Abstract
Natural killer (NK) cells exert antimyeloma cytotoxicity. The balance between inhibition and activation of NK-cells played by the inherited repertoire of killer immunoglobulin-like receptor (KIR) genes therefore may influence prognosis. One hundred eighty-two patients with multiple myeloma (MM) were analyzed for KIR repertoire. Multivariate analysis showed that progression-free survival (PFS) after autologous stem cell transplantation (ASCT) was significantly shorter for patients who are KIR3DS1+ (P = .01). This was most evident for patients in complete or partial remission (good risk; GR) at ASCT. The relative risk (RR) of progression or death for patients with KIR3DS1+ compared with KIR3DS1− was 1.9 (95% CI, 1.3-3.1; P = .002). The most significant difference in PFS was observed in patients with GR KIR3DS1+ in whom HLA-Bw4, the ligand for the corresponding inhibitory receptor KIR3DL1, was missing. Patients with KIR3DS1+KIR3DL1+HLA-Bw4− had a significantly shorter PFS than patients who were KIR3DS1−, translating to a difference in median PFS of 12 months (12.2 vs 24 months; P = .002). Our data show that KIR–human leukocyte antigen immunogenetics represent a novel prognostic tool for patients with myeloma, shown here in the context of ASCT, and that KIR3DS1 positivity may identify patients at greater risk of progression.
Introduction
Multiple myeloma (MM) remains incurable, and the outcome of patients treated with conventional approaches, mainly alkylating agents and glucocorticoids, is unsatisfactory. High-dose therapy and autologous peripheral blood stem cell transplantation (ASCT) has led to improved survival, with a median survival of 2 to 3 years for older and 5 to 6 years in younger patients.1-3 Recently, 2 classes of drugs, the immunomodulators (thalidomide and lenalidomide) and the proteasome inhibitors (bortezomib), have been shown to increase response rates in patients with MM.4-8 These responses are enhanced by subsequent ASCT.5,6 However, with relatively short follow-up, it is unclear if these responses will be durable or indeed lead to better overall survival, and ASCT remains the current standard of care.9,10
Allogeneic stem cell transplantation (allo-SCT) remains the only potentially curative treatment, at least in part because of a graft-versus-myeloma effect.11,12 However, high transplantation-related mortality remains a barrier to more widespread acceptance and use of allografting.13,14 Attempts to reduce the transplantation-related mortality using reduced intensity conditioning regimens have led to a reduction in mortality but at the expense of higher rates of relapse.8,10,13,15,16 However, the graft-versus-myeloma effect supports a role for the immune system in the outcome of myeloma. Thus, the identification of biologic factors that can help to predict the outcome of current standard therapies and hence direct the use of more intensive or novel treatments would be valuable.
Natural killer (NK) cells are an important component of the innate immune system, providing first-line defense against altered tissues, notably virally infected cells and tumors. The physiologic functions of NK cells, including cytotoxicity and cytokine release, are governed by a balance between inhibitory and activating receptors. These receptors include the killer immunoglobulin-like receptors (KIRs), which are specific for allotypic determinants shared by different human leukocyte antigen (HLA) class I molecules (referred to as KIR ligands).17-19 Four inhibitory KIRs have been shown to play a main role in NK-cell alloreactivity; KIR2DL1 recognizes group 2 HLA-C molecules, defined as having a lysine residue at position 80, KIR2DL2 and KIR2DL3 bind to group 1 HLA-C molecules that contain asparagine at position 80, HLA-A and -B allotypes with a polymorphic sequence motif at position 80 to 83 (Bw4 motif) are targeted by KIR3DL1, and HLA-A3 and -A11 are recognized by KIR3DL2.20,21 Although the ligands and functions of inhibitory KIR receptors are well documented, this is not the case for activating KIR receptors. The role of NK cells in the graft-versus-leukemia response has been shown in the haploidentical setting21 and in some studies in the matched related donor and unrelated donor settings.22-24 This together with in vitro evidence that NK cells are capable of killing both allogeneic and autologous myeloma cells, suggests that NK cells may also play a role in the outcome of patients with MM.25,26
The aim of this study was to investigate the effect of KIR genotype on the outcome of patients with MM after ASCT.
Methods
Study population
The subjects of this study were 182 consecutive patients with MM who received a first ASCT with peripheral blood–derived stem cells between 1993 and 2006 and for whom archived genomic DNA was available for KIR genotyping. The study was approved by the Imperial College Research Ethics Committee, and informed consent was obtained from all patients in accordance with the Declaration of Helsinki. Patient and disease characteristics at ASCT, including previous chemotherapy, remission status, serum β-2 microglobulin (β2M), and albumin, were routinely collected. Cytogenetics analysis (fluorescent in situ hybridization [FISH] or conventional Giemsa-banding or both) was performed in bone marrow samples. High-risk cytogenetics were defined as t(14; 16), t(4; 14), del 13q (either as a sole abnormality by conventional Giemsa-banding or in combination with other abnormalities), t(11;14), hypodiploidy, and complex abnormalities. β2M and serum albumin levels were not available from diagnosis because most patients were referred from elsewhere to our center for ASCT. Patients undergoing a tandem ASCT or a tandem auto-allo-SCT procedure were excluded from the study.
After ASCT, all patients had serum electrophoresis with immunofixation performed at monthly intervals. Bone marrow trephine biopsies were taken at 3, 6, and 12 months or at suspicion of disease progression and examined for plasma cell percentage as well as κ and λ in situ hybridization.
KIR genotyping
Genomic DNA was typed for the inhibitory KIR genes KIR2DL1, KIR2DL2, KIR2DL5A (alleles 001 and 002), KIR2DL5B (alleles 002-004, 06, and 007), KIR3DL1, KIR3DL3; the activating KIR genes KIR2DS1, KIR2DS2, KIR2DS3, KIR2DS4, KIR2DS5, KIR3DS1; the framework genes KIR2DL3, KIR2DL4, KIR3DL2, KIR3DP1; and the pseudogene KIR2DP1. Briefly, genotyping was performed with polymerase chain reaction (PCR) amplification with 2 locus sequence-specific primers (Invitrogen). The primer sets were amplified alleles described by the International Nomenclature Committee of the World Health Organization (www.ebi.ac.uk). Internal control primers that amplify an 800-base pair (bp) fragment and a 200-bp fragment of the alleles of KIR3DP1 (framework gene expressed in nearly all haplotypes) were used to confirm robust PCR amplifications. Amplification was performed in 10 μL of PCR mix containing 500 ng of genomic DNA and 5 U/μL Taq polymerase (Bioline). Cycling was performed as follows: 30 cycles of 94° for 20 seconds, 63° for 20 seconds, and 72° for 90 seconds. PCR products were then electrophoresed in 2% agarose gels containing ethidium bromide, and the products were visualized under ultraviolet light.
Diverse KIR haplotypes can be broadly simplified into 2 biologically distinct groups, A and B. Persons with group A haplotype have a fixed number of genes that encode only inhibitory receptors with the exception of the activating KIR gene KIR2DS4; persons with the group B haplotype carry additional activating KIR genes. All persons can be categorized as having 1 of 2 KIR genotypes: A/A, which is homozygous for group A haplotypes, or B/x, which contains either 1 (A/B heterozygotes) or 2 (B/B homozygotes) group B haplotypes.
HLA typing
High-resolution HLA typing for HLA-A, -B, -Cw, DRB1 was performed with the use of PCR with sequence-specific primer and reference strand conformational analysis using in-house primers.
Statistical analysis
Survival and progression-free survival (PFS) curves were calculated with the Kaplan-Meier method, and groups were compared with the log-rank test. PFS was defined according to the international uniform response criteria.27 KIR genotypes that were significant at the level of P less than .20 in the univariate analyses were entered into a multivariate analysis with the use of a backward stepping procedure that also included previously identified prognostic factors, including disease state at ASCT (GR vs poor risk), number of lines of previous chemotherapy regimens (1 vs > 1), β2M (< 3.5 vs > 3.49 mg/L), and albumin (< 3.5 vs > 3.49 g/dL). Variables were tested for proportional hazards with the use of a time-dependent covariate method. P values less than .05 were considered significant.
Results
Patient characteristics
Genomic DNA for KIR genotyping was available on 182 patients, 111 (61%) of whom were male. The median age at ASCT was 58.5 years (range, 26.7-72.4 years). Patient and disease characteristics are summarized in Table 1. All patients were sero-negative for HIV1 and HIV2 and hepatitis B and hepatitis C viruses. Response to induction therapy before ASCT was defined according to the European Group for Blood and Marrow Transplantation criteria.28 Induction chemotherapy regimens are shown in Table 1. None of the patients received bortezomib or lenalidomide before ASCT in accordance with national (United Kingdom) guidelines on the use of such agents as first-line therapy. At ASCT 149 patients were defined as having chemosensitive disease (complete or partial remission) and 33 had progressive disease (PD) or chemo-refractory disease, thus defining GR and poor-risk groups, respectively. One hundred forty-one patients (77%) underwent ASCT after first line of chemotherapy and 41 (32%) had received more than 1 line of therapy. Cytogenetics results were available in 76 patients; Giemsa-banding alone, FISH alone, and Giemsa-banding plus FISH were performed in 25, 24, and 27 patients, respectively. Poor risk cytogenetics were found in 33 of 76 patients (43%). Three patients had del13q as the sole abnormality; in all 3 cases this was identified by Giemsa-banding. β2M and serum albumin results were available in 171 of 182 (94%) and 167 of 182 (92%) of patients before ASCT, respectively. Median β2M and serum albumin levels at ASCT were 2.0 mg/L (range, 0.9-13.0 mg/L) and 3.7 g/dL (range, 2.4-4.8 g/dL), respectively. Transplantation conditioning used melphalan 200 mg/m2 in 126 patients and 140 mg/m2 in 56 patients older than 65 years or with preexisting renal dysfunction. In all patients the stem cell source was peripheral blood stem cells collected after cyclophosphamide (4 g/m2) or etoposide (1.6 g/m2) and granulocyte–colony stimulating factor mobilization. The median CD34+ stem cell dose infused was 3.4 × 106/kg (range, 1.7-19.2 × 106/kg). Of note, none of the patients received maintenance therapy after ASCT.
Patient characteristics
| . | Value . |
|---|---|
| Male-to-female ratio | 111:71 |
| Median age at ASCT, y (range) | 58.5 (26.7-72.4) |
| Disease status at ASCT | |
| Chemosensitive disease, n | 149 |
| Chemo-refractory disease/progressive disease, n | 33 |
| Induction chemotherapy | |
| VAD, n | 97 |
| CTD, n | 47 |
| Z-Dex, n | 34 |
| ABCM, n | 1 |
| Melphalan, n | 1 |
| Cyclophosphamide/prednisolone, n | 2 |
| Disease subtype | |
| IgG, n | 102 |
| IgA, n | 35 |
| IgD, n | 13 |
| Light chain myeloma, n | 21 |
| Nonsecretory, n | 11 |
| Median β2M level at ASCT, mg/L (range) | 2.0 (0.9-13) |
| Median serum albumin level at6 ASCT, g/L (range) | 3.7 (2.4-4.8) |
| Median plasma cell percentage at ASCT, %(range) | 10 (0-90) |
| Median CD34 cell dose, × 106/kg (range) | 3.4 (1.7-19.2) |
| . | Value . |
|---|---|
| Male-to-female ratio | 111:71 |
| Median age at ASCT, y (range) | 58.5 (26.7-72.4) |
| Disease status at ASCT | |
| Chemosensitive disease, n | 149 |
| Chemo-refractory disease/progressive disease, n | 33 |
| Induction chemotherapy | |
| VAD, n | 97 |
| CTD, n | 47 |
| Z-Dex, n | 34 |
| ABCM, n | 1 |
| Melphalan, n | 1 |
| Cyclophosphamide/prednisolone, n | 2 |
| Disease subtype | |
| IgG, n | 102 |
| IgA, n | 35 |
| IgD, n | 13 |
| Light chain myeloma, n | 21 |
| Nonsecretory, n | 11 |
| Median β2M level at ASCT, mg/L (range) | 2.0 (0.9-13) |
| Median serum albumin level at6 ASCT, g/L (range) | 3.7 (2.4-4.8) |
| Median plasma cell percentage at ASCT, %(range) | 10 (0-90) |
| Median CD34 cell dose, × 106/kg (range) | 3.4 (1.7-19.2) |
ASCT indicates autologous stem cell transplantation; VAD, vincristine, Adriamycin, dexamethasone; CTD, cyclophosphamide, thalidomide, dexamethasone; Z-Dex, idarubicin, dexamethasone; ABCM, Adriamycin, carmustine, cyclophosphamide, and melphalan; IgG, immunoglobulin G; and β2M, β-2 microglobulin.
KIR genotypes and PFS
The KIR gene frequencies are summarized in Table 2. The frequencies of KIR haplotypes and individual KIR genes in our patient cohort were comparable to published data in healthy controls.29,30
Association between individual KIRs and progression-free survival on univariate analysis
| KIR gene . | Frequency in patient sample (%) . | P* . |
|---|---|---|
| KIR2DL1 | 171 (94) | .999 |
| KIR2DL2 | 98 (53.6) | .97 |
| KIR2DL3 | 151 (89) | .046† |
| KIR2DL4 | 177 (97) | .999 |
| KIR2DL5A (Alleles 001/005) | 55 (30) | .019† |
| KIR2DL5B (Alleles 002-004, 006/007 | 45 (25) | .93 |
| KIR3DL1 | 167 (92) | .999 |
| KIR3DL2 | 177 (97) | .999 |
| KIR3DL3 | 173 (95) | .999 |
| KIR2DS1 | 51 (28) | .041† |
| KIR2DS2 | 95 (52) | .32 |
| KIR2DS3 | 47 (26) | .13 |
| KIR2DS4 (alleles: 0010101-0010103, 00102/002) | 82 (45) | .38 |
| KIR2DS4 (alleles 003/004/006/007) | 139 (76) | .385 |
| KIR2DS5 | 55 (30) | .31 |
| KIR3DS1 | 62 (34) | .01† |
| KIR2DP1 | 180 (99) | .999 |
| KIR3DP1 | 180 (99) | .999 |
| KIR gene . | Frequency in patient sample (%) . | P* . |
|---|---|---|
| KIR2DL1 | 171 (94) | .999 |
| KIR2DL2 | 98 (53.6) | .97 |
| KIR2DL3 | 151 (89) | .046† |
| KIR2DL4 | 177 (97) | .999 |
| KIR2DL5A (Alleles 001/005) | 55 (30) | .019† |
| KIR2DL5B (Alleles 002-004, 006/007 | 45 (25) | .93 |
| KIR3DL1 | 167 (92) | .999 |
| KIR3DL2 | 177 (97) | .999 |
| KIR3DL3 | 173 (95) | .999 |
| KIR2DS1 | 51 (28) | .041† |
| KIR2DS2 | 95 (52) | .32 |
| KIR2DS3 | 47 (26) | .13 |
| KIR2DS4 (alleles: 0010101-0010103, 00102/002) | 82 (45) | .38 |
| KIR2DS4 (alleles 003/004/006/007) | 139 (76) | .385 |
| KIR2DS5 | 55 (30) | .31 |
| KIR3DS1 | 62 (34) | .01† |
| KIR2DP1 | 180 (99) | .999 |
| KIR3DP1 | 180 (99) | .999 |
Distribution of KIR in the study population is presented.
P for significance on PFS on univariate analysis.
Values statistically significant.
The median overall survival (OS) and PFS for the whole group were 62 months (95% CI, 50-75 months) and 18.8 months (95% CI, 16-21 months), respectively. Factors found to be significantly associated with shorter PFS on univariate analysis included PD or chemo-refractory disease at time of transplantation (poor-risk disease; P < .001), adverse cytogenetics (P = .026), low serum albumin level at transplantation (< 3.5 g/dL; P = .002), low serum β2M level (< 3.5 mg/L; P = .07), KIR haplotype Bx (P = .036), the presence of the activating KIRs KIR2DS1 (P = .041) and KIR3DS1 (P = .01), and the presence of 3 or more activating KIRs (P = .046). Absence of the inhibitory KIR KIR2DL5 alleles 001 or 005 was associated with a longer PFS (P = .019). No significant influence of the number of previous chemotherapy regimens or the melphalan dose on PFS was found.
Association between KIR3DS1 genotype and PFS according to disease status at ASCT
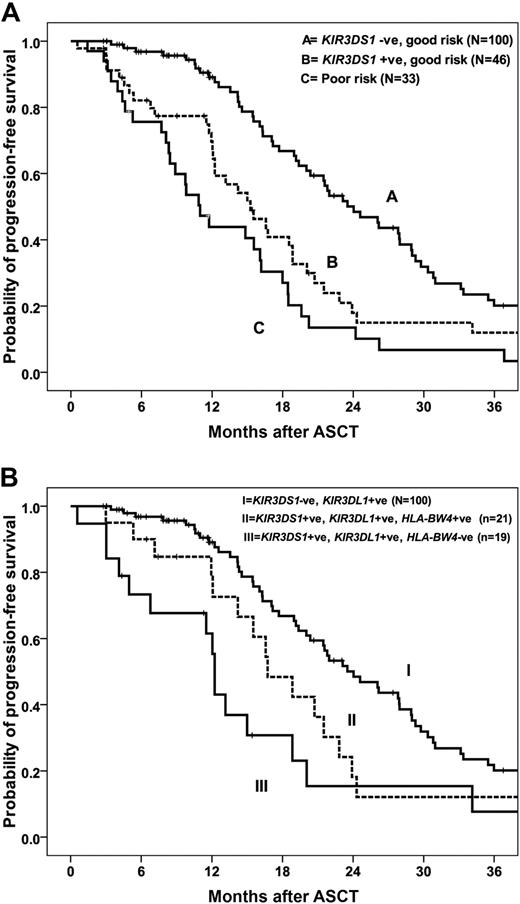
As expected, disease status at ASCT was the most significant predictive factor for outcome in all patients.1-3 Inclusion of KIR2DS1, KIR3DS1, and KIR2DL5 genotypes and disease status in a multivariate analysis resulted in KIR3DS1 being identified as the most significant factor for PFS in patients with GR (complete or partial remission at ASCT) but not in those with poor-risk disease. GR patients who are KIR3DS1− have a median PFS of 24 months (95% CI, 20-28 months) compared with 15.3 months (95% CI, 11-19 months) for those who are KIR3DS1+ (P = .003; Figure 1A). The PFS was reduced for patients with poor-risk disease irrespective of KIR3DS1 status, with median PFS of 9.7 months (95% CI, 0-21 months) and 11 months (95% CI, 8-13 months) for patients who were KIR3DS1− and KIR3DS1+, respectively (P = .31) Importantly, the median PFS for patients with GR disease who were KIR3DS1+ was not significantly different from those with PD (15.4 months; 95% CI, 11-20 months) or chemo-refractory disease (11.1 months; 95% CI, 8.4-13.8 months, P = .061; Figure 1A), underscoring the predictive value of KIR3DS1 in determining outcome. The presence of KIR2DS1, KIR2DL3, and KIR2DL5A were not significantly associated with PFS in multivariate analysis.
Association between KIR3DS1 genotype and PFS. (A) PFS for GR patients who were KIR3DS1+ was significantly shorter than for GR patients who were KIR3DS1− (P = .003). The median PFS for poor-risk patients was not significantly different from GR, KIR3DS1+ patients (P = .061). (B) Among patients with GR disease at ASCT, those with KIR3DS1+/KIR3DL1+/HLA-Bw4− have significantly reduced PFS than patients with KIR3DS1+/KIR3DL1+/HLA-Bw4+ and patients with KIR3DS1− irrespective of HLA-Bw4 (P = .002).
Association between KIR3DS1 genotype and PFS. (A) PFS for GR patients who were KIR3DS1+ was significantly shorter than for GR patients who were KIR3DS1− (P = .003). The median PFS for poor-risk patients was not significantly different from GR, KIR3DS1+ patients (P = .061). (B) Among patients with GR disease at ASCT, those with KIR3DS1+/KIR3DL1+/HLA-Bw4− have significantly reduced PFS than patients with KIR3DS1+/KIR3DL1+/HLA-Bw4+ and patients with KIR3DS1− irrespective of HLA-Bw4 (P = .002).
Patients were stratified into 3 groups on the basis of on disease status at ASCT and KIR3DS1 genotype; group 1: GR, KIR3DS1−; group 2: GR, KIRDS1+; and group 3: poor risk, KIR3DS1+ or KIR3DS1−. The RR of disease progression or death was 1.0, 1.9 (95% CI, 1.3-3.1; P = .002), and 3.0 (95% CI, 1.9-4.8; P = .001), respectively (Table 3). Adjusting for factors shown on univariate analysis to be associated with PFS after ASCT, including albumin and β2M levels, the RR of progression for patients with GR KIR3DS1−, GR KIR3DS1+; and poor risk were 1.0, 1.8 (95% CI, 1.2-2.8; P = .008), and 2.7 (95% CI, 1.6-4.3; P = .001), respectively (Table 3). Cytogenetic results were available for 76 patients only, and, when cytogenetics was incorporated into the model, the RRs for the same 3 groups were 1.0, 2.7 (95% CI, 1.2-6.2; P = .021), and 5.3 (95% CI, 2.4-11.7; P < .001), respectively (Table 3).
Multivariate analysis of factors associated with PFS after ASCT for myeloma
| Patient group . | No. . | RR (95%CI) . | P . |
|---|---|---|---|
| GR KIR3DS1− | 100 | 1.0 | |
| GR KIR3DS1+ | 46 | 1.9 (1.3-3.1) | .002 |
| Bad risk KIR3DS1+ or KIR3DS1− | 33 | 3.0 (1.9-4.8) | < .001 |
| Adjusted for albumin, β2M | |||
| GR KIR3DS1− | 89 | 1.0 | |
| GR KIR3DS1+ | 41 | 1.8 (1.2-2.8) | .008 |
| Bad risk KIR3DS1+ or KIR3DS1− | 33 | 2.7 (1.6-4.3) | .001 |
| Adjusted for albumin, β2M, and cytogenetics | |||
| GR KIR3DS1− | 36 | 1.0 | |
| GR KIR3DS1− | 12 | 2.7 (1.2-6.2) | .021 |
| Bad risk KIR3DS1+ or KIR3DS− | 20 | 5.3 (2.4-11.7) | < .001 |
| Patient group . | No. . | RR (95%CI) . | P . |
|---|---|---|---|
| GR KIR3DS1− | 100 | 1.0 | |
| GR KIR3DS1+ | 46 | 1.9 (1.3-3.1) | .002 |
| Bad risk KIR3DS1+ or KIR3DS1− | 33 | 3.0 (1.9-4.8) | < .001 |
| Adjusted for albumin, β2M | |||
| GR KIR3DS1− | 89 | 1.0 | |
| GR KIR3DS1+ | 41 | 1.8 (1.2-2.8) | .008 |
| Bad risk KIR3DS1+ or KIR3DS1− | 33 | 2.7 (1.6-4.3) | .001 |
| Adjusted for albumin, β2M, and cytogenetics | |||
| GR KIR3DS1− | 36 | 1.0 | |
| GR KIR3DS1− | 12 | 2.7 (1.2-6.2) | .021 |
| Bad risk KIR3DS1+ or KIR3DS− | 20 | 5.3 (2.4-11.7) | < .001 |
Patients were stratified into 3 groups based on (1) disease status at ASCT; (2) KIR3DS1 genotype; and (3) good risk KIR3DS1−, good risk KIR3DS1+, poor-risk patients. The RR after further adjustment for albumin level, β2M level, and bone marrow cytogenetics are also shown.
RR indicates relative risk; 95% CI, 95% confidence interval; GR, good risk; and β2M, β-2 microglobulin.
To ascertain that any association between KIR3DS1 genotype and PFS was not due to a differential response to remission induction chemotherapy, we compared the KIR genotype of GR and poor-risk patients at ASCT. No significant difference was observed in the proportion of patients with KIR3DS1+ between the 2 groups; 31% of GR patients were KIR3DS+ compared with 42.4% of poor-risk patients (P = .23). Similarly, no difference was found for KIR3DS1 genotype and the number of lines of chemotherapy regimens before ASCT; 19% of KIR3DS1+ and 25% of KIR3DS1− patients had received more than 1 line of chemotherapy regimens (P = .42).
Presence of both KIR3DS1 and KIR3DL1 in the absence of the inhibitory ligand (HLA-Bw4) is associated with significantly worse PFS and OS
The activating receptor KIR3DS1 is encoded as an allele of KIR3DL1 and shares greater than 97% sequence homology in its extracellular domain with the KIR3DL1 receptor. KIR3DL1 binds HLA-B allotypes that have the Bw4 epitope. The ligand for KIR3DS1 has not been determined, although the presence of this gene along with the presence of alleles encoding Bw4 have been shown to have an epistatic protective effect on progression of AIDS,31 suggesting that like KIR3DL1, KIR3DS1 may recognize at least some of the Bw4 allotypes in persons seropositive for HIV.
It is possible that KIR-mediated activation of NK cells, a phenomenon that increases both with the presence of certain activating KIR and with the absence of ligand for inhibitory KIR, may have an effect on PFS. Because KIR3DL1 interaction with the Bw4 ligand on target cells would theoretically inhibit the drive to cytotoxicity from an activating signal mediated by KIR3DS1, we hypothesized that an effect of KIR3DS1 would be greatest among persons who are missing the Bw4 ligand for KIR3DL1. In our cohort, 167 of 182 patients (92%) were positive for KIR3DL1. We subdivided these patients into 3 groups on the basis of on KIR3DS1 and KIR3DL1 genotype and HLA-Bw4 status to include group I: KIR3DS1+, KIR3DL1+, and Bw4− (n = 26); group II: KIR3DS1+, KIR3DL1+, and Bw4+ (n = 22); and group III: KIR3DS1−, KIR3DL1+ (n = 119). The median PFS of patients in group I was 13.2 months (95% CI, 10-16 months) compared with 16.7 months (95% CI, 13-21 months) for group II and 20.4 months (95% CI, 18-23months) in group III (P = .042; Table 4). This effect is more marked in patients with GR disease. Within the GR patients, the median PFSs were significantly different between patients in groups I, II, and III at 12.2 months (95% CI, 11.9-12.6 months), 16.7 months (95% CI, 12-21months), and 24 months (95% CI, 20-28), respectively (P = .002; Table 4; Figure 1B). A trend toward improved OS was seen for patients in group III (KIR3DS1−, KIR3DL1+), although it did not reach statistical significance (P = .10). These data suggest that the association between the activating gene KIR3DS1 and PFS is even more significant when Bw4, the HLA ligand for the closely related inhibitory receptor KIR3DL1, is absent.
Association between KIR3DS1, KIR3DL1, and HLA-Bw4
| Patient group . | Median PFS, mo (95%CI) . | No. . | P . |
|---|---|---|---|
| All patients regardless of disease status at ASCT | .042 | ||
| KIR3DS1+/KIR3DL1+HLA-Bw4− | 13.2 (10-16) | 26 | |
| KIR3DS1+/KIR3DL1+HLA-Bw4+ | 16.7 (13-21) | 22 | |
| KIR3DS1−/KIR3DL1+ | 20.4 (18-23) | 119 | |
| Good risk patients | .002 | ||
| Good risk KIR3DS1+/KIR3DL1+HLA-Bw4− | 12.2 (11.9-12.6) | 19 | |
| Good risk KIR3DS1+/KIR3DL1+HLA-Bw4+ | 16.7 (12-21) | 21 | |
| Good risk KIR3DS1−/KIR3DL1+ | 24 (19.7-28) | 100 |
| Patient group . | Median PFS, mo (95%CI) . | No. . | P . |
|---|---|---|---|
| All patients regardless of disease status at ASCT | .042 | ||
| KIR3DS1+/KIR3DL1+HLA-Bw4− | 13.2 (10-16) | 26 | |
| KIR3DS1+/KIR3DL1+HLA-Bw4+ | 16.7 (13-21) | 22 | |
| KIR3DS1−/KIR3DL1+ | 20.4 (18-23) | 119 | |
| Good risk patients | .002 | ||
| Good risk KIR3DS1+/KIR3DL1+HLA-Bw4− | 12.2 (11.9-12.6) | 19 | |
| Good risk KIR3DS1+/KIR3DL1+HLA-Bw4+ | 16.7 (12-21) | 21 | |
| Good risk KIR3DS1−/KIR3DL1+ | 24 (19.7-28) | 100 |
Patients were divided into three groups: (1) KIR3DS1+/KIR3DL1+HLA-Bw4−, (2) KIR3DS1+/KIR3DL1 HLA-Bw4−, and (3) KIR3DS1−/KIR3DL1+.
PFS indicates progression-free survival; and ASCT, autologous stem cell transplantation.
We determined the various combinatorial frequencies of KIR2DS1/KIR2DL1 with group 2 HLA-C alleles (Lys80) and KIR2DS2/3/KIR2DL2/3 and HLA-Cw group 1 (Asn80) alleles. The absence of ligand for inhibitory KIR occurs when persons are homozygous for either HLA-Cw group 1 (ligands for KIR2DL2/3) or group 2 (ligands for KIR2DL1). We found no significant association of these KIRs and their missing ligands on PFS (P = .31 and P = .28, respectively). Similarly, we found no effect of KIR3DL2 and HLA-A3/11 status on PFS and OS (P = .72).
Discussion
The association between KIR genotype and outcome after allo-SCT has been shown by a number of groups.21,24 However, the relationship between KIR genotype and outcome in the setting of autologous transplantation is unknown. With the use of a large homogeneous cohort of patients with MM, we have shown that patients carrying activating KIR genotypes, specifically the activating KIR receptor KIR3DS1, have a shorter PFS than persons who lack this receptor. This effect was more marked in patients who received an ASC transplant in complete or partial remission and who lacked HLA-Bw4, the inhibitory ligand for KIR3DL1.
On univariate analysis, we found that the presence of haplotype Bx, more than 3 activating KIRs, and the activating NK receptors, KIR2DS1 and KIR3DS1, are associated with significantly worse PFS. These results contrast with a recent study by Cooley et al29 who showed that with the use of a donor with a KIR Bx compared with an A/A haplotype resulted in a 30% improvement in the RR of relapse-free survival in patients receiving an unrelated allo-SC transplant for acute myelogenous leukemia. Interestingly, another study showed that the use of HLA-matched unrelated donors with haplotype A/A was associated with lower RR of relapse in patients with myeloid malignancies after a myeloablative allo-SCT.32 In keeping with our findings, they reported that the use of a donor with more than 3 activating KIRs or donors who are positive for KIR3DS1, KIR2DS1, or KIR2DS5 genes was associated with shorter PFS. The discrepancy between our findings and that of Cooley et al29 could in part be explained by differences between the patient groups for the underlying diagnosis (lymphoid vs myeloid), transplantation conditioning (total body irradiation based vs chemotherapy based, and different stem cell source (predominantly bone marrow in the allogeneic setting compared with peripheral blood mononuclear cells only in the autologous setting). The clearest difference between the 2 studies is however the graft type. Our study was performed in the autologous rather than an allogeneic setting and therefore without any interference of an allo-reactive phenomenon or immunosuppressive graft-versus-host disease therapies. All of these factors may contribute significantly to the different KIR effects observed in these studies.
In our patient cohort, KIR3DS1 genotype was associated with a significantly worse outcome, most evident in patients who received a transplant in complete or partial remission after induction chemotherapy, translating into a significant reduction in the median PFS (8.7 months). KIR3DS1 has been shown to be involved in other human diseases. KIR3DS1 appears to have a protective role in patients with HIV and hepatitis C. The presence of KIR3DS1 genotype has been implicated in lower HIV infection rates and in slowing disease progression.31,33 KIR3DS1 has also been shown to confer protection against the development of hepatocellular carcinoma in patients chronically infected with the hepatitis C virus.34 However and in keeping with our results, a meta-analysis of 3 large independent studies on cervical carcinoma showed that in women with high-grade cervical intraepithelial neoplasia, KIR3DS1 is associated with increased risk of progression to invasive cervical cancer.35 Furthermore, contrary to the idea that activating NK receptors protect against malignancy, an analysis of more than 300 cases of nasopharyngeal carcinoma showed that increasing numbers of activating receptors are associated with an increased risk of nasopharyngeal carcinoma in persons seropositive for Epstein-Barr virus.36 These findings suggest that KIRs and the innate immune system may be involved in the pathogenesis and/or progression of some cancers.35
The greatest effect on PFS was observed in KIR3DS1+ patients with GR disease in whom the ligand for the corresponding inhibitory KIR3DL1 receptor, HLA-Bw4, was missing. GR patients who had the genotype: KIR3DS1+KIR3DL1+HLA-Bw4− had a significantly shorter PFS (P = .002) than patients with GR disease who were KIR3DS1−, translating into a difference in median PFS of 12 months (12.2 vs 24 months). This suggests that, in the presence of the HLA ligand, the corresponding inhibitory KIR neutralizes the effect of the activating KIR. It is possible that KIRs and HLA class I alleles may have either beneficial or deleterious consequences, depending on the type of disease.
In contrast to previously published studies we failed to demonstrate an improved outcome with increased numbers of inhibitory KIR-ligand mismatches. Leung et al37 recently reported reduced relapse rates after ASCT for non-Hodgkin lymphoma or solid tumors in pediatric patients with an inhibitory KIR–HLA mismatch. Similarly, in a larger study of 169 patients with neuroblastoma treated with ASCT, a survival advantage was shown in patients lacking HLA class I ligands for autologous inhibitory KIRs.38 Another study, however, failed to show any effect of missing KIR-ligand interactions in 67 recipients of ASCT who had solid tumors or lymphoma.39 All these data support an association between KIR genotype and outcome in the setting of ASCT, although a better understanding of the biologic complexity responsible for these effects in different disease settings is needed.
The mechanism by which KIR3DS1 affects outcome in patients after ASCT is unclear. The finding that a similar proportion of GR and poor-risk patients were KIR3DS1+ at ASCT suggests that KIR3DS1 positivity did not affect response to induction chemotherapy but that any effect is more likely to be related to the peritransplantation or posttransplantation period. It is possible that KIR3DS1+ NK clones exert an immunomodulatory effect on antitumor responses. Any effect on antitumor responses is likely to have a greater effect after ASCT, at a time of lymphopenia-driven homeostasis and minimal tumor burden. Such potential immune modulation could be effected through the production of immunomodulatory cytokines, affecting T-cell function or through a direct NK effect on antigen presentation by dendritic cells,21 thereby indirectly affecting T-cell activation. Furthermore, it is also possible that T-cell populations expressing KIR are involved in this process.
NK cells undergo rapid expansion after ASCT, consistent with their role in defense against viruses. Interestingly, recent studies have shown that proliferating NK cells expressing activating receptors secrete cytokines such interleukin-10 (IL-10)40 and transforming growth factor β-1 (TGFβ-1)41 that have inhibitory effects. This is believed to be a homeostatic protective mechanism to “dampen down” a proinflammatory response and to prevent autoimmunity. IL-10 has direct and indirect inhibitory effects on several T-cell responses, including CD8 T-cell cytotoxicity and CD4 T-cell proliferation, and results in selective expansion of regulatory T cells.42 IL-10 also appears to be an important growth factor for malignant plasma cells.43 Recently NK cells expressing the activating KIR receptors KIR2DS1 and KIR2DS2 were also shown to secrete TGFβ1 with inhibitory effect on NK-mediated target lysis and proinflammatory cytokine production.41 TGFβ1 can modulate antigen-presenting cell function and lymphocyte differentiation, lead to regulator T-cell expansion and the suppression of autoreactive T cells.44 Furthermore, TGFβ1 is elevated in the serum of patients with MM.45 Therefore, it is possible that in patients with more than 3 activating KIRs, and more specifically in persons with KIR3DS1+, the expansion of NK cells in the early period after ASCT might be associated with secretion of anti-inflammatory cytokines that either directly promote tumor growth or affect disease control indirectly by abrogating antitumor immune responses. Interestingly, Venstrom et al46 have recently shown that the risk and severity of graft-versus-host disease is significantly reduced in patients with myeloid malignancies receiving allogeneic stem cells from KIR3DS+ donors. It is, however, possible that, rather than being directly involved in the disease process, KIR3DS1 may be simply a surrogate marker for another neighboring gene that is directly involved in pathogenesis of myeloma.
In conclusion, we report that KIR genotype may have a significant effect on the outcome of autologous transplantation for myeloma and that KIR3DS1, a gene with a frequency of greater than 30% in the general population, is also a negative prognostic marker in this disease. The mechanism by which the KIR gene products exert their effect is not yet clear, although an immune-mediated effect seems most probable. Functional and phenotypic studies to determine the expression of KIR3DS1 on the surface of NK cells and to assess the role of the cytokine milieu and the NK phenotype on outcome are currently under way. In vitro data support NK cytotoxicity against both allogeneic and autologous myeloma,25,26 and the use of NK cells in adoptive cell therapy in hematologic malignancies is an attractive therapeutic strategy. Within a person, a diverse NK-cell repertoire exists, and individual NK clones expressing a specific array of receptors have been shown to respond differently to diverse viral infections.24,47,48 It is therefore conceivable that in myeloma, depending on receptor expression and ligand engagement, different NK clones may exert divergent responses against the tumor. Selection of specific NKclones may therefore improve the beneficial effect of NK-adoptive therapy.
We propose that combining KIR-HLA genotyping with known factors associated with outcome in patients with MM, such as cytogenetics and serum albumin and β2M levels, may improve the prediction of outcome after ASCT and select patients who may benefit from more-intensive or novel treatment strategies. It is important to note, however, that the use of novel agents may affect the prognostic value of KIR3DS1; therefore, these data require validation in patients treated with such agents at induction.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Professor Danny Altmann for critical review of the manuscript.
This work was supported by Leuka registered charity 286231 and the National Institute for Health Research (NIHR) Biomedical Research Center.
Only the authors participated in study design, interpretation of the data, and preparation of the manuscript.
Authorship
Contribution: I.H.G. performed the experiments and participated in the design and interpretation of the analysis. and wrote the paper; K.R. designed and directed the study and wrote the manuscript; A.R., C.G., and J.F.A. provided the patient samples and information and commented on the manuscript; E.K., D. Marin, D. Milojokovic, M.B., and D. MacDonald participated in patient care and commented on the paper; R. Sergeant provided advice on the genotyping of the DNA samples and commented on the paper; H.d.L., N.C., L.F., A.K., J.D., and A.A. participated in the design of the study and revised the manuscript; and R. Szydlo and D. Marin performed the statistical analyses and helped write the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Katayoun Rezvani, Department of Hematology, Imperial College, Hammersmith Campus, 4th Fl Commonwealth Bldg, DuCane Rd, London W12 0NN, United Kingdom; e-mail: k.rezvani@imperial.ac.uk.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal