Abstract
T-cell immunotherapy that targets minor histocompatibility (H) antigens presented selectively by recipient hematopoietic cells, including leukemia, could prevent and treat leukemic relapse after hematopoietic cell transplantation without causing graft-versus-host disease. To provide immunotherapy that can be applied to a majority of transplantation recipients, it is necessary to identify leukemia-associated minor H antigens that result from gene polymorphisms that are balanced in the population and presented by common human leukocyte antigen alleles. Current approaches for deriving minor H antigen–specific T cells, which provide essential reagents for the molecular identification and characterization of the polymorphic genes that encode the antigens, rely on in vivo priming and are often unsuccessful. We show that minor H antigen–specific cytotoxic T lymphocyte precursors are found predominantly in the naive CD8+ T-cell subset and provide an efficient strategy for in vitro priming of native T cells to generate T cells to a broad diversity of minor H antigens presented with common human leukocyte antigen alleles. We used this approach to derive a panel of stable cytotoxic T lymphocyte clones for discovery of genes that encode minor H antigens and identify a novel antigen expressed on acute myeloid leukemia stem cells and minimally in graft-versus-host disease target tissues.
Introduction
Allogeneic hematopoietic stem cell transplantation (HCT) can cure many patients with leukemia in part because of a graft-versus-leukemia (GVL) effect mediated by donor T cells that recognize recipient minor histocompatibility (H) antigens.1-7 Unfortunately, the GVL effect is impeded by immunosuppression administered after transplantation to prevent graft-versus-host disease (GVHD) and frequently fails to eradicate leukemia.7 A strategy that could be used to prevent or treat relapse is to administer cytotoxic T lymphocytes (CTLs) that target one or more minor H antigens expressed selectively on leukemic cells.7-9 The adoptive transfer of minor H antigen–specific donor T cells can eliminate leukemia in humans but may cause on-target toxicity to recipient tissue cells that express the target antigen.10 Several human minor H antigens have been molecularly characterized, and some of these are expressed preferentially on hematopoietic cells and would appear to be candidates to target for a selective GVL effect.11-24 However, less than 30% of recipients of human leukocyte antigen (HLA) identical allogeneic HCT grafts would currently be eligible for intervention directed against these known minor H antigens because the donor and recipient must be appropriately disparate for expression of the minor H antigen and express the correct HLA-restricting alleles.25 Thus, the discovery of additional minor H antigens expressed on leukemic cells would facilitate targeted immunotherapy.
Current approaches to discover minor H antigens use T-cell clones as reagents for molecular identification. CD8+ minor H antigen–specific CTL clones have been isolated from patients after allogeneic HCT, which allows in vivo priming and expansion of donor T cells specific for recipient minor H antigens.26 This strategy is not uniformly successful and could bias the selection of T cells toward immunodominant minor H antigens. In vitro priming has been used previously to isolate a few minor H antigen–specific CTL clones, but the feasibility of this approach for deriving T cells that can be used to discover potentially therapeutically relevant minor H antigens has not been established.27-29 Here, we show that stimulation of donor CD8+ T cells in microcultures with recipient monocyte-derived dendritic cells (DCs) can be used to isolate minor H antigen–specific CD8+ T cells and derive T-cell clones that can be stably propagated for antigen discovery. Derivation of minor H antigen–specific T cells was most efficient if cultures were initiated with sort-purified naive T cells (TN) and interleukin 12 (IL-12), and then supplemented with IL-15 to promote T-cell expansion. A diverse panel of CTL clones was obtained from granulocyte colony-stimulating factor (G-CSF) mobilized stem cell products of 10 HCT donors, and this panel included clones specific for minor H antigens presented in association with common HLA alleles, encoded by polymorphisms with a balanced phenotype frequency (between 26% and 78% of the population), and expressed on hematopoietic cells, including leukemia. We demonstrate that such CTL clones can provide a resource to identify the genetic polymorphisms responsible for minor H antigens expressed on human leukemia initiating cells (LICs).
Methods
Human subjects
Blood and skin biopsies were obtained from normal HLA-identical volunteer siblings and HCT-related donors and recipients who provided written informed consent in accordance with the Declaration of Helsinki to participate in research protocols approved by the Institutional Review Board of the Fred Hutchinson Cancer Research Center. HCT donors and recipients consented to the donor providing an aliquot of G-CSF–mobilized donor peripheral blood stem cells (G-PBSCs).
Cell lines
Monocyte-derived DCs were used as antigen-presenting cells and generated by a modified fast DC protocol.30 CD14+ monocytes were selected from peripheral blood mononuclear cells after labeling with αCD14 monoclonal antibody (mAb)–conjugated immunomagnetic beads (Miltenyi Biotec). The monocytes were cultured in 6-well plates in AIM V serum-free medium (Invitrogen) with 800 U/mL granulocyte-macrophage colony-stimulating factor and 1000 U/mL IL-4 for 48 hours, and then matured by incubation with tumor necrosis factor-α 10 ng/mL, IL-1β 2 ng/mL, IL-6 1000 U/mL, and prostaglandin E2 1000 ng/mL for 48 hours before use.
Epstein-Barr virus (EBV) transformed lymphoblastoid cell lines (B-LCL) and phytohemagglutinin-stimulated T-cell blasts (PHA blasts) were prepared and maintained in RPMI 1640, 10% fetal calf serum (LCL medium), or RPMI 1640 supplemented with 25mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid, 10% human serum, 1% penicillin/streptomycin, and 3mM l-glutamine (CTL medium), respectively, as described.26,31,32 Fibroblast lines were generated from skin biopsy specimens and cultured as described.33 Center d'Etude du Polymorphisme Humain (CEPH) B-LCL registered to the 13th International Histocompatibility Working Group were provided by Paul Martin or purchased from the Coriell Institute for Medical Research.
Limiting dilution analysis of antigen-specific T cells in CD8+ TN and TM subsets
Highly purified CD8+ naive (TN) and memory (TM) T cells were obtained by positive selection of CD8+ cells from volunteers using αCD8 mAb-conjugated immunomagnetic beads (Miltenyi Biotec) followed by fluorescence-activated cell sorter (FACS) sorting of TN (CD45RO− CD45RA+ CD62L+ CD8+) and TM (CD45RO+ CD45RA− CD62L+/− CD8+) cells. To analyze the frequency of minor H antigen–specific T cells in each subset, TN and TM were plated in a minimum of 60 replicate wells of 96-well plates at various concentrations (1 × 104 to 12 × 104) with a fixed T-cell to DC ratio of 30:1 in CTL medium. The cultures were supplemented with IL-12 (10 ng/mL) at initiation and IL-15 (10 ng/mL) at day 7. In some experiments, IL-12 was omitted and/or alternative γ-chain cytokines were used. Split-well cytotoxicity assays were performed on days 12 or 13 by removing 100-μL aliquots from each well, diluting 1:1 with medium, and distributing to 2 parallel sets of 96-well plates containing donor (responder) and recipient (stimulator) target cells. A well was scored positive if it exhibited more than 10% specific lysis of recipient cells and lysis of recipient cells was more than or equal to 5 times greater than lysis of donor cells.
The frequency of virus-specific T cells in the TM subset was determined by plating replicate wells with 1.5 × 103 to 1.5 × 104 TM and autologous DC (T-cell/DC ratio 30:1) pulsed with a pool of cytomegalovirus, EBV, and influenza peptides (2 μg/mL; PANATecs). IL-12 (10 ng/mL) was added at culture initiation, and IL-15 (10 ng/mL) was added at day 7. Individual wells were evaluated at day 12 in a split-well cytotoxicity assay using peptide pulsed and unpulsed autologous DC as target cells.
Statistical evaluation was performed with extreme limiting dilution analysis (http://bioinf.wehi.edu.au/software/elda/index.html).34
Generation of minor H antigen–specific T-cell lines and clones
CD8+ minor H antigen–specific T-cell lines were generated from HCT donors by stimulating TN obtained from an aliquot of G-PBSCs by depleting other cell types using a cocktail of biotin-conjugated antibodies against CD4, CD15, CD16, CD19, CD34, CD36, CD56, CD123, TCR-γ/δ, and CD235a (glycophorin A) and αbiotin beads, and then depleting CD45RO+ T cells using an αCD45RO bead (Miltenyi Biotec). CD45RA+ TN were plated in 96-well plates at 1.5 × 104 T cells per well with 5 × 102 DCs per well derived from the HLA-identical sibling (transplantation recipient) and supplemented with IL-12 and IL-15 as in the previous subsection. Wells were tested for lysis of donor and recipient B-LCL at days 12 to 14 after stimulation, and those wells that lysed recipient but not donor B-LCLs were expanded with αCD3 mAb and/or cloned by limiting dilution as described.26,35
Cytotoxicity assays
Cytotoxicity was measured using 51Cr-labeled target cells that included PHA blasts, DCs, or B-LCLs derived from the T-cell donor (“donor”) or monocyte/DC donor (“recipient”).26 Specific lysis was calculated using the standard formula.33 Peptides for epitope reconstitution studies were synthesized using standard Fmoc chemistry (Genscript) and pulsed onto 51Cr-labeled donor B-LCL for 30 minutes at 37°C before addition of CTLs. CEPH B-LCLs were transfected with a recombinant vaccinia virus encoding HLA-B*0801 (termed vac/HLA-B8, a generous gift of J. Yewdell, National Institute of Allergy and Infectious Diseases) or wild-type vaccinia at a multiplicity of infection of 5:1 and were used in cytotoxicity assays performed for linkage analysis as described.36
A modified version of a flow cytometry–based cytotoxicity assay was used to evaluate in vitro killing of leukemia by CTL clones.37 Primary acute myeloid leukemia (AML) cells were obtained from bone marrow (BM) or peripheral blood of patients with leukemia at presentation or after relapse and cryopreserved. AML samples were subsequently thawed, labeled with carboxyfluorescein diacetate succinimidyl ester (CFSE, 0.05μM; Cell Trace, Invitrogen), and centrifuged over Ficoll-Hypaque to remove dead cells. Viable CFSE-labeled AML cells were plated in a 48-well plate in LCL medium at a concentration of 0.5 × 106/mL, and CTL clones were added at a fixed ratio of 20:1. After 12 hours of incubation at 37°C and 5% CO2, the cells were harvested, washed, and stained with phycoerythrin-conjugated αCD34, allophycocyanin-conjugated αCD33, and propidium iodide (PI)/RNAase (10 μL/100 μL) staining buffer (BD Biosciences PharMingen) for 10 minutes at 4°C, washed and analyzed on a FACSCalibur cytometer (BD Biosciences). A total of 50 μL of Flow-Count Fluorospheres (Coulter) was added just before sample acquisition, and 5000 microbead events were acquired. The data were analyzed with FlowJo software (TreeStar). Viable leukemia cells were defined as CFSE+ PI− cells expressing CD33 and/or CD34 according to the phenotype of the leukemic blasts in each sample. The number of viable leukemia cells was determined for each sample in duplicate, and percentage cytotoxicity was calculated with the formula: 100 − ([mean absolute number of viable CFSE+ target cells in wells plated with the minor H antigen–specific CTL/mean absolute number of viable CFSE+ target cells in control wells plated with an irrelevant control CTL] × 100).
Linkage analysis
Genotypes for the CEPH B-LCLs were obtained from the CEPH database V9.0/V10.0 (http://www.cephb.fr/cephdb/),38,39 and for a subset were supplemented by microsatellite data (generated by the laboratory of M.F.L.). CEPH B-LCLs that were lysed by a CTL clone were scored as expressing the gene encoding the minor H antigen. A dominant autosomal gene model was assumed for the minor H antigen trait based on the CTL-defined segregation pattern in the CEPH families. Pairwise 2-point linkage analysis was conducted using the MLINK subroutine of the FASTLINK (v4.1p) program as described.36,40 The population frequency of the allele encoding the minor H antigen was estimated at 0.5, and the penetrance was set at 0.9. Resulting logarithm of odds scores were examined for regions of highly significant linkage. The University of California Santa Cruz genome browser (http://genome.ucsc.edu/) and the Map Viewer (http://www.ncbi.nlm.nih.gov/mapview/) were used to search for candidate genes in the region deduced from the linkage analysis.
NOD/SCID assay of leukemia engraftment
The expression of the HEATR1 minor H antigen on LIC was evaluated using a nonobese diabetic/severe combined immune-deficient mice (NOD/SCID) engraftment assay as described.41 Briefly, primary leukemia cells were thawed, washed, and then cultured for 16 hours at 37°C in CTL medium supplemented with 25 U/mL IL-2 in the absence or presence of HEATR1-specific CTL or an irrelevant CTL clone at a T-cell/leukemia ratio of 10:1. Leukemia cells and T-cell/leukemia cell mixtures were washed once, resuspended in 400 μL phosphate-buffered saline, and injected via the tail vein into cohorts of 5 to 10 sublethally irradiated (325 cGy) NOD/SCID mice. In some experiments, mice were pretreated with 200 μg of TMβ1 mAb (a generous gift of Dr T. Tanaka) by intraperitoneal injection 24 hours before inoculation with leukemia.42 BM aspirates were obtained at 3 and 6 weeks after inoculation and at 8 to 10 weeks when mice were killed for analysis of leukemia engraftment. BM mononuclear cells and splenocytes were incubated with phycoerythrin-conjugated αCD45, fluorescein isothiocyanate–conjugated αCD33, peridinin chlorophyll protein complex-conjugated αCD3, allophycocyanin-conjugated αCD19 (BD Biosciences PharMingen), and PI, and analyzed on a FACSCalibur cytometer (BD Biosciences). The data were analyzed with FlowJo software.
Quantitative PCR analysis of HEATR1 expression
First-strand cDNA from poly (A)+ RNA from normal human tissues (Human Multiple Tissue cDNA panels I and II; Clontech) was analyzed for expression of CD45, HEATR1, HA-1 P2X5, and IIp45 by polymerase chain reaction (PCR) of duplicate samples. Data were normalized to expression of porphobilinogen deaminase (PBGD). Amplifications were performed on an ABI Prism 7900 (Applied Biosystems) with a 50-μL reaction mixture consisting of 2.5 ng of cDNA, 25 μL of TaqMan Gene Expression Master Mix (Applied Biosystems), 300nM gene-specific forward and reverse primers, and 250nM gene-specific TaqMan probe. The following gene-specific primers and TaqMan minor groove binding probes were used: PBGD-F 5′-CATCTTGGATCTGGTGGGTGTG-3′, PBGD-R 5′-GCTGTATGCACGGCTACTGG-3′, PBGD-probe 5′-(VIC)-CAGCCTCCTTCCAGGTGCCTC AGG; CD45-F 5′-CATCAGTACAGACGCCTCACC-3′, CD45-R 5′-TGGGGTAGGGTTGAGTTTTGC-3′, CD45-probe 5′-(TET)ACGCACGCAGACTCGCAGACGC; HEATR1-F 5′-TCCTTTTTGATACCCAGCATTTTAT-3′, HEATR1-R 5′-TGATCCACCAGAGGCATCATC-3′, and HEATR1-probe 5′-(6FAM)-AGAGAGCAGAAGCCT; HA-1-F 5′-CCATCATGTTCTCCAGGAAGAAA-3′, HA1-R 5′-CCCGCGCGGTTCTTTT-3′, HA1-probe 5′-(6FAM)-AGAGCTCATGAAAACC; P2X5-F 5′-CAGAGTGCTGTCATCACCAAAGTC-3′, P2X5-R 5′-GCCCAAGATCCGAGGTGTT-3′, P2X5-probe 5′-(6FAM)-AGGGCGTGGCCTT; IIp45-F-5′-GAGTTTCGGGAAACCAACAAGG-3′, IIp45-R-5′-CACGGTAACAGTACACGCATTC, IIp45 probe 5′-(6FAM)-CTTCCTCCACGCCGCTGCCACT. The cycle threshold (Ct) was determined by software analysis (Sequence Detection System, Version 2.2.2; Applied Biosystems), and the level of gene expression of test samples was calculated relative to gene expression in B-LCLs using the comparative Ct method [2−(ΔΔCt)].
Genotyping of HEATR1 polymorphism
Genomic DNA was isolated from B-LCL (QIAamp DNA Blood Kit; QIAGEN), and the polymorphic region of HEATR1 was amplified by PCR on a GeneAmp 9700 (Applied Biosystems) using single nucleotide polymorphism (SNP)–specific primers 2275687-F 5′-TCCATTGTGTTCTGCCTTCG-3′ and 2275687-R 5′-GCAAGCTGTCATTTGAGCAG-3′ in a 50-μL reaction mixture consisting of 1 μL of DNA, 45 μL of Platinum PCR SuperMix (Invitrogen), and 200nM forward and reverse primer. PCR products were purified using the QIAquick PCR Purification Kit (QIAGEN), sequenced, and analyzed using Chromas software.
Results
In vitro priming of CD8+ minor H antigen–specific T cells from CD45RA+ CD62L+ TN
We speculated that CD8+ T cells specific for minor H antigens might be isolated and expanded directly from the HLA-identical donor and used subsequently as reagents to discover novel minor H antigens. Studies in animal models have shown that TN have a greater capacity than TM to cause GVHD and that recipient DCs are required for the induction of alloreactive T cells in vivo.43-45 Therefore, we first evaluated whether isolation of human minor H antigen–specific T cells might be facilitated if cultures were initiated with the TN fraction and stimulated with HLA-identical DCs as antigen-presenting cells. We performed experiments in 3 volunteer HLA-identical sibling pairs that had never been pregnant or had a prior blood transfusion that could prime a TM response to minor H antigens. We separated CD8+ T cells from leukapheresis products of one sibling into purified (> 90%) CD45RAhiCD62L+CD45RO− TN and CD45ROhiCD62L+/−CD45RA− TM subsets (supplemental Figure 1A, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). DCs were derived from CD14+ monocytes of the other sibling by culture in granulocyte-macrophage colony-stimulating factor and IL-4, and matured with a mixture of cytokines before use as antigen-presenting cells. DCs prepared in this fashion had a mature CD80+CD83+CD86+CD11c+CD40+CD14− phenotype and were HLA class I and II+, CD1a 20%+, CD54+, and CD58+ (data not shown). Purified TN and TM were then plated in limiting dilution cultures at various concentrations with DCs from the HLA-matched sibling. IL-12 is required for efficient priming of murine cytolytic T cells46 ; therefore, we examined whether the addition of IL-12 at culture initiation was necessary for priming of TN, and compared the later addition of IL-2 or IL-15 for expansion of activated T cells. The addition of IL-12 at the initiation of the cultures was required for the development of cytolytic responses to recipient cells, and adding IL-7 at culture initiation could not compensate for the absence of IL-12 (supplemental Figure 1B). In addition to IL-12, a γ-chain cytokine was required for efficient expansion of CTLs (data not shown), and IL-15 provided the most robust T-cell growth of CTLs (supplemental Figure 1C) and was used in all subsequent experiments.
Using the conditions determined to be optimal in preliminary experiments, we then performed a frequency analysis of minor H antigen–specific T cells in TN and TM subsets obtained from each of the 3 HLA-matched sibling pairs. The proportion of wells that specifically recognized recipient target cells and the calculated cytotoxic T lymphocyte precursor (CTLp) frequency for recipient minor H antigens was significantly higher for TN compared TM cells, and only rare wells of the TM cultures exhibited cytotoxicity of more than 10% (Figure 1A-B). To confirm the results, we expanded each individual well that exhibited specific cytotoxicity of more than 10% with αCD3 and IL-2, and retested the reactivity of the expanded T-cell lines against autologous and HLA-identical sibling target cells using a constant and high effector-to-target (ET) ratio of 10:1. The majority of the CTL lines generated from TN showed cytolytic activity after expansion, whereas none of the cultures from wells initiated with TM exhibited cytolytic responses (Figure 1C). These data demonstrate that, in the absence of prior in vivo priming, human CD8+ T cells specific for minor H antigens are found predominantly in the TN subset and can be differentiated to cytolytic effector cells in vitro in cultures containing IL-12 and a γ-chain cytokine.
A higher frequency of CD8+ T cells specific for minor H antigens is present in the TN subset than in the TM subset. (A) Lysis of PHA blasts prepared from the HLA-identical sibling/monocyte donor (“recipient”) and from the from the T-cell donor (“donor”). The symbols represent the percentage lysis of recipient cells by individual wells plated with TN (○) or TM (△) that demonstrated both ≥ 10% lysis of recipient PHA blasts and ≥ 5× the lysis of donor PHA blasts. Wells with ≤ 10% lysis of recipient PHA blasts or nonspecific cytotoxicity (lysis of recipient PHA blasts that was not ≥ 5× the lysis of recipient blasts) are not shown. Data are shown for assays from 3 volunteer HLA-identical sibling donors (donors 1, 2, and 3, left to right). (B) Comparison of the minor H antigen–specific T-cell precursor frequency (f) between the TN and TM population for HLA-identical siblings. For TN, f was 1/485 794, 1/1 619 886, and 1/225 188 (broken lines) for donors 1, 2, and 3, respectively; and for TM, f was 1/2 377 182, 1/5 817 135, and 1/2 803 498 (dotted lines) for donors 1, 2, and 3, respectively. Bold lines represent the frequency estimate; and the nonbold lines, 95% confidence intervals. The P values for the difference in f between TN and TM for the 3 pairs were 4.13 × 10−9, 5.65 × 10−4, and 2.86 × 10−16, respectively. Each limiting dilution analysis was performed with a minimum of 60 replicates per cell concentration. (C) Repeat analysis of wells with alloreactivity after expansion. Each well plated with TN (○) or TM (△) that was positive for specific lysis of recipient cells in the initial assay was expanded and retested for lysis of recipient and donor PHA blasts at an ET ratio of 10:1. The x-axis indicates the level of cytotoxicity detected before expansion; and y-axis, the level of cytotoxicity detected after expansion. The validation assay was performed on expanded cultures from donors 1 and 2.
A higher frequency of CD8+ T cells specific for minor H antigens is present in the TN subset than in the TM subset. (A) Lysis of PHA blasts prepared from the HLA-identical sibling/monocyte donor (“recipient”) and from the from the T-cell donor (“donor”). The symbols represent the percentage lysis of recipient cells by individual wells plated with TN (○) or TM (△) that demonstrated both ≥ 10% lysis of recipient PHA blasts and ≥ 5× the lysis of donor PHA blasts. Wells with ≤ 10% lysis of recipient PHA blasts or nonspecific cytotoxicity (lysis of recipient PHA blasts that was not ≥ 5× the lysis of recipient blasts) are not shown. Data are shown for assays from 3 volunteer HLA-identical sibling donors (donors 1, 2, and 3, left to right). (B) Comparison of the minor H antigen–specific T-cell precursor frequency (f) between the TN and TM population for HLA-identical siblings. For TN, f was 1/485 794, 1/1 619 886, and 1/225 188 (broken lines) for donors 1, 2, and 3, respectively; and for TM, f was 1/2 377 182, 1/5 817 135, and 1/2 803 498 (dotted lines) for donors 1, 2, and 3, respectively. Bold lines represent the frequency estimate; and the nonbold lines, 95% confidence intervals. The P values for the difference in f between TN and TM for the 3 pairs were 4.13 × 10−9, 5.65 × 10−4, and 2.86 × 10−16, respectively. Each limiting dilution analysis was performed with a minimum of 60 replicates per cell concentration. (C) Repeat analysis of wells with alloreactivity after expansion. Each well plated with TN (○) or TM (△) that was positive for specific lysis of recipient cells in the initial assay was expanded and retested for lysis of recipient and donor PHA blasts at an ET ratio of 10:1. The x-axis indicates the level of cytotoxicity detected before expansion; and y-axis, the level of cytotoxicity detected after expansion. The validation assay was performed on expanded cultures from donors 1 and 2.
A possible but unlikely explanation for the failure to detect minor H antigen–specific T cells among the TM population was that the cytokine conditions did not permit the expansion of antigen-specific TM. Thus, control experiments were performed in which purified TM cells were stimulated under identical culture conditions with autologous DCs pulsed with cytomegalovirus, EBV, or flu peptides, and individual wells were tested on day 12 for cytolytic activity against peptide pulsed and unpulsed autologous DC targets. In each of 3 experiments, we detected a high frequency of cytolytic T cells for each of these pathogens in the TM population (supplemental Figure 2).
Minor H antigen–specific CD8+ CTL can be isolated from G-CSF–mobilized PBSCs
It would be ideal if minor H antigen–specific T cells could be isolated directly from HCT donors at the time of stem cell donation to facilitate their potential therapeutic use to promote a GVL effect. G-CSF is administered to mobilize stem cells for HCT but has been suggested to suppress T-cell responses through poorly defined mechanisms.47 To determine whether minor H antigen–specific CD8+ T cells that recognize leukemia could be isolated from G-PBSCs by in vitro priming, we obtained an aliquot of the G-PBSC product from 10 persons who served as HCT donors for their HLA-identical siblings (Table 1). CD8+ CD45RA+ TN were enriched from G-PBSCs to more than 90% purity using immunomagnetic isolation, plated at 1.5 × 104 cells/well in replicate cultures with recipient DCs, and supplemented with IL-12 on day 0 and IL-15 on day 7. Each well was assayed for lysis of donor and recipient B-LCL after 12 to 14 days. Multiple wells (median, 12; range, 2-41) that contained T cells that lysed recipient but not donor B-LCLs were identified from the TN cells of all 10 donors (Table 1).
Minor H antigen-specific CTL lines generated by primary in vitro stimulation for 10 recipient donor pairs
| ID . | Sex, (patient/donor) . | Age, y (patient/donor) . | Class I HLA antigens (patient and donor) . | Patient diagnosis . | CD8+ TN cells stimulated . | No. of CTL lines with cytotoxicity greater than 20% (ET 10:1) . |
|---|---|---|---|---|---|---|
| 1 | Female/female | 18/22 | A1, A2 | AML | 13 × 106 | 13 |
| B8, B55 | ||||||
| C7, C3 | ||||||
| 2 | Male/female | 43/39 | A2, A2 | RAEB | 20 × 106 | 14 |
| B38, B40 | ||||||
| C7, C7 | ||||||
| 3 | Male/male | 40/45 | A1, A2 | T-ALL | 16.8 × 106 | 6 |
| B8, B18 | ||||||
| C7, C7 | ||||||
| 4 | Male/male | 52/58 | A2, A2 | PLL | 20 × 106 | 2 |
| B7, B7 | ||||||
| C7, C7 | ||||||
| 5 | Male/female | 47/45 | A24, A32 | AML | 31.5 × 106 | 11 |
| B35, B44 | ||||||
| C4,C4 | ||||||
| 6 | Female/male | 51/52 | A1, A3 | B-ALL | 31 × 106 | 5 |
| B8, B49 | ||||||
| C7, C7 | ||||||
| 7 | Male/female | 23/25 | A1, A68 | AML | 50 × 106 | 24 |
| B44, B55 | ||||||
| C5,C3 | ||||||
| 8 | Male/female | 30/32 | A2, A3 | Hodgkin disease | 18 × 106 | 4 |
| B7, B18 | ||||||
| C7, C7 | ||||||
| 9 | Male/male | 53/48 | A1, A2 | AML | 30 × 106 | 41 |
| B8, B14 | ||||||
| C7, C8 | ||||||
| 10 | Female/female | 42/50 | A1, A2 | RA | 22 × 106 | 20 |
| B7, B27 | ||||||
| C7, C2 |
| ID . | Sex, (patient/donor) . | Age, y (patient/donor) . | Class I HLA antigens (patient and donor) . | Patient diagnosis . | CD8+ TN cells stimulated . | No. of CTL lines with cytotoxicity greater than 20% (ET 10:1) . |
|---|---|---|---|---|---|---|
| 1 | Female/female | 18/22 | A1, A2 | AML | 13 × 106 | 13 |
| B8, B55 | ||||||
| C7, C3 | ||||||
| 2 | Male/female | 43/39 | A2, A2 | RAEB | 20 × 106 | 14 |
| B38, B40 | ||||||
| C7, C7 | ||||||
| 3 | Male/male | 40/45 | A1, A2 | T-ALL | 16.8 × 106 | 6 |
| B8, B18 | ||||||
| C7, C7 | ||||||
| 4 | Male/male | 52/58 | A2, A2 | PLL | 20 × 106 | 2 |
| B7, B7 | ||||||
| C7, C7 | ||||||
| 5 | Male/female | 47/45 | A24, A32 | AML | 31.5 × 106 | 11 |
| B35, B44 | ||||||
| C4,C4 | ||||||
| 6 | Female/male | 51/52 | A1, A3 | B-ALL | 31 × 106 | 5 |
| B8, B49 | ||||||
| C7, C7 | ||||||
| 7 | Male/female | 23/25 | A1, A68 | AML | 50 × 106 | 24 |
| B44, B55 | ||||||
| C5,C3 | ||||||
| 8 | Male/female | 30/32 | A2, A3 | Hodgkin disease | 18 × 106 | 4 |
| B7, B18 | ||||||
| C7, C7 | ||||||
| 9 | Male/male | 53/48 | A1, A2 | AML | 30 × 106 | 41 |
| B8, B14 | ||||||
| C7, C8 | ||||||
| 10 | Female/female | 42/50 | A1, A2 | RA | 22 × 106 | 20 |
| B7, B27 | ||||||
| C7, C2 |
CTL indicates cytotoxic T lymphocyte; RAEB indicates refractory anemia with excess blasts; T-ALL, T-cell acute lymphoblastic leukemia; PLL, prolymphocytic leukemia; B-ALL, B-cell acute lymphoblastic leukemia; and RA, refractory anemia.
We expanded the CTL lines from 9 donors for whom a skin fibroblast line was available from the recipient and assayed each for recognition of fibroblasts to determine whether any of the minor H antigens recognized might be preferentially expressed on recipient hematopoietic cells. Forty-two of 119 CTL lines from 8 of the 9 donors exhibited minimal (< 10%) or no lysis of recipient fibroblasts (Table 2). These 42 CTL lines were then tested against a panel of more than 20 B-LCLs from unrelated donors that shared at least one HLA allele with the recipient to define the HLA-restricting allele and estimate the phenotype frequency of the minor H antigen. The HLA-restricting allele was determined for 35 of the 42 T-cell lines but could not be defined for the remaining 7 because the phenotype frequency of the minor H antigen was low and a larger panel of partially matched unrelated B-LCLs would be required (Table 2). Each of the 35 T-cell lines recognized a distinct minor H antigen based on a different HLA-restricting allele or a different pattern of lysis of unrelated B-LCLs with the same restricting allele. These data demonstrate that in vitro priming of CD8+ TN from G-PBSCs can generate a panel of T cells specific for multiple distinct recipient minor H antigens, including many that are not presented by dermal fibroblasts.
CTL lines with preferential cytolytic activity against hematopoietic target cells
| ID . | Class I HLA antigens (patient and donor) . | CTL line . | Percentage lysis of recipient B-LCL . | Percentage lysis of donor B-LCL . | Percentage lysis of recipient fibroblasts . | HLA restricting allele . | Estimated phenotype frequency of minor H antigen, percentage . |
|---|---|---|---|---|---|---|---|
| 1 | A1, A2, B8, B55, C7, C3 | 4 | 40 | 4 | 2 | B55 | 40 |
| 6 | 34 | 6 | 2 | A2 | 30 | ||
| 8 | 45 | 7 | 6 | B55 | 80 | ||
| 13 | 58 | 5 | 7 | B8 | 50 | ||
| 3 | A1, A2, B8, B18, C7, C7 | 3 | 27 | 0 | 6 | A1 | < 10 |
| 5 | 24 | 0 | 8 | A1 | 13 | ||
| 5 | A24, A32, B35, B44, C4,C4 | 1 | 47 | 6 | 3 | ND | < 10 |
| 4 | 21 | 0 | 1 | B35 | 30 | ||
| 5 | 46 | 0 | 3 | A32 | < 10 | ||
| 8 | 58 | 0 | 3 | B44 | 28 | ||
| 10 | 28 | 1 | 2 | B44 | 36 | ||
| 6 | A1, A3, B8, B49,C7, C7 | 3 | 25 | 5 | 1 | B8 | 11 |
| 4 | 57 | 4 | 5 | A1 | 50 | ||
| 7 | A1, A68, B44, B55, C5, C3 | 1 | 22 | 1 | 1 | A1 | 17 |
| 2 | 33 | 0 | 2 | B55 | 60 | ||
| 3 | 43 | 0 | 1 | A1 | 33 | ||
| 6 | 41 | 0 | 2 | ND | < 10 | ||
| 7 | 39 | 0 | 6 | A68 | 27 | ||
| 8 | 29 | 0 | 0 | B55 | 40 | ||
| 10 | 29 | 0 | 7 | A68 | 55 | ||
| 11 | 40 | 0 | 2 | A1 | 14 | ||
| 13 | 29 | 0 | 0 | B44 | 18 | ||
| 14 | 27 | 0 | 2 | B44 | 11 | ||
| 15 | 22 | 0 | 4 | ND | < 10 | ||
| 16 | 38 | 2 | 0 | B44 | 26 | ||
| 19 | 31 | 1 | 8 | C5 | 67 | ||
| 21 | 40 | 0 | 4 | ND | < 10 | ||
| 22 | 45 | 0 | 1 | A1 | 44 | ||
| 8 | A2, A3, B7, B18, C7, C7 | 2 | 24 | 1 | 0 | B7 | 78 |
| 4 | 77 | 1 | 3 | B7 | 56 | ||
| 9 | A1, A2, B8, B14, C7, C8 | 1 | 42 | 0 | 2 | B8 | 10 |
| 8 | 59 | 1 | 2 | B8 | 20 | ||
| 12 | 51 | 1 | 2 | ND | < 10 | ||
| 14 | 60 | 0 | 2 | ND | < 10 | ||
| 10 | A1, A2, B7, B27, C7, C2 | 1 | 47 | 7 | 0 | B7 | 28 |
| 3 | 26 | 4 | 1 | A1 | 47 | ||
| 4 | 46 | 1 | 0 | B27 | 25 | ||
| 7 | 37 | 1 | 0 | ND | < 10 | ||
| 8 | 37 | 4 | 0 | A2 | 40 | ||
| 9 | 25 | 4 | 3 | B27 | 33 | ||
| 10 | 39 | 7 | 8 | B27 | 20 | ||
| 13 | 54 | 2 | 1 | A2 | 63 | ||
| 4 | A2, A2, B7, B7, C7, C7 | All CTL lines killed fibroblasts | |||||
| ID . | Class I HLA antigens (patient and donor) . | CTL line . | Percentage lysis of recipient B-LCL . | Percentage lysis of donor B-LCL . | Percentage lysis of recipient fibroblasts . | HLA restricting allele . | Estimated phenotype frequency of minor H antigen, percentage . |
|---|---|---|---|---|---|---|---|
| 1 | A1, A2, B8, B55, C7, C3 | 4 | 40 | 4 | 2 | B55 | 40 |
| 6 | 34 | 6 | 2 | A2 | 30 | ||
| 8 | 45 | 7 | 6 | B55 | 80 | ||
| 13 | 58 | 5 | 7 | B8 | 50 | ||
| 3 | A1, A2, B8, B18, C7, C7 | 3 | 27 | 0 | 6 | A1 | < 10 |
| 5 | 24 | 0 | 8 | A1 | 13 | ||
| 5 | A24, A32, B35, B44, C4,C4 | 1 | 47 | 6 | 3 | ND | < 10 |
| 4 | 21 | 0 | 1 | B35 | 30 | ||
| 5 | 46 | 0 | 3 | A32 | < 10 | ||
| 8 | 58 | 0 | 3 | B44 | 28 | ||
| 10 | 28 | 1 | 2 | B44 | 36 | ||
| 6 | A1, A3, B8, B49,C7, C7 | 3 | 25 | 5 | 1 | B8 | 11 |
| 4 | 57 | 4 | 5 | A1 | 50 | ||
| 7 | A1, A68, B44, B55, C5, C3 | 1 | 22 | 1 | 1 | A1 | 17 |
| 2 | 33 | 0 | 2 | B55 | 60 | ||
| 3 | 43 | 0 | 1 | A1 | 33 | ||
| 6 | 41 | 0 | 2 | ND | < 10 | ||
| 7 | 39 | 0 | 6 | A68 | 27 | ||
| 8 | 29 | 0 | 0 | B55 | 40 | ||
| 10 | 29 | 0 | 7 | A68 | 55 | ||
| 11 | 40 | 0 | 2 | A1 | 14 | ||
| 13 | 29 | 0 | 0 | B44 | 18 | ||
| 14 | 27 | 0 | 2 | B44 | 11 | ||
| 15 | 22 | 0 | 4 | ND | < 10 | ||
| 16 | 38 | 2 | 0 | B44 | 26 | ||
| 19 | 31 | 1 | 8 | C5 | 67 | ||
| 21 | 40 | 0 | 4 | ND | < 10 | ||
| 22 | 45 | 0 | 1 | A1 | 44 | ||
| 8 | A2, A3, B7, B18, C7, C7 | 2 | 24 | 1 | 0 | B7 | 78 |
| 4 | 77 | 1 | 3 | B7 | 56 | ||
| 9 | A1, A2, B8, B14, C7, C8 | 1 | 42 | 0 | 2 | B8 | 10 |
| 8 | 59 | 1 | 2 | B8 | 20 | ||
| 12 | 51 | 1 | 2 | ND | < 10 | ||
| 14 | 60 | 0 | 2 | ND | < 10 | ||
| 10 | A1, A2, B7, B27, C7, C2 | 1 | 47 | 7 | 0 | B7 | 28 |
| 3 | 26 | 4 | 1 | A1 | 47 | ||
| 4 | 46 | 1 | 0 | B27 | 25 | ||
| 7 | 37 | 1 | 0 | ND | < 10 | ||
| 8 | 37 | 4 | 0 | A2 | 40 | ||
| 9 | 25 | 4 | 3 | B27 | 33 | ||
| 10 | 39 | 7 | 8 | B27 | 20 | ||
| 13 | 54 | 2 | 1 | A2 | 63 | ||
| 4 | A2, A2, B7, B7, C7, C7 | All CTL lines killed fibroblasts | |||||
CTL indicates cytotoxic T lymphocyte; B-LCL, Epstein-Barr virus–transformed lymphoblastoid cell line; and ND, not determined.
Stable CD8+ minor H antigen–specific T-cell clones can be derived from TN for gene discovery
The identification of polymorphic genes that encode minor H antigens can assist in defining candidates that might be targeted to induce a selective GVL effect, and in genotyping to identify disparate donor/recipient pairs. Strategies for gene identification often require repeated screening of target cells with antigen-reactive T cells.11,12,14,16,17,19,21 To avoid the possibility that nonspecific T cells might overgrow during expansion of T-cell lines for screening, we derived T-cell clones from 10 CTL lines, which recognized minor H antigens that were presented by a common HLA allele, had an estimated allele frequency of 0.26 to 0.78, and were not presented by fibroblasts. T-cell clones that could be expanded long-term and mediate stable effector function, including HLA-restricted cytotoxicity and interferon-γ secretion in response to B-LCLs, were isolated from all 10 CTL lines (supplemental Figure 3; Table 3). These T-cell clones also lysed leukemia obtained from patients who shared the HLA-restricting allele with the recipient (Figure 2). Thus, derivation of minor H antigen–specific T-cell clones from TN precursors in vitro can provide the cellular reagents to identify genes that encode leukemia-associated minor H antigens.
Characterization of minor H antigen–specific CTL clones derived by primary in vitro stimulation
| CTL clone . | HLA-restricting allele . | Frequency of minor H antigen phenotype, percentage . | Clonal expansion more than 12 days, log10 . | Interferon-γ secretion after stimulation with B-LCL, pg/mL . | Cytotoxicity versus B-LCL, percentage (ET 20:1) . | Cytotoxicity versus fibroblasts, percentage (ET 20:1): recipient . | ||
|---|---|---|---|---|---|---|---|---|
| Recipient . | Donor . | Recipient . | Donor . | |||||
| J 47 | B0801 | 50 | 3.01 | 600 | < 50 | 76 | 0 | 3 |
| J 7 | B0801 | 30 | 2.95 | 100 | < 50 | 77 | 0 | 6 |
| L 3.2 | B7 | 28 | 2.94 | > 5000 | < 50 | 40 | 0 | 0 |
| L 17.4 | A0201 | 35 | 3.01 | 450 | < 50 | 59 | 0 | 0 |
| R 7 | B35/C0401 | 30 | 2.90 | 200 | < 50 | 95 | 0 | 1 |
| R 29 | B4402,4403 | 45 | 2.96 | 750 | < 50 | 100 | 0 | 1 |
| K 9.3 | B0702 | 52 | 3.09 | > 5000 | < 50 | 91 | 0 | 12 |
| I 19.2 | A2406 | 56 | 2.87 | 1100 | < 50 | 70 | 0 | 0 |
| M 56 | A0201 | 50 | 2.93 | 150 | < 50 | 64 | 1 | 8 |
| M 37.6 | B2705 | 40 | 2.94 | 100 | < 50 | 74 | 0 | 0 |
| CTL clone . | HLA-restricting allele . | Frequency of minor H antigen phenotype, percentage . | Clonal expansion more than 12 days, log10 . | Interferon-γ secretion after stimulation with B-LCL, pg/mL . | Cytotoxicity versus B-LCL, percentage (ET 20:1) . | Cytotoxicity versus fibroblasts, percentage (ET 20:1): recipient . | ||
|---|---|---|---|---|---|---|---|---|
| Recipient . | Donor . | Recipient . | Donor . | |||||
| J 47 | B0801 | 50 | 3.01 | 600 | < 50 | 76 | 0 | 3 |
| J 7 | B0801 | 30 | 2.95 | 100 | < 50 | 77 | 0 | 6 |
| L 3.2 | B7 | 28 | 2.94 | > 5000 | < 50 | 40 | 0 | 0 |
| L 17.4 | A0201 | 35 | 3.01 | 450 | < 50 | 59 | 0 | 0 |
| R 7 | B35/C0401 | 30 | 2.90 | 200 | < 50 | 95 | 0 | 1 |
| R 29 | B4402,4403 | 45 | 2.96 | 750 | < 50 | 100 | 0 | 1 |
| K 9.3 | B0702 | 52 | 3.09 | > 5000 | < 50 | 91 | 0 | 12 |
| I 19.2 | A2406 | 56 | 2.87 | 1100 | < 50 | 70 | 0 | 0 |
| M 56 | A0201 | 50 | 2.93 | 150 | < 50 | 64 | 1 | 8 |
| M 37.6 | B2705 | 40 | 2.94 | 100 | < 50 | 74 | 0 | 0 |
CTL indicates cytotoxic T lymphocyte; B-LCL, Epstein-Barr virus–transformed lymphoblastoid cell line; and ET, effector-to-target.
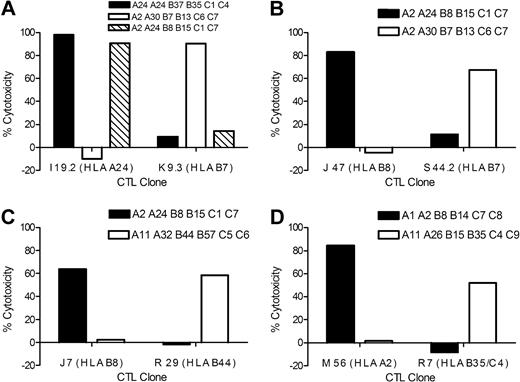
HLA-restricted lysis of AML by minor H antigen–specific T-cell clones. AML samples that were matched and mismatched at the HLA-restricting allele with individual minor H antigen–specific T-cell clones were labeled with CFSE and incubated for 12 hours with minor H antigen–specific CTL clones at an ET ratio of 20:1. Lysis of CFSE-labeled AML cells was determined by FACS analysis to allow analysis of leukemia cells. (A) Lysis of AML by HLA-A24–restricted CTL clone I 19.2 and HLA-B7–restricted CTL clone K 9.3. (B) Lysis of AML by HLA-B8–restricted CTL clone J 47 and HLA-B7–restricted CTL clone S 44.2. (C) Lysis of AML by HLA-B8–restricted CTL clone J 7 and HLA-B44–restricted CTL clone R 29. (D) Lysis of AML by HLA-A2–restricted CTL clone M 56 and by HLA-B35/C4–restricted CTL clone R 7. The data are representative of multiple independent experiments for each CTL clone.
HLA-restricted lysis of AML by minor H antigen–specific T-cell clones. AML samples that were matched and mismatched at the HLA-restricting allele with individual minor H antigen–specific T-cell clones were labeled with CFSE and incubated for 12 hours with minor H antigen–specific CTL clones at an ET ratio of 20:1. Lysis of CFSE-labeled AML cells was determined by FACS analysis to allow analysis of leukemia cells. (A) Lysis of AML by HLA-A24–restricted CTL clone I 19.2 and HLA-B7–restricted CTL clone K 9.3. (B) Lysis of AML by HLA-B8–restricted CTL clone J 47 and HLA-B7–restricted CTL clone S 44.2. (C) Lysis of AML by HLA-B8–restricted CTL clone J 7 and HLA-B44–restricted CTL clone R 29. (D) Lysis of AML by HLA-A2–restricted CTL clone M 56 and by HLA-B35/C4–restricted CTL clone R 7. The data are representative of multiple independent experiments for each CTL clone.
Molecular characterization of a novel HLA-B8–restricted leukemia-associated minor H antigen
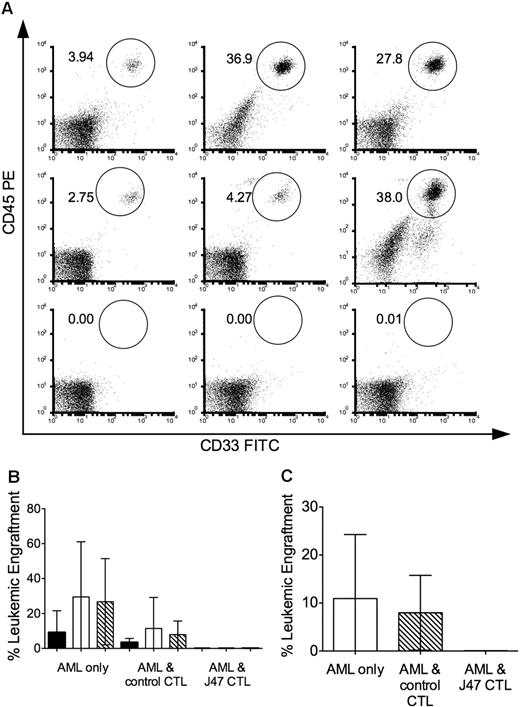
A goal of our studies is to identify genes that encode minor H antigens that are expressed on LICs. We focused on CTL clone J 47 derived from donor 1 that lysed 2 different primary HLA-B8+ AML samples for which we had sufficient aliquots to evaluate engraftment in NOD/SCID mice (Figure 2). Cohorts of NOD/SCID mice were inoculated with 5 × 106 AML cells alone, after coculture with a control minor H antigen–specific CTL clone that did not lyse the leukemic cells in vitro, and after coculture with J 47 CTL. All mice inoculated with both AML samples alone or after coculture with the control CTL clone exhibited leukemic engraftment in the BM as early as week 3 after inoculation and had significant levels of leukemia engraftment in BM and spleen at 6 and 8.5 weeks after inoculation (Figure 3). By contrast, engraftment was undetectable at both sites in all mice inoculated with AML samples cocultured with J 47 (Figure 3). These results are consistent with the minor H antigen recognized by J 47 being expressed on the AML LICs.41
The minor H antigen recognized by J 47 is expressed on AML LICs. Cohorts of sublethally irradiated NOD/SCID mice were inoculated with 5 × 106 AML cells alone or with AML cells that had been cocultured overnight with clone J 7 (specific for a minor H antigen not expressed on the AML) or with clone J 47 at an E/T ratio of 10:1. (A) FACS analysis of BM obtained at 6 weeks and stained for human CD45+CD33+ cells in individual mice that received AML alone (top panel); AML and the control clone (middle panel); and AML and J 47 (bottom panel). Data are shown for 3 representative mice (of 5-10 in each cohort). The mouse on the righthand side of the middle panel died just before the 6-week evaluation, and the BM was obtained postmortem, accounting for higher nonspecific staining in the lower right of the flow plot. (B) Leukemic engraftment in the BM at 3 (■), 6 (□), and 8.5 (▧) weeks after transplantation evaluations in mice in each cohort. Bars represent SD. (C) Leukemic engraftment in the spleen killed at 8.5 weeks after transplantation in mice in each cohort. (A-C) The data are representative of 2 experiments with 2 different AML samples that express the minor H antigen recognized by J 47.
The minor H antigen recognized by J 47 is expressed on AML LICs. Cohorts of sublethally irradiated NOD/SCID mice were inoculated with 5 × 106 AML cells alone or with AML cells that had been cocultured overnight with clone J 7 (specific for a minor H antigen not expressed on the AML) or with clone J 47 at an E/T ratio of 10:1. (A) FACS analysis of BM obtained at 6 weeks and stained for human CD45+CD33+ cells in individual mice that received AML alone (top panel); AML and the control clone (middle panel); and AML and J 47 (bottom panel). Data are shown for 3 representative mice (of 5-10 in each cohort). The mouse on the righthand side of the middle panel died just before the 6-week evaluation, and the BM was obtained postmortem, accounting for higher nonspecific staining in the lower right of the flow plot. (B) Leukemic engraftment in the BM at 3 (■), 6 (□), and 8.5 (▧) weeks after transplantation evaluations in mice in each cohort. Bars represent SD. (C) Leukemic engraftment in the spleen killed at 8.5 weeks after transplantation in mice in each cohort. (A-C) The data are representative of 2 experiments with 2 different AML samples that express the minor H antigen recognized by J 47.
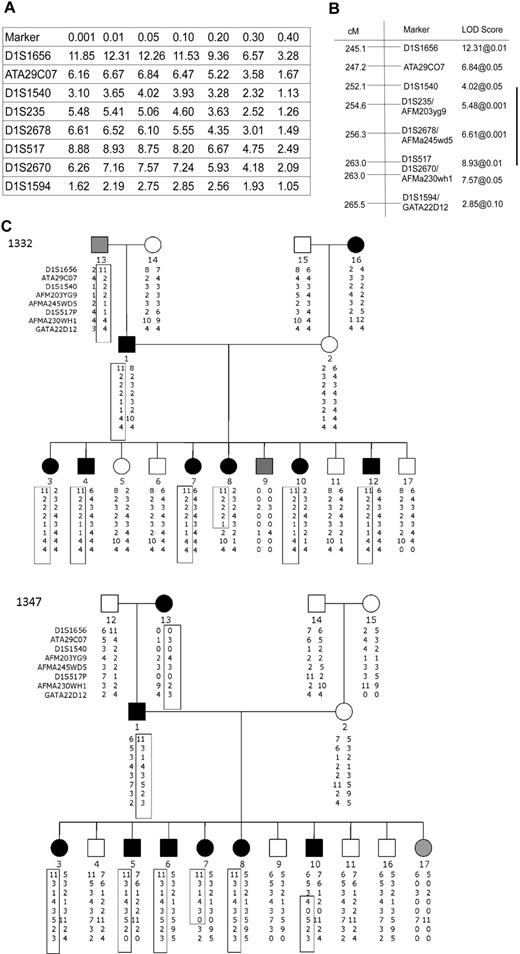
To identify the chromosomal region and candidate genes encoding the minor H antigen recognized by J 47, we performed genetic linkage analysis using B-LCL on a total of 8 CEPH families that were transfected to express HLA-B8. A genome-wide scan using available genotypic data from 478 microsatellite polymorphic markers was carried out initially on 5 families. These families identified a region of significant linkage on chromosome 1q, whereas no other chromosomal regions demonstrated significant linkage. To fine map this region on 1q, we analyzed all 8 CEPH families using CEPH V9.0/10.0 genotypes. The analysis on a total of 8 CEPH families defined a 9.87-Mb region of interest between 1q42.2 and 1q43. Haplotype analysis identified flanking recombinants in 3 of the 8 families, including pedigrees 1332 and 1347, narrowing the region to a 4.7-Mb region (Figure 4).
Identification of the locus encoding the minor H antigen recognized by J 47 using genetic linkage analysis. Two-point linkage analysis localized the coding sequence to chromosome 1q. (A) Genotypic data were extracted for chromosome one from the CEPH V9.0/V10.0 database, and a LOD table was constructed using 8 CEPH families. Markers on chromosome 1q identify a 20.4-cM region of interest with significant linkage to the locus encoding J 47. (B) According to the University of California Santa Cruz Genome Browser (http://genome.ucsc.edu/), this region corresponds to a physical map distance of 9.87 Mb. (C) Haplotype analysis identified informative recombinants in pedigrees 1332 and 1347 that implicate a 4.7-Mb region between D1S1540 and D1S517 as encoding the minor H antigen recognized by J 47. In family 1332, individual 8 who is minor H antigen+ shares the involved haplotype only proximal to marker D1S517 with the minor H antigen+ father. In family 1347, individual 10 is minor H antigen+ and shares the haplotype on chromosome 1q with the minor H antigen+ father only distal to D1S1540. Black, white, and gray circles (female) or squares (male) indicate a person who was scored as positive, negative, or indeterminate (B-LCLs not available or marginal lysis level), respectively, for the minor H antigen phenotype.
Identification of the locus encoding the minor H antigen recognized by J 47 using genetic linkage analysis. Two-point linkage analysis localized the coding sequence to chromosome 1q. (A) Genotypic data were extracted for chromosome one from the CEPH V9.0/V10.0 database, and a LOD table was constructed using 8 CEPH families. Markers on chromosome 1q identify a 20.4-cM region of interest with significant linkage to the locus encoding J 47. (B) According to the University of California Santa Cruz Genome Browser (http://genome.ucsc.edu/), this region corresponds to a physical map distance of 9.87 Mb. (C) Haplotype analysis identified informative recombinants in pedigrees 1332 and 1347 that implicate a 4.7-Mb region between D1S1540 and D1S517 as encoding the minor H antigen recognized by J 47. In family 1332, individual 8 who is minor H antigen+ shares the involved haplotype only proximal to marker D1S517 with the minor H antigen+ father. In family 1347, individual 10 is minor H antigen+ and shares the haplotype on chromosome 1q with the minor H antigen+ father only distal to D1S1540. Black, white, and gray circles (female) or squares (male) indicate a person who was scored as positive, negative, or indeterminate (B-LCLs not available or marginal lysis level), respectively, for the minor H antigen phenotype.
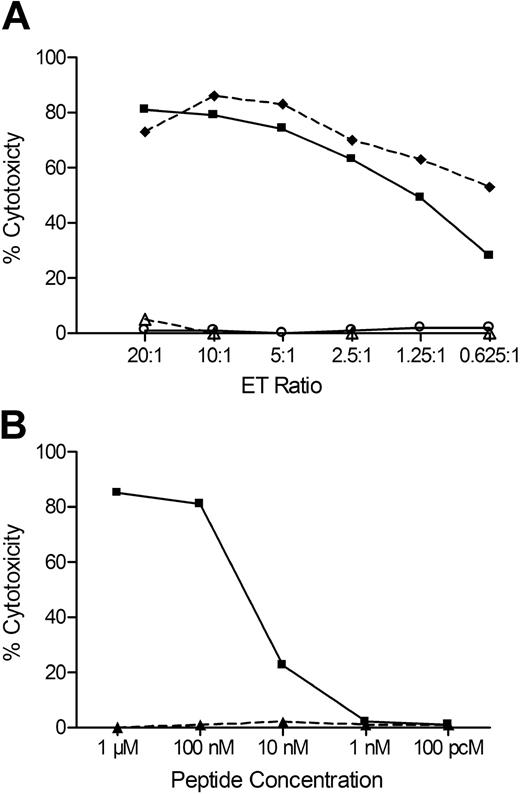
Molecular databases containing human genome sequence and SNP data were searched for known genes and SNPs in the region. We identified 6 candidate genes that contained one or more common exonic nonsynonymous SNPs. Genotype data for these SNPs were available for a subset of the CEPH B-LCL through the NCBI Hap-Map SNP database, and analysis of these data revealed a 100% correlation between lysis of the CEPH B-LCL and the genotype data for 7 SNPs within the HEATR1 gene and 3 SNPs within the adjacent LGALS8 gene. Amino acid sequences encompassing these SNPs were examined for predicted binding to HLA-B8 using the SYFPEITHI program,48 and a 9-mer peptide (ISKERAEAL) with a high binding score was identified in HEATR1. PCR sequencing of the region of HEATR1 encoding this epitope in DNA prepared from recipient and donor B-LCL showed that the recipient genotype for the SNP rs2275687 was A/A encoding ISKERAEAL, and donor genotype was G/G encoding ISKERAGAL. Sequencing of DNA from an additional 29 HLA-B8+ B-LCL lines, 15 of which were lysed by J 47, showed a complete correlation between lysis and the A/A or A/G genotype at rs2275687. J 47 also lysed donor B-LCL donor pulsed with ISKERAEAL but not with ISKERAGAL, confirming this sequence of HEATR1 as the antigenic epitope (Figure 5).
A peptide encoded by a SNP in HEATR1 is the minor H antigen recognized by J 47. (A) Lysis of recipient B-LCLs (♦), donor B-LCLs (○), donor B-LCLs pulsed with 1μM of peptide ISKERAEAL (■), and donor B-LCLs pulsed with 1μM of peptide ISKERAGAL (△) by the CTL clone J 47. (B) Lysis of donor B-LCLs pulsed with various concentrations of ISKERAEAL (■) or ISKERAGAL (▲) peptides. Data are at an ET of 20:1.
A peptide encoded by a SNP in HEATR1 is the minor H antigen recognized by J 47. (A) Lysis of recipient B-LCLs (♦), donor B-LCLs (○), donor B-LCLs pulsed with 1μM of peptide ISKERAEAL (■), and donor B-LCLs pulsed with 1μM of peptide ISKERAGAL (△) by the CTL clone J 47. (B) Lysis of donor B-LCLs pulsed with various concentrations of ISKERAEAL (■) or ISKERAGAL (▲) peptides. Data are at an ET of 20:1.
The HEATR1 minor H antigen is a candidate target for GVL therapy
The HEATR1 gene is a multiply spliced 7-kb gene that encodes BAP28, a protein of unknown function. The reported genotype frequency of rs2275687 in the white population is A/A 10%, A/G 45%, and G/G 45%, which results in a calculated donor recipient disparity for the HEATR1+ (A) allele of 13.9% for HLA-identical sibling recipient donor pairs, and 24.7% for unrelated recipient donor pairs.25 Adjusting for the 22.5% frequency of the HLA-B8 allele in the white population, immunotherapy targeting the HEATR1+ (A) allele would be applicable to 3.1% and 5.6% of HLA-identical sibling and unrelated recipient donor pairs, respectively.
HEATR1 is expressed in hematopoietic cells, including AML cells, but the potential for HEATR1-specific T cells to recognize target tissues of GVHD is not known. It is not feasible to directly evaluate functional recognition of most human nonhematopoietic cells by minor H antigen–specific CTL; thus, expression of the gene encoding the minor H antigen was used as a surrogate to infer CTL recognition. We evaluated the expression of HEATR1 in a panel of nonhematopoetic cells using TaqMan PCR, and compared the expression of these genes with HA-1 and P2X5, which encode previously defined minor H antigens that are generally considered to be restricted in their expression to hematopoietic cells and to be excellent candidates to target for GVL therapy.11,16,23,24 The expression of HEATR1 in nonhematopoietic tissues was very low in target tissues of GVHD, including liver, colon, small intestine, and lung, and was comparable with the expression of HA-1 and P2X5 (Figure 6).
The HEATR1 gene is expressed at levels comparable with HA-1 and P2X5 in GVHD target tissues. Expression of CD45, HEATR1, HA-1, P2X5, and IIp45 (a representative ubiquitously expressed gene) in cDNA tissue panels was evaluated by real-time quantitative PCR. The level of expression of each gene is shown relative to gene expression measured in B-LCLs, which are susceptible to lysis by the minor H antigen specific CTL after normalizing to the expression of a housekeeping gene PBGD. Data are representative of multiple experiments for each gene.
The HEATR1 gene is expressed at levels comparable with HA-1 and P2X5 in GVHD target tissues. Expression of CD45, HEATR1, HA-1, P2X5, and IIp45 (a representative ubiquitously expressed gene) in cDNA tissue panels was evaluated by real-time quantitative PCR. The level of expression of each gene is shown relative to gene expression measured in B-LCLs, which are susceptible to lysis by the minor H antigen specific CTL after normalizing to the expression of a housekeeping gene PBGD. Data are representative of multiple experiments for each gene.
Discussion
We show here that primary in vitro stimulation of purified CD8+ TN in microcultures with DCs from an HLA-identical sibling in the presence of IL-12 and a γ-chain cytokine is an effective strategy for isolating polyclonal CD8+ T cells and T-cell clones specific for human minor H antigens that can be used to discover novel leukemia-associated minor H antigens. We found that CD8+ minor H antigen–specific CTLp directed against both ubiquitously and selectively expressed minor H antigens were found nearly exclusively within the TN subset in nonparous, untransfused donors. The TM subset is predominantly specific for pathogens, and the paucity of minor H antigen–specific T cells in this subset is consistent with the lack of reported cross reactivity of virus-specific T cells with minor H antigens, and direct measurements showing that recognition of unrelated peptides by virus-specific T cells is very rare.49 The predominance of alloreactive T cells in the TN subset is also consistent with studies in murine models demonstrating that this subset has the greatest propensity to cause severe GVHD,43,44 and suggests the possibility that selective depletion of this subset might reduce GVHD in human HCT.
The culture conditions defined here enabled the isolation of a large panel of minor H antigen–specific CTL lines from 10 of 10 G-PBSC donors for patients undergoing HCT. Stable T-cell clones that were specific for multiple distinct recipient minor H antigens were readily derived from the T-cell lines and retained function over repeated cycles of in vitro expansion. The panel of T-cell clones included several that recognized minor H antigens expressed on recipient hematopoietic cells but not by dermal fibroblasts, and these CTL clones also lysed primary leukemia cells in vitro. Thus, primary in vitro stimulation could provide a feasible strategy for the generation of minor H antigen–specific CTLs for adoptive immunotherapy from an aliquot of the G-PBSC product. This strategy would provide minor H antigen–specific CTLs that, if specific for a hematopoietic-restricted minor H antigen, could be used early after HCT for the prophylaxis or treatment of leukemic relapse.
A focus of our work was to develop primary in vitro stimulation as a method to expand the number of T-cell clones available to identify novel minor H antigens expressed on leukemia cells. As proof of principle, we used linkage analysis to identify a novel polymorphic minor H antigen encoded by the HEATR1 gene and recognized by one of the CTL clones generated by primary in vitro stimulation. This novel minor H antigen is presented in association with HLA-B*0801 and results from a polymorphism with a balanced distribution in the population. Leukemia cells present the HEATR1 minor H antigen, and HEATR1-specific CTL abrogated engraftment of human leukemia in NOD/SCID mice demonstrating that the antigen is expressed on LICs. The HEATR1 gene has a similar level of mRNA expression in GVHD target tissues as the HA-1 and P2X5 minor H antigens. Significant numbers of CTLs specific for HA-1 and P2X5 have been identified in the peripheral blood of patients after allogeneic HCT or donor lymphocyte infusion in the absence of GVHD. Thus, the novel minor H antigen encoded by HEATR1 should be considered as a candidate target antigen for evaluation in clinical trials of immunotherapy for HLA-B8+ patients who are discordant with their donor for the antigenic HEATR1 allele and experience posttransplantation leukemic relapse. The genes encoding 4 additional novel minor H antigens have been identified using CD8+ CTL clones generated by primary in vitro stimulation, and these are currently being characterized for expression in LICs and target tissues of GVHD.
A further potential application of this work is in the study of GVHD. Prior studies have linked the development of GVHD in humans with disparity of broadly expressed minor H antigens encoded by the Y-chromosome or by the UGT2B17 gene that is subject to copy number variation as a consequence of gene deletion.24,50 Many of the minor H antigen–specific CTLs that were isolated in this study readily lysed dermal fibroblasts as well as several hematopoietic cell types, indicating recognition of a broadly expressed minor H antigen. Thus, minor H antigen–specific CTLs that are derived from the TN repertoire of the donor are probably valuable reagents for identifying the molecular targets of GVHD and for the development of reagents to track the development and migration of T cells specific for ubiquitously expressed minor H antigens in patients experiencing GVHD after HCT.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Toshiyuki Tanaka for his kind gift of the TMβ1 hybridoma; Dr Jon Yewdell for the HLA-B8 vaccinia viral vector; Juli Armstrong, Pavani Maragowni, and Michelle Brown for technical assistance; Cynthia Nourigat, Melissa Comstock, and Amanda Egge for technical assistance with immunodeficient mice; David Yadock and Shelly Heimfeld for advice with cell selection; and Jerry Davidson for help with ELDA graphics.
This work was supported by the Leukemia & Lymphoma Society (M.B.), the Amy Strelzer Manasevit SuperGen Post-Doctoral Fellowship Program of the National Marrow Donor Program (M.B.), the National Blood Foundation (M.B.), the American Society for Blood and Marrow Transplantation and Millennium Foundation (M.B.), National Institutes of Health (CA18029; S.R.R.), the W. H. Keck Foundation, and the George S. and Delores Doré Eccles Foundation (M.F.L.).
National Institutes of Health
Authorship
Contribution: M.B. designed and performed research, analyzed data, and wrote the paper; B.E.O., J.L.R., A.D.M., M.H., and T.N. performed research and analyzed data; C.N.C. recruited subjects and coordinated samples; E.H.W. and M.F.L. provided reagents and expertise; and S.R.R. designed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Marie Bleakley, Fred Hutchinson Cancer Research Center, Program in Immunology, D3-100, 1100 Fairview Ave N, PO Box 19024, Seattle, WA 98109; e-mail: mbleakle@fhcrc.org.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal