Abstract
Phosphorylated signal transducer and activator of transcription 5 (STAT5) is a biomarker and potential molecular target for hematologic malignancies. We have shown previously that lethal myeloproliferative disease (MPD) in mice mediated by persistently activated STAT5 (STAT5aS711F) requires the N-domain, but the mechanism was not defined. We now demonstrate by retrovirally complementing STAT5abnull/null primary mast cells that relative to wild-type STAT5a, STAT5a lacking the N-domain (STAT5aΔN) ineffectively protected against cytokine withdrawal-induced cell death. Both STAT5a and STAT5aΔN bound to a site in the bcl-2 gene and both bound near the microRNA 15b/16 cluster. However, only STAT5a could effectively induce bcl-2 mRNA and reciprocally suppress miR15b/16 leading to maintained bcl-2 protein levels. After retroviral complementation of STAT5abnull/null fetal liver cells and transplantation, persistently active STAT5aS711F lacking the N-domain (STAT5aΔNS711F) was insufficient to protect c-Kit+Lin−Sca-1+ (KLS) cells from apoptosis and unable to induce bcl-2 expression, whereas STAT5aS711F caused robust KLS cell expansion, induction of bcl-2, and lethal MPD. Severe attenuation of MPD by STAT5aΔNS711F was reversed by H2k/bcl-2 transgenic expression. Overall, these studies define N-domain–dependent survival signaling as an Achilles heel of persistent STAT5 activation and highlight the potential therapeutic importance of targeting STAT5 N-domain–mediated regulation of bcl-2 family members.
Introduction
Phosphorylation of signal transducer and activator of transcription 5 (STAT5) is prevalent downstream of many oncogenic receptor and nonreceptor tyrosine kinases. Examples include Flt3–internal tandem duplication (ITD)1 in human acute myeloid leukemia or activated mutants of Janus kinase 2 (JAK2)2 or MPL3 in human myeloproliferative disease (MPD). The dosage of STAT5 activity has been suggested as a major mechanism for the different disease pathology reported for Flt3-ITD compared with Flt3–tyrosine kinase domain (TKD) mutant.4,5 The double mutant Flt3-ITD-TKD is capable of inducing hyperactivation of STAT56 and overexpression of bcl-XL.6 A major role of STAT5 in Flt3-ITD–induced disease was also demonstrated by Y589 and Y591 mutations, where loss of the STAT5-binding sites was sufficient to ablate disease.1 Two transgenic mouse models of JAK2V617F showed that high dosage of JAK2 (and STAT5 activation) was associated with erythroid expansion, whereas lower dosage of JAK2V617F was associated with megakaryocytic disease.7,8 Therefore, although no activating mutations have been reported for STAT5 in humans, STAT5 is highly activated via upstream kinases in myeloid malignancies.
Kotecha et al9 demonstrated by flow cytometry that phosphorylated STAT5 can be viewed as an important biomarker that can inform diagnosis and clinical outcome in juvenile myelomonocytic leukemia and other forms of acute myeloid leukemia. The most direct evidence that STAT5 plays an essential role in oncogenic transformation has come from mouse models, which showed that STAT5-deficient hematopoietic cells are resistant to transformation by oncogenic tyrosine kinases, such as TEL-JAK210 or BCR-ABL.11 Therefore, the oncogenic role of STAT5 has been recognized. However, although up-regulation of genes such as bcl-2, bcl-XL, mcl-1, cyclin D, and myc by activated oncogenic tyrosine kinases has been demonstrated, the specific mechanisms by which STAT5 can critically control preleukemic expansion in the myeloid lineage have not been defined.
STAT5a with a mutation in the C-terminal transactivation domain (STAT5aS711F) can confer lethal MPD in mice and is a useful tool for study of persistently active STAT5 signaling.10,12 We previously reported that the binding site preferences for STAT5a and STAT5aS711F determined by systematic evolution of ligands by exponential environment (SELEX) sequence motifs were not different and that retroviral overexpression of STAT5aS711F, but not STAT5a lacking the first 136 amino acids (STAT5aΔNS711F), in mice that underwent transplantation was sufficient for lethal MPD.13 Strikingly, the N-domain played a major role in STAT5aS711F-mediated leukemogenesis,13 although the mechanism for this N-domain function has been undefined but of potential high therapeutic significance. Previously, we also showed that STAT5abΔN/ΔN mast cells had a reduced level of both bcl-2 and bcl-XL.14 However, STAT5ΔN protein level was only 10% to 30% of that in wild-type mast cells, and the direct versus indirect connection between the STAT5 N-domain and bcl-2 was not defined. The Flt3Y591 duplication is associated with high levels of bcl-2 expression15 and the connection between Flt3-ITD, STAT5, and downstream survival targets such as bcl-2 is important to understand due to the potential clinical significance. In these studies, we set out to test the hypothesis that STAT5 mediates prosurvival signaling through the N-domain to drive MPD progression and to determine mechanisms for this survival activity.
Methods
Animals
STAT5ab+/null mice on the C57BL/6 background have been described.16 Mice were housed in a specific pathogen–free environment. H2k/bcl-2 transgenic mice express bcl-2 from the H2k promoter and Moloney murine leukemia virus enhancer.17 All studies were approved by the Case Institutional Animal Care and Use Committee.
Fetal liver cell collection and mast cell culture
Fetal liver (FL) cells from embryonic day 14.5 (E14.5) and E15.5 STAT5abnull/null mice were collected and retrovirally transduced as previously described.18 The producer cells contained murine stem cell virus (MSCV)–based bicistronic retrovirus expressing green fluorescent protein (GFP) from an internal ribosomal entry sequence (IRES). GFP+ cells were sorted using a BD FACSAria (BD Biosciences). The sorted cells were cultured in complete RPMI 1640 medium (Invitrogen Life Technologies) with 10% fetal bovine serum, 2mM l-glutamine, 100 U/mL penicillin, 100 μg/mL streptomycin, 1mM sodium pyruvate, and 1mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid, supplemented with interleukin-3 (IL-3; 5 ng/mL) and stem cell factor (SCF; 50 ng/mL). Four to 6 weeks later, the cells matured into c-Kit+ CD34+ FcϵR1+ mast cells as previously described.14 Recombinant murine IL-3, IL-6, and SCF were purchased from R&D Systems and Peprotech. Mast cells in normal culture and following IL-3 or SCF withdrawal were washed twice with phosphate-buffered saline. Cells were cultured in IL-3 or SCF at 5 × 105/mL at the indicated factor concentrations overnight. Cells were then assessed for subdiploid DNA content by Hoechst 33342 (Molecular Probes) staining and analyzed by flow cytometry.
In vivo complementation and transplantation
STAT5abnull/null FL cells were derived and transduced with concentrated retrovirus-containing supernatants from GP+E86 producer cell lines (MSCV-IRES-GFP, MSCV-STAT5aS711F-IRES-GFP, and MSCV-STAT5aΔNS711F-IRES-GFP) as described.19 For bcl-2 add-back, STAT5ab+/null mice were crossed to H2k/bcl-2 transgenic mice.17 The STAT5abnull/null FL cells expressing H2k/bcl-2 were transduced with either MSCV-IRES-GFP or MSCV-STAT5aΔNS711F-IRES-GFP retroviral supernatant and transplanted into lethally irradiated recipient mice.
Flow cytometric assay for hematopoietic stem cell survival and bcl-2 expression
Mice that received a transplant were killed 15 days after transplantation. Bone marrow (BM) cells were counted and stained with biotin-conjugated lineage markers, Ly-6G (Gr-1), CD11b (Mac-1) CD45R/B220, CD4 (L3T4), CD8 (Ly2), and Ter119/Ly76. Lineage-depleted cells were isolated per the manufacturer's instructions (magnetic-activated cell sorting; Miltenyi Biotec) and 5 × 105 cells were stained with the lineage markers plus c-Kit–allophycocyanin (APC) and Sca-1–phycoerythrin (PE)–cyanin 7 (Cy7). The lineage markers were then conjugated to APC-Cy7 by secondary streptavidin-APC-Cy7. The cells were stained with annexin V–PE (BD Biosciences) and DAPI (4,6 diamidino-2-phenylindole; Invitrogen). To measure the bcl-2 expression in KLS cells, the Lin− fraction was stained with the lineage markers c-Kit–APC and Sca-1–PE-Cy7. The cells were then fixed and permeabilized with BD Cytofix/Cytoperm Fixation and Permeabilization solution (BD Biosciences), followed by anti–bcl-2–PE staining. All flow cytometric analyses were performed on a BD LSR II (BD Biosciences).
Chromatin immunoprecipitation and PCR
Mast cells and BaF3 cells were cross-linked with a 1.6% formaldehyde solution (Fisher Scientific) in phosphate-buffered saline for 10 minutes at room temperature. Cross-linking was stopped by the addition of glycine to a final concentration of 0.125 M. Cell pellets were resuspended in 1 mL of radioimmunoprecipitation assay (RIPA) buffer as described.20 Lysates were sonicated in three 15-second bursts, clarified by centrifugation, and protein concentration was determined. Then, 2 μg of STAT5 antibody L-20 (Santa Cruz) or recombinant immunoglobulin G (rIgG; Sigma) was added to 2 mg of protein lysate for specific and isotype-control immunoprecipitations, rocked overnight at 4°C, and washed twice with ice-cold RIPA buffer, 4 times with ice-cold wash buffer (100mM Tris [tris(hydroxymethyl)aminomethane], pH 8.5, 500mM LiCl, 1% Nonidet P-40, 1% deoxycholic acid), and twice more with ice-cold RIPA buffer. Cross-links were reversed by heating at 99°C for 30 minutes in Crosslinking Reversal Buffer (125mM Tris, pH 6.8, 10% β-mercaptoethanol, 4% sodium dodecyl sulfate). DNA was isolated as described.20 Polymerase chain reactions (PCRs) to detect the putative STAT5 consensus binding sites were performed in a final concentration of 50mM KCl, 20mM Tris, pH 8.4, 1.5mM MgCl2, 0.2mM deoxynucleotide triphosphates, 0.2μM each primer, and 1.25 U Platinum Taq (Invitrogen). The PCR cycling conditions were 1 cycle at 95°C for 5 minutes, 32 cycles at 95°C for 30 seconds, 60°C for 45 seconds, 72°C for 30 seconds, followed by 1 cycle at 72°C for 5 minutes. Sequences for PCR primers for bcl-2 and miR sites are available upon request.
Quantitative RT-PCR
Mast cells were cultured in either IL-3 (5 ng/mL) or IL-3 (5 ng/mL) and SCF (50 ng/mL) for 24 hours. RNA was prepared with Trizol reagent (Invitrogen). cDNA was prepared using SuperScript III First Strand Synthesis system for reverse-transcription (RT)–PCR (Invitrogen) and analyzed in triplicate with FastStart Universal SYBR Green Master (Roche). Primers specific for bcl-2, bcl-XL, cis, and osm have been described.21 gapdh served as an internal control.21 Real-time RT-PCR was performed using a 7500 FAST Real-time PCR system (Applied Biosystems). For mature miR quantification, N-code microRNA First-strand cDNA synthesis and quantitative RT-PCR Kits were used (Invitrogen). Small nuclear RNA U6 was used as endogenous control. Reactions were run in triplicate for each experiment.
Results
The STAT5 N-domain controls bcl-2 expression and survival in primary hematopoietic cells
Studies with primary cells from STAT5abΔN/ΔN mice have shown that STAT5 can induce genes in mast cells that are essential for proliferative and antiapoptotic functions.14 However, the gene-targeting strategy used to generate STAT5abΔN/ΔN mice resulted in variable expression of STAT5ΔN protein in mast cells and questionable expression in hematopoietic stem cells (HSCs).19,22 Therefore, it has been unclear to what extent the N-terminal domain of STAT5 could promote proliferation and survival of mast cells and HSCs in particular. To explore the potential effects of the N-domain of STAT5, we retrovirally transduced MSCV-IR-GFP (empty vector), MSCV-STAT5a, and MSCV-STAT5aΔN into E14.5 and E15.5 STAT5abnull/null FL cells and cultured mature mast cells expressing c-Kit, CD34, and FcϵR1. Growth rates of primary retrovirally transduced mast cells were variable, especially IR-GFP controls where integration-site selection could have an impact. No average significant growth differences were observed (data not shown).
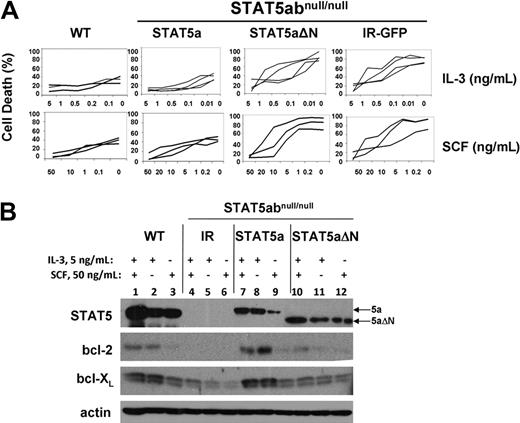
To examine potential survival functions of retrovirally expressed STAT5a or STAT5aΔN, STAT5abnull/null mast cells were complemented with IR-GFP, STAT5a, or STAT5aΔN vectors. In regular culture with IL-3 + SCF, no significant differences were measured between wild-type and STAT5abnull/null mast cells (P > .36). When we stressed the cells by culture in limiting concentration of IL-3 or SCF as done previously to induce apoptosis,14 STAT5a- and IR-GFP–expressing mast cells showed significantly different dose-response curves (Figure 1A). Add-back of STAT5a (P < .01 for either IL-3 or SCF only) but not STAT5aΔN (P = .63 for IL-3 only; P = .09 for SCF only) was sufficient to protect STAT5abnull/null mast cells from cytokine withdrawal–induced cell death. Importantly, the lack of survival protection by STAT5aΔN was not due to different protein expression level. There was no difference in response to cytokine withdrawal between STAT5abnull/null mast cells expressing IR-GFP or STAT5aΔN, indicating that the N-terminal domain is responsible for survival function of STAT5. Add-back of bcl-2 or bcl-XL to STAT5abnull/null mast cells also restored survival as expected (data not shown). Transduction-induced protein expression of STAT5a and STAT5aΔN was comparable (Figure 1B).
The STAT5 N-domain is required for STAT5-mediated mast cell survival and bcl-2 expression. (A) STAT5abnull/null mast cells transduced with MSCV-based bicistronic retrovirus expressing either vector control (IR) wild-type STAT5a or STAT5aΔN were cultured in the indicated concentrations of either IL-3 or SCF for 16 to 24 hours, and compared with wild-type mast cells. Apoptosis was assessed by the presence of subdiploid DNA after Hoechst 33342 DNA staining. Each line in the plot represents an independent experiment (IL-3 withdrawal: IR-GFP vs STAT5a, P < .01; IR-GFP vs STAT5aΔN, P = .64; STAT5a vs STAT5aΔN, P < .01; SCF withdrawal: IR-GFP vs STAT5a, P < .01; IR-GFP vs STAT5aΔN, P = .87; STAT5a vs STAT5aΔN, P < .01; Student t test for all data points in each line). (B) Mast cells as in panel A were cultured in either IL-3 (5 ng/mL), SCF (50 ng/mL), or both IL-3 (5 ng/mL) plus SCF (50 ng/mL) for 48 hours, and total cell lysates were subjected to Western blot analysis for STAT5, bcl-2, and bcl-XL, and actin expression on the same membrane.
The STAT5 N-domain is required for STAT5-mediated mast cell survival and bcl-2 expression. (A) STAT5abnull/null mast cells transduced with MSCV-based bicistronic retrovirus expressing either vector control (IR) wild-type STAT5a or STAT5aΔN were cultured in the indicated concentrations of either IL-3 or SCF for 16 to 24 hours, and compared with wild-type mast cells. Apoptosis was assessed by the presence of subdiploid DNA after Hoechst 33342 DNA staining. Each line in the plot represents an independent experiment (IL-3 withdrawal: IR-GFP vs STAT5a, P < .01; IR-GFP vs STAT5aΔN, P = .64; STAT5a vs STAT5aΔN, P < .01; SCF withdrawal: IR-GFP vs STAT5a, P < .01; IR-GFP vs STAT5aΔN, P = .87; STAT5a vs STAT5aΔN, P < .01; Student t test for all data points in each line). (B) Mast cells as in panel A were cultured in either IL-3 (5 ng/mL), SCF (50 ng/mL), or both IL-3 (5 ng/mL) plus SCF (50 ng/mL) for 48 hours, and total cell lysates were subjected to Western blot analysis for STAT5, bcl-2, and bcl-XL, and actin expression on the same membrane.
We next examined the expression level of bcl-2 and bcl-XL expression in all 4 groups of mast cell cultures. Mast cells were cultured in both IL-3 and SCF, IL-3 alone, or SCF alone at 3 to 5 × 105 cells/mL for 48 hours to measure changes in protein expression. In the concentrations of cytokine used, viability was more than 80% for all groups at the time of analysis. As seen in Figure 1B, there was dramatic reduction of bcl-2 expression in IR-GFP–expressing STAT5abnull/null mast cells compared with wild-type mast cells. A modest reduction in bcl-XL expression was also observed. Reconstitution of STAT5abnull/null mast cells with full-length STAT5a restored bcl-2 and bcl-XL expression, whereas STAT5aΔN-expressing cells had a slight induction of either bcl-2 or bcl-XL expression, compared with the vector control cells. Notably, IL-3 response was most responsible for the difference of bcl-2/bcl-XL expression among all 4 groups (Figure 1B lanes 2, 5, 8, 11). This striking difference in survival-related target gene expression was dependent on the N-terminal domain of STAT5 and was consistent with the cytokine response curve.
STAT5 interacts with conserved binding sites in bcl-2 intron 2 and up-regulates bcl-2 mRNA
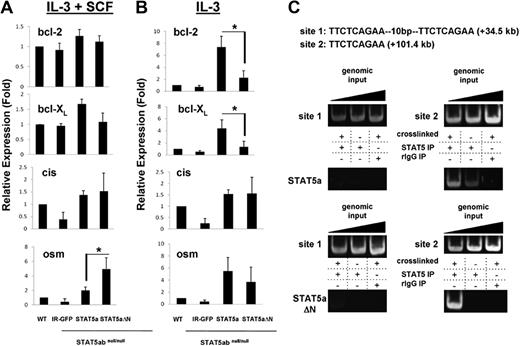
Because the protein level of bcl-2 was STAT5 N-domain dependent, it was important to determine whether bcl-2 might be a direct STAT5 transcriptional target gene and whether STAT5-dependent bcl-2 regulation might occur through a transcriptional and/or posttranscriptional mechanism. The relative levels of mRNA for bcl-2 were determined by real-time RT-PCR in wild-type or STAT5abnull/null mast cells reconstituted with vectors expressing IR-GFP, STAT5a, or STAT5aΔN (Figure 2A). Because we observed that IL-3 has the most effect on bcl-2 protein, we focused on conditions involving IL-3. In the presence of IL-3 and SCF, bcl-2 mRNA level was similar among all 4 groups. When cultured in IL-3 alone, bcl-2 mRNA level in STAT5abnull/null IR-GFP–transduced mast cells was modestly reduced to 50% to 70% of wild type, indicating that other pathways may also be involved in regulation of bcl-2. Nevertheless, complementation with STAT5a increased bcl-2 mRNA 10-fold higher than IR-GFP control, compared with STAT5aΔN, which increased bcl-2 mRNA by only 2- to 3-fold (Figure 2B). Therefore, STAT5aΔN was less effective in inducing bcl-2 mRNA. In contrast, regulation of cis and osm, 2 other well-characterized STAT5 target genes, was not dependent on the N-domain.
STAT5 regulates bcl-2 mRNA by binding a conserved site in intron 2 of the bcl-2 gene as determined by chromatin immunoprecipitation. (A) Mast cells as in Figure 1 were incubated in IL-3 (5 ng/mL) plus SCF (50 ng/mL), or IL-3 (5 ng/mL) alone (B) for 24 hours. mRNA level of bcl-2, bcl-XL, cis, or osm was quantitated by RT-PCR. Values obtained from wild-type mast cells were set to 1 (for IL-3 + SCF: osm group, *P = .03; for IL-3 alone: bcl-2 group, *P = .01 and bcl-XL group, *P = .03; Student t test). Error bars represent SD of the mean. (C) STAT5 was immunoprecipitated from formaldehyde–cross-linked lysates prepared from STAT5abnull/null mast cells transduced with either wild-type STAT5a (WT) or STAT5aΔN (ΔN). Binding of STAT5 to putative STAT5 consensus binding sites 1 and 2 in intron 2 of the bcl-2 gene was determined by PCR amplification of immunoprecipitated DNA. Nonspecific binding was assayed by immunoprecipitating STAT5 from non–cross-linked lysates and rIgG isotype-control immunoprecipitation of cross-linked lysates. Addition of increasing amounts of genomic input DNA ensured PCRs were performed in the linear range. Results are representative of 2 independent experiments.
STAT5 regulates bcl-2 mRNA by binding a conserved site in intron 2 of the bcl-2 gene as determined by chromatin immunoprecipitation. (A) Mast cells as in Figure 1 were incubated in IL-3 (5 ng/mL) plus SCF (50 ng/mL), or IL-3 (5 ng/mL) alone (B) for 24 hours. mRNA level of bcl-2, bcl-XL, cis, or osm was quantitated by RT-PCR. Values obtained from wild-type mast cells were set to 1 (for IL-3 + SCF: osm group, *P = .03; for IL-3 alone: bcl-2 group, *P = .01 and bcl-XL group, *P = .03; Student t test). Error bars represent SD of the mean. (C) STAT5 was immunoprecipitated from formaldehyde–cross-linked lysates prepared from STAT5abnull/null mast cells transduced with either wild-type STAT5a (WT) or STAT5aΔN (ΔN). Binding of STAT5 to putative STAT5 consensus binding sites 1 and 2 in intron 2 of the bcl-2 gene was determined by PCR amplification of immunoprecipitated DNA. Nonspecific binding was assayed by immunoprecipitating STAT5 from non–cross-linked lysates and rIgG isotype-control immunoprecipitation of cross-linked lysates. Addition of increasing amounts of genomic input DNA ensured PCRs were performed in the linear range. Results are representative of 2 independent experiments.
Because we found that STAT5 could regulate bcl-2 mRNA level, we then investigated whether STAT5-binding sites could be found within the bcl-2 gene. From a total of 71 potential STAT5-binding sites in the mouse bcl-2 gene, 7 were conserved between mice and humans (supplemental Figure 1, available on the Blood website; see the Supplemental Materials link at the top of the online article). We therefore performed chromatin immunoprecipitation (ChIP) using mast cells grown in IL-3 + SCF and expressing reconstituted STAT5a or STAT5aΔN (Figure 2C). Using the chromatin fraction pulled down by anti-STAT5 antibody as template, a PCR fragment containing the binding site 2 in bcl-2 intron 2 was detected. Background levels of nonspecific binding were defined using isotype control IgG or no cross-linking as described.23 In contrast, no STAT5 binding to site 1 (tandem STAT5-binding sites) or to any of the other 5 sites (data not shown) was observed. Although both STAT5a and STAT5aΔN were able to bind the conserved site 2 in bcl-2 intron 2, regulation of bcl-2 mRNA by STAT5 may require an N-domain–mediated coactivator function. As additional controls, STAT5 bound to known bcl-XL and il-2R sites and STAT5abnull/null cells as a third control gave similar background on bcl-2 site 2 (supplemental Figure 2A). To study persistently active STAT5 binding of bcl-2 site 2 in BaF3 cells, the STAT5aS711F mutant was used and both STAT5aS711F and STAT5aΔNS711F bound to this site (supplemental Figure 2B).
STAT5 represses miR-15/16 to promote bcl-2 expression at a posttranscriptional level
The observation that in the presence of both IL-3 and SCF the mRNA level of bcl-2 was comparable between wild-type and STAT5abnull/null mast cells whereas the protein level was significantly reduced suggested additional posttranscriptional regulation of bcl-2. miR-15b and miR-16-2 form a cluster on chromosome 3 and have recently been shown to regulate bcl-2.24 We therefore investigated expression levels of miR-15/16 in mast cells by real-time PCR. miR-15a, miR-15b, and miR-16 were increased approximately 2-fold in STAT5abnull/null mast cells compared with wild-type mast cells (Figure 3A). Interestingly, add-back of STAT5a into STAT5abnull/null mast cells reduced miR-15b and miR-16 expression to the level of wild-type cells, whereas STAT5aΔN had no effect. Consistent with reports in other cell types, we found that transfection of miR15b or miR16 was sufficient to reduce bcl-2 protein levels in myeloid cells (supplemental Figure 3). To study persistently active STAT5 regulation of microRNA in BaF3 cells, the STAT5aS711F mutant was used. Corroborating the data with wild-type STAT5a, the expression of miR-15b and miR-16 was suppressed by STAT5aS711F transduction into BaF3 cells, whereas transduction with STAT5aΔNS711F was less effective (Figure 3B).
STAT5 regulates bcl-2 posttranscriptionally through the mIR-15/16 cluster. (A) Level of miR-15a, miR-15b, and miR-16 was quantitated by RT-PCR from wild-type or STAT5abnull/null mast cell cultures that were transduced with either IR-GFP–, STAT5a-, or STAT5aΔN-expressing vectors (*IR vs STAT5a, P < .05; **STAT5a vs STAT5aΔN, P < .05; Student t test). (B) The level of miR-15a, miR-15b, and miR-16 was assayed in BaF3 cells expressing IR-GFP, STAT5aS711F, or STAT5aΔNS711F; *STAT5aS711F vs STAT5aΔNS711F, P < .05. Error bars represent SD of the mean. (C) Shown is the conserved STAT5-binding site in the promoter of miR-15b/16-2. The numbers indicate the distance upstream from the start site for miR-15b. (D) ChIP data display STAT5aS711F and STAT5aΔNS711F bound to the conserved binding site near the miR15/16 cluster in BaF3 cell extracts. The nonconserved site is approximately 8 kb upstream of the starting site of miR-15b and is included as a control.
STAT5 regulates bcl-2 posttranscriptionally through the mIR-15/16 cluster. (A) Level of miR-15a, miR-15b, and miR-16 was quantitated by RT-PCR from wild-type or STAT5abnull/null mast cell cultures that were transduced with either IR-GFP–, STAT5a-, or STAT5aΔN-expressing vectors (*IR vs STAT5a, P < .05; **STAT5a vs STAT5aΔN, P < .05; Student t test). (B) The level of miR-15a, miR-15b, and miR-16 was assayed in BaF3 cells expressing IR-GFP, STAT5aS711F, or STAT5aΔNS711F; *STAT5aS711F vs STAT5aΔNS711F, P < .05. Error bars represent SD of the mean. (C) Shown is the conserved STAT5-binding site in the promoter of miR-15b/16-2. The numbers indicate the distance upstream from the start site for miR-15b. (D) ChIP data display STAT5aS711F and STAT5aΔNS711F bound to the conserved binding site near the miR15/16 cluster in BaF3 cell extracts. The nonconserved site is approximately 8 kb upstream of the starting site of miR-15b and is included as a control.
We next used ChIP to detect the presence of STAT5 at the promoter region of the miR-15b/16-2 cluster in BaF3 cells, because we noted a STAT5-binding site conserved between mice and humans (Figure 3C). As shown in Figure 3D, STAT5 specifically immunoprecipitated a region including the conserved STAT5-binding site near the miR15b/16-2 cluster. BaF3 extracts expressing STAT5aS711F or STAT5aΔNS711F both pulled down miR15b/16-2 by ChIP, whereas either non–cross-linked or IgG control could not. As an additional control, neither STAT5 mutant was able to bind to a nonconserved binding site approximately 8 kb upstream of the miR-15b/16-2 cluster.
The STAT5 N-domain is essential for bcl-2 expression, survival, and expansion of KLS cells
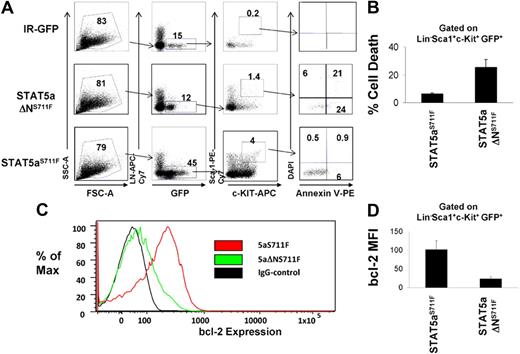
Because we found that the STAT5 N-domain was responsible mainly for mast cell survival and bcl-2 expression in vitro, we next tested the hypothesis that STAT5aΔNS711F is deficient at MPD induction because of an intrinsic inability to maintain HSC survival in vivo. To test this, we transduced STAT5abnull/null FL cells with IR-GFP control, STAT5aS711F, or STAT5aΔNS711F, and transplanted these cells into lethally irradiated recipient mice. The retroviral ecotropic producer cells used were not significantly different in terms of retroviral transduction efficiency and yielded initial copy number of 1 to 3 per cell (supplemental Figure 4A) and comparable STAT5a mRNA levels (supplemental Figure 4B). On day 15 after transplantation when all mice that received a transplant were still without signs of MPD, the KLS fraction from recipient mice was analyzed for survival and bcl-2 expression. As seen in Figure 4A, STAT5abnull/null FL expressing the IR-GFP vector gave rise to very few (GFP+c-Kit+Lin−Sca-1+) GFP+ KLS cells, whereas reconstitution with FL expressing STAT5aS711F showed a robust KLS cell expansion, roughly 25-fold higher than the IR-GFP control. Meanwhile, STAT5aΔNS711F expression expanded the KLS cells 3-fold less than full-length STAT5aS711F, but still 8-fold higher than vector control. We found that cell death in STAT5aΔNS711F-expressing KLS cells, as measured by the annexin V+ DAPI− cell population, was 4-fold higher compared with the STAT5aS711F group, which was only approximately 5% (Figure 4B; P < .01). The STAT5 N-terminus thus is rate limiting for KLS cell expansion.
KLS cells expressing STAT5aΔNS711F have reduced survival and bcl-2 expression. (A) STAT5abnull/null FL cells at E14.5 and E15.5 were transduced with either MSCV/IRES-GFP vector control (IR), STAT5aS711F, or STAT5aΔNS711F, followed by transplantation into lethally irradiated recipient mice. Fifteen days later, apoptosis was assayed on the GFP+c-Kit+Lin−Sca-1+ (GFP+KLS) fraction of BM cells from each group by annexin V–DAPI assay. Early apoptotic cells were defined as annexin V+ DAPI−. A representative flow cytometric plot shows annexin V and DAPI gating on GFP+ KLS cells among 3 groups. (B) Percentage of annexin V+ DAPI– GFP+ KLS cells from 3 independent mice among each group (STAT5aS711F vs STAT5aΔNS711F, P < .01; Student t test). Error bars represent SD of the mean. (C) A representative histogram showing bcl-2 expression in GFP+KLS cells. (D) Bcl-2 expression was determined by mean fluorescence intensity minus IgG background after intracellular staining (STAT5aS711F: n = 4, STAT5aS711FΔN: n = 3; STAT5aS711F vs STAT5aΔNS711F, P < .01; Student t test). Error bars represent SD of the mean.
KLS cells expressing STAT5aΔNS711F have reduced survival and bcl-2 expression. (A) STAT5abnull/null FL cells at E14.5 and E15.5 were transduced with either MSCV/IRES-GFP vector control (IR), STAT5aS711F, or STAT5aΔNS711F, followed by transplantation into lethally irradiated recipient mice. Fifteen days later, apoptosis was assayed on the GFP+c-Kit+Lin−Sca-1+ (GFP+KLS) fraction of BM cells from each group by annexin V–DAPI assay. Early apoptotic cells were defined as annexin V+ DAPI−. A representative flow cytometric plot shows annexin V and DAPI gating on GFP+ KLS cells among 3 groups. (B) Percentage of annexin V+ DAPI– GFP+ KLS cells from 3 independent mice among each group (STAT5aS711F vs STAT5aΔNS711F, P < .01; Student t test). Error bars represent SD of the mean. (C) A representative histogram showing bcl-2 expression in GFP+KLS cells. (D) Bcl-2 expression was determined by mean fluorescence intensity minus IgG background after intracellular staining (STAT5aS711F: n = 4, STAT5aS711FΔN: n = 3; STAT5aS711F vs STAT5aΔNS711F, P < .01; Student t test). Error bars represent SD of the mean.
We next checked by intracellular flow cytometry the endogenous bcl-2 protein expression in the GFP+ KLS cells among all 3 groups. As expected, STAT5aS711F was able to drive a significantly higher bcl-2 expression than STAT5aΔNS711F (Figure 4C-D; P < .01) in transplanted FL cells, indicating that the N-domain–mediated regulation of bcl-2 expression by STAT5 may be responsible for the KLS cell survival under conditions where MPD follows STAT5aS711F expression.
Transgenic bcl-2 expression restores lethal MPD in the absence of the STAT5 N-domain
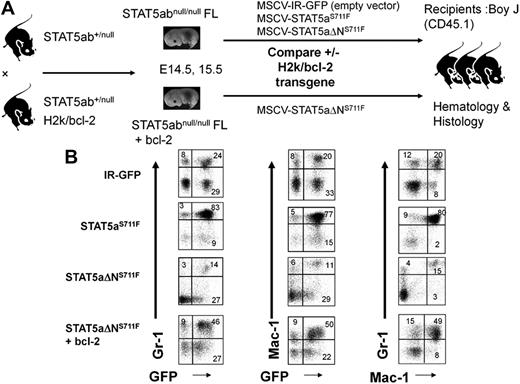
Having uncovered an important survival function mediated by the STAT5 N-domain, we next set out to determine whether deficient survival function could be restored by adding back a bcl-2 transgene. H2k/bcl-2 transgenic mice do not normally develop MPD and remain generally healthy, although a few mice can develop B-cell lymphomas at more than 9 months of age. The bcl-2 transgene was crossed onto the STAT5abnull/null background and FL cells from these mice or STAT5abnull/null controls were used for retroviral transduction with STAT5aΔNS711F or IR-GFP control vector (Figure 5A). STAT5abnull/null FL cells transduced with IR-GFP conferred a normal percentage of Gr-1/Mac-1–positive myeloid cells in the peripheral blood (Figure 5B). In contrast, mice expressing STAT5aS711F showed increases in the percentage of Gr-1/Mac-1–positive cells. As expected, STAT5aΔNS711F conferred a normal percentage of Gr-1/Mac-1–positive cells in most mice when STAT5aS711F-expressing mice already exhibited MPD. Interestingly, mice expressing both bcl-2 and STAT5aΔNS711F demonstrated a high frequency of Gr-1/Mac-1–positive cells, indicating that MPD was restored by bcl-2 add-back.
Bcl-2 transgenic add-back increases the contribution of transplanted HSCs to the myeloid lineage. (A) STAT5abnull/null FL cells at E14.5 and E15.5 were transduced with either MSCV/IRES-GFP vector control (IR), STAT5aS711F, or STAT5aΔNS711F and transplanted into lethally irradiated recipient mice. For the bcl-2 add-back, the STAT5abnull/null mice were crossed with bcl-2 transgenic mice as stated in “In vivo complementation and transplantation.” STAT5abnull/null bcl-2 transgenic FL cells were transduced with either IR-GFP or STAT5aΔNS711F. (B) Representative flow cytometric plots show the accumulation of myeloid cells in the peripheral blood. The panels show the Gr-1+ or Mac-1+ cells co-gated for expression of GFP. The right panels show the percentage of Gr-1+Mac-1+ double-positive myeloid cells. The flow cytometry was performed 20 to 24 days after transplantation for all 4 groups.
Bcl-2 transgenic add-back increases the contribution of transplanted HSCs to the myeloid lineage. (A) STAT5abnull/null FL cells at E14.5 and E15.5 were transduced with either MSCV/IRES-GFP vector control (IR), STAT5aS711F, or STAT5aΔNS711F and transplanted into lethally irradiated recipient mice. For the bcl-2 add-back, the STAT5abnull/null mice were crossed with bcl-2 transgenic mice as stated in “In vivo complementation and transplantation.” STAT5abnull/null bcl-2 transgenic FL cells were transduced with either IR-GFP or STAT5aΔNS711F. (B) Representative flow cytometric plots show the accumulation of myeloid cells in the peripheral blood. The panels show the Gr-1+ or Mac-1+ cells co-gated for expression of GFP. The right panels show the percentage of Gr-1+Mac-1+ double-positive myeloid cells. The flow cytometry was performed 20 to 24 days after transplantation for all 4 groups.
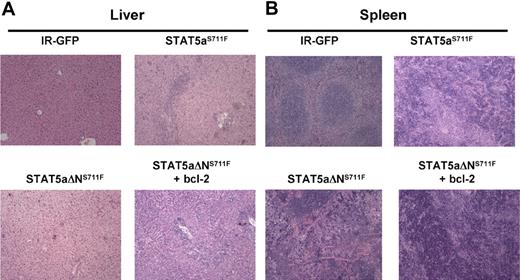
Histopathology analysis of spleen and liver was compared among all 4 groups at the time of killing. In the liver of the STAT5aΔNS711F mice, the hepatic architecture was largely intact with scattered small foci of extramedullary hematopoietic components in the sinusoids (supplemental Figure 5A). However, in the liver of mice expressing STAT5aΔNS711F plus H2k/bcl-2, the hepatic lobular architecture was markedly distorted by dense infiltrate of predominately myeloid cells at different stages of differentiation in the hepatic lobules or portal triads (Figure 6A). This showed that STAT5aΔNS711F and bcl-2 could phenocopy STAT5aS711F-induced MPD. In the spleen, the mice expressing STAT5aΔNS711F plus bcl-2 also recapitulated the disease of STAT5aS711F, and showed markedly distorted splenic architecture (Figure 6B). The red and white pulps were diffusely effaced by myeloid cells at different stages of differentiation. Spleens of STAT5aΔNS711F-expressing mice without bcl-2 were enlarged and had evidence of panmyelosis with some mice showing focal areas of megakaryocytes (supplemental Figure 5B) in contrast to STAT5aS711F, which provoked a more aggressive myelomonocytic phenotype.
STAT5aΔNS711F-expressing cells confer a milder form of MPD, which is restored to aggressive disease by transgenic human bcl-2 expression. STAT5abnull/null FL cells at E14.5 and E15.5 were transduced with either MSCV/IRES-GFP vector control (IR), STAT5aS711F, or STAT5aΔNS711F and transplanted into lethally irradiated recipient mice. Tissues from mice that underwent transplantation were collected and formalin-fixed, and histologic sections were stained with hematoxylin and eosin. Representative sections of liver (A) and spleen (B) are shown. Histologic sections were prepared for mice expressing IR-GFP, STAT5aS711F, and STAT5aΔNS711F + bcl-2 groups at < 30 days after transplantation. For mice that received STAT5aΔNS711F-expressing BM cells that survived long term, the histology was performed at the end of the survival curve (80-90 days). Images were taken using an Olympus BX41 microscope equipped with PanAchromatic objectives (20×) and a mounted Spot In-Sight digital camera (Diagnostic Instruments Inc). The software for image acquisition was Spot Advance. Image processing was done in Adobe Photoshop.
STAT5aΔNS711F-expressing cells confer a milder form of MPD, which is restored to aggressive disease by transgenic human bcl-2 expression. STAT5abnull/null FL cells at E14.5 and E15.5 were transduced with either MSCV/IRES-GFP vector control (IR), STAT5aS711F, or STAT5aΔNS711F and transplanted into lethally irradiated recipient mice. Tissues from mice that underwent transplantation were collected and formalin-fixed, and histologic sections were stained with hematoxylin and eosin. Representative sections of liver (A) and spleen (B) are shown. Histologic sections were prepared for mice expressing IR-GFP, STAT5aS711F, and STAT5aΔNS711F + bcl-2 groups at < 30 days after transplantation. For mice that received STAT5aΔNS711F-expressing BM cells that survived long term, the histology was performed at the end of the survival curve (80-90 days). Images were taken using an Olympus BX41 microscope equipped with PanAchromatic objectives (20×) and a mounted Spot In-Sight digital camera (Diagnostic Instruments Inc). The software for image acquisition was Spot Advance. Image processing was done in Adobe Photoshop.
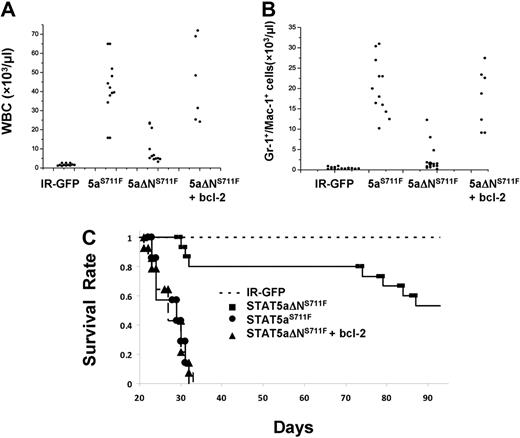
The most striking evidence for an important role of bcl-2 expression in MPD was obtained in peripheral white blood cell (WBC) and myeloid cell counts among all groups. As expected, IR-GFP control mice had generally low WBC counts (1-2 × 109/L) and no elevation in Gr-1/Mac-1 (5 × 108/L)–positive myeloid cells, as shown in Figure 7A and B. Expression of STAT5aS711F increased WBC and myeloid cells to an average 4 × 1010/L and 2 × 1010/L, respectively. Elevation of peripheral WBC and myeloid counts was ablated in STAT5aΔNS711F-expressing mice, which averaged only approximately 6 × 1010/L and 1 × 1010/L. However, 3 mice had a higher WBC count and developed MPD, indicating that the N-terminal domain although important for MPD was not essential upon overexpression. Nevertheless, overall there was a significant 8-fold difference in WBC and myeloid expansion between STAT5aS711F and STAT5aΔNS711F mice (P < .01). Add-back of transgenic bcl-2 restored the WBC and myeloid cell expansion, which increased to 4 × 1011/L and 1.5 × 1011/L, respectively. Notably, mice expressing STAT5aS711F died within 5 weeks after transplantation, whereas more than half of the mice expressing STAT5aΔNS711F survived up to the latest analysis at 90 days (Figure 7C) and at terminal analysis had normal peripheral blood counts (data not shown). Add-back of bcl-2 greatly shortened the life span of the mice, which showed an overlapping survival curve with those that received a transplant of STAT5aS711F. Bcl-2 transgene expression alone was not associated with any disease in mice monitored for more than 6 months (data not shown). Overall, these data demonstrate that the N-domain of STAT5 is responsible mainly for survival function to promote lethal MPD and that bcl-2 expression can restore the lethality of the disease.
Transgenic expression of human bcl-2 in STAT5aΔNS711F-expressing HSCs restores peripheral myeloid expansion and lethality in transplant recipients. Mice that received a transplant as described in Figure 5 were analyzed 3 weeks after transplantation. Total WBC counts (A) and the absolute number of Gr-1+/Mac-1+ cells (B) are plotted for mice from 4 groups (for WBC: STAT5aS711F vs STAT5aΔNS711F, P < .01; STAT5aS711F vs STAT5aΔNS711F + bcl-2, P = .58; STAT5aΔNS711F vs STAT5aΔNS711F + bcl-2, P < .01; for Gr-1+/Mac-1+ cells: STAT5aS711F vs STAT5aΔNS711F, P < .01; STAT5aS711F vs STAT5aΔNS711F + bcl-2, P = .64; STAT5aΔNS711F vs STAT5aΔNS711F + bcl-2, P < .01; Student t test). Each dot represents an individual mouse. Horizontal bars indicate the average for all mice analyzed per group. (C) The survival of mice in each group was determined daily and deaths were recorded. The number of mice per group for all 3 panels was as follows: IR-GFP: n = 12; STAT5aS711F: n = 12; STAT5aΔNS711F: n = 12; STAT5aΔNS711F + bcl-2: n = 12. IR denotes IRES-GFP vector and STAT5aS711F denotes STAT5aS711F-IRES-GFP vector. The white blood cell counts and flow cytometry were performed 20 to 24 days after transplantation for IR-GFP, STAT5aS711F, and STAT5aΔNS711F + bcl-2. Terminal analyses of STAT5aΔNS711F-expressing mice were performed 2 to 3 months after transplantation.
Transgenic expression of human bcl-2 in STAT5aΔNS711F-expressing HSCs restores peripheral myeloid expansion and lethality in transplant recipients. Mice that received a transplant as described in Figure 5 were analyzed 3 weeks after transplantation. Total WBC counts (A) and the absolute number of Gr-1+/Mac-1+ cells (B) are plotted for mice from 4 groups (for WBC: STAT5aS711F vs STAT5aΔNS711F, P < .01; STAT5aS711F vs STAT5aΔNS711F + bcl-2, P = .58; STAT5aΔNS711F vs STAT5aΔNS711F + bcl-2, P < .01; for Gr-1+/Mac-1+ cells: STAT5aS711F vs STAT5aΔNS711F, P < .01; STAT5aS711F vs STAT5aΔNS711F + bcl-2, P = .64; STAT5aΔNS711F vs STAT5aΔNS711F + bcl-2, P < .01; Student t test). Each dot represents an individual mouse. Horizontal bars indicate the average for all mice analyzed per group. (C) The survival of mice in each group was determined daily and deaths were recorded. The number of mice per group for all 3 panels was as follows: IR-GFP: n = 12; STAT5aS711F: n = 12; STAT5aΔNS711F: n = 12; STAT5aΔNS711F + bcl-2: n = 12. IR denotes IRES-GFP vector and STAT5aS711F denotes STAT5aS711F-IRES-GFP vector. The white blood cell counts and flow cytometry were performed 20 to 24 days after transplantation for IR-GFP, STAT5aS711F, and STAT5aΔNS711F + bcl-2. Terminal analyses of STAT5aΔNS711F-expressing mice were performed 2 to 3 months after transplantation.
Discussion
STAT5 is activated by a wide range of growth factors and cytokines that are critical for hematopoiesis. Therefore, understanding STAT5 biologic function and mapping functional domains is generally important for understanding hematopoiesis and cancer development. Previous studies showed that STAT5 is a central regulator of IL-3 and SCF signaling with overt effects on mast cell biology in vitro and mice lacking expression of full-length STAT5 do not have detectable mast cells in peripheral tissues.14 Retroviral transduction into the STAT5abnull/null background provides a powerful tool for further characterization of STAT5 structure-function both in vitro and in vivo. In this system, we also documented initial transduction efficiency by our ecotropic producer cells and found relatively low transduction (1-3 copies per cell), which importantly did not differ among constructs. Of course, after the initial transduction, selective pressure in vitro or in vivo could be expected to bias copy number or expression. STAT5abnull/null mast cells were deficient in both survival and bcl-2 or bcl-XL expression, compared with wild-type mast cells, and permitted analysis of complementation of function in the absence of endogenous STAT5.
STAT5-mediated regulation of bcl-2 in hematopoietic cells has been described in mouse25 and human26 hematopoietic cells and is induced in a STAT5 dosage–dependent manner.27 STAT5 has been reported to bind a nonconserved site in the human bcl-2 promoter by electrophoretic mobility shift assay.28 However, whether bcl-2 is a direct STAT5 target gene has not been tested on species-conserved sites in native chromatin by ChIP. A STAT5-binding site search found 7 sites in bcl-2 that were conserved between humans and mice. Our ChIP analysis demonstrated direct binding of STAT5 to bcl-2 at site 2 within intron 2, indicating that bcl-2 is a bona fide direct target of STAT5. It has been previously demonstrated that up to 50% of STAT5-binding sites reside in introns.29 Although we did not observe STAT5 bound to site 1 that contains 2 tandem dimer-binding sites, we cannot rule out that binding site selection could be cell type specific. Alternatively, other STAT family members could bind and modify bcl-2 expression at this site or it may be differentially bound in HSCs. It is experimentally difficult to examine ChIP on HSCs, especially in our retroviral model system, due to the limiting cell numbers. Nevertheless, we found that both STAT5a and STAT5aΔN bound to site 2 and both STAT5a and STAT5aΔN were able to induce bcl-2 and bcl-XL mRNA expression 7-fold and 2-fold, respectively, under limiting cytokine conditions. This demonstrated that STAT5 can regulate bcl-2 and bcl-XL, at least in part through a direct transcriptional mechanism. Interestingly, we observed that STAT5aΔN was less efficient in induction of bcl-2 and bcl-XL expression compared with STAT5a under limiting cytokine conditions, but not for expression of the known direct target genes cis and osm. Therefore, the defects in survival were not accounted for by a general hypomorphic transcriptional activity in all target genes.
Interestingly, we found that the mRNA level of bcl-2 was not changed in STAT5abnull/null mast cells in the presence of IL-3 and SCF compared with wild-type mast cells, although the protein level was dramatically reduced. To test for posttranscriptional regulation of bcl-2, microRNAs (miRs) were considered to be candidates. miRs are small, noncoding, single-stranded RNAs of approximately 22 nucleotides that negatively regulate gene expression at the posttranscriptional level primarily through targeting the 3′-UTR of target mRNAs. miR expression can be regulated by transcription factors such as STAT3, which was recently demonstrated to be involved in regulation of miR-21 and the miR-17/92 cluster.30,31 However, there are no current reports of STAT5-mediated miR expression. We used computational screening to determine whether conserved STAT5-binding sites might exist in the promoter of the current known miRs that can target bcl-2. Several miRs, including miR-1, miR-15, miR-16, miR-182, miR-143,24,32-34 and miR-182a,35 have been reported to regulate bcl-2 expression. However, only miR-15b and miR-16-2, which form a cluster at chromosome 3, have a putative conserved STAT5-binding site, which was located approximately 5 kb upstream of the miR-15b/16-2 cluster. miR15/16 can regulate bcl-2 expression in megakaryocytic,36 gastric,34 and primary hepatic stellate33 cells. We now add myeloid 32D cl.3 and primary mast cells. Real-time PCR demonstrated that miR-15b was significantly increased when STAT5 was absent, whereas overexpression of STAT5a reduced miR-15b levels. Interestingly, the STAT5 N-domain was essential for the inhibition. Our ChIP data, however, found that the N-domain is not essential for promoter binding, suggesting that its role might be of repressive nature, which might involve remodeling the chromatin structure. For instance, interaction of STAT5 with SMRT through the N-terminal coiled coil domain represses STAT5 transcriptional activity.37 On the other hand, regulation of bcl-2 and bcl-XL mRNA through direct STAT5 binding required N-domain–mediated transcription, likely due to cofactor binding. This reciprocal regulation would be expected to coordinately regulate bcl-2 in a novel manner that has not been described previously.
Although we did not find a conserved STAT5-binding site in the vicinity of the miR-15a/16-1 cluster, we cannot rule out that STAT5 still may regulate these through an indirect mechanism. Recently negative regulation of miR-15a/16-1 by the STAT5 target gene myc has been reported.38 myc can also be regulated by many transcription factors apart from STATs. Human miR-15a/16-1 is located in the 13q14 region where deletion or point mutations occur at high frequency in chronic lymphocytic leukemia, lymphoma, and prostate cancer,39,40 but has not been described in myeloid malignancies. Validated gene targets of miR-15a/16-1 have been identified including wnt3a, cyclin D1, ets1, jun, and other cancer-associated genes.36,41 Therefore, aberrant STAT5 activation, often observed in human chronic myeloid leukemia and acute myeloid leukemia, might regulate the miR-15/16 gene cluster to broadly control cancer-relevant genes.
Our data are consistent with a previous report of the importance of HSCs in MPD development mediated by STAT5.12 We observed that STAT5aS711F supported KLS cell expansion and survival. STAT5aS711F was much more efficient at inducing MPD than STAT5aΔNS711F, which was devoid of survival function. KLS cell survival correlated very well with endogenous bcl-2 expression in HSCs. Bcl-2 and bcl-XL have a well-documented role in leukemogenesis. Therefore, we reasoned that restoration of bcl-2 expression in STAT5ΔNS711F-expressing mice could recapitulate the MPD by STAT5aS711F. Indeed, the disease obtained after add-back of bcl-2 was indistinguishable from STAT5aS711F, indicating that attenuation of MPD by loss of the STAT5 N-domain was due mainly to the impaired survival function. We cannot rule out that the STAT5 N-domain could regulate other survival genes and that bcl-2 is generically replacing a defective survival function. We did not observe changes in mRNA for survival-related genes in mast cell cultures containing IL-3 and SCF (supplemental Figure 6). It is likely that many STAT5 direct and indirect target genes are reduced due to the generally reduced transcriptional activity of STAT5aΔN. Certainly our retroviral transplant model relied largely on overexpression, which could account for higher activation of STAT5aΔNS711F. However, we observed striking dependency on survival function in both mast cells and in the MPD setting through the STAT5 N-domain.
It is interesting that in the absence of the N-domain we observed mostly a milder and less lethal MPD phenotype characterized by splenomegaly and maintained megakaryopoiesis, in contrast to the aggressive myelomonocytic disease provoked by STAT5aS711F. This milder and more megakaryocytic phenotype observed in some mice is reminiscent of other murine transplant models for MPD42 or BCR/ABL transgenic mice43,44 where lower levels of STAT5 activation were associated with large populations of megakaryocytes. Phospho-STAT5 dosage controls erythroid versus megakaryocyte differentiation45 and may explain differences in pathophysiology observed in MPD mouse models.
N-domain–targeted STAT knockout mice can still retain the potential for significant N-domain–independent activities. Here we have described novel survival mechanisms mediated by the STAT5 N-domain. The identification of binding partners and secondary signaling events mediated by the N-domain of STAT5 is an important area due to the potential therapeutic significance. The glucocorticoid receptor interacts with the STAT5 N-domain, which can control gene expression as either a coactivator or corepressor.46 Glycosylation of Th92 in the N-domain of STAT5 is important for binding to the coactivator of transcription CREB-binding protein47 and eventually p300 and CREB-binding protein/p300 are established coactivators of bcl-2 expression.48 STAT5 might also regulate bcl-2 through direct repression of IRF-8 (ICBSP),49 which is a known direct repressor of the bcl-2 promoter.50 Other N-domain–dependent interactions may also be critical for expression of a subset of target genes through tetramerization13 or through protein-protein interactions to activate unique sets of genes. This study did not focus on tetramer mutants of STAT5 and thus cannot rule out a role for tetramers in bcl-2 regulation. Future studies will focus on the role of the glucocorticoid receptor and tetramers in the N-domain–mediated survival mechanism.
In summary, our study reveals a dynamic relationship between phosphorylated STAT5 and survival signaling, reciprocally regulated through microRNA suppression and bcl-2 family member transactivation. Defining the cofactors associated with the N-domain functions of STAT5 and targeting those interactions or directly targeting bcl-2 and bcl-XL may present therapeutic opportunities for exploiting the Achilles heel of aberrant STAT5 activation.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Jos Domen (Medical College of Wisconsin) for providing the H2k/bcl-2 transgenic mice. We also thank Yu-Chung Yang, Cheng-Kui Qu, and Shigemi Matsuyama for helpful discussions.
This work was supported by National Institutes of Health R01DK059380 (K.D.B), Hematology Training Grant T32HL007147 (K.M.), SFB-F28 (R.M.), and the Flow Cytometry and Radiation Resources Core Facilities of the Case Comprehensive Cancer Center (P30CA43703).
National Institutes of Health
Authorship
Contribution: K.L.M., Z.W., X.Y.X., and J.B. performed research and analyzed data; J.J.R., R.M., and W.T. designed research and analyzed data; and G.L. and K.D.B. designed and performed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Kevin D. Bunting, Department of Medicine, Division of Hematology/Oncology, Case Western Reserve University, 10900 Euclid Ave, WRB 2-131, Cleveland, OH 44106-7284; e-mail: kdb10@case.edu.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal