Abstract
RNA-binding motif protein 15 (RBM15) is involved in the RBM15-megakaryoblastic leukemia 1 fusion in acute megakaryoblastic leukemia. Although Rbm15 has been reported to be required for B-cell differentiation and to inhibit myeloid and megakaryocytic expansion, it is not clear what the normal functions of Rbm15 are in the regulation of hematopoietic stem cell (HSC) and megakaryocyte development. In this study, we report that Rbm15 may function in part through regulation of expression of the proto-oncogene c-Myc. Similar to c-Myc knockout (c-Myc-KO) mice, long-term (LT) HSCs are significantly increased in Rbm15-KO mice due to an apparent LT-HSC to short-term HSC differentiation defect associated with abnormal HSC-niche interactions caused by increased N-cadherin and β1 integrin expression on mutant HSCs. Both serial transplantation and competitive reconstitution capabilities of Rbm15-KO LT-HSCs are greatly compromised. Rbm15-KO and c-Myc-KO mice also share related abnormalities in megakaryocyte development, with mutant progenitors producing increased, abnormally small low-ploidy megakaryocytes. Consistent with a possible functional interplay between Rbm15 and c-Myc, the megakaryocyte increase in Rbm15-KO mice could be partially reversed by ectopic c-Myc. Thus, Rbm15 appears to be required for normal HSC-niche interactions, for the ability of HSCs to contribute normally to adult hematopoiesis, and for normal megakaryocyte development; these effects of Rbm15 on hematopoiesis may be mediated at least in part by c-Myc.
Introduction
RNA-binding motif protein 15 (RBM15) and its fusion partner megakaryoblastic leukemia 1 (MKL1) were cloned from the leukemic blasts of childhood acute megakaryoblastic leukemia (AMkL) patients with t(1;22)(p13;q13).1,2 AMkL is a subtype of acute myeloid leukemia (AML-M7, according to the French-American-British classification system), and AML-M7 with t(1;22) occurs at a very early stage in life, the median age of onset being approximately 4 months.3 The prognosis for these patients is typically quite poor, with a median survival time of only 8 months.3 The t(1;22) is often the sole abnormality found in younger patients, whereas in older patients (> 6 months of age), additional karyotypic abnormalities can be found,3 suggesting that the RBM15-MKL1 (also known as OTT1-MAL) fusion protein generated by t(1;22) plays a crucial role in the pathogenesis of this disease.1
RBM15 is a member of the spen family, a group of proteins with homology to the Drosophila split ends (spen) protein. Rbm15 possesses the motif structure typical of spen family proteins, including 3 RNA-recognition motif domains at its N terminus and a spen paralog and ortholog C-terminal domain at its C terminus.4 Spen family members and spenlike proteins have been shown to be involved in a variety of important signaling cascades, including mitogen-activated protein kinase,5 Wnt,6,7 Notch,8 cyclin E,9 Hox,10 and epidermal growth factor receptor11 pathways. Multilineage hematopoietic abnormalities have been described recently in a Rbm15 knockout (Rbm15-KO) mouse model, including a B-cell differentiation block, myeloid and megakaryocytic expansion in the spleen and bone marrow (BM), and an increase in the Lin−Sca1+c-Kit+ (LSK) population.12 However, the role of Rbm15 in the regulation of hematopoietic stem cell (HSC) function has not been fully characterized, and the molecular mechanisms responsible for the deregulation of HSC and megakaryocyte development in the absence of Rbm15 remain to be elucidated.
c-Myc is a proto-oncogene that has been studied extensively in many tissue types. Nonetheless, the role of c-Myc in the regulation of adult mouse HSCs has been described only recently due to limitations in the analysis of mice lacking the gene because of early embryonic lethality; however, using an inducible Cre-LoxP system, c-Myc was conditionally deleted in the adult hematopoietic system and new, unexpected roles for gene were discovered, roles involving more than just the enhancement of hematopoietic progenitor cell proliferation.13,14 In these studies, Wilson and coworkers found that the increased long-term (LT) HSCs in c-Myc–deficient BM were caused not by alterations in HSC proliferation or survival but rather by an accumulation of LT-HSCs associated with a differentiation block caused by increased HSC-niche adhesion.13,14
In this study, we report that Rbm15 has an important role in regulating HSCs and megakaryocyte development, which may occur partly through its regulation of c-Myc expression. Rbm15-KO LT-HSCs have a differentiation block during the transition to short-term (ST) HSCs, presumably due to increased cell-niche interactions. These findings are highly reminiscent of those observed for the HSCs in c-Myc-KO mice. c-Myc expression is down-regulated in Rbm15-KO LSKs, and can be up-regulated by ectopic RBM15 overexpression in wild-type (WT) HSCs/progenitors. In addition, we found that Rbm15- and c-Myc-KO mice showed similar defects in megakaryocyte development, and the increased megakaryocyte production by Rbm15-mutant megakaryocytic progenitors (Mk-Ps) could be partially reversed by ectopic c-Myc expression, suggesting a possible functional interplay between Rbm15 and c-Myc in the regulation of both HSC and megakaryocyte development.
Methods
Mice
To generate Rbm15flox/flox mice, we constructed a targeting vector in which the entire Rbm15 exon 1 was flanked by 2 loxP sites (supplemental Figure 1, available on the Blood website; see the Supplemental Materials link at the top of the online article). The construct was introduced by homologous recombination into 129SvJ embryonic stem cells and the targeted embryonic stem cells used to produce mouse chimeras. The Rbm15flox/flox mice were subsequently backcrossed and are maintained on a pure C57BL/6 background. To delete Rbm15 conditionally in the hematopoietic system, Rbm15flox/flox mice were crossed with Mx1-Cre transgenic mice (The Jackson Laboratory). By proper mating, we were able to obtain Mx1-Cre–positive Rbm15flox/flox (Rbm15-KO hereafter), Mx1-Cre–positive Rbm15flox/+ (Rbm15-hetero), and Mx1-Cre–negative Rbm15flox/flox or Mx1-Cre–negative Rbm15flox/+ (Rbm15-WT hereafter) mice in the same litter. The mice were genotyped by polymerase chain reaction (PCR) and Southern blot analysis using the probes indicated in supplemental Figure 1B. To induce Cre expression by the Mx1-Cre transgene, both Mx1-Cre–positive Rbm15flox/flox mice and their Mx1-Cre–negative littermate controls were injected i.p. with 250 μg polyinosinic-polycytiaylic acid (Sigma-Aldrich) 5 times at 2-day intervals starting at 3 weeks of age. The generation and experimental use of the genetically engineered mice were approved by the Animal Care and Use Committee of St Jude Children's Research Hospital.
BM preparation, flow cytometric analysis, and culture
The isolation and preparation of BM and spleen cells have been described previously.15 Red blood cells were lysed using red blood cell lysis buffer (Sigma-Aldrich), and mononuclear cells were stained with fluorescein isothiocyanate (FITC)–lineage (Lin+) markers (CD8, CD3, B220, CD19, Gr1, and Ter119), phycoerythrin (PE)-Sca1, allophycocyanin (APC)-c-Kit, and biotin-CD135 (Flk2), followed by streptavidin-peridinin chlorophyll protein for HSC analysis on a BD Calibur Cytometer. For 5-color flow analysis on a BD LSR II, FITC-CD45.2, PE-Cy7-Lin+ markers, PE-Sca1, APC-c-Kit, PE-Cy5-CD135 were used. PE-Cy7-Sca1, PE-labeled CD49d, CD49e, CD49f, CD184, CD18, CD11a, CXC chemokine receptor (CXCR)4, and biotin-CD29 (β1 integrin), followed by streptavidin-PE, FITC-Lin+, and APC-c-Kit were used for adhesion molecule staining. All of the antibodies listed above were sourced from eBiosciences and BD Biosciences. The N-cadherin antibody was purchased from IBL-America, and FITC goat anti–rabbit IgG (Jackson ImmunoResearch Laboratories) secondary antibody was used for flow analysis. Fluorescence-activated cell sorting (FACS) was performed on BD Vantage DiVa or Aria High-Speed Cell Sorters. Sorted LT-HSCs were cultured in StemSpan serum-free expansion medium (StemCell Technologies) supplemented with 10 ng/mL recombinant murine (rm) interleukin (IL)–3 (R&D Systems), 50 ng/mL rmIL-6 (StemCell Technologies), and 100 ng/mL stem cell factor (rmSCF; R&D Systems) to differentiate cells. Apoptosis of HSCs was analyzed using the Annexin V:FITC Apoptosis Detection Kit I (BD Biosciences).
