The GFI1 gene encodes a transcriptional repressor, which regulates myeloid differentiation. In the mouse, Gfi1 deficiency causes neutropenia and an accumulation of granulomonocytic precursor cells that is reminiscent of a myelodysplastic syndrome. We report here that a variant allele of GFI1 (GFI136N) is associated with acute myeloid leukemia (AML) in white subjects with an odds ratio of 1.6 (P < 8 × 10−5). The GFI136N variant occurred in 1806 AML patients with an allele frequency of 0.055 compared with 0.035 in 1691 healthy control patients in 2 independent cohorts. We observed that both GFI1 variants maintain the same activity as transcriptional repressors but differ in their regulation by the AML1/ETO (RUNX1/RUNX1T1) fusion protein produced in AML patients with a t(8;21) translocation. AML1/ETO interacts and colocalizes with the more common GFI136S form in the nucleus and inhibits its repressor activity. However, the variant GFI136N protein has a different subnuclear localization than GFI136S. As a consequence, AML1/ETO does not colocalize with GFI136N and is unable to inhibit its repressor activity. We conclude that both variants of GFI1 differ in their ability to be regulated by interacting proteins and that the GFI136N variant form exhibits distinct biochemical features that may confer a predisposition to AML.
Introduction
In acute myeloid leukemia (AML), malignant blasts of myeloid origin accumulate in the bone marrow.1,2 It has been shown for different mouse models that deficiency of myeloid transcription factors such as CCAAT/enhancer-binding protein alpha (CEBPA) and PU.1 can promote the development of AML.3,–5 In addition, mutations in certain transcription factors, such as CEBPA, are not only causative for AML in experimental models but also influence the prognosis of patients with AML.6,7 Similar to CEBPA and PU.1, Growth Factor Independence 1 (GFI1) is a hematopoietic transcription factor.8,9 The GFI1 protein consists of a N-terminal Snail/Growth factor independence 1 domain and 6 C-terminal zinc fingers.10,11 Gfi1 is involved in: T-cell lymphomagenesis; maturation and activation of B, T, and dendritic cells; regulation of alternative splicing of the CD45 gene in T cells; development of sensory epithelial cells in the inner ear, development of neuroendocrine lung, and Purkinje cells.10,,,,,,,,,–20 In addition, Gfi1−/− mice show reduced self-renewal of hematopoietic stem cells and a block in the development of granulocytes causing severe neutropenia.8,9,21,22
Several patients with congenital neutropenia have mutations in the GFI1 gene, generating a dominant-negative loss of function.23 The combination of a severe neutropenia and the accumulation of atypical monocytes8 in Gfi1-deficient mice is reminiscent of myelodysplastic diseases and thus suggestive of a role of Gfi1 in myeloid leukemia. These observations prompted us to investigate whether mutations or single nucleotide polymorphisms (SNPs) exist that may play a role in the pathogenesis of AML. In this study we report a nonsynonymous SNP, which leads to the replacement of serine by asparagine in the N-terminal part of the coding region of GFI1. The frequency of this SNP has been determined in 2 different cohorts of patients and control patients in Germany and The Netherlands. Our studies suggest that this variation is associated with AML and has a different biochemical function and subnuclear localization compared with the more common variant.
Methods
Patients and control patients
AML patients were identified on the basis of their clinical-pathologic1,2 presentations. Peripheral blood and bone marrow samples were collected before initiation of treatment. The study was approved by the Institutional Review Boards of the participating institutions, and all patients provided written informed consent in accordance with federal and institutional guidelines and the Declaration of Helsinki.
Patients
Patients from Germany.
All AML patients from Germany were recruited by the Study Alliance Leukemia (AML96; AML2003) and by the University Hospitals of Dresden, Marburg, and Essen between 1993 and 2003. Patients were white. The AML96 and AML2003 group recruited their patients from more than 80 hospitals all over Germany. The mean age (± SD) of the German patient cohort was 53.56 years (± 16.57 years; range, 19-86 years) with 54% male patients. More than 90% of the eligible AML patients presenting to the University Hospitals of Dresden, Essen, and Marburg were recruited for genetic studies. The study documentation did not record what percentage of eligible patients took part in the other centers. Because most AML patients in these centers are treated according to study protocols, the participation rate is expected to be also greater than 90%. Smoking status, duration, and intensity of smoking were not recorded consistently and were only available for patients from Essen and Marburg.
Patients from The Netherlands.
All AML patients from The Netherlands were recruited from the European Organization for Treatment and Research of Cancer (EORTC) study group by the University Hospitals at Nijmegen and from the Dutch Hemato-Oncology Cooperative Group Hemato-Oncologie voor Volwassenen Nederland (HOVON) at Rotterdam between 1989 and 2007. Patients were white. The mean age (± SD) of this patient cohort was 50.21 years (± 14.75 years; range, 14-77 years), with 52% of them being male.
Study protocols.
The details of the study protocols have been published previously6,24,–26 or are registered at the National Cancer Institute (EORTC 06 991, NCT 00004128, AML-12/06 991). Patients first register and are later randomized, if at all, when their cytogenetic risk profile is known. At this time, patients had already agreed to provide material for scientific analyses. Thus, this should not affect the composition of the cohort or GFI1 allele frequency.
Patients with t(8;21) translocations.
Besides the t(8;21) patients from the study groups in Germany and The Netherlands, additional white t(8;21) patients were recruited from the City of Hope (COH), the Cancer and Leukemia Group B (CALGB), and the Munich Leukemia Laboratory to correlate relapse-free survival with the presence of a variant GFI136N allele. These patients were recruited between 1992 and 2004. Only those patients were taken into account for whom complete follow-up was available and who were treated in a comparable way regarding induction, consolidation, and maintenance therapy (eg, excluding autologous or allogenic stem cell transplantation).
Smoking status.
Because there is some controversy about the possible role of smoking in AML, which is not yet fully established,27,28 we also compared the smoking status between patients and control persons. The study groups do not record smoking status regularly because it is not an established risk factor for AML. For 80% of a control group in Germany and for 70% and 90% of patients from Essen and Marburg, respectively, the smoking status was determined. The odds ratio for smokers carrying the GFI136N allele to develop AML was 3.4 (Table 1) and 4.3 for nonsmokers (Table 1). Thus, we conclude that smoking does not affect the association between GFI136N and AML
Distribution of GFI136N in AML patients and healthy control patients
| Population . | AA . | GA . | GG . | Frequency allele A . | OR . | P* . | 95% CI . |
|---|---|---|---|---|---|---|---|
| German and Dutch patients and control patients | |||||||
| Patients | 1 | 195 | 1610 | 0.054 ± 0.005 | 1.6 | < 8 × 10−5 | 1.3-2 |
| Control patients | 1 | 118 | 1572 | 0.035 ± 0.003 | |||
| Patients by location | |||||||
| Germany patients | 1 | 134 | 1129 | 0.053 ± 0.004 | 1.6 | < 2 × 10−3 | 1.2-2 |
| Germany control patients | 1 | 87 | 1162 | 0.035 ± 0.004 | |||
| The Netherlands patients | 0 | 61 | 481 | 0.056 ± 0.007 | 1.6 | .02 | 1.1-2.6 |
| The Netherlands control patients | 0 | 31 | 410 | 0.035 ± 0.006 | |||
| According to smoking status† | |||||||
| Smokers AML | 0 | 9 | 39 | 0.09 ± 0.028 | 3.4 | .04 | 1.1-11.8 |
| Smokers control patients | 0 | 4 | 59 | 0.03 ± 0.015 | |||
| Nonsmokers AML | 0 | 13 | 41 | 0.12 ± 0.016 | 4.3 | .003 | 1.5-12.2 |
| Nonsmokers control patients | 0 | 6 | 82 | 0.03 ± 0.013 |
| Population . | AA . | GA . | GG . | Frequency allele A . | OR . | P* . | 95% CI . |
|---|---|---|---|---|---|---|---|
| German and Dutch patients and control patients | |||||||
| Patients | 1 | 195 | 1610 | 0.054 ± 0.005 | 1.6 | < 8 × 10−5 | 1.3-2 |
| Control patients | 1 | 118 | 1572 | 0.035 ± 0.003 | |||
| Patients by location | |||||||
| Germany patients | 1 | 134 | 1129 | 0.053 ± 0.004 | 1.6 | < 2 × 10−3 | 1.2-2 |
| Germany control patients | 1 | 87 | 1162 | 0.035 ± 0.004 | |||
| The Netherlands patients | 0 | 61 | 481 | 0.056 ± 0.007 | 1.6 | .02 | 1.1-2.6 |
| The Netherlands control patients | 0 | 31 | 410 | 0.035 ± 0.006 | |||
| According to smoking status† | |||||||
| Smokers AML | 0 | 9 | 39 | 0.09 ± 0.028 | 3.4 | .04 | 1.1-11.8 |
| Smokers control patients | 0 | 4 | 59 | 0.03 ± 0.015 | |||
| Nonsmokers AML | 0 | 13 | 41 | 0.12 ± 0.016 | 4.3 | .003 | 1.5-12.2 |
| Nonsmokers control patients | 0 | 6 | 82 | 0.03 ± 0.013 |
Allele frequencies for the GFI136N variant among white AML patients and control patients in Germany and in The Netherlands. GFI136N was enriched 1.6-fold in AML patients (P < 8 × 10−5) compared with the control population, which was confirmed after adjusting for age and sex.
AML indicates acute myeloid leukemia; CI, confidence interval; GFI1, Growth Factor Independence 1; and OR, odds ratio.
The Hardy-Weinberg equilibrium was tested and fit expectation with the exception that the frequency of genotype AA was lower than expected (1 vs 5; P = .01).
With reference to Essen and Marburg.
Control persons
Control persons from Germany.
The control groups were recruited by the University Hospitals of Essen, Marburg, and Dresden and consisted of healthy blood or stem cell donors recruited between 1996 and 2003. All control persons were white. Another control group consisted of samples derived from the Department of Pharmacogenetics, University Hospital Essen, with known smoking status and was randomly derived from mandatory citizen registries in the Ruhr area of Germany. These participants were neither physician- nor self-referred. For 80% of these control patients, the smoking status was known. The median age (± SD) of the German control group was 46.03 years (± 12.27 years; range, 19-86 years) with 55% of them being male. All control persons were white.
Control persons from The Netherlands.
The Dutch control patients were recruited by the University Hospitals of Nijmegen and Rotterdam and consisted of healthy blood donors. Control patients were recruited between 1996 and 2006. The median age (±SD) of Dutch control group was 53.58 years (± 13.86 years; range, 19-87 years) with 50% of all participants being male. All control persons were white.
Probability of control persons and patients attending the same institutions.
The diagnosis and subsequent therapy (including a possible subsequent stem cell therapy) of AML requires a specialized hematologic laboratory and department. This type of expertise can only be provided by university hospitals or university-affiliated hospitals such as the ones providing the samples in Germany and The Netherlands for the present study. It is very likely that the vast majority of all AML patients would visit the same institutions in their local area, which were taking part in our study. This implies in case of Essen, Marburg, Dresden, Rotterdam, and Nijmegen that the control persons would most likely visit these same institutions in the event they developed AML.
Therapeutic regimens of patients
Patients with AML (except FAB M3) were treated according to the published multicenter chemotherapy protocols (Marburg, Deutsche Studieninitiative Leukämie [DSIL], EORTC, CALGB, or COH).6,24,–26 A complete remission was achieved if neutrophils (> 1000 or > 1500 neutrophils/μL) and platelets (> 100 000 platelets/μL) recovered in peripheral blood, no blasts were detected in peripheral blood, no signs for extramedullar tumor masses were found, and less than 5% blasts were detectable in the reconstituted bone marrow. The overall survival analysis was restricted to patients younger than 65 years with de novo AML treated in the DSIL (ie, exclusion of AML cases evolving from a preceding myeloid disorder or related to previous anticancer therapy). Relapse-free survival refers to relapse in patients who have initially attained a complete remission. Disease-free survival is also a clinical end point that we use in complete responders, but here the events taken into account are relapse or death whatever comes first. Overall survival is an analysis for all patients.
Statistical analysis
Odds ratios for the German and Dutch population were calculated in analogy of the common odds ratio described by Sasieni29 by use of the χ2 test. The odds ratio for the smoker and nonsmoker cohorts (consisting of the respective control persons and patients) was calculated by use of the Fisher exact test. The unit of analysis for both approaches was the individual person. Odds ratios were adjusted for age and sex with the use of a logistic regression model with the individual person being the unit of analysis, the outcome being whether the person represents patients or control persons, and the key variant being the presence or absence of the GFI136N allele. The overall odds ratio for the German and Dutch populations was calculated on the basis of the Mantel-Haenszel method because allele frequencies were almost identical and thus the individual odds ratios for the 2 populations also.
The Fisher exact test was used with regard to the different subnuclear localization of GFI136S and GFI136N protein. The Hardy-Weinberg equilibrium was calculated by log likelihood ratio χ2. For the difference between age, sex, platelet, and lymphocyte numbers, number of blast cells and percentage of CD34+ cells in the bone marrow, and the distribution of different cytogenetic aberrations, the Mann-Whitney U test was used. For comparing survival rates of the AML patients, the log-rank test was used. For differences in reporter-assay experiments and the mean values of AML1/ETO patients, an unpaired Student t test was chosen. All P values were calculated 2-sided, and values of P less than .05 were considered significantly different. Statistical analysis was performed with Graph-Pad Prism software (GraphPad software) and SysStat 12 software (SysStat).
SNP, mutational analysis, and quality control
Three different methods were used to genotype patients and control patients. GenBank accession numbers were BC074867 for the GFI1 cDNA and NT032977 for genomic DNA. The technical quality of each sequencing result was validated by the assessment of each individual chromatogram. Sequencing results with poor quality (low raw signal intensity, broad peaks, or high background noise) were retested and rejected if the sequencing quality was still low. As a second approach, 20 ng of genomic DNA was used for polymerase chain reaction (PCR) amplification of Exon 2. PCR product was restricted with BfaI (New England Biolabs) for 24 hours. BfaI restricts the GFI136S allele (CTA⇓GC), whereas the GFI136N allele (CTA AC) is not restricted. Third, genotyping was performed on an ABI PRISM 7900 (ABI) or MX3005 (Stratagene) with the use of genomic GFI136S and GFI136N allele-specific primers designed by ABI. Each call was verified with regard to the time course of the intensity increase of the 2 fluorescence markers. Within this third approach, GFI136N-positive samples were reconfirmed, where possible, by the use of 20 ng of genomic DNA for PCR amplification of Exon 2. For SNP alleles rs6662618, rs1325432, rs2031494, rs10782922, rs186682, and rs177371561, primer sets were purchased from ABI, and genotyping was performed on an ABI PRISM 7900 (ABI) or Mx 3005 from Stratagene.
Cell lines and cell culture
Cos 7, NIH-3T3, and HeLa cells were maintained in Dulbecco modified Eagle medium 10% fetal bovine serum (HyClone) and 1% penicillin-streptomycin (Invitrogen). Kasumi-1 cells were maintained in RPMI 1640, 20% fetal bovine serum, and 1% penicillin-streptomycin.
Nuclear matrix preparation, transient transfections, and reporter gene assays
Nuclear matrix preparation was performed as described.30 Cells were transfected with 400 ng of GFI1 binding reporter and β-Gal reporter (400 ng) with Lipofectamine 2000 Transfection Reagent (Invitrogen). In all cases, DNA amount was added up to 1 μg with empty Flag-N3-plasmid. Promoter activity was determined 30 hours after transfection as previously published.14 All transfection settings were repeated 3 times with new plasmid preparations. For Western blot, α-GFI1 (sc-8558; Santa Cruz Biotechnology) and α-ETO (sc-9737; Santa Cruz Biotechnology) were used. Either 50 ng of or 150 ng of GFI136S or GFI136N plasmid and/or 300 ng of AML1/ETO was used.
Immunofluorescence
NIH-3T3 cells were transfected with 150 ng of GFI136S, 150 ng of GFI136N, and 50 ng of AML1/ETO plasmid as previously described.14 After 30 hours, medium was removed, and cells were washed with phosphate-buffered saline, fixed for 10 minutes with ice-cold methanol, washed twice with phosphate-buffered saline, and equilibrated 30 minutes in solution A (10mM Tris, pH 7.5; 100mM NaCl; 0.05% Tween 20; 1% bovine serum albumin). Cells were stained with primary antibody (Gfi1 N-20) or Flag Antibody (M2; Sigma-Aldrich), which was diluted 1:200 in solution A, for a 1-hour incubation time and secondary labeled (FITC or rhodamine) antibody (Jackson ImmunoResearch Laboratories). Nuclear staining was done with the use of TO-PRO-3 (Invitrogen), and cells were analyzed with the use of confocal microscope (LSM; Zeiss) and LSM Browser 5.0 software. For detecting endogenous GFI1 an α-GFI1 antibody (clone 2.5 D17; Sigma-Aldrich) was used.
Immunoprecipitation
Cos 7 cells were electroporated with 10 μg of GFI136S or GFI136N or AML1/ETO plasmid in Dulbecco modified Eagle medium at 500 μF and 250 V. After 24 hours, cells were disrupted in Flag lysis buffer (25mM Tris, pH 7.4; 150mM NaCl; 1mM CaCl2; 1% TritonX-100; and 3% bovine serum albumin; Sigma-Aldrich). After 2 hours of incubation of either 250 μg of GFI136S or GFI136N with 250 μg of AML1/ETO lysate, complexes were precipitated with α-GFI1 antibodies, bound to Protein-Sepharose G (Sigma-Aldrich), and then subjected to separation by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and detected by immunoblotting using an α-ETO antibody (Santa Cruz Biotechnology).
Results
The SNP GFI136N predisposes to AML
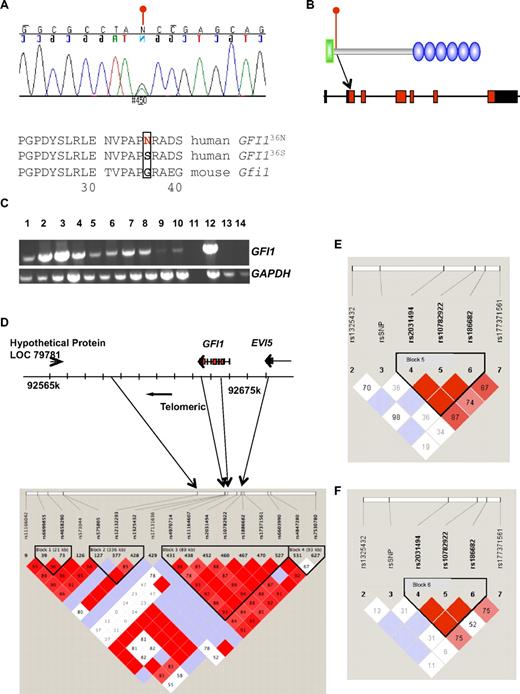
To determine whether GFI1 mutations or SNPs might play a role in the pathogenesis of AML, we sequenced genomic DNA from bone marrow or blood from 92 AML patients treated at the University Hospital Essen. We repeatedly found in 13 cases a heterozygosity caused by a single-base substitution in the GFI1 coding region (c.107G > A), leading to the replacement of a serine (S) by an asparagine (N) residue at amino acid position 36 (Figure 1A-B).GFI1 mRNA was expressed in the blast cells of the patients that were collected at time of diagnosis (Figure 1C). The GFI136N variant allele also was present in the epithelial cell DNA of GFI136N heterozygous AML patients (data not shown), from whom epithelial DNA was available. Finally the GFI136N allele also could be detected in the blood sample of normal control persons (Table 1). These findings indicate that the base substitution at c.107G>A represents an SNP and not a somatic acquired mutation. The fact that GFI136N is indeed a SNP was confirmed by the UCSC genome bioinformatics group denominating it as rs34631763.
Expression, genomic representation, and LD of GFI136N. (A) Chromatogram of the GFI136S/36N cDNA sequence of one of the patients. Change of G to A at position c107 (c107G>A) results in the replacement of serine by asparagine. The neighboring amino acid sequences of the more common human and mouse sequences are shown. (B) Location of the GFI136N variant on genomic and protein level. The SNP is located in exon 2 and replaces a serine by an asparagine at amino acid position 36. Green indicates N-terminal Snail/Growth factor independence 1 repressor domain of GFI1; blue, 6 C2H2 zinc finger domains. (C) GFI1 mRNA expression in different patients. GFI1 expression in different AML patients was semiquantitatively assayed by reverse transcriptase PCR. Lanes 1 to 9 represent bone marrow and peripheral blood samples of AML patients at diagnosis. Lane 10 shows cell from the t(8;21)-positive Kasumi 1 AML cell line. Lane 11 shows the control without reverse transcriptase and lane 12 a peripheral blood aphaeresis sample from an AML patient. Lane 13 shows HeLa cells (human cervical cancer) and lane 14 cells originating from a patient with a chronic lymphocytic leukemia (CLL). (D) Results of LD analysis in the genomic region encompassing the GFI1 and EVI5 loci and neighboring regions. LD block 3 spans part of GFI1 and EVI5. (E) LD as determined after genotyping of 39 GFI136N heterozygous AML patients from Essen, Marburg, and the DSIL study group. The genotypes of 5 SNPs in the proximity of the GFI136N SNP were determined. The results show that GFI136N is not within the LD block that spans part of EVI5. (F) Results of LD of a group consisting of the aforementioned 39 GFI136N heterozygous AML patients and 26 healthy persons homozygous for GFI136S from Essen. Similar to the analysis described previously, GFI136N is not within the LD block that spans part of EVI5.
Expression, genomic representation, and LD of GFI136N. (A) Chromatogram of the GFI136S/36N cDNA sequence of one of the patients. Change of G to A at position c107 (c107G>A) results in the replacement of serine by asparagine. The neighboring amino acid sequences of the more common human and mouse sequences are shown. (B) Location of the GFI136N variant on genomic and protein level. The SNP is located in exon 2 and replaces a serine by an asparagine at amino acid position 36. Green indicates N-terminal Snail/Growth factor independence 1 repressor domain of GFI1; blue, 6 C2H2 zinc finger domains. (C) GFI1 mRNA expression in different patients. GFI1 expression in different AML patients was semiquantitatively assayed by reverse transcriptase PCR. Lanes 1 to 9 represent bone marrow and peripheral blood samples of AML patients at diagnosis. Lane 10 shows cell from the t(8;21)-positive Kasumi 1 AML cell line. Lane 11 shows the control without reverse transcriptase and lane 12 a peripheral blood aphaeresis sample from an AML patient. Lane 13 shows HeLa cells (human cervical cancer) and lane 14 cells originating from a patient with a chronic lymphocytic leukemia (CLL). (D) Results of LD analysis in the genomic region encompassing the GFI1 and EVI5 loci and neighboring regions. LD block 3 spans part of GFI1 and EVI5. (E) LD as determined after genotyping of 39 GFI136N heterozygous AML patients from Essen, Marburg, and the DSIL study group. The genotypes of 5 SNPs in the proximity of the GFI136N SNP were determined. The results show that GFI136N is not within the LD block that spans part of EVI5. (F) Results of LD of a group consisting of the aforementioned 39 GFI136N heterozygous AML patients and 26 healthy persons homozygous for GFI136S from Essen. Similar to the analysis described previously, GFI136N is not within the LD block that spans part of EVI5.
To determine the overall frequency of this SNP, we determined its frequency in 2 independent white cohorts from Germany and The Netherlands and their respective control subjects. In total, approximately 1806 AML (including the initial 92 from Essen) patients treated at hospitals in Germany and The Netherlands (see “Patients”), as well as 1691 healthy control patients from different locations in Germany and The Netherlands, were tested for the frequency of this SNP. The GFI136N variant was observed in Germany and The Netherlands with the same elevated frequency in patients over controls (Table 1, odds ratio 1.6, P = 8 × 10−5 using a Mantel-Haenszel approach). This correlation was confirmed (odds ratio 1.5, P ≤ 1 × 10−3, 95% confidence interval, 1.1-2) after adjusting for age and sex (see “Patients”) and this finding was independent of smoking status (Table 1), which is a controversial risk factor in the development of AML.27,28 Interestingly, the number of detected homozygous carriers was lower in the control group (1 vs expected 2) and significantly lower in the patient cohort (1 observed vs expected 5; P = .01), indicating that homozygous carriers might experience a selective disadvantage (Table 1). We verified also the allele frequency of GFI136N in healthy control patients of other ethnicities such as Tanzania and Nigeria (allele frequency 0.004 in 210 samples) and Chinese (allele frequency 0 in 205 samples). The incidence of GFI136N is lower in these populations, as is the incidence of AML in these populations.31
Linkage disequilibrium of GFI136N
A greater frequency of GFI136N among AML patients could be explained by linkage disequilibrium (LD) of GFI136N with other, unknown, causative genetic variations. To address this, we analyzed patterns of LD in the CEPH sample32 using data from the International HapMap project.33 In the telomeric direction, the most proximal SNP (rs11164607) to GFI136N is 1 kbp away and belongs to the LD block 3 spanning part of GFI1 and EVI5 (Figure 1D; Table 2). The locus of GFI1 is not in LD with any genes, which lie further in the telomeric direction (Figure 1C). With regard to the centromeric direction, we tested 5 SNPs around GFI136N in 28 healthy control patients homozygous for GFI136S and 39 AML cases heterozygous for the GFI136N. We found complete allelic association of alleles (r2 = 0.88) at loci located in the region between rs2031494 and rs186682 but not between these alleles and GFI136N (Figure 1E-F). In conclusion, it is unlikely that variants in EVI5 are associated with AML. However, a population stratification effect cannot be entirely ruled out.34
Position of tested SNPs relative to each other
| Name . | Relative position, in bp . |
|---|---|
| rs6662618 | 0 |
| rs1325432 | 5533 |
| Rs4970714 | 5646 |
| Rs11164607 | 12 327 |
| Rs34631763 (rsSNP = GFI136N) | 13 518 |
| rs2031494 | 30 558 |
| rs10782922 | 39 184 |
| rs186682 | 41 864 |
| rs177371561 | 45 828 |
| Name . | Relative position, in bp . |
|---|---|
| rs6662618 | 0 |
| rs1325432 | 5533 |
| Rs4970714 | 5646 |
| Rs11164607 | 12 327 |
| Rs34631763 (rsSNP = GFI136N) | 13 518 |
| rs2031494 | 30 558 |
| rs10782922 | 39 184 |
| rs186682 | 41 864 |
| rs177371561 | 45 828 |
GFI1 indicates Growth Factor Independence 1; and SNP, single nucleotide polymorphism.
GFI136N is not associated with other established AML markers
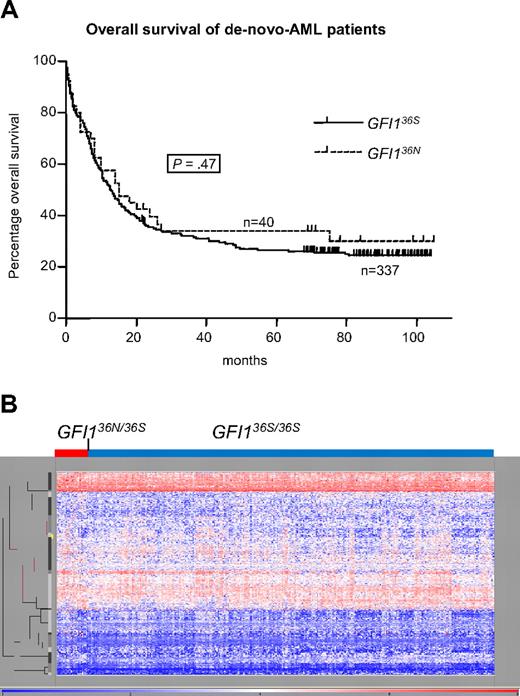
After determining that GFI136N predisposed to AML in these 2 populations, we investigated whether GFI136N also might be associated with prognosis or other known AML factors. Among 377 de novo AML patients recruited by the AML96 study group, the presence of GFI136N did not correlate with any established factors,1,2,6,7 such as age, white blood cell count, lactate dehydrogenase level, frequency of CD34+ cells, morphologic subgroups as defined by the French-American-British classification, cytogenetic aberrations, or mutation status of FLT3, NPM1, or PTPN11 (Tables 3,Table 4–5), nor with 5-year overall survival or relapse-free survival (Figure 2A; data not shown). Valk et al24 have recently published a cluster analysis of genome-wide mRNA expression patterns and mutational analysis that separated the Rotterdam AML patient into subgroups that correlate in many cases with known AML risk factors (eg, t[8;21]; EVI1 or WT1 expression; KRAS, NRAS, or CEBPA mutations). We determined the carrier status of the patients of this population with regard to status of GFI136N and could observe that GFI136N heterozygous patients were not associated within any cluster (Figure 2B) or with any of the aforementioned prognostic factors.
Features of AML96 GFI136S and GFI136N patients
| . | GFI136N (n) . | GFI136S (n) . | P . |
|---|---|---|---|
| Number | 40 | 337 | |
| Median age, y | 53.5 (40) | 57 (337) | .746 |
| Sex, % male | 42.5 (17) | 51 (172) | .307 |
| Leukocytes, per fl | 49 (40) | 39 (337) | .652 |
| Blast percentage | 61 (35) | 62 (303) | .618 |
| LDH, IU | 542 (38) | 691 (323) | .751 |
| Platelets, per fl | 92 (40) | 77 (337) | .225 |
| FLT3 status negative, %* | 90 (40) | 88 (332) | .688 |
| NPM1 mutations, % | 64 (14) | 50 (93) | .64 |
| PTPN11 mutations, % | 16 (3) | 3 (5) | .03 |
| MLL PTD, %† | 6 (1) | 7 (11) | 1 |
| . | GFI136N (n) . | GFI136S (n) . | P . |
|---|---|---|---|
| Number | 40 | 337 | |
| Median age, y | 53.5 (40) | 57 (337) | .746 |
| Sex, % male | 42.5 (17) | 51 (172) | .307 |
| Leukocytes, per fl | 49 (40) | 39 (337) | .652 |
| Blast percentage | 61 (35) | 62 (303) | .618 |
| LDH, IU | 542 (38) | 691 (323) | .751 |
| Platelets, per fl | 92 (40) | 77 (337) | .225 |
| FLT3 status negative, %* | 90 (40) | 88 (332) | .688 |
| NPM1 mutations, % | 64 (14) | 50 (93) | .64 |
| PTPN11 mutations, % | 16 (3) | 3 (5) | .03 |
| MLL PTD, %† | 6 (1) | 7 (11) | 1 |
The characteristics of GFI136 homozygous and carriers of the GFI136N allele with regard to different parameters are described.
AML indicates acute myeloid leukemia; GFI1, Growth Factor Independence 1; LDH, lactate dehydrogenase; MLL, mixed-lineage leukemia; and PTD, partial tandem duplication.
No internal tandem duplication mutation.
PTD mutations.
Distribution of AML96 DSIL patients with the GFI136S or GFI136N form, respectively, in morphological AML subtypes as defined in the FAB classification
| FAB subgroup . | GFI136S% (no. of all cases) . | GFI1S36N% (no. of all cases) . | P . |
|---|---|---|---|
| M0 (17) | 88 (15) | 12 (2) | .69 |
| M1 (79) | 90 (71) | 10 (8) | 1 |
| M2 (132) | 89 (117) | 11 (15) | .723 |
| M4 (48) | 98 (47) | 2 (1) | .046 |
| M4eo (21) | 91 (19) | 10 (2) | 1 |
| M5a (51) | 92 (47) | 8 (4) | .629 |
| M5b (14) | 71 (10) | 29 (4) | .043 |
| M6 (8) | 75 (6) | 25 (2) | .197 |
| M7 (4) | 75 (3) | 25 (1) | .355 |
| RAEB-T (2) | 100 (2) | 0 | 1 |
| Total (377) | 89 | 11 |
| FAB subgroup . | GFI136S% (no. of all cases) . | GFI1S36N% (no. of all cases) . | P . |
|---|---|---|---|
| M0 (17) | 88 (15) | 12 (2) | .69 |
| M1 (79) | 90 (71) | 10 (8) | 1 |
| M2 (132) | 89 (117) | 11 (15) | .723 |
| M4 (48) | 98 (47) | 2 (1) | .046 |
| M4eo (21) | 91 (19) | 10 (2) | 1 |
| M5a (51) | 92 (47) | 8 (4) | .629 |
| M5b (14) | 71 (10) | 29 (4) | .043 |
| M6 (8) | 75 (6) | 25 (2) | .197 |
| M7 (4) | 75 (3) | 25 (1) | .355 |
| RAEB-T (2) | 100 (2) | 0 | 1 |
| Total (377) | 89 | 11 |
By the use of a mosaic analysis, no significant difference could be observed between patients carrying the 2 GFI1 variants.
AML indicates acute myeloid leukemia; DSIL, Deutsche Studieninitiative Leukämie; FAB, French-American-British; and GFI1, Growth Factor Independence 1.
Frequency of cytogenetic aberrations in AML patients from the AML96DSIL study group carrying the GFI136S or GFI136N allele with regard to all aberrations in this group
| Type of aberration . | GFI136S, % (n) . | GFI136N, % (n) . | P . |
|---|---|---|---|
| t(8;21) | 17 (21) | 8 (1) | .62 |
| t(6;9) | 3 (4) | 0 | 1 |
| t(9;11) | 1 (1) | 0 | 1 |
| t(9;22) | 2 (2) | 0 | 1 |
| del(5q) | 10 (12) | 16 (2) | .3 |
| −del(7q) | 8 (9) | 17 (2) | .26 |
| inv(3q) | 1 (1) | 0 | 1 |
| inv(16) | 14 (17) | 16 (2) | .83 |
| −5 | 2 (2) | 8 (1) | .25 |
| −7 | 12 (14) | 16 (2) | .64 |
| −Y | 6 (7) | 8 (1) | .9 |
| −Trisomy 8 | 20 (23) | 25 (3) | .62 |
| Trisomy 11 | 3 (4) | 0 | 1 |
| −Trisomy 13 | 4 (5) | 0 | 1 |
| −Trisomy 21 | 5 (6) | 0 | 1 |
| Trisomy 22 | 4 (5) | 17 (1) | .44 |
| Complex | 32 (38) | 33 (4) | .74 |
| Abn(11q) | 8 (9) | 0 | 1 |
| Abn(12p) | 9 (10) | 8 (1) | 1 |
| Type of aberration . | GFI136S, % (n) . | GFI136N, % (n) . | P . |
|---|---|---|---|
| t(8;21) | 17 (21) | 8 (1) | .62 |
| t(6;9) | 3 (4) | 0 | 1 |
| t(9;11) | 1 (1) | 0 | 1 |
| t(9;22) | 2 (2) | 0 | 1 |
| del(5q) | 10 (12) | 16 (2) | .3 |
| −del(7q) | 8 (9) | 17 (2) | .26 |
| inv(3q) | 1 (1) | 0 | 1 |
| inv(16) | 14 (17) | 16 (2) | .83 |
| −5 | 2 (2) | 8 (1) | .25 |
| −7 | 12 (14) | 16 (2) | .64 |
| −Y | 6 (7) | 8 (1) | .9 |
| −Trisomy 8 | 20 (23) | 25 (3) | .62 |
| Trisomy 11 | 3 (4) | 0 | 1 |
| −Trisomy 13 | 4 (5) | 0 | 1 |
| −Trisomy 21 | 5 (6) | 0 | 1 |
| Trisomy 22 | 4 (5) | 17 (1) | .44 |
| Complex | 32 (38) | 33 (4) | .74 |
| Abn(11q) | 8 (9) | 0 | 1 |
| Abn(12p) | 9 (10) | 8 (1) | 1 |
No significant difference for a specific aberration in association with the 2 GFI1 variants could be observed.
AML indicates acute myeloid leukemia; and GFI1, Growth Factor Independence 1.
Influence of GFI136N allele on progression. (A) Prognostic impact of GFI136N on disease progression of AML patients. GFI136S (homozygous for GFI136S) and GFI136N (heterozygous for GFI136N or homozygous for GFI136N; 1 patient only) de novo AML patients, recruited from the DSIL, had the same prognosis for a 5-year overall survival rate of approximately 27% (GFI136S) and 34% (GFI136N). Also, the 5-year relapse-free survival was not different between both groups (data not shown). (B) Genome-wide mRNA expression pattern of patients heterozygous for the GFI136N allele was not overall different from GFI136S-homozygous patients. Genome-wide RNA expression data were obtained by gene array analysis (Affymetrix) from a patient cohort from the HOVON study group analyzed in The Netherlands (Rotterdam; n = 350).
Influence of GFI136N allele on progression. (A) Prognostic impact of GFI136N on disease progression of AML patients. GFI136S (homozygous for GFI136S) and GFI136N (heterozygous for GFI136N or homozygous for GFI136N; 1 patient only) de novo AML patients, recruited from the DSIL, had the same prognosis for a 5-year overall survival rate of approximately 27% (GFI136S) and 34% (GFI136N). Also, the 5-year relapse-free survival was not different between both groups (data not shown). (B) Genome-wide mRNA expression pattern of patients heterozygous for the GFI136N allele was not overall different from GFI136S-homozygous patients. Genome-wide RNA expression data were obtained by gene array analysis (Affymetrix) from a patient cohort from the HOVON study group analyzed in The Netherlands (Rotterdam; n = 350).
The nuclear localization and repressor activity of the GFI136N variant
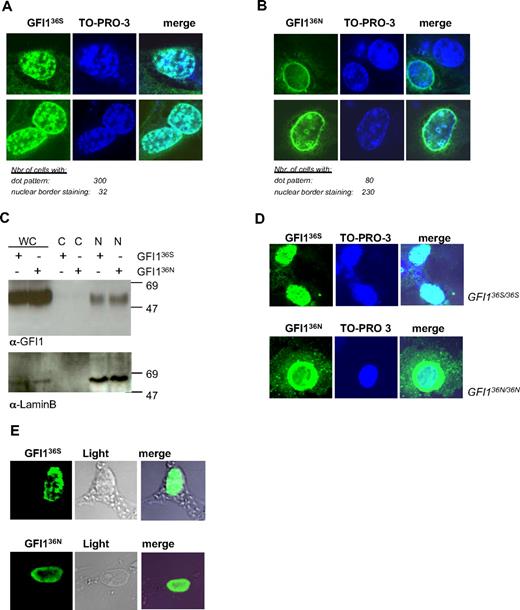
After investigating the association of GFI136N with AML we investigated the possible molecular mechanism behind this observation. We transfected cells with vectors encoding the 2 GFI1 variants and noticed that the 2 forms of GFI1 differed in their subnuclear localization (Figure 3A-B). GFI136N was predominantly localized at the nuclear/cytoplasmic border, whereas GFI136S showed the previously described nuclear dotted pattern (Figure 3A-B).35 However, both GFI1 protein variants could be copurified with components of the nuclear compartment (Figure 3C), indicating that they are both located in the nucleus. This difference was confirmed in more than 300 single cells transfected to express one or the other of the 2 Flag-tagged GFI1 protein variants (Figure 3A-B). The same results were obtained in independent experiments with the use of either an α-Gfi1 antibody (Figure 3A-B) or an α-Flag-tag antibody (data not shown).
