Abstract
The chromosomal translocation t(4;11)(q21;q23) is the most frequent genetic aberration of the human MLL gene, resulting in high-risk acute lymphoblastic leukemia (ALL). To elucidate the leukemogenic potential of the fusion proteins MLL·AF4 and AF4·MLL, Lin−/Sca1+ purified cells (LSPCs) were retrovirally transduced with either both fusion genes or with MLL·AF4 or AF4·MLL alone. Recipients of AF4·MLL- or double-transduced LSPCs developed pro-B ALL, B/T biphenotypic acute leukemia, or mixed lineage leukemia. Transplantation of MLL·AF4- or mock-transduced LSPCs did not result in disease development during an observation period of 13 months. These findings indicate that the expression of the AF4·MLL fusion protein is capable of inducing acute lymphoblastic leukemia even in the absence of the MLL·AF4 fusion protein. In view of recent findings, these results may imply that t(4;11) leukemia is based on 2 oncoproteins, providing an explanation for the very early onset of disease in humans.
Introduction
The mixed lineage leukemia (MLL) gene1,2 participates in a large variety of different chromosomal translocations, resulting in the production of MLL fusion proteins which initiate critical steps of malignant transformation and lead either to the development of acute myeloid (AML) or lymphoblastic (ALL) leukemia.3-5 Currently, the MLL recombinome comprises more than 64 characterized translocation partner genes, but the chromosomal translocation t(4;11)(q21;q23) is the most frequent genetic aberration of the human MLL gene in pediatric and adult ALL patients.6 Analysis of t(4;11) leukemic cells revealed the presence of MLL·AF4 and AF4·MLL fusion alleles in approximately 80% of patients, whereas approximately 20% of t(4;11) leukemia patients bear complex rearrangements, involving MLL, AF4, and a third genomic locus.7 Thus, approximately 20% of t(4;11) patients encode 3 independent fusion alleles. Reciprocal MLL fusion alleles, deriving from complex translocations, are occurring not only in the group of t(4;11) leukemia patients but also in leukemia deriving from other MLL rearrangements (eg, MLL·AF10).6
Several MLL fusion alleles have been shown to mediate myeloid transformation capacity in genetically modified hematopoietic cells.8-12 Most of the tested fusion partners represent nuclear proteins. Certain fusions to cytosolic proteins (eg, MLL·GAS7 or MLL·SEPT6) were also able to induce an AML phenotype,13,14 however, other MLL fusions to cytosolic protein sequences failed to transform primary hematopoietic cells.15 All positively tested MLL fusion alleles led to an ectopic transcriptional activation of Hoxa7, Hoxa9, and Meis1.16,17 A similar deregulation of murine Hoxa genes was observed when the fusion partner conferred dimerization or oligomerization.18-21
Although several attempts were made in the past, the molecular mechanism mediated by the oncogenic function of t(4;11) fusion proteins remained elusive. Recently, 2 murine MLL·AF4 models (knockin and inverter model) were established. In both models, the MLL·AF4 allele caused a B-cell lymphoma phenotype with low penetrance and long latency.22,23 Moreover, MLL·AF4 displayed an instructive role for B-lineage specification.23 Using a slightly different transgenic approach, a genetically engineered MLL·AF4 allele resulted in the development of AML, and less frequently, in pre-B ALL.24 These data verified for the first time that the MLL·AF4 fusion protein is able to provide leukemogenic properties.
By contrast, the overexpression of MLL·AF4 in cell culture conferred resistance to apoptosis and caused cell cycle arrest,25,26 whereas expression of the reciprocal AF4·MLL fusion protein resulted in a loss of contact inhibition, similar to mutant RAS-transformed cells. In addition, the same study demonstrated that the reciprocal AF4·MLL fusion protein is converted into an oncoprotein only after Taspase1-mediated proteolytic cleavage.27
Therefore, we decided to investigate the functions of both t(4;11) fusion proteins in murine hematopoietic precursor cells to unravel their oncogenic potential. For the purpose of our study, retroviral transduction/transplantation (RTT) experiments were conducted in Lin−/Sca1+ purified cells (LSPCs) that were transduced either with MLL·AF4 or AF4·MLL alone, or with both t(4;11) fusion alleles. Mice that received a transplant of genetically modified LSPCs developed pro-B ALL, B/T biphenotypic acute leukemia, or mixed lineage leukemia (MLL). The observed immunophenotypes were stable in secondary recipients, demonstrating that the ectopic expression of the AF4·MLL fusion protein is capable of initiating and maintaining the onset of ALL disease development in mice.
Methods
Cloning procedures and retroviral vectors
The retroviral vector PIDE used in these studies is a derivative of the PINCO retrovirus, where the cytomegalovirus promoter::green fluorescent protein (GFP) cassette was exchanged by an internal ribosome entry site::GFP cassette.28 The reciprocal MLL fusion genes were cloned, resulting in the proviral vectors PIDE::MLL·AF4 and PIDE::AF4·MLL, respectively. The cDNA expression cassettes coding for the MLL·AF4 and AF4·MLL fusion proteins were constructed after reverse-transcription polymerase chain reaction (RT-PCR) amplification of appropriate cDNA fragments using isolated mRNA from the t(4;11) cell line SEM.29,30 To avoid PCR amplification artifacts, all cassettes were completely sequenced on both strands.
Cell lines
The ecotropic Phoenix E packaging cell line was cultured at 37°C, 7.5% CO2 in Dulbecco modified Eagle medium (Gibco) supplemented with 10% fetal calf serum (FCS; heat inactivated), 1% l-glutamine (Gibco), and 1% penicillin/streptomycin (Gibco). BA/F3 cells were cultured in RPMI 1640 (Gibco) supplemented with 10% FCS (PAA), 1% l-glutamine (Gibco), 1% penicillin/streptomycin (Gibco), and 10 ng/mL recombinant murine interleukin-3 (mIL-3; Cell Concepts).
PCR experiments
Genomic DNA was isolated from transduced BA/F3 cells and spleen cells of primary leukemic mice using the DNeasy Blood & Tissue kit (QIAGEN). Genomic DNA was used for titration PCR experiments, touchdown PCR, and long-range PCR experiments. Titration and touchdown PCR experiments were performed with the oligonucleotides used for RT-PCR experiments (see next paragraph). For long-range PCR experiments, 660 ng of genomic DNA and specific oligonucleotides were used to amplify the full-length integrates of MLL·AF4 (3·3 5′-GTGCGAAGTCCCACAAGGTC-3′; der11/Hpa1·RW CCGGTTAACAGGTGTTTTGGTTAATTCTTGTAGC-3′) and AF4·MLL (der4/Eag1·FW 5′-GGCCGATGGCAGCCCAGTCAAGTTT-3′; der4/Cla1·RW 5′-CCATCGATGTTTAGGAACTTCCGGCA-3′).
For RT-PCR analysis, RNA was isolated from bone marrow (BM) of primary leukemic mice with the RNeasy kit (QIAGEN), treated with DNaseI, and reverse transcribed using SuperScript (Invitrogen) and random hexamers (Amersham). The cDNA was then subjected to PCR reaction with specific primers for MLL·AF4 (8·3 5′-CCCAAAACCACTCCTAGTGAG-3′; AF4·5 5′-ACTGTCACTGTCCTCACTGTCA-3′), for AF4·MLL (AF4·3 5′-GTTGCAATGCAGCAGAAGCC-3′; 13·5 5′-CAGGGTGATAGCTGTTTCGG-3′), and for GAPDH (GAPDH*3 5′-CTTCACCACCATGGAGAAGG-3′; GAPDH*5 5′-CCTGCTTCACCACCTTCTTG-3′).
For quantitative RT-PCR (QRT-PCR) experiments, RNA was isolated from bone marrow (BM) samples of primary leukemic mice and from mice that did not develop leukemia after an observation period of 13 months. Expression of the fusion genes was normalized to GAPDH as endogenous control and control BM as sample reference. The following oligonucleotides were used: d4_qpcr·3 5′-TGGTCAAGATCAGGCCCCTA-3′, d4_qpcr·5 5′-TTGTGGGTTTGGTGGGGTAG-3′, d11_qpcr·3 5′-CAGGTCCAGAGCAGAGCAAAC-3′, and d11_qpcr·5 5′-GAGCACTTGGAGGTGCAGATG-3′.
Enrichment of murine hematopoietic stem cells
Bone marrow was collected from tibias and femurs of male C57BL/6 (CD45.2/Ly5.2) mice (Charles River) by flushing the bones with PBS supplemented with 0.5% FCS. Lineage-negative (Lin−) bone marrow cells were isolated using the Lineage Cell Depletion kit (CD3, CD45R [B220], CD11b [Mac1], Gr1 [Ly-6G/C], Ter119) from Miltenyi Biotec. Additional purification of Sca1-positive (Sca1+) cells was performed according to the manufacturers' instruction (Miltenyi Biotec) using magnetic-activated cell separation (MACS) columns. Purified Lin−/Sca1+ cells were prestimulated before further use for 2 days in Dulbecco modified Eagle medium (Invitrogen) supplemented with 10% Hyclone fetal bovine serum, 1% l-glutamine (Invitrogen), 1% penicillin/streptomycin (Invitrogen), 20 ng/mL mIL-3, 20 ng/mL mIL-6, and 100 ng/mL mCSF (Cell Concepts).
Retroviral transduction
Viral particles were generated in ecotropic Phoenix E cells using a standard calcium-phosphate transfection protocol (7.5 μg of vector DNA and 62 μL of 2M CaCl2 in 500 μL of sterile water; 500 μL of 2× HBS pH 7.05). Retroviral supernatant was collected at days 2 and 3 after transfection. RetroNectin-coated (20 μg/mL) non–tissue culture–treated 24-well plates were incubated 4 times with 500 μL of the retroviral stock solution after blocking with 2% BSA in PBS. Target cells (BA/F3 cells or LSPCs) were plated onto the virus-loaded RetroNectin layer and incubated for 24 hours at 37°C. A second infection round was performed by 2 times exposure of the cells to the retroviral stock solution supplemented with 20 ng/mL mIL-3, 20 ng/mL mIL-6, and 100 ng/mL mCSF (Cell Concepts) for 3 hours at 37°C and subsequent centrifugation at 600g for 45 minutes. Coinfections were carried out using only 250 μL of each viral stock solution. Cells were incubated for another 24 hours at 37°C, 7.5% CO2, before infection efficiency was determined by PCR experiments.
Transplantation and monitoring
Genetically modified Lin−/Sca1+ cells were transplanted by retro-orbital injection into sublethally irradiated (8 Gy) syngeneic primary recipients (aged 8-12 weeks). Secondary transplantation experiments were performed using whole spleen cell suspensions (∼5000 cells) of primary leukemic mice (CD45.2/Ly5.2) and subsequent transplantation into C57BL/6 (CD45.1/Ly5.1) mice. All transplantation experiments performed were graphically outlined in supplemental Figure 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Secondary recipients were sublethally irradiated (4.5 Gy) and leukemic cells were retro-orbitally injected. All mice that underwent transplantation were observed daily and moribund recipients were sacrificed for further examination. Diagnoses of hematopoietic neoplasms were made according to the Bethesda proposals for classification of lymphoid neoplasms in mice.31 Mice that did not develop leukemia were analyzed according to the same protocol after an observation period of 13 months. All animal experiments were approved by the Frankfurt state government and performed according to its guidelines.
Hemogram and histopathology
Peripheral blood (PB) was collected, analyzed for PB profile (Hemavet 950FS; Drew Scientific), and spread for blood smears. Cytospins from bone marrow and spleen were made using Shandon Double Cytofunnel (Thermo Scientific). Blood smears and cytospins were stained by Wright-Giemsa (Fisher Scientific). Organ samples (spleen, thymus, liver, ectopic thymic tissue, lymph nodes, and kidney) were either directly embedded in Tissue-Tek OCT Compound (Sakura) or preserved in 10% formalin before being embedded in paraffin, sectioned, and stained with hematoxylin and eosin.
Flow cytometric analysis
Hematopoietic or leukemic cells were obtained by flushing tibias and femurs or smashing the spleen, thymus, ectopic thymic tissue, and lymph nodes. Single-cell suspensions (PBS, 0.5% FCS) were then incubated with fluorescence-conjugated or with biotin-conjugated antibodies for 30 minutes at 4°C and subsequently analyzed by flow cytometry. FACS analyses were performed by collecting data with BD FACSCanto, BD FACSAria, BD FACSCalibur, or CyFlowML (Partec), followed by analysis with FCS Express V3 (De Novo Software).
The following biotin-conjugated antibodies were all purchased from BD: B220-biotin (RA3-6B2), CD3-biotin (145-2C11), Mac-1–biotin (M1/70), Gr-1–biotin (RB6-8C5), and Ter119-biotin (TER119). The fluorescence-conjugated antibodies that were used in this study are as follows: B220–phycoerythrin (PE; RA3-6B2), CD3-PE (145-2C11), Mac-1–PE (M1/70), Gr-1–PE (RB6-8C5), Ter119-PE (TER119), CD43-PE (S7), CD44-PE (IM7), CD45.1-PE (A20), ckit-PE (2B8), sca-1–PE (E13-161.7), B220–fluorescein isothiocyanate (FITC; RA3-6B2), CD3-FITC (145-2C11), Mac-1–FITC (M1/70), Gr-1–FITC (RB6-8C5), Ter119-FITC (TER119), CD43-FITC (S7), CD45.2-FITC, (104) ckit-FITC (2B8), sca-1–FITC (E13-161.7), B220–PE–cyanin 7 (Cy7; RA3-6B2), CD19-PE-Cy7 (1D3), ckit–allophycocyanin (APC; 2B8), streptavidin-PE-Cy5, and streptavidin–peridinin-chlorophyll-protein complex (PerCP; all from BD). CD8-PE-Cy7 (53-6.7), CD25-APC (3C7), and CD4-APC-Cy7 (GK1.5) were from Abcam. To label lineage-positive cells, either an APC-conjugated cocktail (CD3, B220, Mac-1, Gr-1, Ter119) or a biotin-conjugated cocktail (CD3, B220, Mac-1, Gr-1, Ter119) from BD was used. The latter lineage cocktail was developed using streptavidin-PerCP or streptavidin-PE-Cy5 secondary antibodies.
RNA isolation and microarray analysis
Leukemic blasts of all 3 histo/immunophenotypes and normal bone marrow were isolated using RNA purification on RNeasy columns (RNeasy Mini Kit; QIAGEN). Gene expression analysis was performed using the Affymetrix Mouse Genome 430 2.0 microarrays. All Affymetrix experiments were performed at the NGFN II Affymetrix working platform in Essen (L. Klein-Hitpass) after passing the quality control. The microarray data are found in the GEO public database under accession number GSE20519.32
Results
Transduction experiments with retrovirally encoded MLL·AF4 and AF4·MLL alleles
Both PIDE vectors (PIDE::MLL·AF4 and PIDE::AF4·ML) were used for transient transfection experiments into Phoenix E cells to produce viral stock solutions of both MLL fusion alleles. Because the cloned MLL·AF4 and AF4·MLL expression cassettes have sizes of 6684 bp (MLL exons 1-9::AF4 exons 4-20) and 8738 bp (AF4 exons 1b-3::MLL exons 11-37), respectively, the resulting proviral DNA has a length of 11 344 and 13 281 bp. This is at the limit for efficient packaging in Phoenix E cells, and thus, we expected only low titers of infectious viral particles during stock preparation. Therefore, viral stocks were first tested for their infectivity using BA/F3 cells that were infected 2 times/day on 2 consecutive days. In case of coinfections, both viral stock solutions were used along with the above-mentioned infection schedule.
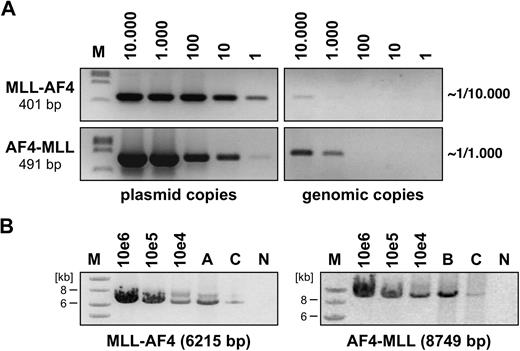
None of the performed transduction experiments in BA/F3 cells resulted in GFP expression, either with PIDE::MLL·AF4 or with PIDE::AF4·MLL. This was presumably because of the length of the transgene cassettes that compromised the use of the internal ribosome entry site::GFP cassette, and prevented us from using flow cytometry to estimate the percentage of infected cells. Therefore, we used an alternative method to approximately calculate the transduction efficiency. As shown in Figure 1A, genomic DNA was isolated from infected BA/F3 cells (1 × 107 genomes) and aliquots were titrated against distinct amounts of PIDE::MLL·AF4 and PIDE::AF4·MLL plasmid copies (1, 10, 102, 103, 104). Using specific oligonucleotides binding to the MLL·AF4 or AF4·MLL cDNA cassettes, we were able to roughly calculate the transduction efficiency in individual experiments (PIDE::MLL·AF4 infected ∼ 1 in 104 cells; PIDE::AF4·MLL infected ∼ 1 in 103 cells).
Semiquantitative assessment of retroviral transduction efficiency. (A) Genomic DNA from PIDE::MLL·AF4- and PIDE::AF4·MLL-transduced BA/F3 cells was isolated and diluted to obtain 1 to 10 000 diploid genome copies. Simultaneously, PIDE::MLL·AF4 and PIDE::AF4·MLL plasmid copies (1 to 10 000) were used to detect MLL·AF4 and AF4·MLL sequences by PCR experiments. Direct comparison revealed that approximately 1 in 10 000 BA/F3 cells were transduced with the MLL·AF4 transgene, whereas 1 in 1000 BA/F3 cells were transduced with the AF4·MLL transgene. M indicates DNA size marker (lambda DNA, ClaI digested). (B) Long-range PCR was performed with infected BA/F3 cells (A: MLL·AF4, B: AF4·MLL, C: MLL·AF4 and AF4·MLL). The indicated plasmid dilutions were amplified in parallel. For the MLL·AF4 transgene, specific oligonucleotides were used that bind to MLL exon 3 and AF4 exon 20. The AF4·MLL transgene was amplified using oligonucleotides specifically binding to AF4 exon 1b and MLL exon 37. The sizes of both amplification products are indicated. M indicates DNA size marker (1-kb ladder); N, negative control.
Semiquantitative assessment of retroviral transduction efficiency. (A) Genomic DNA from PIDE::MLL·AF4- and PIDE::AF4·MLL-transduced BA/F3 cells was isolated and diluted to obtain 1 to 10 000 diploid genome copies. Simultaneously, PIDE::MLL·AF4 and PIDE::AF4·MLL plasmid copies (1 to 10 000) were used to detect MLL·AF4 and AF4·MLL sequences by PCR experiments. Direct comparison revealed that approximately 1 in 10 000 BA/F3 cells were transduced with the MLL·AF4 transgene, whereas 1 in 1000 BA/F3 cells were transduced with the AF4·MLL transgene. M indicates DNA size marker (lambda DNA, ClaI digested). (B) Long-range PCR was performed with infected BA/F3 cells (A: MLL·AF4, B: AF4·MLL, C: MLL·AF4 and AF4·MLL). The indicated plasmid dilutions were amplified in parallel. For the MLL·AF4 transgene, specific oligonucleotides were used that bind to MLL exon 3 and AF4 exon 20. The AF4·MLL transgene was amplified using oligonucleotides specifically binding to AF4 exon 1b and MLL exon 37. The sizes of both amplification products are indicated. M indicates DNA size marker (1-kb ladder); N, negative control.
To exclude recombination events during infection and to ensure the integrity of the cDNA cassettes, we also performed long-range PCR experiments. Using MLL- and AF4-specific oligonucleotides, we were able to demonstrate the presence of intact MLL·AF4 and AF4·MLL alleles in the infected target cells (Figure 1B). Thus, we concluded that transduction experiments were in principle possible, although we were aware that the actual number of infectious particles may compromise the outcome of subsequent RTT experiments.
Retroviral transduction of t(4;11) fusion alleles into LSPCs results in the development of acute leukemia
Bone marrow was prepared from tibias and femurs of C57BL/6 (CD45.2/Ly5.2) donor mice and sorted by magnetic-activated cell separation (MACS) for Lin−/Sca1+ cells. The freshly prepared cells were prestimulated for 72 hours before transduction. For all experiments, 2 × 105 Lin−/Sca1+ cells were cultured on RetroNectin-coated microwell plates, transduced 4 times, harvested, and transplanted by retro-orbital injection into sublethally irradiated recipient mice (CD45.2/Ly5.2; 2 × 105 cells/mouse; 8 Gy). A total of 15 mice received a transplant of mock-transduced cells (PIDE vector), 8 mice received a transplant of PIDE::MLL·AF4-transduced cells, 23 mice received a transplant of PIDE::AF4·MLL-transduced cells, and 8 mice received a transplant of coinfected cells. All mice were observed daily over a period of 381 days (∼ 13 months). Moribund mice were sacrificed and analyzed by molecular methods, immunophenotyping, and histologic analyses.
As shown in Figure 2 (top graph), none of the mice that received a mock transplant or PIDE::MLL·AF4 transplant developed a disease phenotype during the observation period. In contrast, 8 of 23 mice that received a transplant of PIDE::AF4·MLL-transduced LSPCs developed acute leukemia (penetrance of 35%; time to leukemia [TTL]: 233 ± 21 days), whereas 3 of 8 mice that received a transplant of coinfected LSPCs developed acute leukemia (penetrance of 38%; TTL: 266 ± 76 days). All leukemic phenotypes were classified by immunophenotyping.
Kaplan-Meier survival analysis of primary and secondary leukemic mice. (Top graph) Survival data showing the onset of primary leukemia in C57BL/6 mice (CD45.2/Ly5.2) during an observation period of approximately 13 months. A total of 15 mice received a transplant of PIDE-transduced (mock) LSPCs; none developed a disease phenotype. Eight mice received a transplant of either MLL·AF4-tranduced LSPCs or both t(4;11) fusion genes. Three mice that received a transplant of both fusion alleles (MLL·AF4/AF4·MLL) developed acute leukemia. Twenty-three recipients received a transplant of AF4·MLL-transduced LSPCs. Of those, 8 mice developed acute leukemia. y-axis indicates percentage; x-axis, days. (Bottom graph) Survival data showing the onset of secondary leukemia in C57BL/6 mice (CD45.1/Ly5.1). A total of 15 mice received a retransplant of spleen cells deriving from AF4·MLL-mediated leukemic mice. All of these mice developed acute leukemia with a TTL of 25 ± 10 days. Eighteen mice received a retransplant of spleen cells deriving from MLL·AF4/AF4·MLL-mediated leukemic mice. Sixteen of 18 mice developed acute leukemia with TTL of 29 ± 15 days. y-axis indicates percentage; x-axis, days.
Kaplan-Meier survival analysis of primary and secondary leukemic mice. (Top graph) Survival data showing the onset of primary leukemia in C57BL/6 mice (CD45.2/Ly5.2) during an observation period of approximately 13 months. A total of 15 mice received a transplant of PIDE-transduced (mock) LSPCs; none developed a disease phenotype. Eight mice received a transplant of either MLL·AF4-tranduced LSPCs or both t(4;11) fusion genes. Three mice that received a transplant of both fusion alleles (MLL·AF4/AF4·MLL) developed acute leukemia. Twenty-three recipients received a transplant of AF4·MLL-transduced LSPCs. Of those, 8 mice developed acute leukemia. y-axis indicates percentage; x-axis, days. (Bottom graph) Survival data showing the onset of secondary leukemia in C57BL/6 mice (CD45.1/Ly5.1). A total of 15 mice received a retransplant of spleen cells deriving from AF4·MLL-mediated leukemic mice. All of these mice developed acute leukemia with a TTL of 25 ± 10 days. Eighteen mice received a retransplant of spleen cells deriving from MLL·AF4/AF4·MLL-mediated leukemic mice. Sixteen of 18 mice developed acute leukemia with TTL of 29 ± 15 days. y-axis indicates percentage; x-axis, days.
Isolated blasts from the spleen of primary leukemic mice (3 × 104 to 105 cells) were retransplanted into sublethally irradiated secondary C57BL/6 recipient mice (CD45.1/Ly5.1; 4.5 Gy). A total of 15 mice received a retransplant of AF4·MLL-expressing leukemic cells, whereas 18 mice received a retransplant of MLL·AF4/AF4·MLL-expressing leukemic cells. More than 90% of the secondary recipients developed acute leukemia with a short latency (2 mice that received a transplant of leukemic cells bearing both t(4;11) transgenes died disease-free for unknown reasons). Mice that received a transplant of leukemic blasts deriving from AF4·MLL-transduced LSPCs had a TTL of 25 (± 10) days, whereas recipients of coinfected primary leukemic blasts had a TTL of 29 (± 15) days (Figure 2 bottom graph).
Characterization of AF4·MLL and MLL·AF4/AF4·MLL leukemic mice and histopathologic analysis
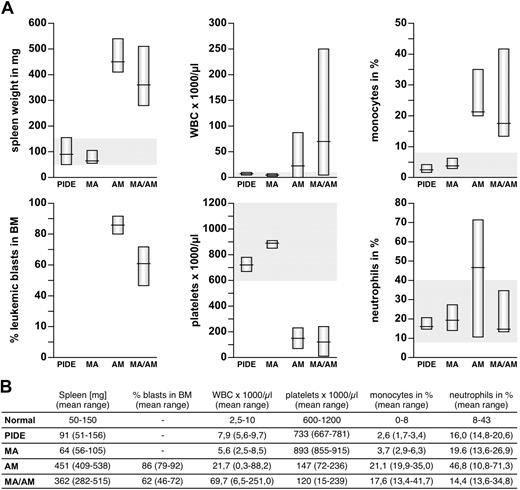
Peripheral blood (PB), bone marrow (BM), and organs samples were investigated from sacrificed mice. Collected peripheral blood was analyzed automatically and spread for blood smears. Single-cell suspensions of bone marrow and spleen were used for cytospins and subjected to flow cytometry experiments. Tissues were dissected and analyzed for leukemic blast infiltration. All diseased animals had a significantly enlarged spleen (282-538 mg, mean AF4·MLL: 451 mg, mean MLL·AF4/AF4·MLL: 362 mg), in contrast to control (51-156 mg, mean control: 91 mg) and animals that received a transplant of MLL·AF4 (56-105 mg, mean MLL·AF4: 64 mg; Figures 3–4). In addition, the majority of diseased animals presented with a strongly enlarged thymus and inclusions of ectopic thymic tissue near the thyroid. Leukemogenesis was always accompanied by massive infiltration of the bone marrow (mean AF4·MLL: 86%, mean MLL·AF4/AF4·MLL: 62%), elevated white blood counts (WBCs; up to 21.7 × 109/L for AF4·MLL and 69.7 × 109/L for MLL·AF4/AF4·MLL), increased monocytes (mean AF4·MLL: 21.1%, mean MLL·AF4/AF4·MLL: 17.6%) and neutrophils (mean AF4·MLL: 46.8%, MLL·AF4/AF4·MLL: not significantly increased), and decreased platelets (mean AF4·MLL: 147 × 109/L, mean MLL·AF4/AF4·MLL: 120 × 109/L; Figure 3).
Characterization of the pathologic parameters for all investigated mice. (A) Charts of spleen weight, blasts in BM, WBC (white blood cell counts), platelets, monocytes, and neutrophils are shown. Values for recipients of PIDE- and MLL·AF4 (MA)–transduced LSPCs are located in the average range. In contrast, recipients of AF4·MLL (AM)– and co(MA/AM)-transduced LSPCs developed typical signs of leukemia, with splenomegaly, increased WBC, monocytes, and neutrophils, reduced platelets, and infiltration of the BM with leukemic blasts. (B) Table summarizing all leukemic characteristics.
Characterization of the pathologic parameters for all investigated mice. (A) Charts of spleen weight, blasts in BM, WBC (white blood cell counts), platelets, monocytes, and neutrophils are shown. Values for recipients of PIDE- and MLL·AF4 (MA)–transduced LSPCs are located in the average range. In contrast, recipients of AF4·MLL (AM)– and co(MA/AM)-transduced LSPCs developed typical signs of leukemia, with splenomegaly, increased WBC, monocytes, and neutrophils, reduced platelets, and infiltration of the BM with leukemic blasts. (B) Table summarizing all leukemic characteristics.
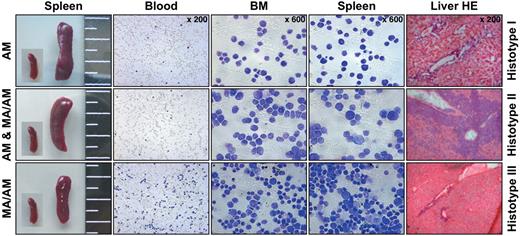
Histopathologic characterization revealed 3 histotypes associated with leukemia. All 3 histotypes are shown. Panels from left to right: spleen sizes (with an embedded picture of a control spleen), blood smears, cytospins of bone marrow (BM) and spleen, and liver sections after hematoxylin and eosin (HE) staining.
Histopathologic characterization revealed 3 histotypes associated with leukemia. All 3 histotypes are shown. Panels from left to right: spleen sizes (with an embedded picture of a control spleen), blood smears, cytospins of bone marrow (BM) and spleen, and liver sections after hematoxylin and eosin (HE) staining.
Histopathology of all diseased mice displayed extensive leukemic infiltrations in analyzed organs including spleen, thymus, liver, ectopic thymic tissue, lymph nodes, and kidney. As indicated in Figure 4, we obtained 3 different histopathologic phenotypes in leukemic mice. Histotype 1 was observed in AF4·MLL-mediated leukemia and characterized by lymphoblast-like leukemic cells (AF4·MLL: 5 of 8 diseased animals). The second histotype, mediated by expression of the AF4·MLL fusion protein alone or by the expression of both fusion proteins (MLL·AF4/AF4·MLL), resembled large granular lymphoblasts (AF4·MLL: 3 of 8 diseased animals, MLL·AF4/AF4·MLL: 2 of 3 diseased animals). Finally, histotype 3 appeared only in MLL·AF4/AF4·MLL-mediated leukemia and was characterized by leukemic blasts of lymphoid and myeloid type (MLL·AF4/AF4·MLL: 1 of 3 diseased animals).
Flow cytometric analysis of AF4·MLL and MLL·AF4/AF4·MLL leukemic mice
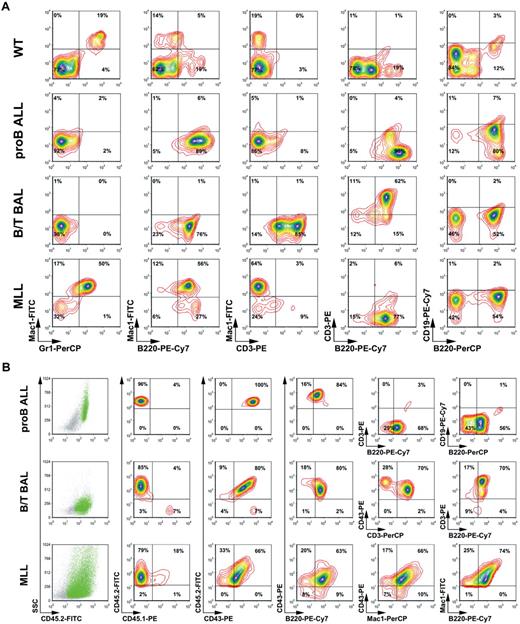
Leukemic blasts isolated from bone marrow and spleens of all primary leukemic mice were analyzed by flow cytometry. Bone marrow of C57BL/6 wild-type mice was analyzed in parallel with AF4·MLL- and AF4·MLL/MLL·AF4-transduced leukemic blasts. Using different surface marker combinations (staining 1: Mac1-FITC, Gr1-PerCP, B220-PE-Cy7, CD3-PE; staining 2: CD43-PE; CD3-FITC; CD4-APC-Cy7; CD8-PE-Cy7; c-kit–APC; staining 3: CD43-PE; B220-PerCP; CD19-PE-Cy7; c-kit–FITC; CD25-APC), we were able to classify 3 different types of acute leukemia. As summarized in Figure 5A, these analyses revealed that AF4·MLL-mediated leukemia can be classified as either pro-B ALL (B220+, CD19−; histotype 1) or as B/T biphenotypic acute leukemia (B/T BAL; B220+, CD3+; histotype 2), although the presence of both t(4;11) transgenes resulted in B/T BAL (B220+, CD3+; histotype 2) or a mixed lineage leukemia (MLL; B220+, Mac1+; histotype 3).
Confirmation of the different types of acute leukemia by flow cytometric analyses. (A) Primary leukemic cells of all 3 histotypes were analyzed by flow cytometric analyses. Different antibody combinations and their conjugated fluorochromes are indicated. Leukemias are classified as pro-B ALL, B/T BAL, and mixed lineage leukemia (MLL). (B) CD45.2+ cells were used for the analysis and classification of secondary leukemias. The different antibody combinations are indicated. These analyses demonstrate engraftment and recapitulation of the primary disease phenotypes.
Confirmation of the different types of acute leukemia by flow cytometric analyses. (A) Primary leukemic cells of all 3 histotypes were analyzed by flow cytometric analyses. Different antibody combinations and their conjugated fluorochromes are indicated. Leukemias are classified as pro-B ALL, B/T BAL, and mixed lineage leukemia (MLL). (B) CD45.2+ cells were used for the analysis and classification of secondary leukemias. The different antibody combinations are indicated. These analyses demonstrate engraftment and recapitulation of the primary disease phenotypes.
Similar flow cytometric experiments were performed for all secondary leukemic mice. Therefore, the staining setup was designed as follows: staining 1: Mac1-FITC, Gr1-PerCP, B220-PE-Cy7, CD3-PE; staining 2: CD45.2-FITC, CD43-PE, B220-PECy7, CD3-PerCP, staining 3: CD45.2-FITC, CD43-PE, Mac1-PerCP; staining 4: CD45.2-FITC, CD45.1-PE, B220-PerCP, CD19-PECy7. As exemplarily shown in Figure 5B, where only the CD45.2+ cells were displayed, a successful engraftment of all transplants could be demonstrated. Moreover, the distribution of CD45.2+ cells in the side scatter/CD45.2 blots also suggests 3 different leukemic types. AF4·MLL-mediated leukemia displayed bone marrow engraftment in the range of 83% (± 9%; means ± SD), whereas the presence of both transgenes resulted in an engraftment of 57% (± 35%). Cells were analyzed by the antibody combinations CD45.1/CD45.2, CD45.2/CD43, and CD43/B220. Subsequently, the 3 different subtypes were further characterized by CD3/B220 and CD19/B220 (histotype 1), CD43/CD3 and CD3/B220 (histotype 2), and CD43/Mac1 and Mac1/B220 (histotype 3). These more precise analyses revealed again that AF4·MLL-mediated leukemia can be classified as either pro-B ALL (CD43+, B220+, CD3−, CD19−) or B/T BAL (CD43+, B220+, CD3+), although the presence of both t(4;11) transgenes resulted in B/T BAL or a mixed lineage leukemia (CD43+, B220+, Mac1+). All analyzed leukemic cells displayed a high percentage (> 50%) of c-kit–positive cells (data not shown). Differences in the percentages were because different antibodies were used for cell staining during immunologic classification. We observed no significant changes of the immunophenotype, indicating that disease phenotypes remained stable in secondary recipients.
Molecular analysis of leukemic cells
To verify the expression of all transgenes, we analyzed primary (and secondary) leukemic cells for the presence of the proviral DNA and the transcriptional activity of all transgenes by PCR experiments. As exemplarily shown in Figure 6, genomic DNA of primary leukemic mice was isolated and tested with primer combinations that specifically identify the transgene cassettes. All PCR experiments were performed with proper positive and negative controls. This type of analysis revealed the presence of all transgenes as expected from the initial transduction experiments. Next, transcriptional activity of the transgenes was monitored by RT-PCR experiments. Besides the appropriate controls, cDNA prepared from isolated total RNA revealed the presence of transcripts deriving from the integrated transgenes. This indicated that leukemic cells maintained the transcription of the transduced transgenes. The same experiments have been performed for secondary leukemic mice with identical results (data not shown). Moreover, we investigated the transcript levels of the transduced t(4;11) fusion genes by QRT-PCR experiments in 9 of 11 primary leukemic mice (supplemental Figure 2A). Based on these data, the leukemic phenotypes were always accompanied by active transcription of the transduced t(4;11) fusion allele. We also tested all primary mice that did not develop a disease (MLL·AF4, AF4·MLL, or both fusion alleles). Transcripts of MLL·AF4 were detected in 3 of 8 recipient mice (transduced with the MLL·AF4 fusion allele) and in 2 of 5 mice (transduced with both fusion alleles). Transcripts of the AF4·MLL fusion allele could not be detected in any of the nondiseased recipients, indicating that the presence of the AF4·MLL transcript was always associated with disease development (supplemental Figure 2B).
Molecular analyses revealed the presence and transcription of transgenes in leukemic cells. (Top) Scheme of both proviruses encoding the MLL·AF4 and AF4·MLL alleles. (Left panels) Genomic PCR experiments of 4 primary leukemic mice, displaying the results of leukemic cells that either encode the AF4·MLL transgene and developed a pro-B or a B/T BAL disease phenotype or that exhibit both t(4;11) fusion alleles and developed a B/T BAL or mixed lineage leukemia (MLL). Neg indicates negative control; Pos, positive control; and gDNA, genomic DNA isolated from the leukemic cells of indicated mice. (Right panels) RT-PCR analysis of 3 mice representing the pro-B, B/T BAL, and MLL leukemia disease phenotype. Appropriate controls (−RT) demonstrated that only cDNA derived from total RNA was investigated (+RT). Transcription of all transgenes was demonstrated. GAPDH indicates positive control for isolated RNA; P, PIDE::MLL·AF4 or PIDE::AF4·MLL plasmids served as positive controls; N, negative control; BM, bone marrow, wild type; and M, DNA size marker (lambda DNA, ClaI digested).
Molecular analyses revealed the presence and transcription of transgenes in leukemic cells. (Top) Scheme of both proviruses encoding the MLL·AF4 and AF4·MLL alleles. (Left panels) Genomic PCR experiments of 4 primary leukemic mice, displaying the results of leukemic cells that either encode the AF4·MLL transgene and developed a pro-B or a B/T BAL disease phenotype or that exhibit both t(4;11) fusion alleles and developed a B/T BAL or mixed lineage leukemia (MLL). Neg indicates negative control; Pos, positive control; and gDNA, genomic DNA isolated from the leukemic cells of indicated mice. (Right panels) RT-PCR analysis of 3 mice representing the pro-B, B/T BAL, and MLL leukemia disease phenotype. Appropriate controls (−RT) demonstrated that only cDNA derived from total RNA was investigated (+RT). Transcription of all transgenes was demonstrated. GAPDH indicates positive control for isolated RNA; P, PIDE::MLL·AF4 or PIDE::AF4·MLL plasmids served as positive controls; N, negative control; BM, bone marrow, wild type; and M, DNA size marker (lambda DNA, ClaI digested).
Gene expression profiling experiments of the 3 histo/immunotypes
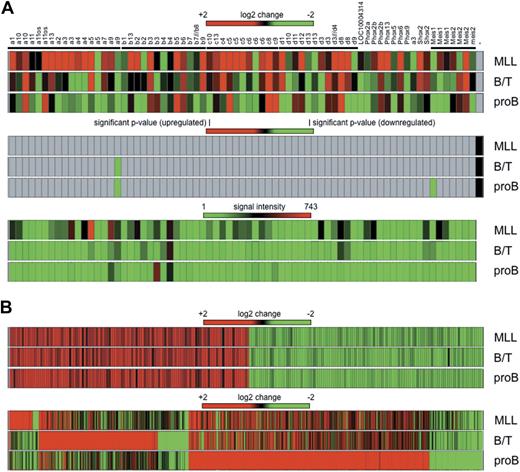
Total RNA was isolated from bone marrow of 3 primary leukemic mice displaying the 3 different histo/immunotypes. Global gene expression was analyzed by hybridization of Affymetrix Mouse Genome 430 2.0 microarrays. First, we analyzed transcriptional deregulation of Hox genes. As summarized in Figure 7A, fold changes (top panel), significant P values (middle panel), and signal intensities (bottom panel) were displayed for the investigated probe sets (primary data are also summarized in supplemental Table 1). Based on the presented data and the fact that all displayed genes were classified as absent calls, no significant transcriptional activation of Hox genes was observed compared with normal bone marrow samples. Slight changes were observed for single probe sets of Hoxa9 (log change −1.8 for B/T BAL and −2.2 for pro-B disease phenotype) and Meis1 (log change −2.3 for the pro-B disease phenotype). Signal intensities of the investigated probe sets were very low and near the noise level. Stronger signals were observed only for Hoxa5 (694 arbitrary light units [alu]) and Hoxa9 (512 alu) in the MLL disease phenotype, and Hoxb3 (490 alu) in the pro-B ALL disease phenotype, however, their P values were not significant.
Gene expression profiling experiments. (A) Hox genes from 4 Hox gene clusters and the Meis gene family are shown as heat maps for the 3 leukemia subtypes (MLL, B/T BAL, and pro-B ALL) after comparison with bone marrow of mock-transduced mice. (Top panel) Log2 changes. (Middle panel) Significant P values. (Bottom panel) Signal intensities in arbitrary light units. With the exception of down-regulated Hoxa9 and Meis1 in B/T BAL and pro-B ALL, none of the observed changes was calculated as significant (gray areas in middle panel). (B) Common and unique signatures of all 3 disease phenotypes. (Top panel) Commonly deregulated genes in all 3 disease phenotypes (regardless of whether only AF4·MLL or both transgenes were present). (Bottom panel) Unique signatures for each subtype. Genes were listed only if the observed log2 change was more than ± 2 (equals ± 4-fold) and arbitrary light units were > 500.
Gene expression profiling experiments. (A) Hox genes from 4 Hox gene clusters and the Meis gene family are shown as heat maps for the 3 leukemia subtypes (MLL, B/T BAL, and pro-B ALL) after comparison with bone marrow of mock-transduced mice. (Top panel) Log2 changes. (Middle panel) Significant P values. (Bottom panel) Signal intensities in arbitrary light units. With the exception of down-regulated Hoxa9 and Meis1 in B/T BAL and pro-B ALL, none of the observed changes was calculated as significant (gray areas in middle panel). (B) Common and unique signatures of all 3 disease phenotypes. (Top panel) Commonly deregulated genes in all 3 disease phenotypes (regardless of whether only AF4·MLL or both transgenes were present). (Bottom panel) Unique signatures for each subtype. Genes were listed only if the observed log2 change was more than ± 2 (equals ± 4-fold) and arbitrary light units were > 500.
The reason for presenting these data at this preliminary point is the identification of a common signature in the 3 types of induced leukemia (MLL, B/T, and pro-B). Based on plus or minus 4-fold changes and arbitrary light units of more than 500, a significant signature of 230 genes was identified (Figure 7B; supplemental Table 2). In all 3 disease types, 116 genes were up-regulated (2- to 35-fold), whereas 114 genes were down-regulated (2- to 25-fold). This common signature clusters together with the “core signature” identified in human t(4;11) leukemic patients (n = 229).33 A total of 189 shared “core signature genes” were identified in human and mouse leukemias, with an overlap of 82.5% and a confidence interval of 95%. Thus, human and mouse t(4;11) leukemias share a set of approximately 190 genes that are equally deregulated. These data have to be confirmed to validate their importance.
In addition, each leukemia disease phenotype displayed a unique signature. Mixed lineage leukemic mice displayed 63 up-regulated (+4- to +979-fold) and 17 down-regulated (−4- to −12-fold) genes. B/T BAL leukemic mice displayed 316 up-regulated (+4- to +1058-fold) and 83 down-regulated (−4- to −100-fold) genes. Finally, pro-B leukemic mice displayed 642 up-regulated (+4- to +676-fold) and 140 down-regulated (−4- to 100-fold) genes (supplemental Table 3).
Rearrangements of immunoglobulin and T-cell receptor genes
We also investigated somatic recombination events because all leukemia disease phenotypes displayed lymphoid character. For this purpose, we established a PCR-based method to investigate Igh and Tcrβ/γ gene rearrangements. Genomic DNA was isolated from leukemic cells and used for direct and inverse PCR experiments. In 1 mouse with B/T BAL, we observed an unique D·J rearrangement (D2-4::J3), whereas 2 pro-B ALL mice displayed a single D·J rearrangement (D2-4::J4) and a highly unusual V·J rearrangement (V2-3::J4). None of the other investigated mice displayed any Igh rearrangement other than germline alleles. We also investigated Tcrβ and Tcrγ gene rearrangements; however, all investigated leukemic cells displayed germline configuration for both investigated genes. A detailed description of the applied method and all data can be accessed in supplemental Figures 3 to 7.
Discussion
We present experimental evidence that the AF4·MLL fusion protein is capable of initiating acute lymphoblastic leukemia (ALL). Our conclusion is based on retroviral transduction experiments using MACS-enriched murine Lin−/Sca1+ cells (LSPCs) in combination with transplantation experiments in sublethally irradiated C57BL/6 mice. This is the first time that a reciprocal AF4·MLL fusion protein has been tested in an experimental animal model system. Retroviral stocks for MLL·AF4 and AF4·MLL transgenes were prepared and used for single and coinfection experiments. Because of the length of both expression cassettes, a low production of infectious particles was observed. Based on PCR titration experiments, using genomic DNA isolated from infected Ba/F3 cells, we calculated that 20 of 2 × 105 transplanted cells carried the MLL·AF4 transgene, whereas approximately 200 of 2 × 105 transplanted cells carried the AF4·MLL transgene (Figure 1A). An explanation of why the 2-kb shorter MLL·AF4-containing retrovirus resulted in 10-fold lower titers may be explained by the effects of overexpressed MLL·AF4 fusion protein that result in cell cycle arrest.25,26 All our transplantation experiments were carried out under highly competitive conditions due to a high percentage of cotransplanted but noninfected LSPCs. This may explain in part the latency of disease onset in mice that underwent primary transplantation (∼ 6 months), or, may argue for secondary mutations that are required for disease onset. By contrast, all secondary recipients displayed a rapid engraftment, a short time to leukemia (2-7 weeks), and a high penetrance (> 90%). Similar data were obtained for tertiary and quaternary transplantation experiments, in which we observed even shorter latencies for disease development (data not shown).
The AF4·MLL transgene alone was able to initiate the onset of acute leukemia, displaying a pro-B ALL (63%) or a B/T biphenotypic acute leukemia (BAL; 37%). The presence of the additional MLL·AF4 allele resulted again in B/T BAL (67%) and the development of mixed lineage leukemia (MLL; 33%). Based on QRT-PCR data obtained from 9 of the 11 primary mice (supplemental Figure 2A), there is no correlation between transcript level and the resulting immunophenotype of the leukemia. By contrast, when we tested all mice that did not develop a disease, we identified several animals that still transcribed the MLL·AF4 fusion allele (3 of 8 transduced with MLL·AF4 fusion allele, and 2 of 5 that were transduced with both alleles; supplemental Figure 2B). This indicated that MLL·AF4-transduced LSPCs were, in principle, able to engraft and to maintain their presence during a period of 13 months, however, seemed to be unable to initiate disease development.
The 3 observed immunophenotypes resemble developmental stages that are closely linked to each other. The MLL disease phenotype, observed only in the presence of both t(4;11) fusion proteins, is presumably a precursor cell type that is able to develop into the myeloid and lymphoid lineage. The B/T biphenotypic acute leukemia is presumably the next step in direction to the lymphatic lineage, whereas the pro-B immunophenotype resembles early B-cell commitment (Figure 5). The analysis of Igh or T-cell receptor ß/γ genes revealed specific gene rearrangements of the Igh gene locus in 2 of 5 pro-B ALL and 1 of 5 B/T BAL leukemic mice. All others leukemic mice displayed nonrearranged germline alleles. Although all observed leukemia disease phenotypes displayed lymphoid surface markers, the rearrangement of immunoglobulin or T-cell receptor genes appears to be rare event. This may indicate that leukemic cells are arrested at very early time points of lymphoid development.
Published data about gene expression profiling experiments performed with MLL-rearranged leukemic patients and with murine model systems expressing specific MLL fusion proteins indicate that the ectopic activation of certain HOXA genes is of great importance. By contrast, a recent study performed by our group—and independently confirmed by another group—demonstrated that a significant portion of t(4;11) leukemic patients does not up-regulate HOXA genes, raising questions about the HOX concept for t(4;11) leukemia.33,34 In particular, transcripts of MLL·AF4 alone or of both t(4;11) fusion alleles were found in both the “HOXA high” and the “HOXA low” group of t(4;11) patients. This may indicate that the presence or absence of elevated HOXA gene transcription is presumably linked to different cell types that were malignantly transformed (eg, fetal liver vs bone marrow cells). Because none of the observed leukemic phenotypes (pro-B, B/T BAL, and MLL) in our murine model resulted in the ectopic activation of Hoxa genes (Figure 7 and supplemental Table 1), it may indicate that the malignant transformation occurred in a murine cell type, where Hox genes were not transcriptionally activated.
What kind of pathologic properties may derive from the reciprocal AF4·MLL fusion protein that would explain our experimental observations? The MLL protein is the essential component of a nuclear machinery that methylates the lysine 4 residue of histone H3 proteins (H3K4), and thus, influences chromatin properties at promoter regions to enable and maintain transcriptional processes.35-38 AF4 is part of a nuclear machinery that activates RNA polymerase II by P-TEFb and the transfer of DOT1L to preinitiating RNA polymerase II complexes.39 Concerning the pathologic functions of the AF4·MLL fusion protein, we have only limited data. According to data from our group, the AF4·MLL fusion protein becomes hydrolyzed by Taspase1 and subsequently forms a high-molecular-weight protein complex. First results with affinity-purified AF4·MLL fusion protein complex indicate that the purified complex exhibits a bona fide H3K4 and H3K79 histone methyltransferase activity. Moreover, the AF4·MLL complex is able to activate P-TEFb kinase, and thus, to influence the elongation process.
Based on the data obtained with an MLL·AF4 allele in a transgenic mouse model system24 and our own data obtained with a transduced AF4·MLL allele, we pose the hypothesis that the pathologic mechanism in t(4;11) leukemia is based on 2 independent oncoproteins. This presumably renders t(4;11) leukemia different from many other MLL-rearranged leukemias.16 However, expression levels of both t(4;11) fusion proteins and their diverse effects on cell physiology in addition to yet unknown events may account for differences in disease onset and immunophenotype.
In conclusion, we successfully established a t(4;11) mouse model, based on a retrovirally transduced AF4·MLL fusion allele or the presence of both t(4;11) fusion alleles. The resulting disease phenotypes displayed a commitment for the lymphoid lineage, with a differentiation block at an early stage of B-cell development. This new model system will allow us to further investigate interesting questions concerning the pathology of t(4;11) leukemia.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Sabrina Böhme, Jennifer Merkens, and Silvia Bracharz for technical assistance, and Marco Giordan for his bioinformatic analyses.
This work was supported by grants 102362, 107819, and 108400 from the Deutsche Krebshilfe (R.M.). This work has been conducted in the framework of the integrated research group “Pathological fusion genes and their disease mechanisms.”
Authorship
Contribution: A.B., K.S., B.R., R.H., and M.R. performed the purification of murine hematopoietic stem cells, infection with retroviral stocks, transplantation into mice, and subsequent analysis (histology, immunology); A.B. and K.S. performed all molecular analyses; R.M. and T.D. analyzed the global gene expression data; and R.M. and A.B. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Rolf Marschalek, Institute of Pharmaceutical Biology, Goethe-University of Frankfurt, Max-von-Laue-Str 9, D-60438 Frankfurt/Main, Germany;e-mail: rolf.marschalek@em.uni-frankfurt.de.
References
Author notes
A.B. and K.S. contributed equally to this work.