Quantitative and semiquantitative reverse transcription–PCR
RNA from FACS-sorted cell populations from the BM of either Rbm15-KO or -WT control mice was extracted using the RNeasy micro kit (QIAGEN). cDNA was generated using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). TaqMan real-time PCR was performed on an Applied Biosystems 7900HT real-time PCR system using a standard program. TaqMan primer/probe sets are listed in supplemental Table 2.
BM transplantation
BM cells were collected from 8- to 10-week-old Rbm15-KO or -WT littermate control mice (CD45.2+) and transplanted by intravenous injection with or without competitor marrow (B6.SJL; 002014, The Jackson Laboratory; CD45.1+) at the ratios indicated into B6.SJL recipients that had received 1100 rad.
In vivo 5-bromo-2′-deoxyuridine incorporation
Mice received an intraperitoneal injection of 2 mg of 5-bromo-2′-deoxyuridine (BrdU; BD Biosciences) and were analyzed 12 hours thereafter. BM cells were isolated and stained with PE-Cy7-labeled Lin+ mixture, c-Kit APC, and Sca1 PE antibodies. The analysis of BrdU incorporation was performed using the FITC BrdU flow kit (BD Biosciences) on a 5-color flow cytometer.
In vitro megakaryocyte culture and colony-forming assays
Lin− cells separated from total mouse BM using the EasySep Mouse Hematopoietic Progenitor Cell Enrichment Kit (StemCell Technologies) were cultured for 3 days in a serum-free expansion media containing rmSCF, rmIL-3, and 1 U/mL erythropoietin with Nutridoma serum-free media supplement (Roche) in RPMI 1640/Dulbecco modified Eagle medium (1:1 mixture; Lonza and CellGro, respectively). Cells were then differentiated in the absence of rmIL-3, but with rmSCF (10 ng/mL) and thrombopoietin (rmTPO; 10 ng/mL; R&D Systems) for differentiation toward megakaryocytes or with a high concentration of erythropoietin (10 U/mL) and rmTPO toward erythrocytes. The cells were allowed to differentiate for 4 days before analysis. The megakaryocyte colony-forming units (CFUs) were cultured by using MegaCult-C (StemCell Technologies). Lin− cells were plated according to the manufacturer's instructions with various concentrations of rmTPO, 10 ng/mL rmIL-3, and 20 ng/mL rmIL-6 (R&D Systems). Slides for histology were dehydrated, fixed, and stained with acetylthiocholiniodide (Sigma-Aldrich) and Harris hematoxylin counterstain. The acetylthiocholiniodide-stained cell numbers and size were quantitated using the Nikon NIS-Element AR system (Nikon).
Megakaryocyte ploidy analysis
Cultured megakaryocytes were fixed with 1% paraformaldehyde before cell surface marker staining using biotin-labeled CD41, followed by streptavidin-APC. The cells were then placed in 70% ethanol overnight at −20°C and stained with propidium iodide (PI) staining buffer (BD Biosciences) for 15 minutes before analysis. The flow data were analyzed using FlowJo software (TreeStar).
Statistical analysis
Paired Student t test was used to assess statistical significance.
Results
Rbm15 expression in hematopoietic cells of adult mice
To determine the expression pattern of Rbm15 in hematopoietic cells, we isolated different stages and lineages of mouse BM cells based on their surface marker expression. This was accomplished by FACS, and analysis of Rbm15 levels was done by semiquantitative reverse transcription–PCR. Rbm15 was found to be expressed in ST-HSC, granulocyte/monocyte progenitor (GMP), and megakaryocytic/erythroid progenitor (MEP) stages, as well as mature B cells and all stages of T-cell maturation (supplemental Figure 1A). These murine Rbm15 expression data closely parallel the expression pattern of human RBM15, which is by far most abundant in the hematopoietic system, is present in committed cells of similar hematopoietic lineages, and is also highly expressed in CD34+ BM cells (http://biogps.gnf.org). Based on these expression data, as well as the role of the RBM15-MKL1 fusion protein in acute megakaryoblastic leukemogenesis, we hypothesized that Rbm15 most likely plays a substantive role in regulating hematopoiesis, including HSC function.
To test this hypothesis, we engineered mice in which Rbm15 could be inactivated on an inducible basis and characterized the hematopoietic system in these animals. To delete Rbm15 conditionally in the hematopoietic system, Rbm15flox/flox mice were crossed with Mx1-Cre transgenic animals. Mx1-Cre-positive Rbm15flox/flox (Rbm15-KO), Mx1-Cre–positive Rbm15flox/+ (Rbm15-hetero), and Mx1-Cre–negative Rbm15flox/flox (Rbm15-WT) mice were obtained from the same litters by proper mating; because Rbm15-hetero and Rbm15-WT mice were phenotypically comparable (data not shown), we used Rbm15-KO and Rbm15-WT mice in the comparative studies reported in this study.
Phenotypic LT-HSCs are increased in Rbm15-KO mice
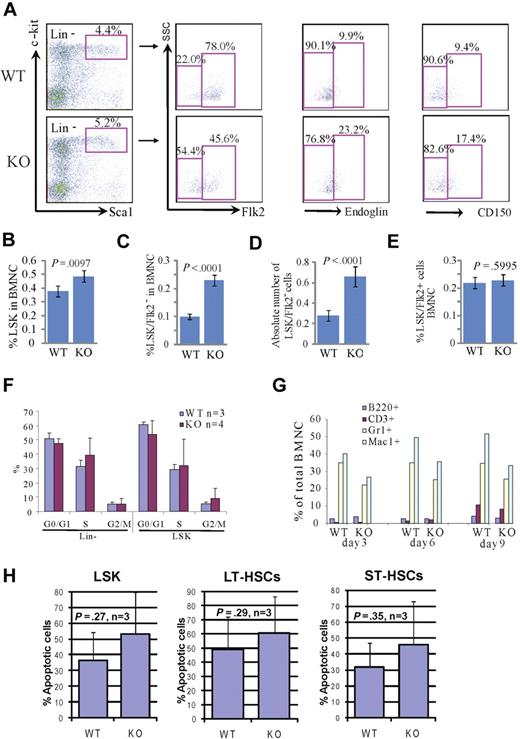
To study the role of Rbm15 in hematopoiesis, we first analyzed the immunophenotype and numbers of HSCs in Rbm15-KO mice by cell surface marker staining. Within the population of total BM nucleated cells (BMNCs), the percentage of LSK cells, which are enriched for HSCs of both LT (LT-HSCs) and ST (ST-HSCs) marrow-reconstituting capabilities, was significantly increased in Rbm15-KO mice compared with Rbm15-WT animals (Rbm15-WT, 0.38 ± 0.18 vs Rbm15-KO, 0.49 ± 0.19, P = .010, n = 18 mice per group; Figure 1A-B). Previous studies have demonstrated that LT-HSCs and ST-HSCs can be distinguished based on Flk2 expression; LT-HSCs are Flk2 negative (LSK/Flk2−), whereas ST-HSCs are Flk2 positive (LSK/Flk2+).16 Therefore, we examined Flk2 expression in the LSK cell population in Rbm15-KO mouse BM. We found that the percentage of LT-HSCs was significantly increased in Rbm15-KO mice (0.23% ± 0.02% of BMNCs, 2.3-fold higher) compared with Rbm15-WT littermate controls (0.10% ± 0.01% BMNCs; P < .001, n = 18 per group; Figure 1A,C). The absolute number of LT-HSCs was also increased significantly in Rbm15-KO mice (Rbm15-WT, 0.28 ± 0.05 × 107 vs Rbm15-KO, 0.66 ± 0.10 × 107, P < .001, n = 17 per group; Figure 1D). By contrast, although the percentage of ST-HSCs was decreased in the LSK population, it was comparable between Rbm15-KO and Rbm15-WT mice compared as a percentage of the total BMNCs (Figure 1E). The increased number of LT-HSCs in Rbm15-KOs was also demonstrated with 2 additional LT-HSC markers, endoglin (CD105)17 and CD150, a SLAM family member.18 Several recent studies have demonstrated that CD10517 and CD15018,19 are expressed by LT-HSCs, but not ST-HSCs. Consonant with our LSK/Flk2 analysis, the percentages of LSK/CD105+ and LSK/CD150+ LT-HSCs were also increased in Rbm15-KO mice compared with Rbm15-WT controls (Figure 1A). This significant increase suggested that Rbm15 might be involved in regulating the program of differentiation of LT-HSCs to ST-HSCs, which correlates with the up-regulation of Rbm15 expression during this process (supplemental Figure 1A).
Increased percentages and absolute numbers of LSK cells and LT-HSCs in Rbm15-KO mice without aberrant proliferation or apoptosis. (A) Representative flow analysis of LSK (Lin−Sca+c-Kit+), LT-HSC, and ST-HSC cells in Rbm15-KO mice and littermate control animals. Lin− cells were gated and analyzed for Sca1 and c-Kit, and the LSK cells were then gated to identify LT-HSCs and ST-HSCs based on Flk2, endoglin, or CD150 expression. Increased percentages of LT-HSCs (indicated as LSK/Flk2−, endoglin+, or CD150+ cells) were documented by studies using all 3 of these markers. (B-E) Quantitation of the percentages of LSK cells (B), LT-HSCs (LSK/Flk2−; C), and ST-HSCs (LSK/Flk2+; E) in total BMNCs, and the absolute number of LT-HSCs (D) in mice BM (n = 18 WT and KO each). (F) In vivo proliferation was assessed by BrdU incorporation. Mice received an intraperitoneal injection of 2 mg of BrdU and were analyzed 12 hours thereafter. BM cells were isolated and stained with a Lin+ mixture, c-Kit, and Sca1 antibodies, and analysis of BrdU incorporation in conjunction with 7-aminoactinomycin D was performed using a 5-color flow cytometer. (G) Rbm15-KO HSCs can terminally differentiate in vitro. FACS-sorted LSK/Flk2− cells from WT and Rbm15-KO BM were bulk cultured in stem cell culture medium containing murine SCF and murine IL-3 to allow the cells to differentiate. After 3, 6, and 9 days, the expression of lineage markers Mac1 (macrophages), Gr1 (granulocytes), B220 (B lymphocytes), and CD3 (T lymphocytes) was analyzed. The percentages of these cells as part of the total nucleated cells are shown. (H) The apoptotic responses of freshly isolated HSCs from Rbm15-KO mice and WT littermate controls were assessed by costaining with stem cell markers and annexin V/PI. The percentages of annexin V+/PI− apoptotic cells in LSK, LT-, and ST-HSC populations were analyzed. Mean ± SD and P values from 3 independent experiments are shown.
Increased percentages and absolute numbers of LSK cells and LT-HSCs in Rbm15-KO mice without aberrant proliferation or apoptosis. (A) Representative flow analysis of LSK (Lin−Sca+c-Kit+), LT-HSC, and ST-HSC cells in Rbm15-KO mice and littermate control animals. Lin− cells were gated and analyzed for Sca1 and c-Kit, and the LSK cells were then gated to identify LT-HSCs and ST-HSCs based on Flk2, endoglin, or CD150 expression. Increased percentages of LT-HSCs (indicated as LSK/Flk2−, endoglin+, or CD150+ cells) were documented by studies using all 3 of these markers. (B-E) Quantitation of the percentages of LSK cells (B), LT-HSCs (LSK/Flk2−; C), and ST-HSCs (LSK/Flk2+; E) in total BMNCs, and the absolute number of LT-HSCs (D) in mice BM (n = 18 WT and KO each). (F) In vivo proliferation was assessed by BrdU incorporation. Mice received an intraperitoneal injection of 2 mg of BrdU and were analyzed 12 hours thereafter. BM cells were isolated and stained with a Lin+ mixture, c-Kit, and Sca1 antibodies, and analysis of BrdU incorporation in conjunction with 7-aminoactinomycin D was performed using a 5-color flow cytometer. (G) Rbm15-KO HSCs can terminally differentiate in vitro. FACS-sorted LSK/Flk2− cells from WT and Rbm15-KO BM were bulk cultured in stem cell culture medium containing murine SCF and murine IL-3 to allow the cells to differentiate. After 3, 6, and 9 days, the expression of lineage markers Mac1 (macrophages), Gr1 (granulocytes), B220 (B lymphocytes), and CD3 (T lymphocytes) was analyzed. The percentages of these cells as part of the total nucleated cells are shown. (H) The apoptotic responses of freshly isolated HSCs from Rbm15-KO mice and WT littermate controls were assessed by costaining with stem cell markers and annexin V/PI. The percentages of annexin V+/PI− apoptotic cells in LSK, LT-, and ST-HSC populations were analyzed. Mean ± SD and P values from 3 independent experiments are shown.
The competitive repopulation ability of Rbm15-KO HSCs is compromised
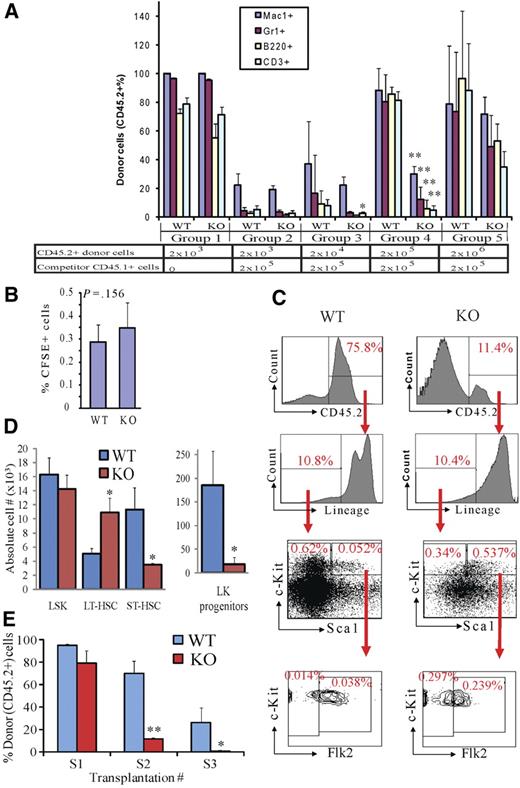
To test whether the increased LT-HSCs in Rbm15-KO BM are functional HSCs, we transplanted limiting-diluted BM from either Rbm15-KO or Rbm15-WT mice (both CD45.2+) into lethally irradiated WT host animals (CD45.1+) with a constant number (2 × 105) of WT competitor BM cells (CD45.1+; Figure 2A). Six weeks after transplantation, the reconstitution of lymphoid (B220+ and CD3+) and myeloid (Mac1+ and Gr1+) lineages was assessed in the peripheral blood (PB).
Rbm15-KO HSCs are less competitive in reconstitution of lethally irradiated recipient mice and compromised in serial transplantation due to a LT-HSC differentiation block. (A) Serially diluted donor BM cells (CD45.2+) were transplanted into lethally irradiated CD45.1+ recipient mice with (groups 2-5) or without (group 1) 2 × 105 competitor WT BM cells (CD45.1+). Six weeks after transplantation, PB was analyzed by FACS to examine the donor-derived (CD45.2+) monocyte/macrophage (Mac1+), granulocyte (Gr1+), B-lymphoid (B220+), and T-lymphoid (CD3+) cells in the recipient animals. The bars represent the averages in each group of mice; error bars show SD. (B) Homing of Rbm15-WT and -KO BM cells. BM cells were labeled with the fluorescent dye CFSE, then transplanted into lethally irradiated (10 Gy) recipient mice. CFSE+ cells homing to the BM of the recipient mice were assessed 5 hours after transplantation by flow analysis. (C) Representative BM analysis of competitive transplanted recipient mice (group 5 in A) 5 months after transplantation. The percentages of Lin− cells, LSKs, LT-HSCs, ST-HSCs, and Lin−c-Kit+Sca1− (LK) progenitors in the total donor-derived cell populations are shown. *P < .05, **P < .01, n = 4. (D) Absolute numbers of different HSC and progenitor subsets derived from CD45.2+ donor cells in the BM of the chimeras shown in (C). Note that, whereas Rbm15-KO LT-HSC numbers are increased approximately 3-fold, the numbers of Rbm15-KO ST-HSCs and LK progenitors are reduced markedly compared with the WT counterparts, indicating an inability of LT-HSCs that lack Rbm15 to normally differentiate. *P < .05, **P < .01, n = 2. (E) BMNCs (2 × 106) from either Rbm15-WT or Rbm15-KO mice (CD45.2+) were transplanted into lethally irradiated (10 Gy) WT recipient mice (CD45.1+) (S1). BMNCs were taken 3 months later from these recipient mice, and the same procedure was applied in secondary (S2) and tertiary (S3) recipients. PB from 3 to 6 mice in each group was analyzed 6 weeks and 3 months after transplantation for CD45.2+ donor cell contribution. The data shown were collected from 4 independent experiments using PB samples taken at 6 weeks after transplantation. *P < .05, **P < .01.
Rbm15-KO HSCs are less competitive in reconstitution of lethally irradiated recipient mice and compromised in serial transplantation due to a LT-HSC differentiation block. (A) Serially diluted donor BM cells (CD45.2+) were transplanted into lethally irradiated CD45.1+ recipient mice with (groups 2-5) or without (group 1) 2 × 105 competitor WT BM cells (CD45.1+). Six weeks after transplantation, PB was analyzed by FACS to examine the donor-derived (CD45.2+) monocyte/macrophage (Mac1+), granulocyte (Gr1+), B-lymphoid (B220+), and T-lymphoid (CD3+) cells in the recipient animals. The bars represent the averages in each group of mice; error bars show SD. (B) Homing of Rbm15-WT and -KO BM cells. BM cells were labeled with the fluorescent dye CFSE, then transplanted into lethally irradiated (10 Gy) recipient mice. CFSE+ cells homing to the BM of the recipient mice were assessed 5 hours after transplantation by flow analysis. (C) Representative BM analysis of competitive transplanted recipient mice (group 5 in A) 5 months after transplantation. The percentages of Lin− cells, LSKs, LT-HSCs, ST-HSCs, and Lin−c-Kit+Sca1− (LK) progenitors in the total donor-derived cell populations are shown. *P < .05, **P < .01, n = 4. (D) Absolute numbers of different HSC and progenitor subsets derived from CD45.2+ donor cells in the BM of the chimeras shown in (C). Note that, whereas Rbm15-KO LT-HSC numbers are increased approximately 3-fold, the numbers of Rbm15-KO ST-HSCs and LK progenitors are reduced markedly compared with the WT counterparts, indicating an inability of LT-HSCs that lack Rbm15 to normally differentiate. *P < .05, **P < .01, n = 2. (E) BMNCs (2 × 106) from either Rbm15-WT or Rbm15-KO mice (CD45.2+) were transplanted into lethally irradiated (10 Gy) WT recipient mice (CD45.1+) (S1). BMNCs were taken 3 months later from these recipient mice, and the same procedure was applied in secondary (S2) and tertiary (S3) recipients. PB from 3 to 6 mice in each group was analyzed 6 weeks and 3 months after transplantation for CD45.2+ donor cell contribution. The data shown were collected from 4 independent experiments using PB samples taken at 6 weeks after transplantation. *P < .05, **P < .01.
We found that, when transplanted without competitor cells, Rbm15-KO HSCs could almost completely reconstitute hematopoiesis in recipient mice (group 1, Figure 2A), indicating that Rbm15-KO HSCs can proliferate and terminally differentiate in vivo. However, when the same numbers of donor and competitor BM cells were transplanted (2 × 105; group 4, Figure 2A), the Rbm15-KO HSCs contributed to lymphomyelopoiesis in recipient mice significantly less than did Rbm15-WT HSCs. Furthermore, even when transplanted at a 10:1 ratio of Rbm15-KO donor (2 × 106) vs competitor (2 × 105) BM cells, the lymphomyeloid reconstitution by Rbm15-KO cells failed to reach levels comparable with that of WT donor cells (group 5, Figure 2A). Similar results were observed when we performed the same PB analysis 3 months after transplantation (data not shown). These data indicate that the hematopoietic reconstitution ability of Rbm15-KO HSCs is impaired compared with Rbm15-WT HSCs.
The hematopoietic reconstitution capacity of HSCs is correlated in part with their BM engraftment (residing) ability, which can be evaluated by their BM homing and lodging capacities, and their growth and differentiation potential after engrafting in the BM niche. We first eliminated the possibility that the reconstitutive impairment we observed was caused by a homing defect of Rbm15-KO BM cells. Specifically, comparable percentages of 5- (and 6-)carboxyfluorescein diacetate succinimidyl ester (CFSE)–labeled cells from Rbm15-KO and Rbm15-WT control donor cells were found in recipients' BM, suggesting that the mutant HSCs possess normal homing capability (Figure 2B).
We then tested whether the reconstitutional disadvantage of Rbm15-mutant HSCs was the result of a reduction of growth or differentiation potential. Five weeks after transplantation, donor-derived HSC (CD45.2+) reconstitution was analyzed in the BM of group 5 recipients (Figure 2A) because not enough Rbm15-KO donor-derived cells could be obtained from other groups. We found that significantly fewer Rbm15-KO HSCs contributed to recipient BM hematopoiesis than those from Rbm15-WT controls, as shown by the percentage of CD45.2+ donor-derived cells (Rbm15-WT donor derived, 75.8%; Rbm15-KO donor derived, 11.4%; Figure 2C). This result was consistent with our findings from analysis of the PB (Figure 2A). Interestingly, whereas we found that the Rbm15-WT donor-derived BM cells yielded a normal percentage of HSCs, the percentage of phenotypic HSCs derived from Rbm15-KO cells was significantly increased in the recipient BM. As shown in Figure 2C, the percentage of donor-derived LSK-HSCs within the total donor-derived cell population was found to be increased by 10.3-fold in the Rbm15-KO transplantation group compared with that observed in the littermate control-transplanted group (0.537% vs 0.052%, respectively). The percentage of the LT-HSC population derived from Rbm15-KO donor cells was increased by an even more dramatic 21.2-fold (0.297% vs 0.014%). The percentage of donor-derived ST-HSCs was also increased, but not to the same extent as the LT-HSCs, in Rbm15-KO–transplanted recipients compared with Rbm15-WT recipients (6.3-fold increase; 0.239% vs 0.038%, respectively), whereas the percentages of lineage-negative (Lin−) cells were the same (10.4% vs 10.8%, respectively), and the percentage of Lin−c-Kit+Sca1− (LK) populations was decreased (1.8-fold decrease; 0.34% vs 0.62% in Rbm15-KO–transplanted recipients compared with Rbm15-WT recipients; Figure 2C). The absolute numbers of Rbm15-KO LT-HSCs in the BM of recipient mice were also increased significantly, whereas those of the ST-HSC and LK progenitor populations were significantly decreased (Figure 2D). This increase in mutant LT-HSCs in the recipient BM, with diminished contribution to LK progenitor and PB cell regeneration, further supported our impression that Rbm15 is required for the differentiation of LT- to ST-HSCs.
Furthermore, these transplantation studies suggest that the defects associated with Rbm15-KO HSCs are cell intrinsic because we were able to reproduce all of the phenotypic abnormalities in the normal BM environment provided by WT recipient mice. Consistent with these competitive repopulation assay results, serial transplantation studies using Rbm15-KO BM also revealed a marked deficiency in the ability of marrow lacking the gene to contribute to hematopoiesis (Figure 2E).
The proliferation, in vitro differentiation, and survival of Rbm15-KO HSCs are comparable with those from WT controls
HSC numbers in the BM of adult mice are relatively stable during normal conditions of homeostasis. Alterations with several underlying mechanisms can result in increased HSC numbers; for example, enhanced HSC proliferation, blocks to HSC differentiation, and protection of HSCs from apoptosis can all result in expanded HSC numbers.
To determine whether the proliferation of Rbm15-deficient HSCs is altered, we analyzed the cell-cycle status of Rbm15-KO HSCs using an in vivo BrdU pulse-labeling assay. We found that the percentages of the cell-cycle phases distinguished by BrdU labeling and 7-aminoactinomycin D staining of both Lin− and LSK populations from Rbm15-KO mice were comparable within like cell populations from littermate controls (Figure 1F). These data indicate that the increase in LT-HSCs in Rbm15-deleted mice is most likely not caused by an alteration in stem cell proliferation.
To investigate whether HSC differentiation is altered in Rbm15-KO mice, we tested the maturation ability of HSCs by bulk sorting of LT-HSCs and incubation using liquid culture conditions.20 As shown in Figure 1G, the Rbm15-KO LT-HSCs could differentiate to all of the lineages tested, thus indicating that these cells are capable of normal maturation ex vivo.
To study whether apoptosis is deregulated in Rbm15-deleted HSCs, we assessed HSC survival by annexin V/PI staining of freshly isolated BM cells. As shown in Figure 1H, the baseline apoptotic rates in LT-HSCs, ST-HSCs, and LSK populations from Rbm15-KO mice were similar to those from WT controls, indicating that the increase in LT-HSC numbers in Rbm15-KO mice is not likely due to reduced apoptotic death.
Up-regulation of expression of adhesion molecules on Rbm15-KO HSCs
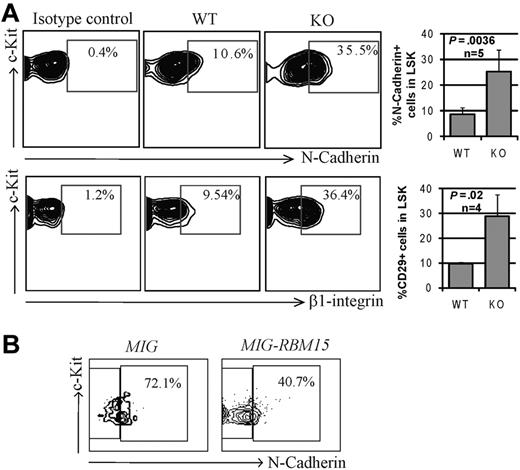
Because the functional impairment of Rbm15-deficient HSCs was observed only in vivo, we speculated that this might be caused by a stem cell niche interaction defect. We therefore compared the expression patterns of selected adhesion molecules known to be important for HSC-niche functions. As shown in Figure 3A, the expression levels of N-cadherin and β1 integrin (CD29) were found to be increased substantially (3.0 ± 0.52-fold, n = 5 and 3.0 ± 0.94-fold, n = 4, respectively) in Rbm15-KO HSCs compared with littermate control HSCs. αL (CD11a), α4 (CD49d), and α5 (CD49e) integrins were shown to be slightly increased in Rbm15-KO HSCs as well, whereas β2 (CD18) and α6 (CD49f) integrins and CXCR4 levels were not altered (supplemental Figure 3). Overexpression of RBM15 by retroviral transduction decreased N-cadherin expression (1.8-fold decrease) in WT LSK cells (Figure 3B), suggesting that the gene may regulate expression of the adhesion molecule and, in turn, can alter HSC-niche interactions. The increase in major adhesion molecules in Rbm15-deficient HSCs suggests that these cells may interact more strongly with their niche, preventing the efficient transition of LT- to ST-HSCs.
Increased adhesion molecule expression by Rbm15-KO HSCs. (A) Representative flow cytometric analysis of the expression of N-cadherin and β1 integrin (CD29) on Rbm15-KO and -WT littermate control LSK HSCs (left panels). Bar graphs showing the mean ± SD and P values for results from multiple independent experiments are also included (right panels). (B) N-cadherin expression in RBM15-overexpressing LSK cells. WT Lin− BM cells were transduced with MSCV-IRES-GFP (MIG) vector control or MSCV-IRES-GFP-RBM15 (MIG-RBM15), and then GFP-positive LSKs were gated and the N-cadherin expression was analyzed after 4 days in culture.
Increased adhesion molecule expression by Rbm15-KO HSCs. (A) Representative flow cytometric analysis of the expression of N-cadherin and β1 integrin (CD29) on Rbm15-KO and -WT littermate control LSK HSCs (left panels). Bar graphs showing the mean ± SD and P values for results from multiple independent experiments are also included (right panels). (B) N-cadherin expression in RBM15-overexpressing LSK cells. WT Lin− BM cells were transduced with MSCV-IRES-GFP (MIG) vector control or MSCV-IRES-GFP-RBM15 (MIG-RBM15), and then GFP-positive LSKs were gated and the N-cadherin expression was analyzed after 4 days in culture.
Rbm15-deficient megakaryocytic progenitors produce increased, but abnormally small and low-ploidy megakaryocytes
RBM15 was discovered due to its involvement in the AMkL fusion gene RBM15-MKL1. The critical role of RBM15-MKL1 in the pathogenesis of acute megakaryocytic leukemia suggested to us that RBM15 might be functionally involved in normal megakaryocyte development.
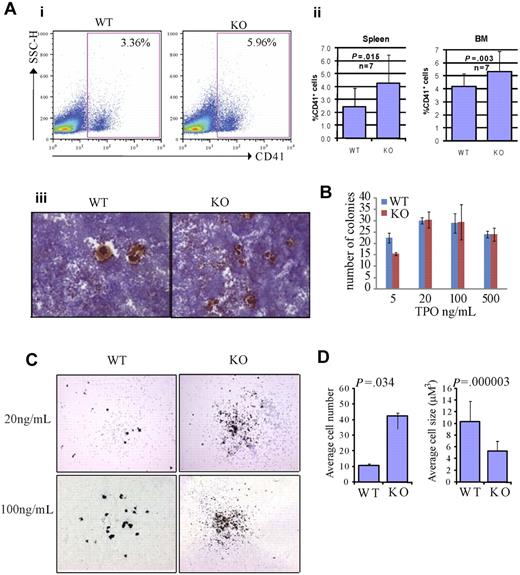
We first investigated megakaryocytes in the BM and spleens of Rbm15-KO mice, and found that the percentage of CD41+ megakaryocytes was increased compared with Rbm15-WT animals (Figure 4Ai-ii). Quantitation of the differences in percentages of CD41+ megakaryocytes based on genotype revealed in the BM is as follows: Rbm15-WT, 4.16% plus or minus 1.00% versus Rbm15-KO, 5.31% plus or minus 1.54% of Mac1−Gr1− cells, P = .003, n = 7 per group; in the spleen: Rbm15-WT, 2.43% plus or minus 1.47% versus Rbm15-KO, 4.26% plus or minus 2.20% of Mac1−Gr1− cells, P = .015, n = 7 per group. Histopathologic examination of the spleens and BM from each group of mice revealed a statistically significant increase in the numbers of morphologically identified megakaryocytes present in the spleens (average of 2.8-fold higher in KO vs WT spleens, P = .005), but not the marrows from Rbm15-KO animals (supplemental Figure 4A-B). This increase of megakaryocyte numbers was also confirmed by acetylcholinesterase (AchE) staining in the spleens of the KO animals (Figure 4Aiii). To further explore the role of Rbm15 in megakaryocyte development, we quantitated megakaryocytic progenitors by CFU assays (CFU-Mk). We found that Rbm15-deleted cells generated similar numbers of colonies compared with cells from WT controls (Figure 4B), suggesting that megakaryocytic colony-forming progenitor numbers do not increase in the absence of Rbm15. However, although the total number of Rbm15-KO progenitor-generated colonies was not different from WT controls, increased numbers of smaller-sized megakaryocytes were observed in each colony derived from Rbm15-KO progenitors (Figure 4C-D and supplemental Figure 5).
Rbm15-deficient HSC/Ps favor megakaryocytic differentiation in vivo and in vitro. (A) Increased megakaryocytes in Rbm15-KO mice. A representative flow analysis of megakaryocytes in the spleens of Rbm15-KO and -WT control mice using megakaryocyte cell surface marker CD41 is shown (i). The percentages of the CD41+ cells in the Mac1−Gr1− splenic and BM populations from 4 independent experiments are shown as bar graphs (ii). Cryosections of Rbm15-WT and -KO spleens were stained for AchE. Representative images are shown (iii). (B) Quantitation of CFU-Mk colony numbers. Lin− BM cells were plated in MegaCult-C medium with IL-3, IL-6, and different concentrations of TPO for CFU-Mk colony formation. The colony numbers were quantitated after a 7-day culture. The bar graph represents results from 2 independent experiments. (C) Morphology of AchE-stained CFU-Mk colonies. Representative pictures of colonies cultured with 20 ng/mL and 100 ng/mL TPO are shown. (D) Quantitation of cell number and size of AchE-positive cells in each CFU-Mk colony. Average cell number in each colony (left) and average cell size (μM3; right) are graphed. Data were collected from 2 independent experiments. Error bars indicate SD.
Rbm15-deficient HSC/Ps favor megakaryocytic differentiation in vivo and in vitro. (A) Increased megakaryocytes in Rbm15-KO mice. A representative flow analysis of megakaryocytes in the spleens of Rbm15-KO and -WT control mice using megakaryocyte cell surface marker CD41 is shown (i). The percentages of the CD41+ cells in the Mac1−Gr1− splenic and BM populations from 4 independent experiments are shown as bar graphs (ii). Cryosections of Rbm15-WT and -KO spleens were stained for AchE. Representative images are shown (iii). (B) Quantitation of CFU-Mk colony numbers. Lin− BM cells were plated in MegaCult-C medium with IL-3, IL-6, and different concentrations of TPO for CFU-Mk colony formation. The colony numbers were quantitated after a 7-day culture. The bar graph represents results from 2 independent experiments. (C) Morphology of AchE-stained CFU-Mk colonies. Representative pictures of colonies cultured with 20 ng/mL and 100 ng/mL TPO are shown. (D) Quantitation of cell number and size of AchE-positive cells in each CFU-Mk colony. Average cell number in each colony (left) and average cell size (μM3; right) are graphed. Data were collected from 2 independent experiments. Error bars indicate SD.
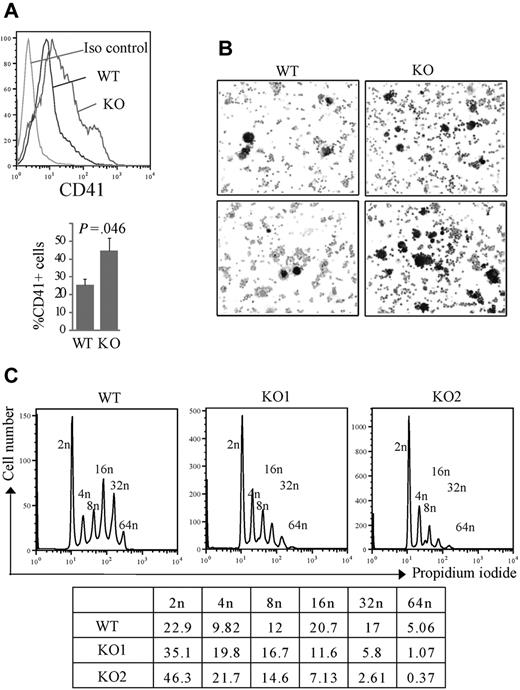
To further characterize the defects of megakaryocytes derived from Rbm15-KO progenitors, we next differentiated Lin− BM cells in liquid cultures and found a significant increase in CD41+ megakaryocyte development from Rbm15-deleted cells (44.63% ± 6.90%) compared with WT cells (25.5% ± 3.15%, P = .046, n = 3; Figure 5A). This megakaryocytic differentiation was also confirmed by AchE staining; consistent with our CFU-Mk culture results, we found much greater numbers of small AchE-positive cells in the cultures of Rbm15-KO cells compared with control cultures (Figure 5B). DNA content analysis showed that Rbm15-KO mice have substantially more low-ploidy (2n-8n), and markedly fewer high-ploidy (16n-64n), CD41+ megakaryocytes than WT animals (Figure 5C). Despite the increased megakaryocytes observed in Rbm15-KO mice, PB platelet counts in these animals were not significantly different from WT littermate controls (supplemental Table 1).
Increased, low-ploidy megakaryocytes develop from Rbm15-KO HSC/Ps. (A-B) Liquid culture of lin− marrow cells. Lin− cells isolated by magnetic-activated cell sorting were cultured in hematopoietic stem cell expansion media for 2 to 3 days, and then allowed to differentiate toward the megakaryocytic lineage, as described.21 (A, top panel) Representative flow analysis of the CD41 surface marker specific for megakaryocytes of the cells after a 4-day induction. (Bottom panel) Average percentages of CD41+ cells from 3 independent experiments are graphed. Error bars show SD. (B) The differentiated megakaryocytes were cytospun onto slides and stained for AchE. AchE-positive megakaryocytes are darkly stained brown. The original magnification was ×100. (C) Ploidy analysis of in vitro cultured megakaryocytes. Ploidy of CD41+ cells from liquid-cultured Lin− cells from Rbm15-KO mice and WT littermates was analyzed by flow cytometry using PI to determine DNA content. The percentages of cells of different ploidy status are shown in the table below. Data from 2 representative experiments using Rbm15-KO cells are shown.
Increased, low-ploidy megakaryocytes develop from Rbm15-KO HSC/Ps. (A-B) Liquid culture of lin− marrow cells. Lin− cells isolated by magnetic-activated cell sorting were cultured in hematopoietic stem cell expansion media for 2 to 3 days, and then allowed to differentiate toward the megakaryocytic lineage, as described.21 (A, top panel) Representative flow analysis of the CD41 surface marker specific for megakaryocytes of the cells after a 4-day induction. (Bottom panel) Average percentages of CD41+ cells from 3 independent experiments are graphed. Error bars show SD. (B) The differentiated megakaryocytes were cytospun onto slides and stained for AchE. AchE-positive megakaryocytes are darkly stained brown. The original magnification was ×100. (C) Ploidy analysis of in vitro cultured megakaryocytes. Ploidy of CD41+ cells from liquid-cultured Lin− cells from Rbm15-KO mice and WT littermates was analyzed by flow cytometry using PI to determine DNA content. The percentages of cells of different ploidy status are shown in the table below. Data from 2 representative experiments using Rbm15-KO cells are shown.
Gene expression profile changes in Rbm15-KO HSCs
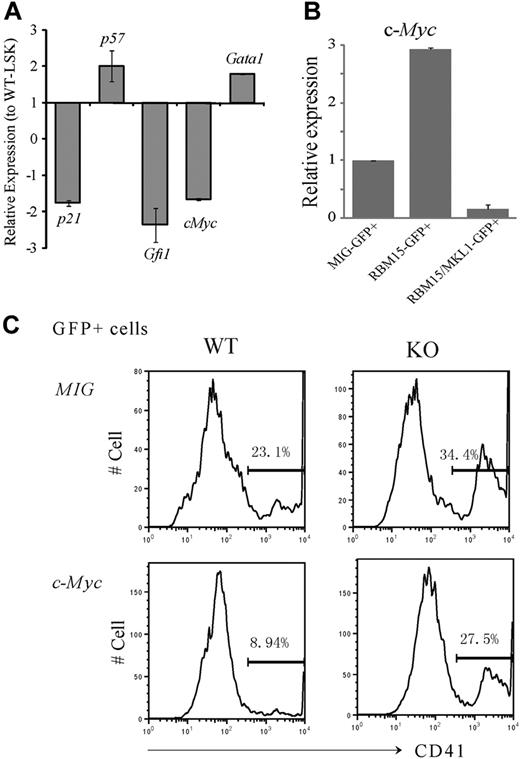
The deletion of a select few genes has been reported to increase LT-HSC numbers. For example, as is the case with Rbm15-KO mice, the Gfi1 zinc finger transcription factor,22 p21cip1/waf cyclin-dependent kinase inhibitor,23 and c-Myc13 KO mice all exhibit increased HSC numbers and defective marrow reconstitution capability. By real-time PCR analysis, we found that the expression levels of p21, c-Myc, and Gfi1 were all down-regulated in Rbm15-KO compared with WT HSCs (Figure 6A).
c-Myc is a downstream target of Rbm15 in the regulation of HSC and megakaryocyte development. (A-B) c-Myc expression levels are regulated by Rbm15 and RBM15-MKL1 in HSCs. (A) Expression of selected genes in Rbm15-KO HSCs. RNA was isolated from FACS-sorted LSK-HSCs from both Rbm15-deleted and WT littermate control mice, reverse transcribed, and analyzed by TaqMan real-time PCR. Normalized expression of each gene in Rbm15-deleted cells relative to that in WT control LSK cells was calculated. Results represent the mean ± SD of 3 independent experiments. (B) c-Myc levels in HSCs with ectopic RBM15 and RBM15-MKL1 expression. LSK-HSCs were FACS sorted and transduced with MSCV-IRES-GFP (MIG) vector control, MSCV-IRES-GFP-RBM15 (MIG-RBM15), or MSCV-IRES-GFP-RBM15-MKL1 (MIG-RBM15-MKL1) retrovirus supernatant. After a 2-day transduction, GFP-positive cells were sorted for real-time PCR analysis (mean ± SD, n = 2). (C) c-Myc overexpression rescues the megakaryocyte increase in ex vivo cultures. Lin− cells were isolated by magnetic-activated cell sorting from Rbm15-KO and -WT littermate control animals and transduced with either MSCV-IRES-GFP (MIG) or MSCV-IRES-GFP-c-Myc (c-Myc) retroviruses before culturing ex vivo using the same conditions as described in Figure 5. GFP+ cells were gated and megakaryocytes were analyzed using the specific cell surface marker CD41. The results shown are representative of 3 independent experiments.
c-Myc is a downstream target of Rbm15 in the regulation of HSC and megakaryocyte development. (A-B) c-Myc expression levels are regulated by Rbm15 and RBM15-MKL1 in HSCs. (A) Expression of selected genes in Rbm15-KO HSCs. RNA was isolated from FACS-sorted LSK-HSCs from both Rbm15-deleted and WT littermate control mice, reverse transcribed, and analyzed by TaqMan real-time PCR. Normalized expression of each gene in Rbm15-deleted cells relative to that in WT control LSK cells was calculated. Results represent the mean ± SD of 3 independent experiments. (B) c-Myc levels in HSCs with ectopic RBM15 and RBM15-MKL1 expression. LSK-HSCs were FACS sorted and transduced with MSCV-IRES-GFP (MIG) vector control, MSCV-IRES-GFP-RBM15 (MIG-RBM15), or MSCV-IRES-GFP-RBM15-MKL1 (MIG-RBM15-MKL1) retrovirus supernatant. After a 2-day transduction, GFP-positive cells were sorted for real-time PCR analysis (mean ± SD, n = 2). (C) c-Myc overexpression rescues the megakaryocyte increase in ex vivo cultures. Lin− cells were isolated by magnetic-activated cell sorting from Rbm15-KO and -WT littermate control animals and transduced with either MSCV-IRES-GFP (MIG) or MSCV-IRES-GFP-c-Myc (c-Myc) retroviruses before culturing ex vivo using the same conditions as described in Figure 5. GFP+ cells were gated and megakaryocytes were analyzed using the specific cell surface marker CD41. The results shown are representative of 3 independent experiments.
c-Myc expression is regulated by ectopically expressed RBM15 and the RBM15-MKL1 fusion
Although the expression levels of several HSC-important genes were found to be altered in Rbm15-KO HSCs, the phenotype of these cells most closely resembles that of HSCs from c-Myc-KO mice,13,14 including LT-HSC accumulation, normal cell proliferation and survival, intact differentiation ability in vitro, and greater accumulation of LT-HSCs in mixed chimeras. In addition, Guo et al have made the important observation that, as in mice lacking Rbm15, megakaryocyte development is also significantly altered in c-Myc-KO mice.24 These c-Myc-KO megakaryocyte abnormalities are phenotypically similar to those observed in our Rbm15-deleted mice, including increased CD41+ megakaryocytes in the spleens and reduced size of the cells.
These findings prompted us to explore whether c-Myc expression might be regulated by Rbm15. As shown in Figure 6B, overexpression of RBM15 in WT LSK-HSCs was associated with increased c-Myc expression levels (∼ 3-fold higher), whereas the RBM15-MKL1 fusion significantly decreased c-Myc expression to nearly undetectable levels, suggesting that c-Myc may be a major target of the RBM15 and chimeric RBM15-MKL1 proteins. Thus, together with the shared abnormalities of HSC and megakaryocyte development in Rbm15-KO and c-Myc-KO mice, our data suggest that down-regulation of c-Myc by the RBM15-MKL1 fusion protein may play an important role in the pathogenesis of AMkL harboring the t(1;22).
Ectopic expression of c-Myc partially rescues the increased megakaryocyte numbers observed in in vitro Rbm15-KO cell cultures
To further explore whether c-Myc and Rbm15 might potentially be functionally interrelated, Lin− cells were isolated from Rbm15-KO and -WT littermate control mice and retrovirally transduced with murine stem cell virus (MSCV)–internal ribosome entry site (IRES)–green fluorescent protein (GFP)–c-Myc or with MSCV-IRES-GFP vector as a control. The transduced cells were expanded and induced toward megakaryocytic differentiation using the same conditions as described for the liquid cultures illustrated in Figure 5. GFP+ cells were gated and subjected to CD41 analysis. Consistent with the analysis shown in Figure 5, CD41+ megakaryocytes were increased in Rbm15-KO cells compared with WT control cells when both were transduced with vector alone (34.4% vs 23.1%, respectively, in a representative experiment; Figure 6C). By contrast, we found c-Myc overexpression in WT cells to decrease the percentage of CD41+ megakaryocytes compared with cells transduced with the vector alone (8.94% vs 23.1%, respectively). More importantly, c-Myc overexpression could partially reverse the megakaryocyte increase of Rbm15-deleted cells transduced with vector only (27.5% vs 34.4%, respectively; Figure 6C). Whereas these findings do not establish an epistatic interplay between c-Myc and Rbm15, they do demonstrate that the expression of c-Myc (independent of Rbm15 status, as shown in Figure 6C) affects megakaryocyte development, consistent with the data reported by Guo and colleagues in the companion paper to this article.24 Thus, because the functional status of Rbm15 affects c-Myc expression levels (Figure 6A-B), we posit that the effects of Rbm15 on megakaryocyte development may be mediated in part via c-Myc; formal proof that this is indeed the case will require additional future investigation.
Discussion
In this study, we report that Rbm15, the murine counterpart of the RBM15 gene involved in the RBM15-MKL1 fusion generated by t(1;22) in pediatric acute megakaryocytic leukemia, plays a critical role in the regulation of HSC and megakaryocyte development. We found that phenotypic LT-HSCs accumulate significantly in the BM of Rbm15-KO mice, most likely due to a differentiation defect caused by increased niche adhesion. Furthermore, we determined that Rbm15-KO Mk-Ps produce more megakaryocytes than their WT counterparts, but of smaller size and lower ploidy. These phenotypic alterations are reminiscent of the hematopoietic abnormalities observed in c-Myc-KO mice,13,14 although they are not as severe.24
We found that c-Myc expression is significantly lower in Rbm15-KO HSCs compared with their WT counterparts, and that the increased production of megakaryocytes by Rbm15-KO Mk-Ps can be at least partially reversed by ectopic expression of c-Myc. We conclude, therefore, that c-Myc is one of the important downstream targets that mediate Rbm15 function in HSC/progenitors (HSC/Ps).
Rbm15 may regulate the differentiation of LT-HSCs to ST-HSCs in a BM niche-dependent manner
The BM niche provides a harbor of sorts to protect HSCs from unfavorable stimulation, yet at the same time allowing for HSC self-renewal. In this study, we show that the proliferation and apoptosis of LT-HSCs are not substantively altered in Rbm15-KO mice, and that the differentiation of these mutant HSCs is comparable with their WT counterparts in in vitro cultures. We therefore speculate that the phenotypic LT-HSC accumulation in the BM of Rbm15-KO mice is most likely due to an inefficient differentiation of LT-HSCs to ST-HSCs, presumably caused by HSC-niche interaction defects. Although we have not formally proven the existence of HSC-niche interaction abnormalities directly, we did find that, compared with HSCs from WT mice, HSCs lacking Rbm15 show a marked increase in the expression of N-cadherin (which helps to mediate binding of HSCs to osteoblastic niche cells),15 as well as up-regulation of various integrins, including α5β1 (CD49e/CD29) and α4β1 (CD49d/CD29) integrins. Both CD49d/CD29 and CD49e/CD29, together with other integrins and the chemokine receptor CXCR4, are known to be expressed on human HSCs and serve to promote their adhesion to the stroma.21,24-26 Interestingly, hematopoiesis is experimentally impaired by genetic deletion or antibody neutralization of CD49d and CD29.25,26
Previous studies have demonstrated that LT-HSCs accumulate in the BM of c-Myc-KO mice; this has been attributed to increased expression of many adhesion molecules, including N-cadherin, as well as α5 and β1 integrins, by the mutant HSCs.13 Like c-Myc, we show in this study that Rbm15 overexpression can down-regulate the expression of N-cadherin by HSCs, demonstrating that the expression of this adhesion molecule is indeed regulated both by c-Myc as well as Rbm15. The increase in N-cadherin and various integrins in Rbm15-KO and c-Myc-KO HSCs, together with the highly similar phenotypes exhibited by both mice lines, suggest that the 2 genes may regulate HSC behavior through at least partially similar mechanisms. We hypothesize that the increased expression of adhesion molecules on the surface of HSCs lacking either Rbm15 or c-Myc enhances their attachment to the BM niche, thus trapping more of the quiescent LT-HSCs and preventing them from cell-cycle entry and differentiation to ST-HSCs, which would normally be induced by activation signaling. Such a scenario would be consistent with our finding that ST-HSCs, but not LT-HSCs, normally express Rbm15 because expression of the gene would be associated with adhesion molecule down-regulation, which in turn would permit exit from the niche and subsequent maturation. The down-regulation of c-Myc in Rbm15-KO HSCs and the up-regulation of c-Myc by ectopic overexpression of Rbm15 shown in this study suggest that c-Myc might be one of the downstream targets of Rbm15 that mediates Rbm15 function in HSCs.
Rbm15 may control endomitosis versus mitotic division in megakaryocytic pregenitors partially through regulation of c-Myc levels
We found that the percentage of CD41-positive megakaryocytes is significantly increased in Rbm15-KO mouse BM and spleens. In vitro analysis demonstrated that the Mk-Ps from the BM of Rbm15 mutant mice produced more megakaryocytes than WT Mk-Ps. The smaller size and lower ploidy of Rbm15-mutant megakaryocytes suggested that Rbm15 might regulate endomitosis versus mitotic division in these megakaryocytes. Mutant megakaryocytes might be inclined toward mitotic cell division in lieu of their endomitotic program. Interestingly, Guo et al found that, compared with other lineages of hematopoietic cells, megakaryocyte development is less dependent upon c-Myc.24 As a result, HSC/Ps from c-Myc-KO mice are also biased toward megakaryocytic lineage differentiation. However, endomitosis of c-Myc-KO megakaryocytes is significantly disrupted, and the megakaryocytes from c-Myc-KO mice are also smaller in size and lower in ploidy compared with megakaryocytes from WT mice. We propose that c-Myc might mediate an endomitotic function of Rbm15 in megakaryocytes (proof of which will require additional study); consistent with this proposal, ectopic expression of c-Myc is able to at least partially reverse the increased megakaryocyte production by Rbm15-KO hematopoietic progenitors.
Comparison of the phenotype of Rbm15-KO mice (in which c-Myc expression is lower) with that of c-Myc-KO mice (in which c-Myc is completely deleted) reveals both mice to exhibit a significantly increased potential for megakaryocytopoiesis. However, the mechanisms by which these occur appear to be at least somewhat different. In contrast to the significant defects of myelolymphopoiesis and erythropoiesis in c-Myc-KO mice, we found the myelocytes, lymphocytes, and erythrocytes of Rbm15-KO mice to be less affected. In addition, the number of Mk-Ps from Rbm15-KO mice, as indicated by the number of CFU-Mk colonies they generated, is comparable with that from WT controls. However, the number of cells in each CFU-Mk colony is greater in Rbm15-KO mice compared with WT mice. On the contrary, Mk-Ps are significantly expanded in c-Myc-KO mice, but fewer CFU-Mk colonies are produced by c-Myc-KO Mk-Ps probably due to increased apoptosis of c-Myc-KO megakaryocytes,24 which is not seen in Rbm15-KO mice. These data suggest that lower levels of c-Myc might be sufficient for myelolymphopoiesis and erythropoiesis, at least under conditions of homeostasis such as in our Rbm15-KO mice. However, a threshold level of c-Myc might be required for the endomitotic machinery in megakaryocytes, and the level of c-Myc in Rbm15-KO megakaryocytes may be lower than this threshold. As a consequence, the majority of megakaryocytes from Rbm15-KO mice might preferentially undergo mitotic division instead of endomitotic growth, thus producing larger numbers of smaller-sized megakaryocytes of lower ploidy. This low-level expression of c-Myc may also be required for normal megakaryocyte survival. It should be noted that a plausible alternative is that Rbm15-KO Mk-Ps may have a generalized impairment of differentiation or differentiation kinetics, independent or instead of an altered balance between endomitosis and mitosis; further study is warranted to examine these biologic possibilities.
RBM15-MKL1 down-regulates c-Myc levels in HSCs
The molecular mechanism of leukemia pathogenesis by the RBM15-MKL1 fusion protein in t(1;22)-positive AMkL is largely unknown. In this study, we found that whereas WT RBM15 up-regulates the expression of c-Myc in HSCs, the RBM15-MKL1 fusion down-regulates c-Myc in the same cell population. This observation suggests that the fusion protein may repress normal RBM15 function through a dominant-negative mechanism. As such, the functional antagonism of RBM15 by RBM15-MKL1 with respect to the regulation of c-Myc could contribute to the determination of the megakaryocytic lineage of the leukemic cells in t(1;22)-positive AMkL patients, given that Guo et al24 showed that c-Myc is less important for megakaryocyte proliferation than other blood cell lineages. Thus, RBM15-MKL1 may promote megakaryocyte proliferation in patients with t(1;22) in part through a default mechanism. We suggest that a second possible factor operative in RBM15-MKL1 leukemogenesis may concern the microenvironment. Adhesion molecules important for HSC-niche interactions, N-cadherin and β1 integrin, are significantly up-regulated in Rbm15-mutant HSCs. If RBM15-MKL1 does in fact inhibit RBM15 in a dominant-negative fashion in leukemic cells, we speculate that the fusion would also deregulate cell growth and differentiation in a microenvironment-dependent manner by causing altered adhesion molecule expression on leukemia stem cells.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Xiaoli Cui for excellent technical assistance; Tong Xin and Jianmin Pan in the Department of Biostatistics, St Jude Children's Research Hospital, for statistical analysis; and the Flow Cytometry and Cell Sorting Shared Resource, the Animal Resources Center (ARC) and ARC Diagnostic Laboratory, and the Transgenic/Gene Knockout Resource of St Jude Children's Research Hospital for their assistance.
This work was supported in part by National Cancer Institute Cancer Center Core Grant CA21675 and by the American Lebanese Syrian Associated Charities, St Jude Children's Research Hospital.
National Institutes of Health
Authorship
Contribution: C.N. designed and performed the majority of the experiments and analyzed data; J.Z. and P.B. helped with the design and performance of selected experiments, including the retroviral transduction of HSCs with RBM15 or RBM15-MKL1 and the AChE staining of megakaryocytes; M.O. provided hematopathologic consultation and diagnosis; Z.M. assisted with the generation of Rbm15-KO mice; and C.N., J.Z., and S.W.M. conceived the overall study design and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Stephan Wade Morris, Department of Pathology, St Jude Children's Research Hospital, MS #343, Thomas Tower, Rm 4026, 262 Danny Thomas Pl, Memphis, TN 38105-3678; e-mail: steve.morris@stjude.org.