Abstract
Although the potential role of Pim2 as a cooperative oncogene has been well described in lymphoma, its role in leukemia has remained largely unexplored. Here we show that high expression of Pim2 is observed in patients with acute promyelocytic leukemia (APL). To further characterize the cooperative role of Pim2 with promyelocytic leukemia/retinoic acid receptor α (PML/RARα), we used a well-established PML-RARα (PRα) mouse model. Pim2 coexpression in PRα-positive hematopoietic progenitor cells (HPCs) induces leukemia in recipient mice after a short latency. Pim2-PRα cells were able to repopulate mice in serial transplantations and to induce disease in all recipients. Neither Pim2 nor PRα alone was sufficient to induce leukemia upon transplantation in this model. The disease induced by Pim2 overexpression in PRα cells contained a slightly higher fraction of immature myeloid cells, compared with the previously described APL disease induced by PRα. However, it also clearly resembled an APL-like phenotype and showed signs of differentiation upon all-trans retinoic acid (ATRA) treatment in vitro. These results support the hypothesis that Pim2, which is also a known target of Flt3-ITD (another gene that cooperates with PML-RARα), cooperates with PRα to induce APL-like disease.
Introduction
Pim2 is a member of the Pim family of serine threonine kinases and is expressed mainly in the hematopoietic system.1 Functionally, it is described as an antiapoptotic protein, which phosphorylates BAD at serine 112 and reverses its apoptotic effects.2,3 Pim2-deficient mice are phenotypically normal and display minimal to no defects in hematopoiesis.4 However, deletion of all 3 Pim proteins (Pim1, Pim2, Pim3) in mice reduced their growth.4 Transgenic mice overexpressing Pim2 alone do not show any hematopoietic malignancies.5,6 However, we and others have shown that Pim2 can cooperate with other oncogenes to induce malignant transformation.7 For example, Pim2 has been shown to cooperate with c-myc and Akt to induce lymphoid tumors.5,6
Flt3-ITD is a mutated form of Flt3 that is expressed in 30% of patients with acute myeloid leukemia (AML) and induces transformation of myeloid cells in vitro and in vivo.8-10 The expression of Pim2 is transcriptionally induced by Flt3-ITD via signal transducer and activator of transcription 5.11 In addition, many AML patients show increased expression of Pim2 regardless of the presence of Flt3-ITD mutations.11 Moreover, we have shown that Pim2 complements the Flt3-D835Y mutant receptor for myeloid cell transformation,7 suggesting that Pim2 coexpression together with Flt3Wt or partially transforming Flt3D835Y receptor mimics the transformation induced by Flt3-ITD. Together, these data indicate that Pim2 might play an important role in the pathogenesis of leukemia.
The promyelocytic leukemia/retinoic acid receptor α (PML/RARα; PRα) fusion protein is the result of a translocation t(15;17)(q22;q11.2) that is present in the vast majority of acute promyelocytic leukemia (APL) cases.12 It has been proposed that PRα acts in a dominant-negative fashion to suppress the normal function of both PML and RARα.13,14 Mouse models and cell culture experiments have demonstrated that PRα contributes to the leukemic phenotype mainly by inhibiting differentiation of hematopoietic progenitor cells.15-17 Data from mouse models indicate that the PRα fusion protein is necessary but not sufficient to induce AML.18-22 Both transgenic and knockin mouse models of PRα induce an AML-like disease after a long latency.20,23,24 The long latency, and additional cytogenetic changes accompanying disease progression in these mice strongly suggest that additional alterations are required for the development of AML by PRα.25-28
When Flt3-ITD was overexpressed in PRα cells and these cells were transplanted into wild-type recipients, all mice developed APL after a short latency.29 This is in line with the genetic and epidemiologic data that suggest Flt3-ITD as a second event in the pathogenesis of APL.30 We therefore hypothesized that Pim2, being an important target of Flt3-ITD, should be able to induce leukemia in mice if overexpressed in PRα cells. We were also interested to see whether there is a difference in the disease latency, penetrance, and phenotype. To investigate the role of Pim2 as a cooperative oncogene to PRα in the pathogenesis of leukemia, we performed bone marrow transplantation experiments using Pim2-transduced PRα knockin (PRα-ki) cells.
Our data demonstrate that Pim2 cooperates with PRα to induce APL in mice after a short latency. The disease was transplantable into secondary and tertiary recipients. Interestingly, high Pim2 expression was observed in APL patients suggesting a possible cooperative role in inducing leukemia. We propose that high expression of Pim2 in AML cooperates with other oncogenes such as PRα, which may then lead to the development of leukemia.
Methods
Mice
Cathepsin G-PML-RARα (PRα) knockin mice as described earlier were kindly provided by Dr Timothy Ley (Washington University School of Medicine, St Louis, MO).24 Female and male PRα ki/ki (C57Bl/6) mice at 8 to 16 weeks of age were used as donors for retroviral transduction and subsequent bone marrow transplantations. Female B6.SJL-PtprcaPep3b/BoyJ (SJL, CD45.1+) mice (purchased from Charles River) at 8 to 10 weeks of age were used as recipients for the bone marrow transplantation experiments. All animals were housed under pathogen-free conditions, and all experiments were conducted with the ethical approval of the University Hospital of Muenster Animal Experiment facility.
Pim2-PMY retroviral vector and preparation of retroviruses
Pim2 was expressed from a retroviral construct that also contained the enhanced green fluorescence (EGFP) coding sequence expressed from an internal ribosomal entry site in the PMY vector. The EGFP-PMY vector was a kind gift from Dr T. Kitamura from The University of Tokyo.31 The cDNA for murine Pim2 was amplified and cloned into the pEntry vector for the gateway system using the TOPO-TA directional cloning kit following the manufacturer's instructions (Invitrogen). The PMY destination vector was made by cloning a gateway cassette into EGFP-PMY at XhoI and ScaII sites. The Pim2 cDNA was then switched from the pEntry vector into the PMY destination vector by a recombination reaction catalyzed by LR-Clonase (Invitrogen). For preparation of retroviruses, the ecotropic packaging cells PlateE were transiently transfected by PMY or Pim2-PMY using Lipofectamine with Plus reagent (Invitrogen). Between 24 and 96 hours after transfection, the retroviral particle-containing supernatants were collected, filtered through 0.45-μm filters (Sarstedt), and stored at −80°C until use.
Primary BM cell isolation, retroviral transduction, and bone marrow transplantation
Primitive mouse bone marrow cells were isolated from long bones of healthy, age-matched PRα ki mice and were pooled for lineage depletion. Hematopoietic progenitor cells (HPCs) were purified by phenotype-sort using a lineage cell depletion kit for mouse (Miltenyi Biotec) following the manufacturer's instructions. Purified 2 × 106 HPCs/mL were preincubated overnight in 20% fetal calf serum/Iscove modified Dulbecco medium (Gibco Life Technology) supplemented with 10 ng/mL murine interleukin-3, 5 ng/mL murine interleukin-6, and 50 ng/mL murine stem cell factor (Peprotech). Retronectin (Takara) coating (50 μg/mL) was used to concentrate the viral particles in the culture dishes, and prestimulated PRα bone marrow (BM) cells were incubated overnight in these plates. The transduction procedure was repeated 3 times. The transduction efficiency as measured by EGFP fluorescence was 32% and 35% for PMY and Pim2-PMY, respectively. A total of 3 × 105 EGFP-positive cells without any preselection together with 105 wild-type BM cells from B6.SJL mice were injected into the lateral tail vein of lethally irradiated (10 Gy) B6.SJL-CD45.1 mice. Cotrim was given in the drinking water for 2 to 3 weeks after bone marrow transplantation. For secondary and tertiary transplantations, mice were lethally irradiated (10 Gy) and received a transplant of 106 bone marrow or spleen cells from mice that underwent primary or secondary transplantation, respectively, together with 105 wild-type BM cells.
Analysis of leukemia development in mice that underwent transplantation
To determine the incidence of leukemia in the mice that underwent transplantation, cohorts of each transplanted construct were followed over time. To screen for leukemia development, peripheral blood was drawn by retro-orbital plexus bleeding at 4- to 5-week intervals. Animals that became moribund were killed, and blood, BM, and spleen samples were analyzed for evidence of leukemia, using automated complete blood count analyses, morphology, flow cytometry, and histopathologic analysis.
Flow cytometry
Freshly harvested hematopoietic tissue cells (peripheral blood, BM, and spleen) were suspended in 1% fetal calf serum/phosphate-buffered saline. The erythrocytes were lysed by alkaline lysis using AKC lysis buffer (0.15 M NH4Cl, 1 mM KHCO3, 0.1 mM Na–ethylenediaminetetraacetic acid, pH = 7.4). The nucleated cells were then stained using fluorescence-labeled antibodies to Gr1, Mac1, CD34, B220, CD19, CD3, Ter119, Sca1, or isotype controls (Becton Dickinson). The flow cytometric data were collected using a BD FACSCalibur, and data were analyzed using CellQuest software (Becton Dickinson). For costaining of Gr1 and CD34 surface markers, the cells from bone marrow or spleen of diseased mice were incubated with Gr1-phycoerythrin (PE; 1:500 diluted) and CD34-allophycocyanin antibody for 20 minutes in the dark. After a subsequent wash, cells were acquired using a FACSAria instrument and data were analyzed by CellQuest and FlowJo software (TreeStar). For analysis of CD34 and Gr1, live cells (4,6 diamidino-2-phenylindole negative) were gated for CD45.2+ (donor cells), and subsequent percentages of cells positive for CD34 and or Gr1 were calculated.
Real-time quantitative reverse-transcription PCR
Freshly isolated spleen or BM cells from Pim2-PRα or control-PRα mice were used for total RNA purification using TRIzol (Invitrogen). The RNA concentration was quantitated by a spectrophotometer. RNA (1 μg) was transcribed into cDNA using random hexamers and Superscript II reverse transcriptase (Gibco Life Technology). The cDNA was diluted to 200 μL with ddH2O, and 2.5 μL was used for each polymerase chain reaction (PCR) reaction. The quantitation of mRNA levels was performed using a real-time fluorescence detection method as described earlier.32 Relative gene expression levels were calculated using standard curves generated by the serial dilutions of cDNA from mouse myeloid progenitor 32D cells. All samples were independently analyzed at least twice for each gene. The housekeeping gene Gapdh served as an additional control for the cDNA quality. Primer and probe sequences for murine Pim2 were forward primer 5′-CGGAACCGTGTGCTAGGCT-3′, reverse primer 5′-AGCAGCGCAACCTCAAGTG-3′, and probe 5′-CCACCGTGTCAGACTCAGTCACCTGC-3′. For detection of the PRα transcript, primers and probes specific for PML-RARα were used. The sequences were forward primer 5′-TCTTCCTGCCCAACAGCAA-3′, reverse primer 5′-GCTTGTAGATGCGGGGTAGAG-3′, and probe 5′AGTGCCCAGCCCTCCCTCGC-3′. Matrix metalloproteinase-9 (MMP-9) primers were used as described.24
For the detection of Pim2 expression in human cells, the primers and probe sequences were forward primer 5′ CATCAAAGTGATTCCCCGGA-3′, reverse primer 5′-TGGGCATGTGACTGAGTCTGA-3′, and probe 5′ TGTGCTGGGCTGGTCCCCCTT-3′. All probes were FAM and TAMRA labeled. The primary APL samples were from the Dresden study group. The RNA from APL patients and controls were isolated by the TRIzol method and cDNA was synthesized as described before.32 The Pim2 expression levels were calculated by the ddCt method using GAPDH gene as control (2(CtGAPDH−CtPim2)). Informed consent was obtained from all patients and controls.
Statistical analysis of clinical data
The primary expression data from AML patients were provided by P.J.M.V. and R.D. (Erasmus University Medical Center).33 All statistical calculations were done using SPSS.17 software.
Colony assay and ATRA experiment
Freshly isolated BM cells from leukemic or control mice were either seeded (5 × 103 cells) in methylcellulose with cytokines (StemCell Technology) in the presence of 1μM all-trans retinoic acid (ATRA) or ethanol or incubated in liquid culture in the complete Iscove modified Dulbecco medium in the presence or absence of ATRA (1μM). RNA was isolated 48 hours later from liquid cultured cells by the TRIzol method followed by cDNA synthesis and quantitative PCR for MMP-9 expression as described above. Colonies were counted on day 8. For morphology assessment, cytospins from either liquid culture or from colony assays were prepared and the cells subsequently stained using Wright-Giemsa staining.
Southern blot analysis
Genomic DNA (10 μg) isolated from spleens was digested with HindIII or SacI for 3 hours and size separated by agarose gel electrophoresis. DNA was transferred to a Biodyne nylon membrane (Pall Corporation) and hybridized to either a GFP or a cyclinD1 probe radioactively labeled by random priming in the presence of alpha-32P Deoxycytidine triphosphate (α32P-dCTP; 50 mC [100 TBq/mMol]) using the Deca Label kit (MBI Fermentas) following the manufacturer's instructions.
Results
High expression of Pim2 is observed in APL patients
Expression of the Pim2 kinase is increased in AML blasts irrespective of the Flt3 mutation status.11 Therefore, we analyzed whether Pim2 expression is associated with specific morphologic or cytogenetic AML subtypes. For this purpose, we analyzed Pim2 mRNA expression in a large dataset obtained from Affymetrix gene expression arrays performed on primary AML samples.33 AML samples were placed into 4 groups according to the cytogenetic findings (normal/intermediate; poor risk; translocations t(8;21), inv(16), and APL with t(15;17)) and analyzed for Pim2 expression. Interestingly, we observed that Pim2 expression was significantly higher in patients with t(15;17) compared with other karyotypes (Figure 1A, P < .001 by 1-way analysis of variance). On the other hand, Pim2 expression was significantly lower in patients with core binding factor leukemias. Next, we performed real-time reverse-transcription (RT)–PCR on primary APL samples (n = 53) to verify the increased Pim2 expression in APL. In addition, normal CD34+ cells (n = 7) and differentiated cells (n = 6) from healthy donors were analyzed for comparison. We found that Pim2 expression was significantly higher in APL samples compared with normal CD34+ as well as differentiated cells (Figure 1B). This high expression of Pim2 in patients with t(15;17) prompted us to investigate the function of Pim2 as a collaborative oncogene with PML-RARα (PRα). We chose the well-defined PRα knockin mouse model to study the in vivo effects of Pim2 in leukemic transformation in association with PRα.21,24
Pim2 expression in AML bone marrow. (A) Expression of Pim2 in AML according to karyotype. Box plots show the range of Pim2 expression between different AML karyotypes. Data were obtained from Affymetrix gene arrays performed on 220 primary AML samples.33 (Normal/intermediate [n = 123]; poor risk [n = 39]; translocations t(8;21) and inv(16) [n = 41]; and APL with t(15;17) [n = 17].) The significance was calculated between groups using univariate 1-way analysis of variance test (P < .001). The median expression is shown by the horizontal line, the size of the box represents the borders that contain 50% of the values, and the error bars represent the upper and the lower quartile of the values. (B) Pim2 expression in primary human APL samples (n = 53), normal CD34+ cells (n = 7), and differentiated cells (normal granulocytes, monocytes, and lymphocytes; n = 6) analyzed by real-time RT-PCR. The expression levels of Pim2 were calculated by ddCt method using GAPDH as an internal control. The horizontal line represents the median.
Pim2 expression in AML bone marrow. (A) Expression of Pim2 in AML according to karyotype. Box plots show the range of Pim2 expression between different AML karyotypes. Data were obtained from Affymetrix gene arrays performed on 220 primary AML samples.33 (Normal/intermediate [n = 123]; poor risk [n = 39]; translocations t(8;21) and inv(16) [n = 41]; and APL with t(15;17) [n = 17].) The significance was calculated between groups using univariate 1-way analysis of variance test (P < .001). The median expression is shown by the horizontal line, the size of the box represents the borders that contain 50% of the values, and the error bars represent the upper and the lower quartile of the values. (B) Pim2 expression in primary human APL samples (n = 53), normal CD34+ cells (n = 7), and differentiated cells (normal granulocytes, monocytes, and lymphocytes; n = 6) analyzed by real-time RT-PCR. The expression levels of Pim2 were calculated by ddCt method using GAPDH as an internal control. The horizontal line represents the median.
Transplanted Pim2–PML-RARα bone marrow cells induce an APL-like disease in mice
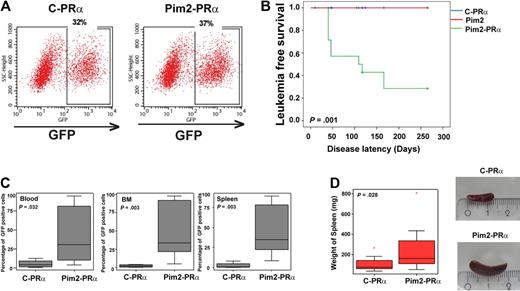
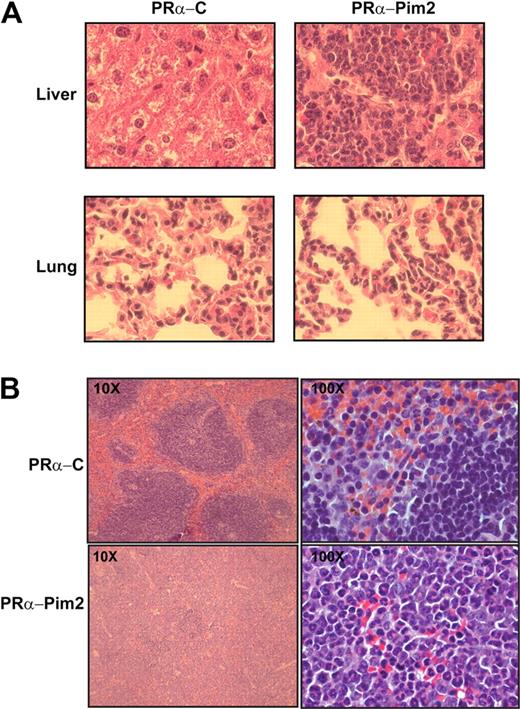
We used PRα knockin (ki) mice to analyze the cooperation of Pim2, because these mice develop disease after a long latency (6-16 months of age) and provide a good congenic system (CD45.1/CD45.2) to differentiate donor from recipient cells. PRα ki/ki lineage-depleted bone marrow (BM) cells were infected with Pim2-PMY or control-PMY retroviruses and transplanted into lethally irradiated C57Bl6 Ly5.1 mice. The transduction efficiency for both constructs was comparable (32%-37%; Figure 2A). In total, 300 000 GFP-positive cells were transplanted into each recipient. In 2 independent experiments, the coexpression of Pim2 in PRα cells (Pim2-PRα) induced an APL-like phenotype in 64% of the mice (9/14). These Pim2-PRα mice developed disease after a short latency (6-18 weeks). None of the mice from the control groups (empty vector–transduced PRα, n = 14; or Pim2 alone, n = 5) developed full disease (Figure 2B). In some mice that received a transplant of PRα bone marrow that had been transfected with GFP, we did see high neutrophil counts and splenomegaly. However, none of them met the criteria for a leukemic phenotype, which we defined on the basis of splenomegaly, infiltration of organs (spleen, liver, and lung), and the presence of CD34+ cells in peripheral blood, spleen, and bone marrow, and that we found in the Pim2-PRα mice. Overexpression of Pim2 led to the expansion of GFP-positive cells in Pim2-PRα mice shortly before the mice became leukemic in comparison with controls (Figure 2C). Splenomegaly was observed only in Pim2-PRα mice (Figure 2D). Donor cells (CD45.2) were detected in peripheral blood (PB), BM, and spleen of recipient mice, regardless of whether they were transfected with Pim2 (Figure 3A). The residual recipient hematopoiesis was largely supplanted by donor hematopoiesis in the Pim2-PRα (leukemic) mice but not in the PRα control mice. In the Pim2-PRα mice, essentially all donor-derived cells were GFP positive, whereas recipients of transplanted control cells (control-PRα) still harbored a significant fraction of CD45.2+ donor cells that were not GFP positive (Figure 3A-B). Immunophenotyping of the transplanted bone marrow by flow cytometry revealed that the overall distribution of mature Mac1+/Gr1+ myeloid cells and of Kit-positive cells was not significantly different between Pim2-PRα– and vector-transduced PRα bone marrow (Figure 3C). However, the lymphoid population (B220+ and CD19+) was significantly decreased in mice that received a transplant of Pim2-PRα compared with control mice (Figure 3D). Interestingly, there was a significant increase in the CD34+ cell population in the BM of Pim2-PRα mice (Figure 3E, P = .030 analyzed using Mann-Whitney U test). Forward and side scatter plots showed that Pim2-PRα diseased mice contained a higher number of large granular cells in the bone marrow compared with control mice (Figure 3F). Histopathologic examination revealed infiltration of myeloid cells into liver, lung, and spleen in recipients of Pim2-PRα bone marrow, whereas no mouse from the control group showed any infiltration (Figure 4).
Effects of Pim2 overexpression in a PML-RARα bone marrow transplantation assay. (A) Flow cytometry for GFP positivity of PML-RARα knock-in bone marrow after retroviral transduction with a GFP-containing vector (C-PRα) or a Pim2-GFP construct (Pim2-PRα) before transplantation. The dot plots show that transduction efficiency for both constructs was comparable. (B) Kaplan-Meier plot shows the probability of leukemia-free survival in animals that underwent transplantation. C-PRα indicates PRα knockin donor bone marrow was retrovirally transduced with a construct containing GFP only before transplantation; Pim2, wild-type donor bone marrow was retrovirally transduced by a construct containing Pim2 and GFP before transplantation; Pim2-PRα, PRα knock-in donor bone marrow was retrovirally transduced with a construct containing Pim2 and GFP. (C) Expression of GFP in recipient animals at the time of leukemia development (for control animals: > 7 weeks after transplantation). Data shown here are from 8 individual mice analyzed for each group for GFP fluorescence by flow cytometry. Significance was calculated by nonparametric Mann-Whitney U test. The median expression is shown by the horizontal line, the size of the box represents the borders that contain 50% of the values, and the error bars represent the upper and the lower quartiles of the values. (D) Weight distribution and photographic image of spleens of the mice that underwent transplantation at the time of leukemia development (for control animals: > 7 weeks after transplantation). Data shown here are from 8 individual mice analyzed for each group. Significance was calculated by nonparametric Mann-Whitney U test. The median expression is shown by the horizontal line, the size of the box represents the borders that contain 50% of the values, and the error bars represent the upper and the lower quartiles of the values.
Effects of Pim2 overexpression in a PML-RARα bone marrow transplantation assay. (A) Flow cytometry for GFP positivity of PML-RARα knock-in bone marrow after retroviral transduction with a GFP-containing vector (C-PRα) or a Pim2-GFP construct (Pim2-PRα) before transplantation. The dot plots show that transduction efficiency for both constructs was comparable. (B) Kaplan-Meier plot shows the probability of leukemia-free survival in animals that underwent transplantation. C-PRα indicates PRα knockin donor bone marrow was retrovirally transduced with a construct containing GFP only before transplantation; Pim2, wild-type donor bone marrow was retrovirally transduced by a construct containing Pim2 and GFP before transplantation; Pim2-PRα, PRα knock-in donor bone marrow was retrovirally transduced with a construct containing Pim2 and GFP. (C) Expression of GFP in recipient animals at the time of leukemia development (for control animals: > 7 weeks after transplantation). Data shown here are from 8 individual mice analyzed for each group for GFP fluorescence by flow cytometry. Significance was calculated by nonparametric Mann-Whitney U test. The median expression is shown by the horizontal line, the size of the box represents the borders that contain 50% of the values, and the error bars represent the upper and the lower quartiles of the values. (D) Weight distribution and photographic image of spleens of the mice that underwent transplantation at the time of leukemia development (for control animals: > 7 weeks after transplantation). Data shown here are from 8 individual mice analyzed for each group. Significance was calculated by nonparametric Mann-Whitney U test. The median expression is shown by the horizontal line, the size of the box represents the borders that contain 50% of the values, and the error bars represent the upper and the lower quartiles of the values.
Immunophenotypic analysis of hematopoiesis of the mice that underwent transplantation. (A) Analysis of mixed chimerism by isotype-specific immunophenotyping. FACS profiles from peripheral blood, bone marrow, and spleen, stained with CD45.1-PE–labeled (host cells) and CD45.2–peridinin-chlorophyll-protein complex (PerCP)–labeled (donor cells) antibodies. (B) Analysis of GFP expression in the host (CD45.1) and donor compartments of peripheral blood, bone marrow, and spleen as indicated of same animals as in panel A. (C-D) Immunophenotypic analysis of bone marrow of animals that underwent transplantation. Box plots show the summarized data for immunophenotypic analyses of bone marrow from mice that received a transplant of empty vector control–PML-RARα (PRα-C; n = 7) or Pim2–PML-RARα (Pim2-PRα; n = 8). Pim2-PRα mice were leukemic at the time of analysis. Bone marrow cells were stained for the indicated surface markers as mentioned in “Flow cytometry” (nonparametric Mann-Whitney U test; B220+P = .045). (E) CD34 expression in the bone marrow of animals that underwent transplantation. The box plot shows the expression of CD34+ cells analyzed in the bone marrow of mice that underwent primary transplantation (PRα-C [n = 7] or Pim2-PRα [n = 8]; nonparametric Mann-Whitney U test; P = .03). (F) FACS profile showing forward and side scatter plots from bone marrow cells isolated from C-PRα and Pim2-PRα mice (leukemic).
Immunophenotypic analysis of hematopoiesis of the mice that underwent transplantation. (A) Analysis of mixed chimerism by isotype-specific immunophenotyping. FACS profiles from peripheral blood, bone marrow, and spleen, stained with CD45.1-PE–labeled (host cells) and CD45.2–peridinin-chlorophyll-protein complex (PerCP)–labeled (donor cells) antibodies. (B) Analysis of GFP expression in the host (CD45.1) and donor compartments of peripheral blood, bone marrow, and spleen as indicated of same animals as in panel A. (C-D) Immunophenotypic analysis of bone marrow of animals that underwent transplantation. Box plots show the summarized data for immunophenotypic analyses of bone marrow from mice that received a transplant of empty vector control–PML-RARα (PRα-C; n = 7) or Pim2–PML-RARα (Pim2-PRα; n = 8). Pim2-PRα mice were leukemic at the time of analysis. Bone marrow cells were stained for the indicated surface markers as mentioned in “Flow cytometry” (nonparametric Mann-Whitney U test; B220+P = .045). (E) CD34 expression in the bone marrow of animals that underwent transplantation. The box plot shows the expression of CD34+ cells analyzed in the bone marrow of mice that underwent primary transplantation (PRα-C [n = 7] or Pim2-PRα [n = 8]; nonparametric Mann-Whitney U test; P = .03). (F) FACS profile showing forward and side scatter plots from bone marrow cells isolated from C-PRα and Pim2-PRα mice (leukemic).
Histopathology of mice receiving transplants of Pim2–PML-RARα or empty vector–PML-RARα HPCs. Histologic sections from liver, lung (A), and spleen (B) were stained with hematoxylin and eosin (×40 magnification for A and B; ×10 and ×100 magnification for B). Note the myeloid infiltrate in the organs of Pim2-PRα mice.
Histopathology of mice receiving transplants of Pim2–PML-RARα or empty vector–PML-RARα HPCs. Histologic sections from liver, lung (A), and spleen (B) were stained with hematoxylin and eosin (×40 magnification for A and B; ×10 and ×100 magnification for B). Note the myeloid infiltrate in the organs of Pim2-PRα mice.
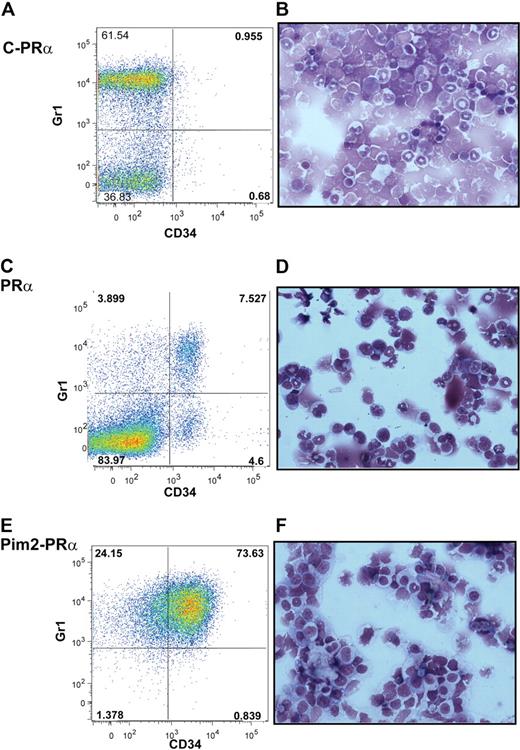
We next analyzed whether the disease induced by Pim2-PRα is recapitulating the previously described APL phenotype in PRα ki mice.24 PRα ki primary mice (which did not undergo transplantation), which became leukemic after 11 months were compared with animals that received a transplant of Pim2-PRα. We performed immunophenotypic analysis to detect coexpression of Gr1 and CD34 on bone marrow and spleen cells. As shown in Figure 5, animals that received a transplant of Pim2-PRα (Figure 5E) contained a high proportion of Gr1/CD34 double-positive cells in their bone marrow. We found a similar population of cells in leukemic PRα ki mice (Figure 5C). In contrast, animals that received a transplant of empty vector–transduced PRα cells did not show this population in their bone marrow (Figure 5A). Obviously, PML-RARα by itself is not sufficient to confer a growth advantage onto the bone marrow transplant and can be efficiently competed out by the recipient hematopoiesis. The morphologic assessment of the bone marrow cells from animals that received a transplant of Pim2-PRα and of leukemic PRα ki mice showed that Pim2-PRα marrow consisted of a relatively homogeneous immature myeloid cell population with some mature myeloid cells, whereas PRα ki diseased marrow contained more mature cells (Figure 5B,D,F). This is consistent with the observed differences in immunophenotype, where the CD34+/Gr1+ population is much more pronounced in the double-transgenic animals that underwent transplantation than in the PRα ki mice. Taken together, these data suggest that the resulting leukemias in mice that received a transplant of Pim2-PRα resemble the APL phenotype observed in PRα ki mice with a more pronounced differentiation block resulting in a higher blast population. This might more closely resemble the situation in human APL.
Immunophenotypic and morphologic characterization of Pim2-PRα–induced leukemias. Costaining of CD34 and Gr1 on BM cells and cytospin morphology from mice that received a transplant of empty vector–transduced PRα bone marrow (A-B), diseased knock-in PRα mice (C-D; that did not undergo transplantation), and mice that received a transplant of Pim2-transduced PRα bone marrow after developing leukemia (E-F) is shown. For analysis of CD34 and Gr1, live cells (4,6 diamidino-2-phenylindole negative) were gated for CD45.2+ (donor cells), and subsequent percentages of cells positive for CD34 and or Gr1 were calculated. Cytospin preparations of BM cells were stained with Wright-Giemsa (original magnification ×60).
Immunophenotypic and morphologic characterization of Pim2-PRα–induced leukemias. Costaining of CD34 and Gr1 on BM cells and cytospin morphology from mice that received a transplant of empty vector–transduced PRα bone marrow (A-B), diseased knock-in PRα mice (C-D; that did not undergo transplantation), and mice that received a transplant of Pim2-transduced PRα bone marrow after developing leukemia (E-F) is shown. For analysis of CD34 and Gr1, live cells (4,6 diamidino-2-phenylindole negative) were gated for CD45.2+ (donor cells), and subsequent percentages of cells positive for CD34 and or Gr1 were calculated. Cytospin preparations of BM cells were stained with Wright-Giemsa (original magnification ×60).
Pim2-PRα–induced leukemias are transplantable in secondary and tertiary recipients
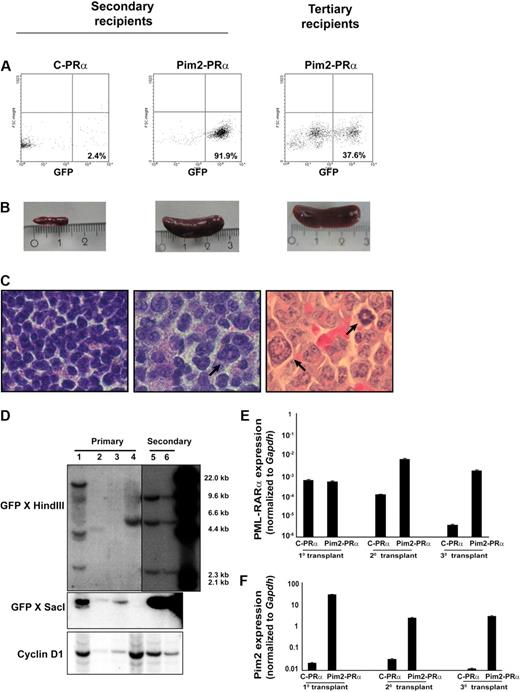
To test directly whether Pim2 coexpression with PRα might confer enhanced leukemogenic potential to bone marrow cells, we assessed the ability of Pim2-PRα–transduced BM cells from preleukemic mice to repopulate secondary recipients. For this purpose, a total of 106 BM or spleen cells (CD45.2+) from diseased primary recipients (Pim2-PRα) were tail-vein injected into lethally irradiated congenic mice (CD45.1+). As already noted, primary control mice (GFP-PRα) did not develop full-blown disease. However, we harvested the bone marrow from primary control mice (that underwent transplantation at the same time as Pim2-PRα mice) and transplanted it into secondary recipients for control purposes. After 9 weeks, all mice that received a transplant of Pim2-PRα cells from primary diseased recipients developed an APL-like disease (Figure 6). Pim2-PRα cells successfully repopulated 100% of secondary recipients (n = 9) in 2 independent experiments. In contrast, control cells (control-PRα) did not repopulate secondary recipients (data not shown). The latency was reduced to 4 weeks when Pim2-PRα cells from secondary leukemic Pim2-PRα mice were transplanted into tertiary recipients (n = 4). GFP-positive cells were detected in BM and spleen of secondary and tertiary recipients (Figure 6A and data not shown). All mice in the Pim2-PRα group developed splenomegaly, but none of the control-PRα mice did (Figure 6B). There was an increase in neutrophils and monocytes and a decrease in lymphocytes and platelets in the PB of Pim2-PRα mice. In addition, in the bone marrow, Pim2-PRα transplants predominantly gave rise to the myeloid lineage (Mac1+, Gr1+) in the spleen, and the amount of B cells (B220+), T cells (CD3+), and erythrocytes (Ter119+) was decreased. An increase in CD34+ immature cells was also observed (data not shown). The Pim2-PRα mice showed splenomegaly and infiltration of myeloid cells into spleen (Figure 6C). These data indicate that Pim2-PRα–induced leukemias are transplantable.
The Pim2–PML-RARα–induced leukemia is transplantable. (A) GFP-positive cells were detected by flow cytometry in the bone marrow and spleen from secondary and tertiary recipients of Pim2–PML-RARα (Pim2-PRα) cells. (B) Photographic image of spleens of the indicated mice. (C) Histologic section of the spleen from secondary and tertiary recipients, stained with hematoxylin and eosin (original magnification ×100). The arrows indicate the infiltration of the spleen of secondary and tertiary recipients by myeloid cells. (D) Southern blot analyses of provirus integration into the bone marrow of primary (lanes 1-4) and secondary (lanes 5-6) transplants. BM cells from lane 1 were used for secondary transplantation. The genomic DNA was digested with HindIII and SacI and probed with GFP sequences. To monitor for loading in the HindIII digest, the blot was reprobed with cyclinD1. (E-F) Real-time RT-PCR was performed on the mRNA samples isolated from spleen of recipient mice. Gapdh was used as an internal control for each experiment. Expression was analyzed in several mice from each group and was repeated independently 2 times. Data shown here are from 1 representative mouse from each group of animals that underwent serial transplantation.
The Pim2–PML-RARα–induced leukemia is transplantable. (A) GFP-positive cells were detected by flow cytometry in the bone marrow and spleen from secondary and tertiary recipients of Pim2–PML-RARα (Pim2-PRα) cells. (B) Photographic image of spleens of the indicated mice. (C) Histologic section of the spleen from secondary and tertiary recipients, stained with hematoxylin and eosin (original magnification ×100). The arrows indicate the infiltration of the spleen of secondary and tertiary recipients by myeloid cells. (D) Southern blot analyses of provirus integration into the bone marrow of primary (lanes 1-4) and secondary (lanes 5-6) transplants. BM cells from lane 1 were used for secondary transplantation. The genomic DNA was digested with HindIII and SacI and probed with GFP sequences. To monitor for loading in the HindIII digest, the blot was reprobed with cyclinD1. (E-F) Real-time RT-PCR was performed on the mRNA samples isolated from spleen of recipient mice. Gapdh was used as an internal control for each experiment. Expression was analyzed in several mice from each group and was repeated independently 2 times. Data shown here are from 1 representative mouse from each group of animals that underwent serial transplantation.
Southern blot analyses showed that the disease in the majority of Pim2-PRα primary recipients was clonal to oligoclonal (Figure 6D lanes 1-4). When used for secondary transplantation experiments (lane 1), one of the subclones grew up as a dominant clone with multiple integration sites (Figure 6D lanes 5-6). These data provide evidence that clonal selection occurred during serial transplantation of the disease.
Spleen cells from diseased mice express Pim2 and PML-RARα
The mRNA expression of Pim2 and PRα was analyzed in the spleen cells from mice that underwent transplantation by real-time RT-PCR. As depicted in Figure 6E, PRα was equally expressed in spleen cells from recipients of either Pim2-PRα or control-PRα cells at primary transplantation. Upon serial transplantation, mice that received a transplant of Pim2-PRα retained PRα expression, whereas control-PRα mice showed decreased expression, possibly due to no further growth of these cells (control-PRα) in mice. High expression of Pim2 was observed in spleen cells from all mice that received a transplant of Pim2-PRα, including secondary and tertiary recipients (Figure 6F).
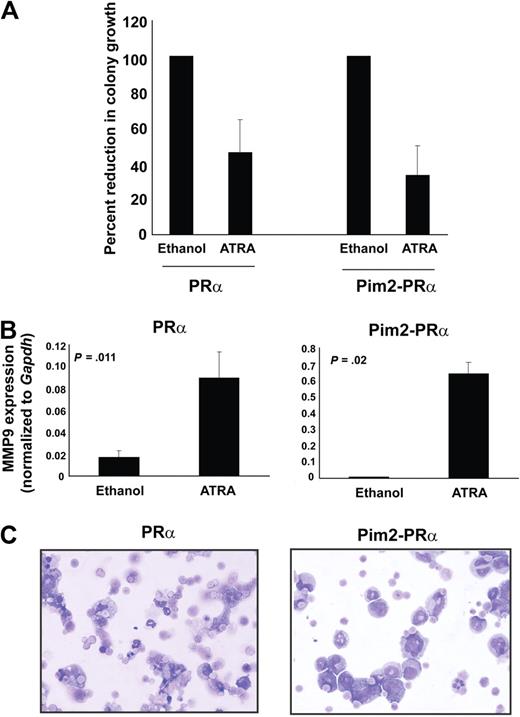
The Pim2-PRα leukemic cells are sensitive to ATRA
Leukemic cells isolated from spleens of diseased Pim2-PRα mice were cultured in vitro in the growth medium containing 1μM ATRA or vehicle control ethanol. Leukemic cells from primary (did not undergo transplantation) PRα-ki diseased mice were used as controls. The effect of ATRA treatment on growth was measured by colony assays. ATRA treatment inhibited growth and induced differentiation of Pim2-PRα as well as PRα cells (Figure 7A). We analyzed MMP-9 expression in these cells after ATRA treatment, as it has been described as a marker of late-stage myeloid differentiation.24 MMP-9 expression was increased in PRα-ki cells after ATRA treatment compared with vehicle-treated control cells, which is in line with a previous report24 (Figure 7B left). Similarly, we also observed an increase of the MMP-9 transcript in Pim2-PRα cells in ATRA-treated cells compared with vehicle control (Figure 7B right). Morphologic analyses showed that ATRA treatment induced differentiation of Pim2-PRα cells into mature myeloid cells to a similar extent as of PRα-ki cells (Figure 7C). These data show that the leukemia induced because of Pim2 overexpression in cooperation with PRα is similar to the previously described PML-RARα–induced APL disease in mice.
Pim2-PRα–induced leukemia is sensitive to ATRA. (A) Colony assays on methylcellulose were performed using freshly isolated BM cells from PRα or Pim2-PRα diseased mice, in the presence or absence of ATRA (1μM). The bars represent the colony growth of PRα and Pim2-PRα cells in the presence of ATRA calculated as percentage of control (ethanol treated). The results shown are the mean of 3 independent experiments ± SD. The significance in growth reduction between 2 groups PRα and Pim2-PRα was calculated by t test (P = .42). (B) MMP-9 expression is increased upon ATRA treatment for 48 hours in PRα as well as Pim2-PRα BM cells assessed by real-time RT-PCR. MMP-9 primers were used as described earlier.24 Results shown here are the mean of 2 independent experiments ± SD. The significance was calculated using t test; P values are indicated in the figure. (C) Cytospin morphology of BM cells incubated with ATRA (1μM) for 48 hours from PRα or Pim2-PRα mice (Wright-Giemsa stain, original magnification ×40).
Pim2-PRα–induced leukemia is sensitive to ATRA. (A) Colony assays on methylcellulose were performed using freshly isolated BM cells from PRα or Pim2-PRα diseased mice, in the presence or absence of ATRA (1μM). The bars represent the colony growth of PRα and Pim2-PRα cells in the presence of ATRA calculated as percentage of control (ethanol treated). The results shown are the mean of 3 independent experiments ± SD. The significance in growth reduction between 2 groups PRα and Pim2-PRα was calculated by t test (P = .42). (B) MMP-9 expression is increased upon ATRA treatment for 48 hours in PRα as well as Pim2-PRα BM cells assessed by real-time RT-PCR. MMP-9 primers were used as described earlier.24 Results shown here are the mean of 2 independent experiments ± SD. The significance was calculated using t test; P values are indicated in the figure. (C) Cytospin morphology of BM cells incubated with ATRA (1μM) for 48 hours from PRα or Pim2-PRα mice (Wright-Giemsa stain, original magnification ×40).
Discussion
In this report, we demonstrate that Pim2 cooperates with PML-RARα (PRα) to induce myeloid leukemia in a bone marrow transplantation assay. The phenotype (splenomegaly, organ infiltration, presence of CD34+/Gr1+ cells) of the disease in this transplantation model is similar to the previously described APL developing in PRα donor mice with a long latency of 6 to 16 months.24,29 However, here we demonstrate that bone marrow from these preleukemic mice does not have the ability to induce disease in bone marrow transplant recipients. In contrast, additional overexpression of Pim2 in the PRα donor bone marrow results in an aggressive transplantable leukemia that develops with a relatively short latency of 6 to 18 weeks.
Pim2 expression has been reported to be up-regulated in various malignancies such as prostate cancer, non-Hodgkin lymphoma, and acute leukemias.11,34-36 Several reports have demonstrated that expression of Pim2 is induced not only by a variety of growth factors and cytokines that regulate blood cell growth and differentiation but also by several oncogenic proteins including Flt3-ITD, Jak2V617F, v-Abl, and Foxp3.11,37-39 Pim kinases are required for the transformation induced by these oncogenes.7,37,40 These studies offer the possibility that Pim proteins might be essential components for the transforming events initiated by other oncogenes.
Although the latency of the disease was comparably short, penetrance was not complete (9/14 animals, Figure 2B). Southern blot analyses showed that the disease was oligoclonal. In addition, the analysis of mice that underwent primary transplantation 4 weeks and 8 weeks after transplantation revealed expansion of the GFP-positive clone only shortly before the mice were leukemic (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Thus, Pim2 expression in the presence of PML-RARα does not induce a rapid polyclonal expansion of leukemic progenitor cells. Rather, we think that Pim2 might act relatively late in the course of leukemic transformation, still requiring a process of clonal evolution.
However, Pim2 enhanced the probability of successful leukemia development and Pim2 strictly required the presence of PRα to induce leukemia. None of the control mice developed a disease, even after prolonged observation. Interestingly, the control transplants expressing PRα, but not Pim2, induced some increase in blood cell counts. However, in these mice the recipient bone marrow was never completely lost, and the mice never became obviously leukemic and were not killed by the increase of their white blood cell counts. In addition, when we analyzed the GFP positivity of these mice late after transplantation, it strongly decreased in 4 of 4 animals (supplemental Figure 1). Taken together, the data imply that Pim2 cooperates with PRα in causing leukemia by enhancing the probability of clonal events that finally induce the leukemic phenotype. In addition, they show that in our model, PRα alone cannot successfully compete out the (normal) recipient hematopoiesis, whereas the double-transgenic progenitor cells can.
Once the leukemia was established, it was readily transplantable into secondary and tertiary recipients (Figure 6). Obviously, it was then advantageous to persistently express both Pim2 and PRα, because the expression of both transgenes was not lost in the course of serial transplantation, although further clonal evolution took place (Figure 6D-F). PRα alone did not induce any Pim2 expression in (aleukemic) recipient mice (Figure 6F). To exclude that this was only because PRα did not induce leukemia in this model, we analyzed Pim2 expression in PRα knockin mice that had become leukemic after a long latency. Here, we did not observe any correlation of Pim2 expression with the leukemic state of these mice (supplemental Figure 2). In addition, in none of these mice was Pim2 expression induced more than 5- to 10-fold in comparison with the other mice. In contrast, in human APL, Pim2 mRNA is highly expressed at levels that exceed the levels in normal CD34+ progenitors by several logs (Figure 1B), well beyond the increase of expression that we achieved by retroviral overexpression in our bone marrow models. Therefore, we think that the observed strong overexpression of Pim2 in human APL is independent of PRα and is recapitulated well in our model system. Our data fit well into a multiple hit model, where PML/RARα has to collaborate with other, independent oncogenes to transform normal progenitors into leukemic progenitor cells. In the human situation, other oncogenes that have been shown to regulate Pim2 expression, for example Flt3-ITD by up-regulating the activity of signal transducer and activator of transcription 5, are possible candidates for such cooperating events.
Immunophenotyping revealed that Pim2-PRα mice harbored a high percentage of mature Mac1+/Gr1+ myeloid cells in the BM and spleen, with a relative loss of B-lymphoid populations (B220+). Intriguingly, there was a significant increase in the CD34+ population in the BM and spleen of Pim2-PRα mice, and these CD34+ cells resembled large granular cells as judged by their forward scatter/side scatter profile. A high proportion of these large granulated cells coexpressed CD34 and lineage markers, for example Gr1, an immunophenotype that well resembles the previously described Gr1+/CD34+ population in the spleen of PRα knock-in mice.21,24 Very recently, several publications provided evidence for a leukemogenic potential of mature committed cells.41,42 Wojiski et al described that promyelocytes from PML-RARα mice possess self-renewal potential.42 It will be interesting in the future to analyze whether subpopulations of the predominant immunophenotype of the disease presented here, showing strong signs of myeloid differentiation, can give rise to leukemias in secondary recipients, or whether we transplanted a rare subpopulation with high leukemia-initiating potential into our secondary and tertiary recipients.
Besides being important for leukemia initiation, does Pim2 overexpression in PRα cells alter sensitivity of the leukemic cells toward ATRA? We analyzed ATRA sensitivity in vitro of APL bone marrow that we obtained from secondary recipients and compared it with the reported ATRA sensitivity of bone marrow of leukemic PRα knock-in mice. As might be expected knowing that human APL cases express high levels of Pim2 and are exquisitely sensitive to ATRA treatment, we did not find any difference in the response of Pim2-PRα bone marrow versus primary leukemic PRα bone marrow to ATRA treatment. Because Pim2 inhibitors are currently being developed preclinically, it will be highly interesting to study their effect on leukemia maintenance in our model in the future.24
In summary, our data show for the first time that Pim2 in cooperation with PML-RARα accelerates leukemia development and provide evidence for a functional role of the observed high Pim2 expression in human APL bone marrow. Our data do not define the mechanism by which Pim2 causes this effect in cooperation with PML-RARα, and further studies are required to investigate the mechanism involved in this. The data indicate that Pim2 might be an important oncogene in AML. It is induced by Flt3 mutations and by other, yet unknown mechanisms. As such, Pim kinases constitute potentially attractive therapeutic targets.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We acknowledge Dr Timothy Ley for PML-RARα ki/ki mice, Dr T. Kitamura for PMY-EGFP vector, Beate Lindtner for creating PMY-gateway vector, and Marion Ziegler for Southern blot analyses.
This work was supported by Deutsche Krebshilfe (106291), Deutsche Krebshilfe Oncogene Networks (108401/Project 108704), Deutsche Jose Carreras Leukämie Stiftung (R05/13; R08/30V), Deutsche Forschungsgemeinschaft (KO2155/2-1), the NGFN-Leukemianet Plus (01GS0873), Wilhelm Sander-Stiftung (2007.048.1), and bmbf; NGFN-Plus (01GS 08188).
Authorship
Contribution: S.A.-S. and H.S. designed the study; S.A.-S. and S.G. performed most of the experiments; S.A.-S., S.K., and C.M.T. analyzed the data; G.K. was involved with histologic analyses; C.S. was involved in Southern blot analysis; M.S. performed fluorescence-activated cell sorting (FACS); C.T., P.J.M.V., R.D., and K.M. provided tools and primary expression data for AML patients; S.A.-S., S.K., C.M.-T., and H.S. wrote the paper; L.T., N.H.T., N.B., K.H., W.E.B., and C.M.-T. provided tools and analysis methods; all authors have checked the final version of the paper.
Conflict of interest disclosure: The authors declare no competing financial interests.
The work of S.K., C.T., W.E.B., C.M.-T., and H.S. was done on behalf of the Study Alliance Leukemia (SAL). A complete list of the members can be found in the supplemental Appendix.
Correspondence: Hubert Serve, Department of Medicine, Hematology and Oncology, University of Frankfurt, Theodor-Stern-Kai 7, 60590 Frankfurt, Germany; e-mail: serve@em.uni-frankfurt.de.

![Figure 1. Pim2 expression in AML bone marrow. (A) Expression of Pim2 in AML according to karyotype. Box plots show the range of Pim2 expression between different AML karyotypes. Data were obtained from Affymetrix gene arrays performed on 220 primary AML samples.33 (Normal/intermediate [n = 123]; poor risk [n = 39]; translocations t(8;21) and inv(16) [n = 41]; and APL with t(15;17) [n = 17].) The significance was calculated between groups using univariate 1-way analysis of variance test (P < .001). The median expression is shown by the horizontal line, the size of the box represents the borders that contain 50% of the values, and the error bars represent the upper and the lower quartile of the values. (B) Pim2 expression in primary human APL samples (n = 53), normal CD34+ cells (n = 7), and differentiated cells (normal granulocytes, monocytes, and lymphocytes; n = 6) analyzed by real-time RT-PCR. The expression levels of Pim2 were calculated by ddCt method using GAPDH as an internal control. The horizontal line represents the median.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/115/22/10.1182_blood-2009-03-210070/4/m_zh89991052320001.jpeg?Expires=1769095745&Signature=bV76QjEVA~-1v4fIBB77SKSL1-9l0zuEw3kAUatmDZrxRSeXjl7gXGyltg475UleRmk0435XxP1sPclm2xpU4llTdx2uMCti8ve0Z8UsRexzL9YpqLbBQ6tMrxvjMHvUaNYM5b1YHzlRTwdI3h8NlT5TJfQAyMqO8qDrk3zeBFLtnmqRJcAKZL1kkRVpdZd47SpmhQUJE-P2BKaaZ57X8T5VDHz-FxNPMGwo284JzBCVpsSlu5Jpw4gAuI3fzO7-qnGwEz15uHz404iYZh0FcGIe2OSByQyDLyDO9RPTEN~rFXGeKfPbwu7pkj9GcHI8mAnyg0D1P4LV4vkYNvpEiA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 3. Immunophenotypic analysis of hematopoiesis of the mice that underwent transplantation. (A) Analysis of mixed chimerism by isotype-specific immunophenotyping. FACS profiles from peripheral blood, bone marrow, and spleen, stained with CD45.1-PE–labeled (host cells) and CD45.2–peridinin-chlorophyll-protein complex (PerCP)–labeled (donor cells) antibodies. (B) Analysis of GFP expression in the host (CD45.1) and donor compartments of peripheral blood, bone marrow, and spleen as indicated of same animals as in panel A. (C-D) Immunophenotypic analysis of bone marrow of animals that underwent transplantation. Box plots show the summarized data for immunophenotypic analyses of bone marrow from mice that received a transplant of empty vector control–PML-RARα (PRα-C; n = 7) or Pim2–PML-RARα (Pim2-PRα; n = 8). Pim2-PRα mice were leukemic at the time of analysis. Bone marrow cells were stained for the indicated surface markers as mentioned in “Flow cytometry” (nonparametric Mann-Whitney U test; B220+ P = .045). (E) CD34 expression in the bone marrow of animals that underwent transplantation. The box plot shows the expression of CD34+ cells analyzed in the bone marrow of mice that underwent primary transplantation (PRα-C [n = 7] or Pim2-PRα [n = 8]; nonparametric Mann-Whitney U test; P = .03). (F) FACS profile showing forward and side scatter plots from bone marrow cells isolated from C-PRα and Pim2-PRα mice (leukemic).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/115/22/10.1182_blood-2009-03-210070/4/m_zh89991052320003.jpeg?Expires=1769095745&Signature=4g4KU3mjNKGzvP9Fc5QGv4KOH55QISF7awjdoNd8HEO8~RiKtBRffl7M16kYeFAojd~rYprtHetCK5lkBVGOvNxdiBKmYQjFov0WqbDbja~A7rv2~~zwOdmLdve-KEJ2XiZSm9asJEfJEwinNppRu7MC8bsHmLF6k-E3A94bnJcrNsO1Wz15JNDYaNzEkYowMBAn66K2kX3KOAdWQXyBBSCdlvF7fFV3O0vyxv-7V84KWoLFaHcOSSTrqUklEpPs-xI3o04kbAoPWG5BluMvvaa3~DuabCp51M-2PyQhqnEV28DMAEmVd~ZQBGTvcN0vd8tYUGQLA6l4oHbr2Litwg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal