Abstract
We performed single-molecule telomere length and telomere fusion analysis in patients at different stages of chronic lymphocytic leukemia (CLL). Our work identified the shortest telomeres ever recorded in primary human tissue, reinforcing the concept that there is significant cell division in CLL. Furthermore, we provide direct evidence that critical telomere shortening, dysfunction, and fusion contribute to disease progression. The frequency of short telomeres and fusion events increased with advanced disease, but importantly these were also found in a subset of early-stage patient samples, indicating that these events can precede disease progression. Sequence analysis of fusion events isolated from persons with the shortest telomeres revealed limited numbers of repeats at the breakpoint, subtelomeric deletion, and microhomology. Array-comparative genome hybridization analysis of persons displaying evidence of telomere dysfunction revealed large-scale genomic rearrangements that were concentrated in the telomeric regions; this was not observed in samples with longer telomeres. The telomere dynamics observed in CLL B cells were indistinguishable from that observed in cells undergoing crisis in culture after abrogation of the p53 pathway. Taken together, our data support the concept that telomere erosion and subsequent telomere fusion are critical in the progression of CLL and that this paradigm may extend to other malignancies.
Introduction
Nonreciprocal translocations (NRTs) are considered to be key mutational events that can drive many types of malignancy.1 The underlying mechanisms that result in these types of events can include, among others, deficiencies in double-strand break repair,2 mitotic checkpoints,3,4 and telomere dysfunction.5 Telomeres play a key role in upholding genomic integrity; in the context of DNA damage checkpoint defects, cells in culture undergo crisis and have extensive telomere erosion, chromosomal fusion, and genomic rearrangements.6,7 NRTs, as well as localized gene amplification,8 can arise as a consequence of cycles of anaphase-bridging, breakage, and fusion initiated by the formation of dicentric chromosomes after telomere fusion.9 This paradigm is exemplified in vivo by telomerase knockout mice, where short telomeres appear to drive the formation of tumors containing NRTs.5 However, evidence for this phenomenon in humans is circumstantial. Numerous malignancies, including breast, prostate, colorectal, and chronic lymphocytic leukemia (CLL),10-15 have been documented to exhibit shorter telomeres compared with normal tissues. These data are consistent with the expected levels of cell division during the progression to malignancy but do not indicate that telomeres become short enough to lose their end-capping function. Telomere fusion, as well as other chromosomal defects, can lead to the formation of anaphase bridges; in situ data show an increase in anaphase bridges, often interpreted as a surrogate marker for telomere fusion, at the adenoma/hyperplasia-carcinoma transition in breast,16 colorectal,17 and squamous cell carcinomas.18 This transition point is also characterized by an up-regulation of telomerase.19,20 In pancreatic carcinomas and osteosarcomas, break points were clustered in terminal regions and were associated with shorter telomeres at specific chromosome arms.21 However, thus far, technical limitations have hampered the detection of critically shortened telomeres and the subsequent telomere fusion events.22 As a consequence, there has not been a direct demonstration that telomeres become short enough during the progression of human cancer to lose their end-capping function, leading to fusion, and genomic instability.
To address this issue, we have developed high-resolution single-molecule approaches both to determine telomere length and to detect telomere fusion events in DNA derived from human tissues.6,23 Single telomere length analysis (STELA) is unique in its ability to detect extremely short telomeres,23 in particular those that are short enough to trigger fusion.6,24 Analysis of cells that have bypassed p53-dependent replicative senescence using STELA, showed that with ongoing cell division, telomeres continue to erode such that as the cells approach crisis the shortest telomeric alleles become completely denuded of telomere repeats and are subjected to telomere-telomere fusion.6 Sequence analysis of telomeres that have been subjected to fusion shows that the mean telomere length is just 5.8 TTAGGG repeats (35 bp) with more than 30% of fusions containing no telomere repeats at all.6,24 The majority of fusion events were also accompanied by the deletion of one or both of the participating telomeres; these deletion events extend into the telomere adjacent DNA up to the limit of the assays (6 kb). In addition, microhomology was observed at the fusion points,6,24 and this distinct profile may be indicative of the repair mechanisms that facilitate the end-joining of short dysfunctional telomeres.
We set out to examine whether the telomere dynamics observed in our in vitro experimental systems are recapitulated during tumor progression in vivo. To do this, we have applied these high-resolution technologies to examine the full extent of telomere erosion and dysfunction at different stages of CLL, the most common form of adult leukemia in Western countries.25 The clinical course of this disease is heterogeneous, with a proportion of patients remaining asymptomatic, never requiring treatment and having similar survival to age-matched persons. In contrast, others have an aggressive illness with a median survival of 2 to 3 years26 ; it is thus possible to compare indolent with more aggressive forms of the disease. Large-scale genomic rearrangements are common events in CLL27 ; NRTs at the 17p and 11q chromosome arms are independent markers of a poor prognosis, and this is related to loss of heterozygosity (LOH) of the p53 (17p13) and ATM (11q22) genes on these chromosome arms.28 CLL B cells express low levels of telomerase,29 but this is insufficient to maintain telomere length, and thus, as observed in many other tumor types,10,13,14 telomeres shorten in CLL.11,12,15 This is consistent with significant cell division during the progression of the disease and, taken together with the deficiencies in p53 and ATM in CLL, indicates the possibility that telomeres may shorten to a length at which they become dysfunctional. Here we show that telomeres in the CLL B cells from poor prognostic CLL patients do have extensive telomere erosion and dysfunction, characteristic of telomeres in cells undergoing crisis in culture. Complete telomere loss was observed in some persons and telomere fusions, some of which were clonal, were detected in persons with the shortest telomere length profiles. Furthermore, telomere dysfunction correlated with large-scale genomic instability, and this was often observed at the telomeric ends of chromosomes. These data are consistent with CLL B cells undergoing a telomere crisis during tumor progression and strongly indicate that this may drive genomic instability in this disease.
Methods
This study was approved by the South East Wales local research ethics committee (LREC# 02/4806).
Patient samples
Peripheral blood samples from 41 CLL patients were obtained after written informed consent had been received in accordance with the Declaration of Helsinki. CLL was defined by clinical criteria as well as cellular morphology and the coexpression of CD19 and CD5 in lymphocytes simultaneously displaying restriction of light-chain rearrangement. Comprehensive clinical information, including treatment histories, was available for all patients. Most of the samples were collected at the time of diagnosis, and none of the previously treated patients had received chemotherapy within 3 months before sample collection. Staging was based on the Binet classification system.30 The clinical characteristics of the CLL patient cohort are presented in supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Isolation of PBMCs
Peripheral blood mononuclear cells (PBMCs) were isolated from ethylenediaminetetraacetic acid venous blood of the 41 CLL patients and from the leukocyte fractions of 6 age-matched healthy donors by density centrifugation using Ficoll-Hypaque (Invitrogen). B cells were subsequently positively isolated using CD19+ Dynabeads (Invitrogen).31 Cells were stored at −20°C as dry pellets before DNA extraction.
DNA extraction and PCR
We extracted DNA from purified B cells using standard proteinase K, RNase A, phenol/chloroform protocols.32 For telomere length analysis at the XpYp and 17p telomeres, we used a modification of the STELA assay previously described.6,23,33 Briefly, genomic DNA was solubilized by digestion with EcoRI, quantified by Hoechst 33258 fluorometry (Bio-Rad), and diluted to 10 ng/μL in 10mM Tris-HCl, pH 7.5. A total of 10 ng of DNA was further diluted to 250 pg/μL in a volume of 40 μL containing 1μM Telorette2 linker and 1mM Tris-HCl, pH 7.5. Multiple polymerase chain reactions (PCRs; typically 6 reactions per sample) were carried out for each test DNA in 10-μL volumes 250 pg of DNA, 0.5μM of the telomere-adjacent and Teltail primers, 75mM Tris-HCl, pH 8.8, 20mM (NH4)2SO4, 0.01% Tween-20, 1.5mM MgCl2, and 0.5 U of a 10:1 mixture of Taq (ABGene) and Pwo polymerase (Roche Molecular Biochemicals). The reactions were cycled with an MJ PTC-225 thermocycler (MJ Research). The DNA fragments were resolved by 0.5% Tris acetate ethylenediaminetetraacetic acid agarose gel electrophoresis, and detected by 2 separate Southern blot hybridizations with random-primed α-33P–labeled (GE Healthcare) TTAGGG repeat probe and a telomere-adjacent probe, together with a probe to detect the 1-kb (Stratagene) and 2.5-kb (Bio-Rad) molecular weight markers. The hybridized fragments were detected by phosphorimaging with a Molecular Dynamics Storm 860 phosphorimager (GE Healthcare). The molecular weights of the DNA fragments were calculated using the Phoretix 1D quantifier (Nonlinear Dynamics).
Telomere variant repeat distributions were determined using an adaptation of the telomere variant repeat (TVR)-PCR technique at the XpYp telomere as previously described.34 To determine the distal extent of the extensive XpYp TVR regions, the XpYpE2 primer was in conjunction with the TVR primers to detect TTAGGG (TAG-TelW), TGAGGG (TAG-TelX), and TCAGGG (TAG-TelY) as detailed previously.34 Products were resolved by 0.5% Tris acetate ethylenediaminetetraacetic acid agarose gel electrophoresis, and detected by Southern hybridization with random-primed α-33P–labeled (GE Healthcare) probe generated by PCR between primers XpYpE2 and XpYpB2.23
Telomere fusion was detected using an adaptation24 of the previously described fusion assay involving the XpYp and 17p telomeres.6 The assay was adapted to detect fusion between additional chromosome ends; to do this, several primers were designed within the subtelomeric repeat regions that are shared among multiple chromosomes detected with the TelBamII probes.35-37 The telomere fusion assay was then carried out as described previously6 with the following modifications. PCR reactions containing 100 ng of DNA were undertaken each containing the XpYpM, 17p6, and 21q1 PCR primers. Fusion molecules were detected and the frequencies quantified by Southern blotting and hybridization with the XpYp telomere-adjacent probes as described previously.6 To determine the chromosomes participating in the fusion events for subsequent sequence characterization, further hybridizations were undertaken with the 17p and 21q telomere adjacent probes; the 21q probe yields additional nonspecific products and thus was not used for quantification. Any fusion products were then reamplified for direct sequence analysis using nested PCR primers (XpYpO, 17p7, and 21qseq1).
Oligonucleotides
Fusion PCR.
XpYpM: 5′-ACCAGGTTTTCCAGTGTGTT-3′; 17p6: 5′-GGCTGAACTATAGCCTCTGC-3′; 21q1: 5′-CTTGGTGTCGAGAGAGGTAG-3′.
Reamplification.
XpYpO:5′-CCTGTAACGCTGTTAGGTAC-3′; 17p7:5′-CCTGGCATGGTATTGACATG-3′; 21qseq1: 5′-TGGTCTTATACACTGTGTTC-3′.
Probes.
21qseq1: 5′-TGGTCTTATACACTGTGTTC-3′; 21qseq1 rev: 5′-AGCTAGCTATCTACTCTAACAGAGC-3′; XpYpO: 5′-CCTGTAACGCTGTTAGGTAC-3′; XpYpB2:5′-TCTGAAAGTGGACC(A/T)ATCAG-3′; 17p7:5′-CCTGGCATGGTATTGACATG-3′; 17p2:5′-GAGTCAATGATTCCATTCCTAGC-3′.
Array CGH analysis
Tumor genomic DNA and reference DNA samples were independently labeled with either Cy3 or Cy5 dyes. Labeled DNA was cohybridized to a NimbleGen comparative genome hybridization (CGH) array (3 × 720 000). The array format included 3 arrays on single slides containing 720 000 probes. Hybridized microarray slides were scanned at 532 nm (for Cy3 dye) and 635 nm (for Cye5 dye) using a 5-μm microarray scanner and quantified using NimbleScan 2.5 software (Roche NimbleGen). The copy number gains and losses were identified using the segMNT algorithm included in NimbleScan software. All microarray data have been deposited into the Gene Expression Omnibus (GEO; National Center for Biotechnology Information) public database under accession number GSE22016.
Telomerase activity
Telomerase activity was detected using the TRAPeze real-time PCR kit (Chemicon International). A total of 1 × 106 CLL B cells were lysed in 200 μL of 3(3-cholamidopropyl) dimethylammonio-1-propane sulfonate buffer, and each lysate was included in the TRAPeze reaction using the Amplifluor primers and Titanium Taq (BD Clontech). Each reaction was carried out in triplicate in 96-well plate format, including positive control extracts, positive PCR controls that generated the standard curves for each plate, heat-treated controls, and a no template control. Telomerase activity was expressed as log copy number calculated using the threshold cycle values and the standard curve for each 96-well plate.
Results
Telomere length correlates with clinical staging and prognostic markers
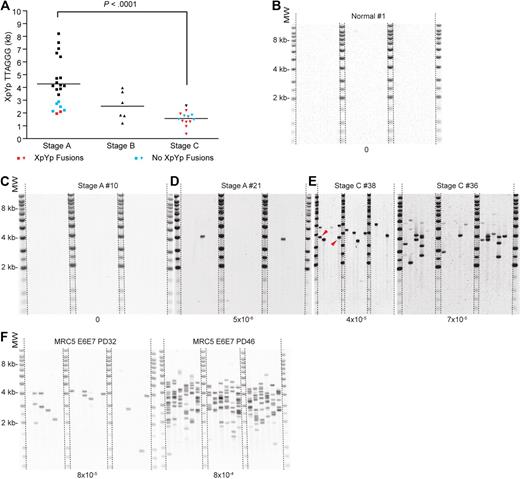
We examined the telomere length distributions using STELA at both the XpYp and 17p telomeres in 41 CLL patients (Figure 1A; supplemental Figure 1; supplemental Table 1). At both telomeres, there was a highly significant difference in mean telomere length between persons with Binet stages A and C (XpYp, 5.110 kb and 2.311 kb P < .001; 17p, 4.832 kb and 3.180 kb, P < .001, Figure 1B-C), with the stage B patients exhibiting intermediate telomere lengths (Figure 1B-C). Despite this stage-related profile, a subset of the stage A samples had similar mean telomere lengths to those found in stage C patients. In addition, there were significant correlations between telomere length and the other prognostic markers currently used for CLL, including ZAP-70, VH gene mutation status, CD38, and β2-microglobulin (Figure 2A-D), but no significant difference was observed between samples derived from previously treated or untreated patients (Figure 2E). It was clear from this analysis that patients with a poor prognosis exhibited very short telomere length distributions. Indeed, the telomere length distributions of many patients were comparable with that observed in replicatively senescent cells in culture.23,33 Moreover, some stage C patients displayed telomeres of lengths similar to that observed in cells undergoing crisis in culture (Figure 1A).6 Strikingly, one person with Binet stage C had a mean telomere length distribution at the XpYp telomere of just 0.959 kb (Figure 1A). At the time of analysis, this represented the shortest telomere length distribution recorded in humans; this person subsequently underwent Richter transformation and died.
Poor-prognosis CLL patients exhibit short telomere length distributions. (A) XpYp STELA gels displaying the telomere length profiles in B cells from normal and CLL patients and MRC5 fibroblast cells entering crisis in culture after the expression of HPV E6E7.6 Binet staging is indicated above the CLL samples. PD is detailed above the MRC5 samples; mean and SD are detailed below. The mean for the MRC5 cells is for the shorter of the 2 XpYp telomeric alleles. At PD53, the short telomere has disappeared. (B-C) Scatter plots of mean length at the XpYp and 17p telomeres plotted against Binet.
Poor-prognosis CLL patients exhibit short telomere length distributions. (A) XpYp STELA gels displaying the telomere length profiles in B cells from normal and CLL patients and MRC5 fibroblast cells entering crisis in culture after the expression of HPV E6E7.6 Binet staging is indicated above the CLL samples. PD is detailed above the MRC5 samples; mean and SD are detailed below. The mean for the MRC5 cells is for the shorter of the 2 XpYp telomeric alleles. At PD53, the short telomere has disappeared. (B-C) Scatter plots of mean length at the XpYp and 17p telomeres plotted against Binet.
Telomere length correlates with prognostic markers, and heterogeneity indicates clonal growth. (A-E) Scatter plots displaying mean length at the XpYp telomere plotted against (A) ZAP-70 expression, (B) VH gene sequence homology, (C) CD38 expression, (D) β2-microglobulin, and (E) whether the patients had undergone treatment at the point of analysis. (F) Telomere length heterogeneity (SD) and clinical staging compared with heterogeneity of clonal fibroblast populations derived from a single cell. Student t test (2-tailed) was used for statistics.
Telomere length correlates with prognostic markers, and heterogeneity indicates clonal growth. (A-E) Scatter plots displaying mean length at the XpYp telomere plotted against (A) ZAP-70 expression, (B) VH gene sequence homology, (C) CD38 expression, (D) β2-microglobulin, and (E) whether the patients had undergone treatment at the point of analysis. (F) Telomere length heterogeneity (SD) and clinical staging compared with heterogeneity of clonal fibroblast populations derived from a single cell. Student t test (2-tailed) was used for statistics.
CLL patients exhibited a reduced heterogeneity in the telomere length distributions compared with normal persons (Figure 1A), with a significant decrease in heterogeneity as a function of clinical staging (P < .001; Figure 2F). Furthermore, telomere length heterogeneity in stage C patients, but not stage A, was indistinguishable from that observed in single-cell fibroblast clones in vitro (P = .42; Figures 1A,2F).23 These data are consistent with the clonal growth of CLL B cells.
Telomerase activity is insufficient to maintain telomere length in CLL B cells
As telomerase can modulate telomere length, we assessed the impact that telomerase activity may have on the telomere dynamics of CLL B cells. We performed a quantitative telomerase activity assay on a subset of the samples; consistent with previous observations,29 we observed no significant correlations between telomerase activity and telomere length. Furthermore, telomerase activity did not appear to be preferentially up-regulated in advanced stage disease and did not correlate with other known prognostic markers (supplemental Figure 2). These data indicate that, although telomerase activity can be detected, it is insufficient to counteract end replication losses during the progression of the disease.
Telomeres are critically eroded in CLL B cells
The proximal regions of human telomere repeat arrays contain variable amounts of TVRs, including among others TGAGGG and TCAGGG repeats interspersed with the canonical telomere repeat TTAGGG.34,38,39 The extreme variability of TVR distributions,34,38 together with the specificity of the telomere repeat DNA binding proteins TRF1, TRF2, and Pot1 for TTAGGG repeats,40,41 indicates that TVRs are not functional. Thus, the start of the functional TTAGGG repeat region is highly variable, ranging from 0 to 3 kb into the telomere repeat array. This can be determined by examining the TVR profiles at the XpYp telomere.34 Although the raw STELA data demonstrate the presence of extremely short telomeres in CLL patients (Figure 1A), it includes the nonfunctional TVR regions and thus represents an overestimate of the functional telomere length. We therefore determined the distal limit of the TVR region at the XpYp telomere using TVR-PCR34 in all of the CLL patients (supplemental Figure 3); this allowed us to calibrate the TTAGGG repeat content of one of the XpYp alleles. In doing so, this revealed the true extent of telomere repeat erosion in CLL patients. The TVR-adjusted STELA data demonstrated a highly significant difference in the TTAGGG repeat length between stage A and C patients (4.281 kb and 1.570 kb, respectively, P < .001, Figure 3A), with stage B patients displaying intermediate length distributions. Once again, the TVR-adjusted STELA data identified a subset of stage A samples (n = 5 of 21) that had telomeres as short as those found in the stage C group, highlighting the potential of this technique to identify patients at risk of disease progression (Figure 3A). Two stage C patients exhibited mean XpYp telomere distributions of a similar length (< 1.0 kb) to telomeres that we have previously described in fibroblasts undergoing crisis in vitro (Figure 1A)6 ; one person had a mean TTAGGG repeat length distribution of 0.360 kb, just 60 TTAGGG repeats (Figure 3A). It was also evident that the telomere length distributions extended to within the length ranges in which we had previously detected fusion6 ; indeed, the mean of the lower 20th percentile of each XpYp distribution was just 0.91 kb in the stage C patients. These data clearly demonstrate that significant telomere erosion occurs during the progression of this disease, and this may be extensive enough to lead to telomeric dysfunction and fusion.
CLL patients displaying short TTAGGG repeat lengths exhibit telomere fusion. (A) Scatter plot displaying XpYp telomere length in terms of TTAGGG repeats distal to the TVR region. Persons marked in color were analyzed for fusion: red represents fusion was detected; and blue, fusion was not detected. (B-C) Telomere fusion assay in one normal person and one stage A patient, both of whom show no evidence of telomere fusion. (D-E) Examples of telomere fusion in stage A and stage C patients. Each band represents a single fusion event. Fusion was detected by Southern hybridization with the XpYp telomere-adjacent DNA probe. Red arrows indicate an example of a clonal fusion event, verified by sequence analysis (Figure 3C). (F) Fusion assay applied to MRC5 fibroblast cells expressing HPVE6E7 entering crisis in culture6 ; PD points are indicated above. Frequencies of fusion of the XpYp telomere with a subset of 13 chromosome ends are indicated below each blot. These frequencies are useful as a comparator of relative fusion frequencies but represent considerable underestimate of the genome-wide frequency of telomere fusion.
CLL patients displaying short TTAGGG repeat lengths exhibit telomere fusion. (A) Scatter plot displaying XpYp telomere length in terms of TTAGGG repeats distal to the TVR region. Persons marked in color were analyzed for fusion: red represents fusion was detected; and blue, fusion was not detected. (B-C) Telomere fusion assay in one normal person and one stage A patient, both of whom show no evidence of telomere fusion. (D-E) Examples of telomere fusion in stage A and stage C patients. Each band represents a single fusion event. Fusion was detected by Southern hybridization with the XpYp telomere-adjacent DNA probe. Red arrows indicate an example of a clonal fusion event, verified by sequence analysis (Figure 3C). (F) Fusion assay applied to MRC5 fibroblast cells expressing HPVE6E7 entering crisis in culture6 ; PD points are indicated above. Frequencies of fusion of the XpYp telomere with a subset of 13 chromosome ends are indicated below each blot. These frequencies are useful as a comparator of relative fusion frequencies but represent considerable underestimate of the genome-wide frequency of telomere fusion.
Short telomeres are subjected to fusion
During crisis in vitro, short telomeres are subjected to fusion with other chromosome ends and nontelomeric loci.6,7 Telomere fusion is considered to represent a significant mutational event that can lead to oncogenic genomic rearrangements5,8 ; we therefore investigated whether the short telomeres observed in CLL were capable of fusion. To do this, we used a modification of the single molecule telomere fusion assay that we had previously developed to detect fusion between the XpYp and 17p telomeres.6 The new assay allowed the detection of fusion between XpYp, 17p, and a family of 13 chromosome ends each sharing the sub-telomeric sequences detected by the TelBam11 probe.24 The increased sensitivity of this assay allowed us to readily detect telomere fusion in the CLL samples (Figure 3). We detected fusion events involving the XpYp telomere in 58% of stage C samples analyzed (Figure 3A). The mean frequency of fusion (2.0 × 10−5) was significantly different from that observed in 4 normal control persons in which, despite analyzing DNA representing a total of 1.8 million diploid genomes, no fusion events were detected (P < .001, Figure 3B). In addition, 7 stage A patients were analyzed and 2 displayed fusion involving the XpYp telomere (P < .03); interestingly, both of these had the shortest XpYp telomere length distributions of the stage A patients (mean, 2.10 and 1.95 kb, Figure 3A,C-D). Strikingly, 2 stage C patients displayed telomere fusion at frequencies (Figure 3E) that were comparable with MRC5 fibroblast cells expressing HPVE6E7 entering crisis in culture with similar XpYp telomere length distributions (Figures 1A, 3F).6,24
Together, these data provide a demonstration of short dysfunctional telomeres occurring in a human neoplastic condition. Our assays examined telomere dysfunction at a limited number of chromosome ends; the fusion assay used here can detect 15 of a possible 1127 different pairwise telomere fusion combinations in the human genome. Furthermore, telomere length at each chromosome end is independently variable.23,42 Thus, we anticipate that there will be additional telomeres that are dysfunctional in these persons.
Telomere fusion displays a characteristic mutational profile
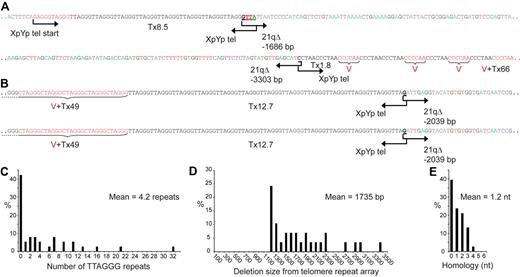
To verify and examine the nature of the telomere fusion events that we detected in CLL patients, a total of 40 events were characterized by DNA sequencing (Figure 4; supplemental Figure 4). The number of TTAGGG repeats at the point of fusion was limited with a mean of 4.2 and a maximum of 32 repeats (Figure 4A-C); no fusion events were detected that contained TTAGGG repeats on either side of the fusion point (supplemental Figure 4). The majority (83%) of fusion events were accompanied by the deletion of the telomere and telomere-adjacent DNA at one or both of the telomeres involved (mean deletion, 1735 bp), this extended up to the limit of the assay (3.3 kb, Figure 4D). Short patches of homology, which were significantly different from that expected by chance, were observed at the fusion points (mean, 1.2 nt; P < .01 by χ2 test; Figure 4E), with additional homology adjacent to the fusion point (supplemental Figure 4). This mutational spectrum was indistinguishable from fusion events observed in cells undergoing crisis in culture, or the rare sporadic fusion events in normal cells in culture.6,24 This implies that the fusion of short dysfunctional telomeres within CLL B cells, normal fibroblasts, and fibroblasts undergoing crisis in culture is mediated by a common end-joining mechanism. The nature of this mechanism is currently unknown; however, it may be indicative of DNA-PK independent, error prone, end-joining processes that use microhomology.6,43 We also detected evidence of clonal fusion events in the form of 2 fusions from the same person that were derived from the same sized fusion molecule and were composed of the same DNA sequence (Figures 3E,4B).
Telomere fusion occurs between very short telomeres and is characterized by extensive deletion and microhomology at the fusion point. (A) DNA sequences of 2 telomere fusion events involving the XpYp and 21q telomeres obtained from CLL B cells. Microhomology at the fusion point is underlined and bold. The amounts of TTAGGG repeats and variants are denoted as T (black) and V (red), respectively. Deletion size (bp) is indicated. (B) DNA sequence of a clonal fusion event. Two identical sequences were from separate fusion molecules obtained from the same patient. (C) Histogram summarizing the amount of contiguous TTAGGG repeats adjacent to the fusion point. (D) Summarizing the amount of deletion into the telomere-adjacent DNA determined from the start of the telomere repeat array. (E) Summarizing the amount of 100% homology between the fusion partners at the fusion junction. Additional homology is observed adjacent to the fusion point (supplemental Figure 3).
Telomere fusion occurs between very short telomeres and is characterized by extensive deletion and microhomology at the fusion point. (A) DNA sequences of 2 telomere fusion events involving the XpYp and 21q telomeres obtained from CLL B cells. Microhomology at the fusion point is underlined and bold. The amounts of TTAGGG repeats and variants are denoted as T (black) and V (red), respectively. Deletion size (bp) is indicated. (B) DNA sequence of a clonal fusion event. Two identical sequences were from separate fusion molecules obtained from the same patient. (C) Histogram summarizing the amount of contiguous TTAGGG repeats adjacent to the fusion point. (D) Summarizing the amount of deletion into the telomere-adjacent DNA determined from the start of the telomere repeat array. (E) Summarizing the amount of 100% homology between the fusion partners at the fusion junction. Additional homology is observed adjacent to the fusion point (supplemental Figure 3).
Complete telomere loss
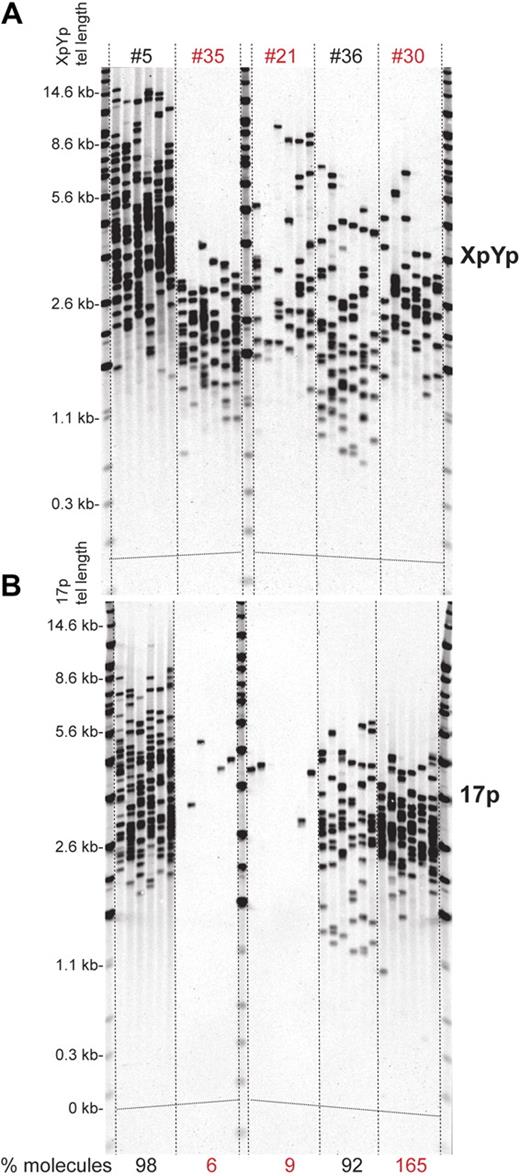
In addition to short telomeres and fusion events, our STELA data showed further evidence of telomeric instability in the form of LOH that was detected by comparison of the single molecule amplification efficiencies between the XpYp and 17p telomeres. The most striking example of this was observed at the 17p telomere in 2 persons where it was apparent that more than 91% of cells had lost both copies of the 17p telomere (Figure 5). Routine FISH analysis showed that both these patients displayed LOH at the p53 locus, array-CGH confirmed that this extended to the telomere (Figure 6).
Complete telomere loss in CLL patients. (A) XpYp and (B) 17p STELA LOH is detected when there is a differential number of amplifiable molecules between the 2 telomeres; the percentage of telomeric molecules at 17p relative to that detected at the XpYp telomere is shown below the 17p STELA blot. Two persons show almost complete LOH at 17p (#35 and #21) and one (#30) shows loss of one XpYp allele, highlighted in red.
Complete telomere loss in CLL patients. (A) XpYp and (B) 17p STELA LOH is detected when there is a differential number of amplifiable molecules between the 2 telomeres; the percentage of telomeric molecules at 17p relative to that detected at the XpYp telomere is shown below the 17p STELA blot. Two persons show almost complete LOH at 17p (#35 and #21) and one (#30) shows loss of one XpYp allele, highlighted in red.
Genomic instability in CLL patients exhibiting telomere dysfunction. Array-CGH (NimbleGen 720k whole genome array) from chromosomes 6-Y in 3 persons that display telomeric dysfunction in the form of telomeric LOH (#21 and #35) and fusion (#21 and #36). One person showed no evidence of telomere dysfunction. Binet stage, XpYp (mean ± SD), and lower 20th percentile are shown to the right of each rainbow CGH view.
Genomic instability in CLL patients exhibiting telomere dysfunction. Array-CGH (NimbleGen 720k whole genome array) from chromosomes 6-Y in 3 persons that display telomeric dysfunction in the form of telomeric LOH (#21 and #35) and fusion (#21 and #36). One person showed no evidence of telomere dysfunction. Binet stage, XpYp (mean ± SD), and lower 20th percentile are shown to the right of each rainbow CGH view.
Telomere dysfunction is associated with genomic instability
Our data demonstrate that short dysfunctional telomeres, complete telomere loss, and telomere fusion are common events in CLL patients and become more common in advanced disease. As telomere fusion can lead to wholesale genomic instability, we tested whether persons displaying evidence of telomeric dysfunction also showed evidence of genomic instability using array-CGH. Using 720k array-CGH, we analyzed 6 persons (5 stage C and 1 stage A) that displayed evidence of telomere dysfunction at 17p and XpYp, and 4 (3 stage A and one stage B) that displayed longer and apparently stable telomeres. Four persons exhibiting telomere dysfunction, including one with stage A disease, displayed clear evidence of large-scale genomic instability that included NRTs resulting in the gain or loss of chromosome arms, which included the telomeres (Figure 6; supplemental Figure 5). In contrast, no large-scale genomic instability was apparent in the samples with no evidence of telomere instability (Figure 6; supplemental Figure 5).
Discussion
Here we have described a high-resolution analysis of telomere dynamics in the commonest adult human leukemia, CLL. Consistent with previous reports, we have observed telomere erosion occurring in CLL patients that is proportional to the stage of disease and prognosis.11,12 However, the resolution of STELA has allowed us to accurately establish the full extent of telomere erosion and provided evidence of telomere dysfunction in CLL B cells.
The mean telomere lengths observed in poor prognostic patients were similar to that observed, using the same technology, in replicatively senescent cells in culture where mean telomere length distributions of less than 1 kb can be readily detected.23,33 Telomerase activity is detectable in CLL B cells; however, this is at comparatively low levels29,44 and does not prevent, but may slow, the rate of telomere erosion. Thus, when similar telomere lengths are observed in CLL B cells and telomerase-negative senescent fibroblasts, they indicate an extended replicative history of the CLL B cells compared with the fibroblasts. Without an accurate definition of the rate of telomere erosion as a function of cell division in CLL B cells, it is not currently possible to use telomere length to quantify the precise amount of cell division in these patients. However, our data do indicate that the CLL B cells in poor prognosis patients have undergone substantial cell division. This is consistent with published studies in vivo using deuterated water incorporation, together with telomere data, that indicate that proliferation is an important aspect of this disease.11,12,15,45 This may also be the case in patients with stable white blood cell counts where significant cell division is counteracted by higher rates of cell death45 ; these patients would also be predicted to have continued telomere erosion. As such, telomere dynamics together with white blood cell counts may provide a marker for the overall kinetics of cell turnover in this condition and thus may also provide a useful marker for clinical progression. Consistent with this, we provide evidence that telomere shortening was found in a subset of stage A patient samples and 2 stage A patients with the shortest telomeres, displayed evidence of telomere fusion. This implies that significant cell division and telomere dysfunction can occur in the earlier stages of the disease, and given the correlation between short telomeres and markers of a poor prognosis, indicate that telomere dysfunction may drive progression in the later stages. Importantly, these data provide further evidence11,46 to indicate that telomere length, particularly when determined with high-resolution methods such as STELA, may provide a useful independent marker of prognosis in CLL.
The in vitro cell culture phenomenon of crisis occurs in cells that, as a consequence of compromised DNA damage checkpoints, have bypassed p53-dependent replicative senescence and have continued to divide to a point at which cell death outweighs cell growth.47,48 The genomic instability observed in cells undergoing crisis in culture is initiated by telomere dysfunction after continued telomere erosion beyond the point of replicative senescence.7 Ongoing telomere erosion as cells enter crisis results in the complete loss of the shortest telomeric alleles and the loss of the end-capping function resulting in fusion with other telomeres or nontelomeric double-strand breaks.6,24 The resulting dicentric chromosomes can then be subjected to cycles of anaphase-bridging, breakage, and fusion that can drive genomic instability in crisis cells. Extending these observations in vivo, it is considered that cells may undergo a telomere driven crisis during the progression to malignancy.5,17 However, thus far, evidence for a telomere crisis during malignant progression has been largely circumstantial; our data now provide direct evidence that the telomere dynamics observed in cells undergoing crisis6,7 occur during the progression of a malignancy in vivo. These dynamics include telomere length distributions that extend to within the length range at which fusion has been observed and evidence of complete clonal telomere loss.6 We also provide evidence that these telomeres are subjected to telomere fusion and that this may lead to large-scale genomic instability. In CLL B cells with evidence of telomere dysfunction, including those from a stage A patient, the types of genomic rearrangements observed in the array-CGH analysis were consistent with the types of rearrangements observed in experimental situations that create dysfunctional telomeres in vitro49 or in mouse models in vivo.5,17 Thus, our data provide direct evidence that CLL B cells undergo a telomere crisis that this can drive genomic instability, which may facilitate progression.
It appears probable that, for telomere erosion to continue to the point that fusion and complete loss are observed, the CLL-B cells in which this occurred may have lost some aspects of checkpoint control. In this regard, it is clear that subsets of CLL patients do display defects in the p53 pathway with mutually exclusive mutations in ATM and p53 identified as markers of a poor prognosis.50-52 We propose the following speculative model whereby, in CLL B cells, subtle defects in the DNA damage checkpoints, or a failure of apoptosis, allows telomeres to erode beyond the length at which replicative senescence or apoptosis would normally be triggered. Erosion continues to the length at which the shortest telomeres lose their end-capping function and telomere fusion is initiated. The distinction between the telomere length at which senescence is triggered and that at which telomere fusion occurs is quite subtle; in vitro data show that telomere fusion can be detected less than 7 cell divisions beyond the point of replicative senescence representing approximately 550 bp of telomere erosion.24 Telomere fusion initiates cycles of anaphase-bridging, breakage, and fusion that cause large-scale genomic rearrangements, including LOH at key tumor suppressor loci, such as p53 or ATM. The complete abrogation of the p53-dependent DNA damage checkpoints as well as other tumor suppressor genes allows for further erosion, telomere dysfunction, and genomic instability, creating the appropriate genomic rearrangements that allows for clonal evolution and disease progression.
Taken together, our data provide a direct demonstration of telomere dynamics in the B-cell populations of CLL patients, which was similar to that observed in cells undergoing crisis in culture. We observed critically shortened telomeres that have the propensity to undergo fusion, complete telomere loss events, fusion events (some of which were clonal), and genomic instability in persons exhibiting telomere dysfunction. These data indicate that telomere dysfunction as a consequence of telomeric erosion to critical lengths may be an important event in driving genomic instability and clonal evolution during the progression of CLL. This instability can lead to the loss of specific chromosome arms, such as 17p, which may in turn drive disease progression. The evidence from mouse models,5 cytogenetic analysis of human tumors,21 and the correlation between telomerase activity and genomic instability in solid tumors,19 together with our own unpublished observations of clonal telomere fusion events in solid tumors (D.M.B. and L. Roger, February 2010), indicate that the telomere dynamics in CLL described here may be an exemplar of the telomere dynamics in other hematologic malignancies and solid tumors.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by Leukaemia & Lymphoma Research (no. 8044) and Cancer Research UK (nos. C17199/A5603 and C1799/A6932). D.M.B. is a Cancer Research UK Senior Fellow.
Authorship
Contribution: T.T.L. carried out the experimental work, analyzed the data, and edited the manuscript; B.T.L. contributed to the experimental work and data analysis; R.E.J. and J.R. contributed to the experimental work; G.P. and S.H. provided clinical samples and data; C.F. provided clinical samples and data and edited the manuscript; C.P. jointly conceived and supervised the study and edited the manuscript; and D.M.B. jointly conceived and supervised the study, analyzed the data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Duncan M. Baird, Department of Pathology, School of Medicine, Cardiff University, Heath Park, Cardiff CF14 4XN, United Kingdom; e-mail: bairddm@cf.ac.uk.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal