Bone marrow transduction and transplantation assay
Bone marrow cells were harvested from donor mice 5 days after they were treated with 150 mg/kg 5-fluorouracil, as described previously.27 Cells were stimulated for 24 hours with interleukin-6 (IL-6), IL-3, and stem cell factor (SCF) before being infected with the indicated viruses in the presence of 2 μg/mL polybrene. Transduced bone marrow cells were intravenously injected into lethally irradiated C57BL/6J.BoyJ mice at 106 cells per recipient mouse. Mice were observed on a daily basis for any signs of disease.
Immunologic phenotyping and flow cytometry
As previously described,28 cells were incubated on ice in blocking buffer containing CD16/32 Fc blocker for 15 minutes, before staining with antibodies on ice for 25 minutes. Subsequently, cells were washed and subjected to flow cytometry analysis using 4-color FACSCalibur flow cytometer (BD Biosciences) following the manufacturer's instruction. All fluorochrome-conjugated antibodies were purchased from BD PharMingen (San Diego, CA), and the applied dilutions of antibodies were optimized in our laboratory. CD45.1–phycoerythrin (PE) and CD45.2–fluorescein isothiocyanate (FITC) were used to distinguish donor and recipient cellular origin. Conjugated antibodies staining for Mac-1, Gr-1, T-cell receptor β (TCR-β), and B220 were used to examine the origin of cell lineages. Data were analyzed using FlowJo 7.0 Software (TreeStar, Ashland, OR).
Cell growth assay
A total of 105 cells per well were seeded into 24-well plates in quadruplicate in acid sphingomyelinase medium19 complemented with 10% WEHI-3–conditioned medium, and cultured at 37°C in a 10% CO2 humidified incubator. An aliquot of 10 μL homogeneous cell suspensions was taken at 0, 24, and 48 hours, respectively, to count the cumulative cell number.
Western blot
Cells were washed with phosphate-buffered saline (PBS) and lysed in Nonidet P-40 (NP-40) lysing buffer.21 Protein concentrations were determined using the Bradford method (Pierce Chemical, Rockford, IL). A total of 40 μg total protein was boiled for 5 minutes before electrophoresis on 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) gels, and subsequently blotted to methanol-activated polyvinylidene difluoride (PVDF) membrane (Bio-Rad, Hercules, CA). The blots were blocked and incubated with the primary antibodies as follows: His probe (1:2000; Santa Cruz Biotechnology, Santa Cruz, CA), rabbit anti–extracellular signal-regulated kinase 1 (ERK1; 1:1500; Santa Cruz Biotechnology), and mouse antibodies against indicated cell-cycle regulatory proteins are available in the Cell Cycle I/II Sampler Kit (BD PharMingen). After incubation with horseradish peroxidase (HRP)–conjugated secondary antibodies, blots were developed by the enhanced chemiluminescence detection system (GE Healthcare, Little Chalfont, United Kingdom) and then exposed to the X-ray films.
Statistics
Student t tests were used to compare means of cell number between different genotypes of immortalized hematopoietic cells. The Fisher exact test was applied to determine whether there was difference in incidence of Bcl11a-targeting PIS between leukemia with wild-type Nf1 and Nf1 deficiency. Differences between groups were considered significant at values of P less than .05.
Results
Identification of CISs of BXH-2 MuLV
We examined genomic DNA specimens derived from BXH-2 strain AML, which are Nf1-intact (n = 25) or Nf1-deficient (n = 55). In 19 Nf1-deficient AMLs, the Nf1 gene was disrupted by murine leukemia virus (MuLV) insertions within the Evi2 locus,12,29 and 36 AMLs were generated using Nf1 knockout mice (Figure 1).12,29 Somatic proviral insertions in these AML were isolated efficiently with STA-PCR and SplinkBlunt-PCR that we have developed.24 We obtained 197 informative PISs, with 2.46 PISs recovered from each tumor on average (Table S1, available on the Blood website; see the Supplemental Materials link at the top of the online article). We expect that this is close to full PIS recovery compared with the number of PISs per BXH-2 tumor detected by Southern blotting assay (data not shown).24,29
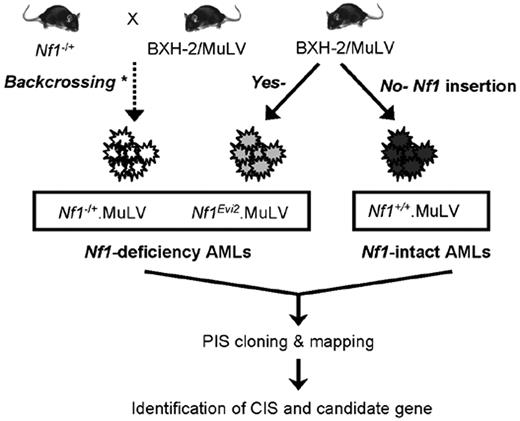
Strategy for collection of leukemia with intact or deficient Nf1 gene for this study. Nf1+/+. MuLV indicates MuLV-infected Nf1-intact leukemia; Nf1−/+.MuLV, MuLV-infected leukemia with engineered null Nf1 allele. Nf1Evi2.MuLV, MuLV-infected leukemia with Nf1 disruption by proviral insertion into Nf1. * indicates that the leukemias were produced previously by the laboratory of Dr Camy I. Brannan (deceased) through multiple rounds of backcrossing Nf1-null mice onto the BXH-2 strain of mice.12,29 PIS indicates proviral insertion site; and CIS, common proviral insertion site.
Strategy for collection of leukemia with intact or deficient Nf1 gene for this study. Nf1+/+. MuLV indicates MuLV-infected Nf1-intact leukemia; Nf1−/+.MuLV, MuLV-infected leukemia with engineered null Nf1 allele. Nf1Evi2.MuLV, MuLV-infected leukemia with Nf1 disruption by proviral insertion into Nf1. * indicates that the leukemias were produced previously by the laboratory of Dr Camy I. Brannan (deceased) through multiple rounds of backcrossing Nf1-null mice onto the BXH-2 strain of mice.12,29 PIS indicates proviral insertion site; and CIS, common proviral insertion site.
Mapping of these PISs using the Ensembl mouse genome database uncovered 12 CISs, including 9 novel CISs (group I CIS in Table 1) and 3 previously reported CISs (group II CIS in Table 1). For some PISs isolated only once in this screen, cross-reference to other PIS profiles deposited in the Retrovirus Tagged Cancer Gene Database (http://rtcgd.abcc.ncifcrf.gov/) resulted in the recognition of 13 CISs (group IV CIS in Table 1). Similarly, 17 CISs were identified where our single PISs hit known CISs (group V CIS in Table 1). Interestingly, many CISs mapped to chromosomal locations that are syntenic to sites of recurrent structural abnormalities in human malignancies (Table S2).
Candidate leukemia genes identified in this MuLV screen
| CIS group*/Gene hit† . | Annotation . | Tumor panel‡ . | ||
|---|---|---|---|---|
| Nf1+ . | Evi2 . | Nf1− . | ||
| I | ||||
| Edg3 | Endothelial differentiation, sphingolipid G protein–coupled receptor, 3 | 0 | 1 | 1 |
| ENS#1 | Unknown | 2 | 1 | 1 |
| FLJ20186 | Differentially expressed in FDCP 8 | 1 | 0 | 2 |
| Ing1 | Inhibitor of growth protein 1 | 1 | 1 | 1 |
| Etnk1 | Ethanolamine kinase 1 | 1 | 0 | 1 |
| II | ||||
| Bcl11a | B-cell CLL/lymphoma 11A | 0 | 3 | 2 |
| Spred2 | Sprouty protein with EVH-1 domain 2, related sequence | 0 | 1 | 4 |
| Pmaip1 | Phorbol-12-myristate-13-acetate–induced protein 1 | 1 | 1 | 0 |
| Jund1 | Jun proto-oncogene–related gene d1 | 1 | 0 | 1 |
| III | ||||
| Ahi1/Myb | WD domain and SH3 domain–containing gene | 6 | 4 | 4 |
| Hoxa7/a9 | Homeobox A7/A9 | 2 | 0 | 2 |
| Meis1 | Myeloid ecotropic viral integration site 1 | 3 | 0 | 2 |
| IV | ||||
| Usp21 | Ubiquitin-specific protease 21 | 1 | 0 | 0 |
| Enah | Enabled homolog | 1 | 0 | 0 |
| Tlr2 | Toll-like receptor 2 | 1 | 0 | 0 |
| Btbe3 | Basic transcription element–binding protein 3 | 0 | 1 | 0 |
| Sar1a | GTP-binding protein Sar1a | 1 | 0 | 0 |
| Fgd3 | Faciogenital dysplasia homolog 3 | 1 | 0 | 0 |
| Pilrb | Paired immunoglobulin-like type 2 receptor beta | 0 | 0 | 1 |
| Foxd1 | Forkhead box protein D1 | 0 | 1 | 0 |
| Gata2 | GATA-binding protein 2 | 1 | 0 | 0 |
| Olig2 | Oligodendrocyte transcription factor 2 | 1 | 0 | 0 |
| Wnt7a | Wingless-type MMTV integration site family, member 7A | 1 | 0 | 0 |
| Ly86 | Lymphocyte antigen 86 | 0 | 1 | 0 |
| ENS#2 | Unknown | 0 | 0 | 1 |
| V | ||||
| ENS#3 | Similar to eosinophil chemotactic cytokine | 0 | 1 | 0 |
| Nfkb1 | Nuclear factor NF-κ-B p105 subunit | 1 | 0 | 0 |
| Centg3 | Centaurin, gamma 3 | 1 | 0 | 0 |
| Dyrk1b | Dual-specificity tyrosine-(Y) phosphorylation-regulated kinase 1b | 1 | 0 | 0 |
| Tbx21 | T cell–specific T-box transcription factor T-bet | 0 | 1 | 0 |
| Lyl1 | Lymphoblastomic leukemia | 1 | 0 | 0 |
| Cbfa2t3h | Core-binding factor, runt domain, alpha subunit 2 | 1 | 0 | 0 |
| PHF5A | PHD finger protein 5A | 0 | 0 | 1 |
| Runx1 | Runt-related transcription factor 1 | 0 | 1 | 0 |
| Tcirg1 | T-cell, immune regulator 1 | 0 | 1 | 0 |
| Lin28 | RNA-binding protein LIN-28 | 0 | 0 | 1 |
| Emid2 | EMI domain–containing protein 2 | 0 | 0 | 1 |
| ATP5G2 | ATP synthase, H+ transporting, mitochondrial complex, subunit C2 | 0 | 1 | 0 |
| Ifngr1 | Interferon-γ receptor 1 | 0 | 0 | 1 |
| Atp5g2 | ATP synthase lipid-binding protein, mitochondrial precursor | 0 | 0 | 1 |
| Vegfb | Vascular endothelial growth factor B | 0 | 0 | 1 |
| Lmo2 | LIM domain only 2 | 1 | 0 | 0 |
| Akt1 | v-akt murine thymoma viral oncogene homolog 1 | 0 | 1 | 0 |
| CIS group*/Gene hit† . | Annotation . | Tumor panel‡ . | ||
|---|---|---|---|---|
| Nf1+ . | Evi2 . | Nf1− . | ||
| I | ||||
| Edg3 | Endothelial differentiation, sphingolipid G protein–coupled receptor, 3 | 0 | 1 | 1 |
| ENS#1 | Unknown | 2 | 1 | 1 |
| FLJ20186 | Differentially expressed in FDCP 8 | 1 | 0 | 2 |
| Ing1 | Inhibitor of growth protein 1 | 1 | 1 | 1 |
| Etnk1 | Ethanolamine kinase 1 | 1 | 0 | 1 |
| II | ||||
| Bcl11a | B-cell CLL/lymphoma 11A | 0 | 3 | 2 |
| Spred2 | Sprouty protein with EVH-1 domain 2, related sequence | 0 | 1 | 4 |
| Pmaip1 | Phorbol-12-myristate-13-acetate–induced protein 1 | 1 | 1 | 0 |
| Jund1 | Jun proto-oncogene–related gene d1 | 1 | 0 | 1 |
| III | ||||
| Ahi1/Myb | WD domain and SH3 domain–containing gene | 6 | 4 | 4 |
| Hoxa7/a9 | Homeobox A7/A9 | 2 | 0 | 2 |
| Meis1 | Myeloid ecotropic viral integration site 1 | 3 | 0 | 2 |
| IV | ||||
| Usp21 | Ubiquitin-specific protease 21 | 1 | 0 | 0 |
| Enah | Enabled homolog | 1 | 0 | 0 |
| Tlr2 | Toll-like receptor 2 | 1 | 0 | 0 |
| Btbe3 | Basic transcription element–binding protein 3 | 0 | 1 | 0 |
| Sar1a | GTP-binding protein Sar1a | 1 | 0 | 0 |
| Fgd3 | Faciogenital dysplasia homolog 3 | 1 | 0 | 0 |
| Pilrb | Paired immunoglobulin-like type 2 receptor beta | 0 | 0 | 1 |
| Foxd1 | Forkhead box protein D1 | 0 | 1 | 0 |
| Gata2 | GATA-binding protein 2 | 1 | 0 | 0 |
| Olig2 | Oligodendrocyte transcription factor 2 | 1 | 0 | 0 |
| Wnt7a | Wingless-type MMTV integration site family, member 7A | 1 | 0 | 0 |
| Ly86 | Lymphocyte antigen 86 | 0 | 1 | 0 |
| ENS#2 | Unknown | 0 | 0 | 1 |
| V | ||||
| ENS#3 | Similar to eosinophil chemotactic cytokine | 0 | 1 | 0 |
| Nfkb1 | Nuclear factor NF-κ-B p105 subunit | 1 | 0 | 0 |
| Centg3 | Centaurin, gamma 3 | 1 | 0 | 0 |
| Dyrk1b | Dual-specificity tyrosine-(Y) phosphorylation-regulated kinase 1b | 1 | 0 | 0 |
| Tbx21 | T cell–specific T-box transcription factor T-bet | 0 | 1 | 0 |
| Lyl1 | Lymphoblastomic leukemia | 1 | 0 | 0 |
| Cbfa2t3h | Core-binding factor, runt domain, alpha subunit 2 | 1 | 0 | 0 |
| PHF5A | PHD finger protein 5A | 0 | 0 | 1 |
| Runx1 | Runt-related transcription factor 1 | 0 | 1 | 0 |
| Tcirg1 | T-cell, immune regulator 1 | 0 | 1 | 0 |
| Lin28 | RNA-binding protein LIN-28 | 0 | 0 | 1 |
| Emid2 | EMI domain–containing protein 2 | 0 | 0 | 1 |
| ATP5G2 | ATP synthase, H+ transporting, mitochondrial complex, subunit C2 | 0 | 1 | 0 |
| Ifngr1 | Interferon-γ receptor 1 | 0 | 0 | 1 |
| Atp5g2 | ATP synthase lipid-binding protein, mitochondrial precursor | 0 | 0 | 1 |
| Vegfb | Vascular endothelial growth factor B | 0 | 0 | 1 |
| Lmo2 | LIM domain only 2 | 1 | 0 | 0 |
| Akt1 | v-akt murine thymoma viral oncogene homolog 1 | 0 | 1 | 0 |
CIS group indicates the way CISs were identified for the corresponding genes. CISs that are composed of at least 2 insertions positioned within 20-kb pairs isolated in this study, and no insertions (I), single insertions (II), or known CISs (III) have previously been reported. IV and V indicate single insertions isolated in this study for which known single insertions (IV) or CISs (V) can be found.
Gene hits indicate genes located nearby CISs, which were determined according to the annotation for the mouse genome (http://www.ensembl.org). ENS#1 indicates ENSMUSG00000019986; ENS#2, ENSMUSG00000030553; and ENS#3, ENSMUSG00000010135.
Tumor panel denotes the genotype of leukemia from which the corresponding proviral insertions were recovered. Nf1+ indicates tumors with intact Nf1 alleles; Evi2, tumors with Nf1 alleles disrupted by Evi2 insertion; and Nf1−, tumors with engineered null Nf1 allele. Values are the number of insertions isolated from each genotype of leukemia.
To explore the biologic significance of the 40 new and known CIS-associated genes (Table 1), the Ingenuity Pathway Analysis program (http://www.ingenuity.com/) was used to produce molecular networks. The NF-κB pathway was predicted with the highest likelihood, organizing 14 (or 39%) of the total genes found in our study. The second most probable pathway involved 9 candidate genes, including Bcl11a. These data suggest that dysregulation of the NF-κB pathway and Bcl11a-associated pathway may contribute to the development of AML.
Analysis of candidate leukemia genes
The Ahi1/Myb CIS represents the most frequent insertion site in our screen. Fourteen proviral insertions with either orientation were recovered immediately downstream to the Ahi-1 gene, also approximately 35 kb downstream to c-Myb (Figure S1), presumably enhancing transcription of Ahi-1 and c-Myb. This finding highlights the potential role of deregulation of Ahi-1 and c-Myb in leukemogenesis.
Bcl11a and Spred2 were hit as often as Meis1 and Hoxa7/a9, which have been strongly implicated in human myeloid leukemogenesis.30 Whereas we observed a similar pattern of insertions in Nf1-deficient and Nf1-intact leukemias at most CISs, the PISs targeting Bcl11a and Spred2 were all isolated from Nf1-deficient AMLs (Table 1). Five PISs with sense or antisense orientation were isolated from the first intron of Spred2, indicating that disruption of normal transcription of Spred2 could cooperate with Nf1 deficiency during formation of leukemia (Figure S2). Consistent with this possibility, SPRED2 are thought to be a negative regulator of RAS/ERK signal transduction,31 and expression of SPRED2 is decreased in some cancers.
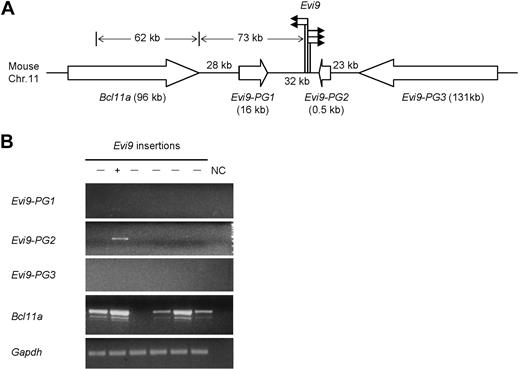
To further explore the putative association between Nf1 deficiency and Bcl11a-targeting insertions, which turn out to overlap with known Evi9 insertions, Southern blot analysis of 40 additional Nf1 intact BXH-2 AMLs with a Bcl11a-specific probe were performed, and we did not detect any evidence of proviral insertions (P < .02; Figure S5). These data suggest that Evi9 insertions are enriched in the Nf1-deficient versus Nf1-intact AML we studied, although they are probably not unique to Nf1 deficiency. In addition to Bcl11a, 3 other genes flanking Evi9 were predicted, tentatively designated as Evi9-PG1, Evi9-PG2, and Evi9-PG3, respectively (http://www.ensembl.org/) (Figure 2A). Strikingly, unlike the majority of reported CISs, our Bcl11a-targeting insertions contain 5 PISs located within a narrow chromosomal region of less than 1 kb, underlining a strong correlation between insertions at Evi9 with leukemogenesis.
Analysis of Bcl11a-targeting CIS. (A) The schematic representation of the CIS-targeting Bcl11a. The solid arrows indicate the relative position and orientation of isolated proviral insertions at Evi9; the open arrows indicate the relative position and transcriptional orientation of Bcl11a and 3 other predicted genes; bracketed numbers, size (in kb) of genes. (B) Expression in BXH-2 leukemia of Bcl11a and 3 other predicted genes nearby Evi9 locus (Evi9-PG1, Evi9-PG2, and Evi9-PG3) was detected by RT-PCR using gene-specific primers. The lanes marked “−” indicate AML samples with no Evi9 insertions; the lane marked “+” indicates 1 of the 5 AML samples with Evi9 insertions. Gapdh was used as a loading control. NC indicates water in place of cDNA as the template for PCR was set up as negative controls.
Analysis of Bcl11a-targeting CIS. (A) The schematic representation of the CIS-targeting Bcl11a. The solid arrows indicate the relative position and orientation of isolated proviral insertions at Evi9; the open arrows indicate the relative position and transcriptional orientation of Bcl11a and 3 other predicted genes; bracketed numbers, size (in kb) of genes. (B) Expression in BXH-2 leukemia of Bcl11a and 3 other predicted genes nearby Evi9 locus (Evi9-PG1, Evi9-PG2, and Evi9-PG3) was detected by RT-PCR using gene-specific primers. The lanes marked “−” indicate AML samples with no Evi9 insertions; the lane marked “+” indicates 1 of the 5 AML samples with Evi9 insertions. Gapdh was used as a loading control. NC indicates water in place of cDNA as the template for PCR was set up as negative controls.
To uncover the gene(s) that is the potential biologic target of proviral insertions at Evi9, we examined expression levels in BXH-2 leukemia of surrounding genes by RT-PCR using gene- specific primers. We could not detect any transcripts from Evi9-PG1 and Evi9-PG3 in these tumors (Figure 2B). Evi9-PG2 is expressed at a higher level in the Evi9 insertion-bearing leukemia than others. Further analysis showed that Evi9-PG2 was only 523 bp in size, harboring a single exon without a predictable coding frame. Thus, Evi9-PG2 could be a pseudogene, or possibly, a noncoding RNA gene. Bcl11a expression could be observed at different levels in 5 of 6 tumors. Sequencing confirmed the specificity of the upper RT-PCR bands shown in Figure 2B (data not shown). As expected, the highest expression level of Bcl11a was detected in the Evi9 insertion-bearing leukemia only for which RNA was available, suggesting that Bcl11a is up-regulated as a consequence of Evi9 insertions.
Expression of Bcl11a in Nf1-deficient bone marrow cells leads to leukemia
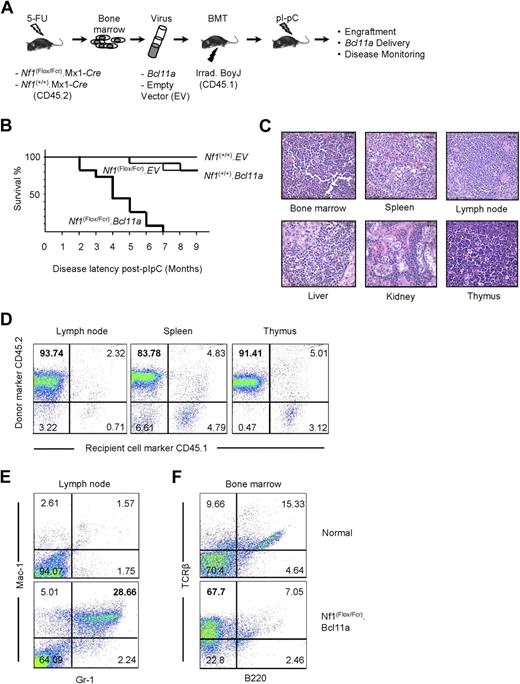
Our findings that (1) Evi9 is a frequent target by MuLV, (2) Evi9 occupies an exceptionally narrow window, (3) Evi9 is enriched in Nf1-deficient tumor panel, and (4) Bcl11a appears to be the primary target of Evi9 established Bcl11a as a strong candidate for cooperation with Nf1 inactivation. In addition, Bcl11a has been implicated in the development of human lymphocytic leukemia.14,15,30 Therefore, we decided to investigate the role of Bcl11a in leukemogenesis using the bone marrow transduction and transplantation assay (BMTT; Figure 3A).27 Donor bone marrow cells were collected from C57BL/6 mice generated via a cross between mice carrying a conditional mutant (floxed) allele of Nf1 and heterozygous Nf1-null (Nf1Fcr/+) mice also transgenic for a Cre transgene driven by the Mx-1 promoter.32 Bone marrow from wild-type Nf1 (Nf1+/+) C57BL/6 mice carrying the Mx1-Cre transgene were used as a control. Wild-type and Nf1 mutant bone marrow cells were transduced with either Bcl11a-expressing or empty vector VSV-G–pseudotyped retrovirus, followed by transplantation into lethally irradiated C57BL/6.BoyJ recipient mice carrying the CD45.1 allele of the cell-surface marker CD45. This allowed us to distinguish the origin of cells in transplant recipients. Therefore, 4 groups of mice were obtained, including mice receiving Nf1-sufficient bone marrow cells, which had been transduced with empty vector packaged retrovirus (Nf1(+/+).EV), Nf1-sufficient bone marrow cells transduced with Bcl11a-expressing retrovirus (Nf1(+/+).Bcl11a), Nf1-deficient bone mar-row cells transduced with empty vector packaged retrovirus (Nf1(Flox/Fcr).EV), and Nf1-engineered bone marrow cells transduced with Bcl11a-expressing retrovirus (Nf1(Flox/Fcr).Bcl11a). We administered polyI:polyC to all mice receiving these donor marrow cells to induce recombination-mediated deletion of the floxed allele by activating the Mx1-Cre transgene.32
Examination of oncogenic effect of Bcl11a overexpression using the BMTT assay in mice. (A) The BMTT scheme used in this study. (B) Survival analysis on the BMTT-recipient mice, which received Bcl11a- or empty vector (used as a negative control)–transduced Nf1-intact (“Nf1(+/+).EV” and “Nf1(+/+). Bcl11a”) or Nf1-deficient bone marrow cells (“Nf1(Flox/Fcr).EV” and “Nf1(Flox/Fcr).Bcl11a”). (C) Histopathologic examination of the Bcl11a-induced leukemia in the recipient mice. Hematoxylin and eosin staining. Representative images with magnification of 60-fold are shown here. Bars represent 25 μm. Slides were viewed with a Nikon Eclipse E600 bright light microscope (Nikon, Tokyo, Japan) using a Nikon Plan Fluor 40×/0.75 objective lens. Images were acquired using a Spot Insight digital camera (model 14.2, Color Mosaic; Diagnostic Instruments, Sterling Heights, MI), and were processed with Spotsoftware Advanced (version 4.6 for Windows; Diagnostic Instruments) and Adobe Photoshop version 7.0 software (Adobe Systems, San Jose, CA). (D) Determination of donor origin of the Bcl11a-induced leukemia developing in the BMTT-recipient mice. Antibodies staining for CD45.1 or CD45.2 cell-surface molecules were used. (E,F) Determination of cell-lineage origin of the Bcl11a leukemia developed in the BMTT-recipient mice. Antibodies against Mac-1, Gr-1, B220, or TCR-β were used to stain lymph nodes and bone marrows harvested from either healthy C57BL/6J mice or mice transplanted with Bcl11a-transduced Nf1-deficient bone marrow cells to collect evidence for acute myeloid leukemia (E) or T-cell acute lymphoid leukemia (F).
Examination of oncogenic effect of Bcl11a overexpression using the BMTT assay in mice. (A) The BMTT scheme used in this study. (B) Survival analysis on the BMTT-recipient mice, which received Bcl11a- or empty vector (used as a negative control)–transduced Nf1-intact (“Nf1(+/+).EV” and “Nf1(+/+). Bcl11a”) or Nf1-deficient bone marrow cells (“Nf1(Flox/Fcr).EV” and “Nf1(Flox/Fcr).Bcl11a”). (C) Histopathologic examination of the Bcl11a-induced leukemia in the recipient mice. Hematoxylin and eosin staining. Representative images with magnification of 60-fold are shown here. Bars represent 25 μm. Slides were viewed with a Nikon Eclipse E600 bright light microscope (Nikon, Tokyo, Japan) using a Nikon Plan Fluor 40×/0.75 objective lens. Images were acquired using a Spot Insight digital camera (model 14.2, Color Mosaic; Diagnostic Instruments, Sterling Heights, MI), and were processed with Spotsoftware Advanced (version 4.6 for Windows; Diagnostic Instruments) and Adobe Photoshop version 7.0 software (Adobe Systems, San Jose, CA). (D) Determination of donor origin of the Bcl11a-induced leukemia developing in the BMTT-recipient mice. Antibodies staining for CD45.1 or CD45.2 cell-surface molecules were used. (E,F) Determination of cell-lineage origin of the Bcl11a leukemia developed in the BMTT-recipient mice. Antibodies against Mac-1, Gr-1, B220, or TCR-β were used to stain lymph nodes and bone marrows harvested from either healthy C57BL/6J mice or mice transplanted with Bcl11a-transduced Nf1-deficient bone marrow cells to collect evidence for acute myeloid leukemia (E) or T-cell acute lymphoid leukemia (F).
Approximately 3 weeks after polyI:polyC treatment, engraftment of donor cells was examined by performing PCR on genomic DNA from peripheral blood cells collected retro-orbitally from recipient mice. Primers specific to the floxed allele of Nf1 were used (Figure S3A). The “WT” PCR band can be amplified from peripheral blood cells with all 4 genotypes, whereas the floxed Nf1 allele–specific (“Flox”) and recombined Nf1 allele–specific (“Del”) products can only be seen in peripheral blood cells derived from the Nf1(Flox/Fcr).EV or Nf1(Flox/Fcr).Bcl11a group of mice, indicating not only engraftment of donor bone marrow but also the occurrence of recombination of the floxed Nf1 allele after polyI:polyC induction. The presence of residual floxed allele may reflect incomplete Nf1 deletion mediated by polyI:polyC-induced Cre expression. Furthermore, with Fcr-null allele-specific primers, the “Fcr” band can be observed in peripheral blood cells from mice receiving Nf1(Flox/Fcr).Cre bone marrow, but not from mice receiving Nf1(+/+).Cre cells (Figure S3), which confirms both engraftment of donor cells and the expected genotype of bone marrow cells received. Importantly, we were able to detect the Bcl11a-expressing retroviral vector that was used for transduction in blood cells from the Nf1(Flox/Fcr).Bcl11a group of mice (Figure S3C), demonstrating successful retroviral delivery of Bcl11a into recipient mice. These results suggest that bone marrow engraftment, recombination-mediated deletion of Nf1, and Bcl11a delivery into recipient mice had been achieved.
By 9 months after polyI:polyC induction, none of 10 mice of the Nf1(+/+).EV group had become sick (Figure 3B). Two mice each had died in the Nf1(+/+).Bcl11a (n = 11) and Nf1(Flox/Fcr).EV (n = 12) cohorts (Figure 3B). By contrast, all 11 recipients that received Nf1(Flox/Fcr) cells transduced with the Bcl11a virus became moribund by 7 months after polyI:polyC treatment, with a median latency of 107.8 days (Figure 3B). A similar trend in latency of disease was observed in an independent BMTT assay (data not shown). Most sick Nf1(Flox/Fcr).Bcl11a mice had enlarged lymph nodes, liver, and spleen (Table S4A). Visible metastases to the liver and kidney were readily observed. Enlarged thymi were seen in 2 mice without apparent abnormalities in other organs (Table S4A). Most mice exhibited high white blood cell counts (Table S4B). Generally, there was a decrease in the levels of hemoglobin and in the number of red blood cells and platelets (Table S4B). Pathologic analysis showed an increase in the number of immature myeloid or lymphoid cells in peripheral blood and bone marrow of Nf1(Flox/Fcr).Bcl11a mice. Spleen touch preps also displayed an excessive infiltration of blast-like cells. Significantly, neoplastic blast cells were found in multiple tissues, including bone marrow, spleen, lymph node, liver, kidney, and thymus (Figure 3C), indicating that normal tissue structures are effaced and replaced by infiltrating tumor cells. In addition, a 3.7-kb band can be detected in the enlarged lymph nodes and thymi by Southern blotting assay using a construct-specific probe (Figure S4), indicating the presence of the Bcl11a-expressing construct in the affected tissues. Furthermore, tumor cells isolated from 3 Nf1(Flox/Fcr).Bcl11a mice were intravenously injected into 9 syngeneic C57BL/6J mice at 106 cells per mouse, 3 mice for each tumor. The recipient mice became moribund within 6 to 10 weeks after inoculation of tumor cells, with an apparent increase in white blood cell counts similar to what has been observed in Nf1(Flox/Fcr).Bcl11a mice, which demonstrates the transplantability of the disease developed in the BMTT assay. Together, these data show that transplantation of Nf1-deficient bone marrow cells overexpressing Bcl11a results in aggressive hematologic malignancies in recipient mice.
Bcl11a-overexpressing, Nf1-deficient leukemia is of myeloid or lymphoid lineage and derived from transduced donor cells
The origin of the tumor cells was determined by immunostaining assay using antibodies specific for CD45.1 or CD45.2. The majority of cells isolated from enlarged lymph node, spleen, and thymus stained positive for CD45.2 (Figure 3D), indicating that these tumor cells are primarily derived from transplanted donor bone marrow. The cell lineage of origin of the leukemia was defined by immunophenotyping with antibodies specific for myeloid or lymphoid lineage hematopoietic cells. Antibodies against Mac-1, Gr-1, B220, or TCR-β were used to profile lymph node and bone marrow cells harvested from either healthy C57BL/6J mice or mice transplanted with Bcl11a-transduced Nf1-deficient bone marrow cells. Although only scarce amounts of Mac-1– and Gr-1–positive cells can be detected in normal lymph node, the number of these cells was increased 11.33-fold plus or minus 4.86-fold in the affected lymph nodes from 5 Nf1(Flox/Fcr).Bcl11a mice (Figure 3E), indicating a myeloid lineage of these cells. Similarly, the bone marrow collected from 4 Nf1(Flox/Fcr).Bcl11a mice displayed 4.60-fold plus or minus 1.82-fold higher cells staining positive for TCR-β than that harvested from normal mice (Figure 3F), suggesting that these mice have developed T-cell leukemia. Therefore, we conclude that Bcl11a-transduced Nf1-deficient bone marrow cells cause leukemia in recipient mice, which is derived from transduced donor cells and is of either myeloid or lymphoid lineage.
Bcl11a down-regulates p21Cip1 and promotes Nf1-deficient cell growth
To investigate the mechanism underlying the cooperation between Bcl11a and Nf1 deficiency in leukemogenesis, myeloid cell cultures with Bcl11a overexpression and/or Nf1 ablation were examined for their growth properties and differentiation potential. Retroviral vectors carrying a His-tagged Bcl11a and an EGFP marker, or EGFP alone, were used to transduce Nf1-intact or Nf1-null Myb-immortalized myeloblast cell line hematopoietic cells12 (Figure 4A). EGFP-positive cells were then sorted to obtain 4 stably transduced populations: Nf1-intact empty vector-transduced cells (Nf1+/+.vector), Nf1-intact Bcl11a-transduced cells (Nf1+/+.Bcl11a), Nf1-null empty vector-transduced cells (Nf1−/−.vector), and Nf1-null Bcl11a-transduced cells (Nf1−/−.Bcl11a). To test functionality of the Bcl11a-carrying construct, 293T cells were transiently transfected and subjected to Western blotting assay using anti-His antibody. Abundant Bcl11a protein was detected approximately at the position of 170 kDa in Bcl11a-transfected cells, but not in vector-transfected cells, indicating specific expression of Bcl11a from the retroviral construct (Figure 4B).
Bcl11a promotes expansion of an Nf1-deficient hematopoietic cell line and p21Cip1 down-regulation. (A) Scheme for generating Nf1(+/+) or Nf1(−/−) cell lines stably transduced with Bcl11a-expressing retroviral construct (Nf1+/+.Bcl11a and Nf1−/−.Bcl11a) or with empty vector (Nf1+/+.vector and Nf1−/−.vector) as negative controls. His6 indicates the protein tag of 6 repeats of histidine. (B) Verification of the Bcl11a-expressing vector by transient transfection of 293T cells followed by Western blotting assay. Anti-His antibody was used. The double arrows indicate the signal for Bcl11a. The single arrow indicates a nonspecific (n.s.) signal. (C) Cumulative growth of the stable Bcl11a- or vector-transduced Nf1(+/+) or Nf1(−/−) cells. (D) The abundance of cell-cycle regulatory proteins in stable Bcl11a or vector transductants. (E) p21Cip1 protein level was determined in stable Bcl11a or vector transductants after doxorubicin treatment (Doxo) for 16 hours. Representative results from 3 independent experiments were shown here.
Bcl11a promotes expansion of an Nf1-deficient hematopoietic cell line and p21Cip1 down-regulation. (A) Scheme for generating Nf1(+/+) or Nf1(−/−) cell lines stably transduced with Bcl11a-expressing retroviral construct (Nf1+/+.Bcl11a and Nf1−/−.Bcl11a) or with empty vector (Nf1+/+.vector and Nf1−/−.vector) as negative controls. His6 indicates the protein tag of 6 repeats of histidine. (B) Verification of the Bcl11a-expressing vector by transient transfection of 293T cells followed by Western blotting assay. Anti-His antibody was used. The double arrows indicate the signal for Bcl11a. The single arrow indicates a nonspecific (n.s.) signal. (C) Cumulative growth of the stable Bcl11a- or vector-transduced Nf1(+/+) or Nf1(−/−) cells. (D) The abundance of cell-cycle regulatory proteins in stable Bcl11a or vector transductants. (E) p21Cip1 protein level was determined in stable Bcl11a or vector transductants after doxorubicin treatment (Doxo) for 16 hours. Representative results from 3 independent experiments were shown here.
After culture for 24 hours, the cumulative cell number of Nf1−/−.Bcl11a cells was only slightly higher than those of 3 other engineered cell lines (Figure 4C). However, this number increased to 1.5-fold those of other cell lines at 48 hours of culture, indicating that Nf1−/−.Bcl11a cells grew significantly faster (P < .05) than control cells (Figure 4C). The enhanced growth potential of Nf1−/−.Bcl11a cells was confirmed by MTS (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt) assay (data not shown). This result suggests that overexpression of Bcl11a can confer a growth advantage to Nf1-deficient cells.
To examine the molecular basis of Bcl11a-stimulated proliferation advantage, we examined the abundance of cell-cycle regulatory proteins in these cell lines. There were no evident changes between Nf1 status and/or Bcl11a transduction in the basal level of 11 proteins examined, including Cdk4, cyclin A, cyclin B, cyclin D1, cyclin D2, cyclin D3, p27, p21Cip1, PCNA, p19Skp1, p36MAT1, Rb, and Erk (Figure 5D). Interestingly, when challenged for 16 hours with 250 ng/mL doxorubicin, a DNA intercalator and damaging agent, both Nf1−/− (Figure 5E lane 3) and Nf1+/+ (lane 7) cells displayed elevated p21Cip1 levels. However, a marked decrease in the induced level of p21Cip1 was observed in Bcl11a-transduced Nf1−/− or Nf1+/+ cells (lane 4 and lane 8). These results suggest that Bcl11a over-expression suppresses elevation of p21Cip1 level induced by DNA damage.
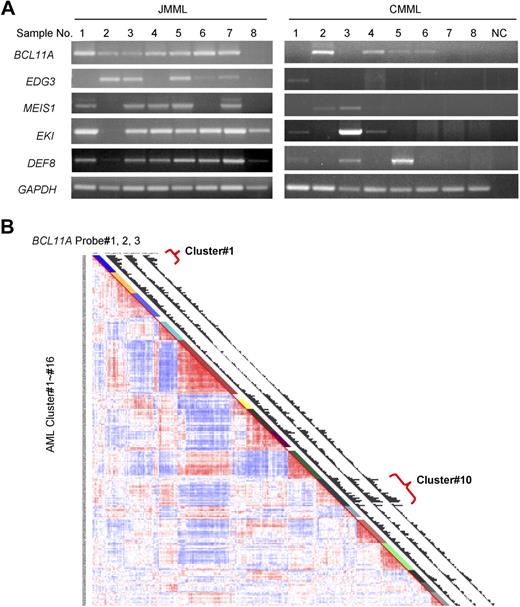
BCL11A expression in human leukemia. (A) BCL11A expression in human chronic myelomonocytic leukemia and juvenile myelomonocytic leukemia was detected by RT-PCR. NC indicates water in place of cDNA as template for PCR. GAPDH was used as a loading control. The JMML samples are found to carry mutations in KRAS (no. 1) or PTPN11 (nos. 2-7), or wild type for both genes (no. 8). (B) Gene expression profiling in a large cohort of human acute myelogenous leukemia. Results using a set of 3 probes for BCL11A are displayed. Probe nos. 1, 2, and 3 indicate probe sets, 210347_s_at, 219497_s_at, and 219498_s_at, respectively. Clusters of AML showing increased level of BCL11A are noted (clusters 1 and 10).
BCL11A expression in human leukemia. (A) BCL11A expression in human chronic myelomonocytic leukemia and juvenile myelomonocytic leukemia was detected by RT-PCR. NC indicates water in place of cDNA as template for PCR. GAPDH was used as a loading control. The JMML samples are found to carry mutations in KRAS (no. 1) or PTPN11 (nos. 2-7), or wild type for both genes (no. 8). (B) Gene expression profiling in a large cohort of human acute myelogenous leukemia. Results using a set of 3 probes for BCL11A are displayed. Probe nos. 1, 2, and 3 indicate probe sets, 210347_s_at, 219497_s_at, and 219498_s_at, respectively. Clusters of AML showing increased level of BCL11A are noted (clusters 1 and 10).
Expression of BCL11A in human leukemia
To determine whether BCL11A could possibly be contributing to human myeloid leukemia, we examined its expression in chronic and acute forms of the disease. BCL11A expression levels in JMML and chronic myelomonocytic leukemia were examined by RT-PCR. As can be seen in Figure 6A, BCL11A expression is detected in most patient samples from both types of diseases, although lower than detectable in a few samples, which is consistent with the possibility that BCL11A expression could make a positive contribution to these diseases. Interestingly, several of the other candidate genes obtained from this insertional mutagenesis screen, including EDG3, MEIS1, EKI, and DEF8, were expressed in these diseases (Figure 5A).
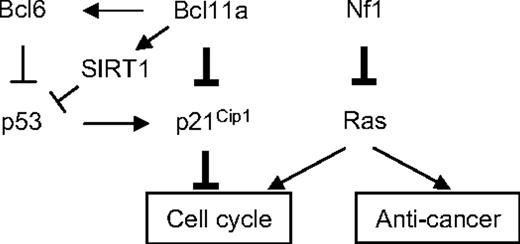
The model for cooperative oncogenesis between Bcl11a and Nf1 deficiency. The thickened lines indicate the major linkage of Bcl11a-Nf1 loss cooperation to leukemogenesis that was identified in this study. The arrows indicate a stimulatory relationship; perpendicular lines, suppression. One major function of Nf1 is to negatively regulate Ras activity, and Nf1 deficiency leads to hyperactivation of Ras.66,67 Ras activation alone can trigger cell growth arrest and senescence.49 However, this effect may be attenuated by an increase in Bcl11a signal, which results in suppression of p21Cip1 induction and, consequently, release of its control over cell- cycle progression, as discovered in this study. Bcl11a may directly repress Cdkn1a transcription or indirectly through p53 mediated by BCL636,68 or SIRT1.64,65,69
The model for cooperative oncogenesis between Bcl11a and Nf1 deficiency. The thickened lines indicate the major linkage of Bcl11a-Nf1 loss cooperation to leukemogenesis that was identified in this study. The arrows indicate a stimulatory relationship; perpendicular lines, suppression. One major function of Nf1 is to negatively regulate Ras activity, and Nf1 deficiency leads to hyperactivation of Ras.66,67 Ras activation alone can trigger cell growth arrest and senescence.49 However, this effect may be attenuated by an increase in Bcl11a signal, which results in suppression of p21Cip1 induction and, consequently, release of its control over cell- cycle progression, as discovered in this study. Bcl11a may directly repress Cdkn1a transcription or indirectly through p53 mediated by BCL636,68 or SIRT1.64,65,69
A large-scale examination of gene expression in a cohort of 285 patients with AML has been performed using Affymetrix-based microarray.33 Based on the overall gene expression profile, these patients were grouped into 16 clusters using an unsupervised cluster approach. Of note, 3 sets of BCL11A-specific probes all generated comparable hybridization signals across these samples (Figure 5B). This microarray analysis revealed that BCL11A displayed variant abundance of transcripts in AML (Figure 5B). More interestingly, patients with high expression levels of BCL11A were mostly segregated into 2 of 16 clusters: cluster 1 and cluster 10 (Figure 6B), whereas other groups and normal bone marrow cells showed low expression of BCL11A. Cluster 10 is associated with a poor outcome, and with monosomy 7, which is also common in JMML and in cases of JMML that transform to AML. The association of BCL11A level with poor outcome of patients in cluster 10 indicates that BCL11A could be up-regulated in this subset of AML and might contribute to these diseases. However, patients in cluster 1 do not show a poor prognosis33 ; even many of patients in this group also carry MLL translocations. Genetic complexity of AML and gene expression–based classification of patients could obscure the effects of certain genetic lesions on the outcome of patients. Other factors, such as monosomy 7 seen in cluster 10, may also contribute to the worse outcome of patients.
Discussion
In this study, we have identified a dozen novel CISs, some of which probably cooperate with Nf1 deficiency in the development of leukemia. We have, for the first time, demonstrated that Bcl11a acts as an oncogene and causes leukemia in the absence of Nf1 in mice, perhaps through suppression of p21Cip1 induction and thereby promotion of cell growth. Our results also suggested that expression of BCL11A may contribute to leukemogenesis in certain groups of AML patients.
An association of BCL11A with hematologic malignancies has been indicated in the literature. Abnormalities involving BCL11A have been detected in a variety of B-cell malignancies in humans.14,15,34 The chromosomal region containing the BCL11A gene is also recurrently rearranged in other human tumors.35 However, it has been controversial whether BCL11A is the real affected gene responsible for the diseases.14,34 Furthermore, in 2 of 209 BXH-2 myeloid leukemia, proviral insertion sites were found to be located within the first intron of Bcl11a and resulted in increased expression, instead of disruption, of Bcl11a.30 Bcl11a has also been reported to transform NIH 3T3 in foci formation assay.36 These data support a role of Bcl11a as a dominantly acting proto-oncogene. On the other hand, by generating Bcl11a knockout mice, Liu et al suggested that Bcl11a is critical for B-cell and alpha/beta T-cell development, but not for macrophage-granulocyte and erythroid lineages.13 Paradoxically, transplantation of Bcl11a knockout mice fetal liver cells resulted in T-cell leukemia originated from recipient mice, suggesting that Bcl11a may be a non–cell autonomous T-cell tumor suppressor gene.13 Moreover, another member of Bcl11 family, Bcl11b, has been suggested to be a tumor suppressor gene, acting via an increase in resistance to DNA damage.37,38 Thus, it has been left unclear whether Bcl11a functions as an oncogene or tumor suppressor gene, under what conditions, and in what manner Bcl11a can cause tumor.
We found that proviral insertions located downstream of Bcl11a concentrated in an Nf1-deficient AML panel. Consequently, expression of Bcl11a was up-regulated, presumably through long-range activation effect of retroviral insertion.39 Given that no other affected genes around these insertions were identified, these data strongly indicate Bcl11a as an oncogene in Nf1-deficient myeloid leukemia. Indeed, our findings of the ability of Bcl11a overexpression in Nf1-deficient bone marrow cells to induce mouse leukemia, combined with our observation of BCL11A dysregulation in certain subsets of human AML, have established that Bcl11a functions as an oncogene in both myeloid and lymphoid lineages, in a cell autonomous manner, and in the absence of Nf1 function. These results indicate that Bcll1a may have a multilineage effect. The tumorigenicity of an oncogene in multiple cell types has been observed previously.40-42 It is possible, however, that the murine stem cell virus–long terminal repeat used to express Bcl11a in our study could create a bias toward T-cell malignancy.43,44 Transgenic models using different promoters to express Bcl11a would provide a different approach to look at the lineage tropism of Bcl11a oncogenicity. Our study also suggests whether Bcl11a is implicated in human neoplasias other than lymphoid diseases will need to be examined.
There are indications that secondary genetic events are needed for the development of leukemia initiated by Nf1 deficiency. Although the cooperation of Nf1 deficiency with other genetic alterations has been proposed or attempted,29,45,46 convincing evidence is still lacking. Our results suggest the existence of a strong synergetic effect of Bcl11a up-regulation and Nf1 deficiency. This cooperation has not been revealed by any other kind of PIS studies, including a recent extensive computational analysis of PISs deposited in the Retrovirus Tagged Cancer Gene Database.47 Recent studies suggested that defects in DNA repair genes influence NF1 tumor progression in mice and humans, presumably by inactivating normal allele of NF1.46,48 We found that formation of leukemia with Nf1 biallelic inactivation and Bcl11a overexpression was markedly accelerated, suggesting that other genetic lesions may contribute more significantly to the observed progression of NF1 tumor than loss of heterozygosity of NF1. In addition, myeloproliferative disorder (MPD) can progress to AML; however, there are no solid experimental data explaining the progression of MPD into AML in the context of JMML or other NF1-associated myeloid diseases. It has been shown that mice with Nf1 deficiency in hematopoietic cells develop an MPD-like disease.11,12,32 Based on these studies, our genetic study has provided a mouse model, clearly demonstrating that Nf1-associated MPD, on acquisition of Bcl11a dysregulation, is capable of progression into AML.
Combined with previously published work, we propose a model of our current understanding of the Bcl11a/Nf1 deficiency cooperation. Dysregulated Ras signaling has been the only known major downstream effect of Nf1 deficiency in cancer so far. Ras activation alone in primary fibroblasts could trigger p53/p16-dependent growth arrest and senescence.49 These cell-defensive responses, however, during the development of leukemia could be overcome through the acquired enhancement of Bcl11a expression, which could result in suppression of p21Cip1 induction. The role of p21Cip1 in multiple aspects of hematopoiesis has been documented.50-59 p21Cip1 induction is correlated with reduced colony formation and cell-cycle arrest of hematopoietic progenitor cells or leukemic cells.50-53 p21Cip1 loss cooperates with AML1-ETO in leukemogenesis and accelerates the onset of mouse mammary tumor virus H-ras–induced tumors.59,60 Interestingly, N-ras oncogene up-regulates p21Cip1 in hematopoietic cells, but Ras does not induce growth arrest in p21Cip1-null fibroblasts, and p21Cip1 loss can rescue Ras-driven anchorage independence and tumorigenesis.56,61,62 The p21Cip1 suppression by Bcl11a could be as a result of direct repression of Cdkn1a transcription by Bcl11a. We have noticed that the Cdkn1a gene has 2 potential Bcl11a-binding sites in its promoter region, which supports the assumption of direct mode of suppression. An experimental validation of Cdkn1a as a direct transcriptional target of Bcl11a is warranted. The suppressive effect of Bcl11a could also be exerted indirectly through other intermediate proteins.36,63-65 Thus, it is probable that Nf1 deficiency in hematopoietic cells may cause the similar effects as Ras activation, which requires cooperation with other genetic abnormalities, such as Bcl11a overexpression, to develop further into aggressive forms of leukemia.
We expect that the establishment of Bcl11a's oncogenic role will stimulate further understanding of its biologic function and molecular mechanisms in disorders and eventually allow for the development of therapeutic strategies in the treatment of Bcl11a-relevant diseases.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Ms Jennifer Jeske-Geurts and Ms Raha Allaei for breeding and genotyping mice, Ms Miechaleen Diers for mouse tail vein injection, Dr Sheri Kuslak for cloning assistance, Dr Xianghua Luo from the Biostatistics Core of the University of Minnesota Cancer Center for help with statistical analyses, the Histopathology Core of the University of Minnesota Cancer Center for tissue sectioning and pathologic examination, the Flow Cytometry Core of the University of Minnesota Cancer Center for assistance in immunophenotyping murine leukemia, Dr Jessica Walrath and Dr Susan Blaydes Ingersoll for providing genomic DNA derived from Nf1-deficient BXH-2 mice, and Dr Luis Parada for providing Nf1 conditional knockout mice and Leukemia & Lymphoma Society of America (LLS 7019-04).
This work was supported by the National Institutes of Health (grant CA84221) and Leukemia & Lymphoma Society of America (LLS 7019-04).
National Institutes of Health
Authorship
Contribution: B.Y. designed, performed, collected, analyzed, and interpreted experimental data, and wrote the manuscript; R.D. and P.J.V. performed research and collected data; M.R.W. and M.L.L. contributed vital new reagents; K.M.S. oversaw the direction of all experimental studies and edited the manuscript; and D.A.L. oversaw the direction of all experimental studies, designed research, evaluated the data, and edited the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: David A. Largaespada, University of Minnesota, 6-160 Jackson Hall, 321 Church Street SE, Minneapolis, MN 55455; e-mail: larga002@tc.umn.edu.
References
Supplemental data
The column "RTCGD CIS" indicates CISs that can be found in the Retrovirus Tagged Cancer Gene Database (http://rtcgd.abcc.ncifcrf.gov/) for the corresponding genes identified in this screen. The column "Human syngeneic pos." describes where the CIS-target genes are located in human cytogenetic positions. The column "Human chromosomal abnormality" denotes those human diseases from which the balanced chromosomal abnormailities are recurrently detected at the corresponding cytogenetic positions according to the Mitelman Database of Chromosome Aberrations in Cancer (http://cgap.nci.nih.gov/Chromosomes/RecurrentAberrations). AML: acute myeloblastic leukemia; ALL: acute lymphoblastic leukemia; CML: chronic myeloid leukemia; CLL: chronic lymphoblastic leukemia.