GFI136S and GFI136N proteins show different subnuclear localization. (A) NIH-3T3 cells were transfected with GFI136S expression plasmid. GFI1 appears green; DNA, blue. The right column represents the merging of both staining. The numbers of cells analyzed for GFI136S are indicated. GFI136S is mainly localized in a dotted pattern. (B) NIH-3T3 cells were transfected with a GFI136N expression plasmid. The numbers of cells analyzed for GFI136N are indicated. GFI136N is localized at the nuclear border. (C) NIH-3T3 cells were transfected with either a GFI136S or GFI136N expression plasmid. Cell lysates were fractionated. Both protein variants are localized in the nuclear cell fraction and cannot be found in the cytosolic fraction. (D) Analysis of a bone marrow sample from the only available homozygous for GFI136N. As in transfected cells, GFI136N was mainly located at the nuclear/cytoplasmic border. As a control, cells from the Kasumi 1 t(8;21)-positive AML cell line were used. This cell line was originally derived from a patient with French-American-British M2 AML and is homozygous for GFI136S. (E) Nuclear matrix preparation of NIH-3T3 cells transiently transfected with expression vectors for GFI136S or GFI136N. Both variants are still attached to nuclear matrix components, although the GFI136N variant appears to be more concentrated at the nuclear membrane, consistent with our observation in regular cell transfection assays.
GFI136S and GFI136N proteins show different subnuclear localization. (A) NIH-3T3 cells were transfected with GFI136S expression plasmid. GFI1 appears green; DNA, blue. The right column represents the merging of both staining. The numbers of cells analyzed for GFI136S are indicated. GFI136S is mainly localized in a dotted pattern. (B) NIH-3T3 cells were transfected with a GFI136N expression plasmid. The numbers of cells analyzed for GFI136N are indicated. GFI136N is localized at the nuclear border. (C) NIH-3T3 cells were transfected with either a GFI136S or GFI136N expression plasmid. Cell lysates were fractionated. Both protein variants are localized in the nuclear cell fraction and cannot be found in the cytosolic fraction. (D) Analysis of a bone marrow sample from the only available homozygous for GFI136N. As in transfected cells, GFI136N was mainly located at the nuclear/cytoplasmic border. As a control, cells from the Kasumi 1 t(8;21)-positive AML cell line were used. This cell line was originally derived from a patient with French-American-British M2 AML and is homozygous for GFI136S. (E) Nuclear matrix preparation of NIH-3T3 cells transiently transfected with expression vectors for GFI136S or GFI136N. Both variants are still attached to nuclear matrix components, although the GFI136N variant appears to be more concentrated at the nuclear membrane, consistent with our observation in regular cell transfection assays.
The dotted nuclear localization of the more common GFI136S variant also was observed in homozygous GFI136S AML cells (Kasumi1), which are derived from a t(8;21) patient (Figure 3D) and in an another AML cell line (HL60 cells; data not shown). By contrast, the same aberrant subnuclear localization of the variant GFI136N form also was observed in a primary tumor sample of the only available homozygous GFI136N AML patient (Figure 3D). This excludes that the aberrant nuclear localization of GFI136N is a particularity of transfected cells. We also found that GFI136N was still able to attach to the nuclear matrix similar to the finding previously reported for the more common GFI136S variant.30 However, it was evident that GFI136N showed a different localization within the nuclear matrix structure than GFI136S, which is consistent with its variant nuclear localization in the cell (Figure 3E).
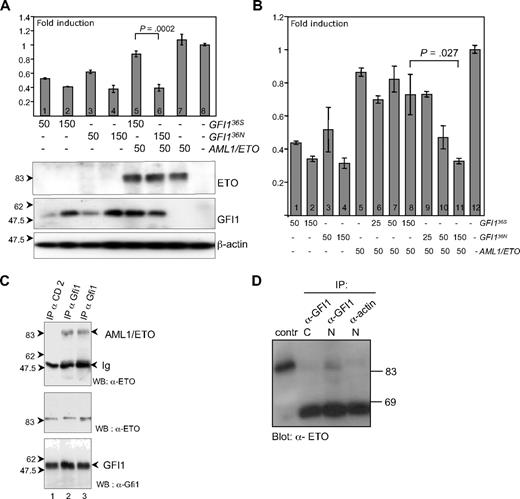
To investigate whether the different subnuclear localization of GFI136N might affect its activity as a transcriptional repressor, we performed a previously described luciferase reporter gene assay in which a promoter containing the GFI1 binding site is used14,36 and that can be repressed by GFI1. We observed that both GFI136S and GFI136N were similarly active as transcriptional repressors (Figure 4A lanes 1-4 and 8; Figure 4B lanes 1-4). This finding suggested that a functional difference of both GFI1 variants might be restricted to specific target genes or alternatively might only become apparent in a particular context for instance in situations in which the function of GFI1 is modulated by other proteins that form complexes with GFI1.
GFI136S and GFI136N maintain similar repressor activity but are differentially regulated by AML1/ETO. (A) Transient transfection of HeLa cells with expression plasmids for GFI136S, GFI136N, AML1/ETO fusion protein, and with a luciferase reporter gene containing Gfi1 binding sequences. Both GFI136S and GFI136N were able to repress the Gfi1 promoter in a concentration-dependent manner (lanes 1–2 and 3–4, respectively), whereas AML1/ETO alone did not influence reporter gene activity (lane 8). In the presence of AML1/ETO, the GFI136S protein lost its repression capacity, but GFI136N retained its repressor activity (lanes 5–6; P < .001). Error bars represent SEM. (B) Same as panel A but with different amounts of expression plasmid transfected as indicated. (C) GFI136S and GFI136N both coprecipitate in transfected cells with AML1/ETO. Cos7 cells were transfected with GFI136S, GFI136N, or AML1/ETO expression plasmids. Immunoprecipitations (IP) were performed with α-GFI1 antibody. AML1/ETO immunoprecipitates in the lysates with GFI136S or the variant GFI136N (lanes 2 and 3, respectively). A control IP was performed with a goat α-CD2 antibody (lane 1). An input control demonstrated the presence of both GFI1 and the AML1/ETO protein. (D) GFI136S coprecipitates with AML1/ETO in extracts from Kasumi1 cells, which is a cell line derived from a t(8;21) AML patient. Nuclear and cytosolic extracts of Kasumi1 cells were prepared and incubated either with the α-Gfi1 (N20) antibody or with a nonspecific α-actin antibody. The cell extracts were immunoblotted and developed with an α-ETO antibody. As a positive control, AML1/ETO transfected Cos7 cells were used.
GFI136S and GFI136N maintain similar repressor activity but are differentially regulated by AML1/ETO. (A) Transient transfection of HeLa cells with expression plasmids for GFI136S, GFI136N, AML1/ETO fusion protein, and with a luciferase reporter gene containing Gfi1 binding sequences. Both GFI136S and GFI136N were able to repress the Gfi1 promoter in a concentration-dependent manner (lanes 1–2 and 3–4, respectively), whereas AML1/ETO alone did not influence reporter gene activity (lane 8). In the presence of AML1/ETO, the GFI136S protein lost its repression capacity, but GFI136N retained its repressor activity (lanes 5–6; P < .001). Error bars represent SEM. (B) Same as panel A but with different amounts of expression plasmid transfected as indicated. (C) GFI136S and GFI136N both coprecipitate in transfected cells with AML1/ETO. Cos7 cells were transfected with GFI136S, GFI136N, or AML1/ETO expression plasmids. Immunoprecipitations (IP) were performed with α-GFI1 antibody. AML1/ETO immunoprecipitates in the lysates with GFI136S or the variant GFI136N (lanes 2 and 3, respectively). A control IP was performed with a goat α-CD2 antibody (lane 1). An input control demonstrated the presence of both GFI1 and the AML1/ETO protein. (D) GFI136S coprecipitates with AML1/ETO in extracts from Kasumi1 cells, which is a cell line derived from a t(8;21) AML patient. Nuclear and cytosolic extracts of Kasumi1 cells were prepared and incubated either with the α-Gfi1 (N20) antibody or with a nonspecific α-actin antibody. The cell extracts were immunoblotted and developed with an α-ETO antibody. As a positive control, AML1/ETO transfected Cos7 cells were used.
GFI136N is refractory to AML1/ETO-mediated regulation
To test whether functional difference of both GFI1 variants was restricted to specific partners, we used AML1/ETO. This protein is a previously described interaction partner of GFI1 that plays a role in AML pathogenesis.30 It is a hallmark protein for AML patients with a t(8;21). The role of AML1/ETO in the initiation of AML has been investigated both in vitro and in vivo.37,,,,–42 In our case coimmuneprecipitation experiments confirmed that both GFI136N and GFI136S were able to bind to AML1/ETO in transfected cells (Figure 4C) and that GFI136S could bind to AML1/ETO at endogenous expression levels in Kasumi1 cells, a AML-blast cell lined derived from a t(8;21) patient (Figure 4D). However, when we tested the repressor activity of both GFI1 variants in the presence of AML1/ETO using the same GFI1-dependent luciferase reporter assay, we observed that the level of repression mediated by the more common GFI136S form was clearly diminished in presence of AML1/ETO (Figure 4A lane 5; Figure 4B lanes 6-8). In contrast, the GFI136N variant maintained its full repressory activity in the presence of AML1/ETO (Figure 4A lane 6; Figure 4B lane 11). This finding suggested that AML1/ETO can negatively regulate the activity of GFI136S but can no longer exert this effect on the GFI136N variant. This example is one in which the 2 GFI1 variants differ in their function in a specific setting.
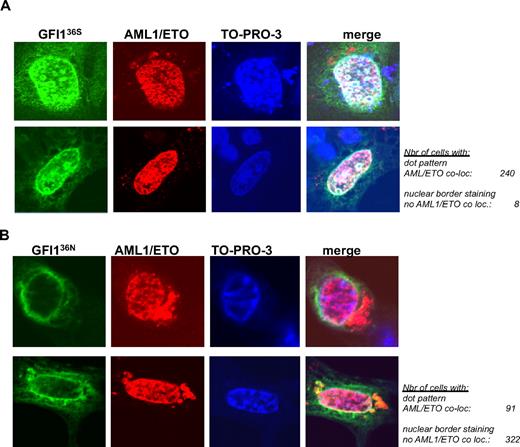
The different repressory activity of GFI136N or GFI136S in the presence of AML1/ETO might be attributable to the different subnuclear localization of both variants. To clarify this, we tested whether GFI136S and GFI136N could colocalize with AML1/ETO. AML1/ETO showed a previously described even nuclear localization40 and colocalized with the GFI136S (Figure 5A) in a statistically significant way in transfected cells (P = .001). However, no areas of overlapping signals were detected in cells coexpressing AML1/ETO and GFI136N. Both GFI136N and AML1/ETO remained separated at the nuclear/cytoplasmic border and in the nucleus (Figure 5B) and did not colocalize in the majority (88%) of transfected cells (Figure 5A-B, P = .001, between GFI136S and GFI136N).
AML1/ETO colocalizes with GFI136S but not with the variant GFI136N. (A) NIH 3T3 cells were transfected with expression vectors for AML1/ETO and GFI136S (original magnification ×100). (B) NIH 3T3 cells were transfected with expression vectors for AML1/ETO and the GFI136N variant form. Expression of GFI1 proteins was revealed by immune-fluorescence with an α-GFI1 antibody and a secondary FITC labeled antibody. The presence of AML1/ETO was revealed by staining with the α-ETO antibody and a secondary rhodamine-labeled antibody. Nuclei were visualized by treatment with the DNA dye TO-PRO-3 (blue). The staining for each protein and DNA is represented in a separate column. Examples of 2 different cells are given for each setting in 2 different rows. The merging of all 3 fluorescence signals is depicted in the last column at the right side and results in a white staining, which can be detected when the signals corresponding to the GFI136S protein and AML1/ETO are merged. White areas or white spots in the nuclei of transfected cells suggest a colocalization of AML1/ETO and the GFI136S protein (A). In contrast, no such white areas can be detected between GFI136N and AML1/ETO (B). The green signal representing the GFI136N protein remained at the nuclear/cytoplasmic border, well separated from the red signal in the nucleus that represents AML1/ETO (B), clearly indicating a lack of colocalization between both. The number of cells analyzed for the subnuclear localization and a colocalization with AML1/ETO are indicated (original magnification ×100).
AML1/ETO colocalizes with GFI136S but not with the variant GFI136N. (A) NIH 3T3 cells were transfected with expression vectors for AML1/ETO and GFI136S (original magnification ×100). (B) NIH 3T3 cells were transfected with expression vectors for AML1/ETO and the GFI136N variant form. Expression of GFI1 proteins was revealed by immune-fluorescence with an α-GFI1 antibody and a secondary FITC labeled antibody. The presence of AML1/ETO was revealed by staining with the α-ETO antibody and a secondary rhodamine-labeled antibody. Nuclei were visualized by treatment with the DNA dye TO-PRO-3 (blue). The staining for each protein and DNA is represented in a separate column. Examples of 2 different cells are given for each setting in 2 different rows. The merging of all 3 fluorescence signals is depicted in the last column at the right side and results in a white staining, which can be detected when the signals corresponding to the GFI136S protein and AML1/ETO are merged. White areas or white spots in the nuclei of transfected cells suggest a colocalization of AML1/ETO and the GFI136S protein (A). In contrast, no such white areas can be detected between GFI136N and AML1/ETO (B). The green signal representing the GFI136N protein remained at the nuclear/cytoplasmic border, well separated from the red signal in the nucleus that represents AML1/ETO (B), clearly indicating a lack of colocalization between both. The number of cells analyzed for the subnuclear localization and a colocalization with AML1/ETO are indicated (original magnification ×100).
We also verified whether the different subnuclear localization of GFI136S or GFI136N might influence survival of t(8;21)-positive patients. In total, 61 t(8;21) patients recruited in Germany, The Netherlands, and the United States were taken in consideration (for selection criteria, see “Patients”). Among the 54 patients homozygous for the more common GFI136S allele, 60% did not show any relapse 5 years after initial remission. In contrast, of 7 t(8;21) patients carrying 1 variant GFI136N allele, only 40% were relapse free 5 years after initial remission, and the median relapse-free survival for GFI136N heterozygous t(8;21) patients was only 5 months.
Discussion
In this study, we have demonstrated that a SNP of GFI1 (GFI136N) was associated with AML in a group of patients and control persons from Germany and that this association could be reproduced in an independent second patient and control cohort from The Netherlands. The association of GFI136N with AML was statistically significant and independent of age, sex, smoking status, and other SNPs in neighboring genes and also was independent of a number of established AML markers (such as FLT3 or NPM mutations and others). Microarray expression data from a large subgroup of AML patients from The Netherlands confirmed this notion. Analysis of clinical data also confirmed that the presence of the SNP coding for the variant GFI136N allele did not affect overall prognosis of AML patients.
It is remarkable that the number of homozygous GFI136N patients with regard to the patient and control patient population was lower than expected. This finding suggests that a selective disadvantage exists for homozygosity at this locus, supporting the view that GFI136N has a pathophysiologic function. A recent case report43 of a patient with severe chronic neutropenia who demonstrated a transient expression of the GFI136N allele (described in this study as a somatic mutation) in hematopoietic cells supports such a pathologic role of the GFI1 variant, in particular because chronic neutropenia is a disease often leading to AML. The physiologic mechanism by which GFI136N predisposes to AML remains to be determined, but it is unlikely that it exerts a dominant-negative effect in contrast to the somatic GFI1 mutations described in neutropenic patients23 ), because the differential blood counts of 2 GFI136N heterozygous AML patients in remission were normal (Table 6) and the GFI136N mRNA was still expressed (data not shown).
Differential blood counts of 2 AML patients in remission that carry the variant GFI136N allele
| . | Patient 1 . | Patient 2 . | Normal range . |
|---|---|---|---|
| Leukocytes, 1/fl | 10.9 | 4.5 | 4.5-11 |
| Band granulocytes, % | 0 | 3 | 0-6 |
| Segmented granulocytes, % | 64 | 55 | 40-75 |
| Eosinophils, % | 2 | 1 | 1-7 |
| Lymphocytes, % | 24 | 30 | 22-40 |
| Monocytes, % | 10 | 11 | 1-10 |
| . | Patient 1 . | Patient 2 . | Normal range . |
|---|---|---|---|
| Leukocytes, 1/fl | 10.9 | 4.5 | 4.5-11 |
| Band granulocytes, % | 0 | 3 | 0-6 |
| Segmented granulocytes, % | 64 | 55 | 40-75 |
| Eosinophils, % | 2 | 1 | 1-7 |
| Lymphocytes, % | 24 | 30 | 22-40 |
| Monocytes, % | 10 | 11 | 1-10 |
The presence of the GFI136N allele did not alter hematological parameters in two patients after achieving remission.
AML indicates acute myeloid leukemia; and GFI1, Growth Factor Independence 1.
One biochemical feature that distinguishes GFI136N from GFI136S was its different subnuclear localization. We and other groups have previously reported that Gfi1 is localized in nuclear dots18 and binds to components of the nuclear matrix.30 We confirmed this for GFI136S in different AML blast cell lines and with regard to binding to the nuclear matrix. In contrast, although GFI136N was still binding to the nuclear matrix in transfected cells, immunofluorescence data on a large number of transfected cells indicated that it is predominantly localized at the nuclear/cytoplasmic border, which was confirmed in blast cells from the only available GFI136N homozygous AML patient. It is unlikely that a defective or incomplete nuclear import of GFI136N is the reason for this because GFI136N was only found in nuclear and not in cytoplasmic extracts.
The different nuclear localization of the 2 GFI1 variants did not interfere with their ability to repress transcription in a reporter gene assay. Although surprising at first, this observation is in agreement with another reported finding44 demonstrating that differences in nuclear localization do not necessarily interfere with the ability of transcription factors to function in reporter assays. This finding also suggests that both GFI1 variant forms do not differ in their capacity to recruit the previously described transcriptional repressor complex consisting of histone modifying enzymes45,46 to target gene promoters. The fact that GFI136S and GFI136N were able to repress expression of a target reporter gene suggests that a functional difference between both may rather be confined to the interaction with specific partner proteins.
We hypothesized that the oncofusion protein AML1/ETO might fall into this category of partner proteins because it is involved in the pathogenesis of AML and it has been reported to form a complex with GFI1.30,37,,,,–42 Our coimmunoprecipitation experiments confirmed this interaction for both forms, GFI136N and GFI136S, in transfected cells. We could even show this interaction at the endogenous level for GFI136S (this was not possible for GFI136N because cells homozygous for GFI136N are not available for such an experiment). This finding suggested that the structure of the GFI136N variant is not altered to a degree that would preclude a complex formation with AML1/ETO. However, immunofluorescence data clearly indicated that GFI136N and AML1/ETO do not overlap in their subcellular localization. It is thus likely that, although a physical interaction of both proteins is still possible in vitro, GFI136N and AML1/ETO do not form a complex in a living cell because they occupy separate subnuclear areas.
Our experiments with a GFI1-dependent reporter gene assay indicated that AML1/ETO dampens the repressor activity of GFI136S significantly in a concentration-dependent manner. AML1/ETO might exert this new regulatory function by displacing corepressor molecules such as histone modifying enzymes from GFI136S while it sits at target gene promoters. Such a mechanism has previously been proposed for the myeloid transcription factor PU.1 (SPI1) that is also inactivated by AML1/ETO.39 Given the different subnuclear localization of GFI136N and its loss of colocalization with AML1/ETO, it is conceivable that GFI136N cannot be regulated in the same manner as the correctly localized GFI136S form. It is therefore possible that GFI136N largely maintains its regular repressor functions under most circumstances, but behaves differently compared with GFI136S under specific conditions, for instance in the presence of AML/ETO.
The different subnuclear localization of GFI136N might lead to a partial and specific loss of those functions of GFI136N that are mediated by specific interaction partners, for instance, by the interaction with the AML1/ETO protein. It was reported recently that a loss of GFI1 function can actually predispose to AML in a study47 demonstrating that Gfi1 deficiency accelerates the development of a KRas-induced myeloproliferative syndrome by up-regulating Hoxa9 in the granulocytic monocytic- and common myeloid progenitor fraction. In light of these findings, our data presented here would be consistent with the hypothesis that the altered function of GFI136N caused by its aberrant subcellular localization might be one of the many factors that predispose for the development of AML.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank all AML patients, whose consent made this work possible. We thank all members of the SAL, EORTC, COH, HOVON, and CALGB for obtaining and processing material. We are indebted to Katja Heydarian, Anja Führer, Inge Spratte, Angelika Warda, Eva Gau, Adriana Parchatka, Marika Karger, and Mathieu Lapointe. We would also like to thank Silke Soucek for statistical analyses, Pierre Lepage and Alexandre Belisle at Genome Québec for providing and genotyping samples from different ethnical backgrounds, and Scott Hiebert, Dan Tenen, and Yoram Groner for kindly providing the respective expression plasmids. Finally, we thank the medical teams of the University Hospitals of Essen and Marburg for providing smoking status information.
The CALGB group was supported in part by CA101140 from the National Cancer Institute and the Leukemia Clinical Research Foundation. B.A.v.d.R. is supported by the Vanderes foundation. This work is supported by a grant from The Cancer Research Society Canada. C.K. is supported by a fellowship from the Cole foundation.
Authorship
Contribution: C.K. performed research, analyzed data, and wrote the manuscript; C.T. contributed to experiments, provided samples, and edited the manuscript; P.J.M.V., H.N., D.L., B.H., W.S., A.N., K.-H.G., C.D.B., G.M., K.M., M.L.S., B.A.v.d.R., J.H.J., H.K.S., S.S., J.K.P., F.K., G.E., and B.L. contributed to experiments, provided samples and clinical data, analyzed data, and edited the manuscript; E.S.-A. and K.A. contributed to experiments and edited the manuscript; U.D. designed research, edited the manuscript, and provided initial funding; and T.M. designed research, oversaw research, analyzed data, wrote the manuscript, and provided funding.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Tarik Möröy, Haematopoiesis and Cancer Laboratory, Institut de Recherches Cliniques de Montréal, 110 Avenue des Pins Ouest, Montréal, QC, H2W 1R7 Canada; e-mail: Tarik.Moroy@ircm.qc.ca.