Abstract
Previous studies in our laboratory demonstrated that the azurophil granule protease neutrophil elastase (NE) cleaves promyelocytic leukemia–retinoic acid receptor (PML-RAR)α (PR), the fusion protein that initiates acute promyelocytic leukemia (APL). Further, NE deficiency reduces the penetrance of APL in a murine model of this disease. We therefore predicted that NE-mediated PR cleavage might be important for its ability to initiate APL. To test this hypothesis, we generated a mouse expressing NE-resistant PR. These mice developed APL indistinguishable from wild-type PR, but with significantly reduced latency (median leukemia-free survival of 274 days vs 473 days for wild-type PR, P < .001). Resistance to proteolysis may increase the abundance of full-length PR protein in early myeloid cells, and our previous data suggested that noncleaved PR may be less toxic to early myeloid cells. Together, these effects appear to increase the leukemogenicity of NE-resistant PR, contrary to our previous prediction. We conclude that NE deficiency may reduce APL penetrance via indirect mechanisms that are still NE dependent.
Introduction
The t(15;17) translocation found in 95% of patients with acute promyelocytic leukemia (APL) generates the promyelocytic leukemia–retinoic acid receptor (PML-RAR)α (PR) fusion gene product. The requirement for this gene product to initiate APL was established by expressing PR under the regulatory sequences of early myeloid-specific promoters in mice.1-4 These mice develop an acute myeloid leukemia with promyelocytic features after a long latency and with incomplete penetrance, suggesting that while PR is the initiating event, additional events are required for the development of leukemia. A number of events associated with the progression of leukemia have been described in these models, including the acquisition of additional cytogenetic abnormalities, activating mutations of FLT3, down-regulation of PU.1, and the dysregulated expression of BCL2.5-8
Our previous work established that neutrophil elastase (NE), an early myeloid-specific serine protease that is maximally expressed in promyelocytes, is important for the development of APL in mice. In our study, NE was shown to cleave PR primarily at positions V420 and V432 in the PML part of the fusion protein. NE deficient mice displayed a decrease in the penetrance of leukemia by approximately 50% compared with wild-type (WT) animals.9 Furthermore, in primary murine bone marrow progenitors, NE activity was found to be relevant for many of the activities of PR, including toxicity to myeloid cells at high doses, increased myeloid proliferation at low doses, and delayed differentiation.10
Because NE directly cleaves PR, we originally proposed that PR cleavage fragments may possess novel gain-of-function activities that are relevant for AML pathogenesis, and that a cleavage-resistant PR may be less likely to cause disease. In this paper, we characterize a NE-resistant form of PR (ie, both V420 and V432 are mutated to arginine). We expressed this mutant PR in the early myeloid cells of mice to determine whether the cleavage resistant form would be less likely to develop APL, and to determine whether the characteristics of the leukemia created by the cleavage-resistant form differ from WT PR. Remarkably, APL penetrance was increased, not decreased, as we originally proposed. This suggests that NE has alternative activities that are relevant for the pathogenesis of APL.
Methods
PML-RARα cleavage assays
In vitro translated protein (TnT System, Promega) derived from plasmids containing PR cDNAs (PRWT vs double mutant V420R and V432R allele PR2VR)2,10 in the pcDNA 3.1 vector were incubated with bone marrow extracts obtained from neutrophil elastase–deficient11 or WT C57Bl/6 mice (Taconic Farms) in 1M NaCl, 25mM Tris pH 7.5, 0.1% Triton X-100 at 37°C for 20 minutes.
Transient transfections
Transfection of pEGFP-PRWT and pEGFP-PR2VR in either U937 or K562 cell lines was performed by electroporation (BTX, Amaxa). For apoptosis assays and PML immunofluorescence analysis, transfected cells were affixed to glass slides by centrifugation, fixed in methanol and air dried. Cells were blocked with 2% goat serum and stained with rabbit anti-PML (PG-M3, Santa Cruz Biotechnology) diluted in Tris-buffered saline Tween-20, detected by fluorescein isothiocyanate–conjugated goat anti–rabbit secondary antibody. Cells were counterstained with DAPI (4′,6-diamidino-2-phenylindole) and visualized by immunofluorescence microscopy. Cell death in U937 transfected cells was assayed using an annexin V apoptosis detection kit (BD Biosciences).
Western blotting
Western blotting was performed using either a using a commercial polyclonal rabbit anti-RARα antibodies (C-20, Santa Cruz Biotechnology) or a rabbit polyclonal antiserum raised against a glutathione S-transferase fusion protein encompassing amino acids 420 to 462 of the human RARα protein with a goat anti–rabbit HRP conjugate secondary antibody12 (a kind gift from S. Kogan, University of California, San Francisco) and visualized using an enhanced chemolumninescent immunodetection system (Amersham). Protein bands were quantified by densitometry using National Institutes of Health ImageJ software Version 1.4.2.
Gene expression analysis
RNA was prepared from transfected green fluorescent protein (GFP)+ cells using a high speed sorter (MoFlo, DAKO-Cytomation) that were then purified, quantified, and qualitatively assessed as described previously.13 RNA samples were labeled and hybridized to Affymetrix Human Genome U133 Plus 2.0 microarrays (Affymetrix) using standard protocols from the Siteman Cancer Center Multiplexed Gene Analysis Core Facility, as described previously, except that the standard amplification protocol was used.
To perform inter-array comparisons, the raw data for each array were scaled to an average intensity of 1500 with Affymetrix Microarray Suite software and the (MAS5 statistical algorithm. Probesets were filtered to remove those that did not have at least 2 present calls in at least 1 of the groups (GFP, GFP-PRWT, GFP-PR2VR). The remaining probesets were subjected to analysis of variance, with differentially expressed genes defined at a significance level of P < .01. MAS5-generated signal intensities were normalized to z-scores for hierarchical clustering and heatmap creation. Statistical analysis, hierarchical clustering, and creation of heat maps were performed with Spotfire software Version 9.1.2 (Spotfire). The data have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus and are accessible through Gene Expression Omnibus Series accession number GSE21550 (www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE21550).
Protease-resistant PML-RARα mice
Mice carrying the human PR cDNA targeted to the murine cathepsin G (mCG) locus, and designated mCG-PML-RARα (mCG-PRWT), have been previously described.14 An identical procedure was employed to create a targeting vector with the PR2VR allele to generate protease cleavage–resistant PML-RARα mice (mCG-PR2VR). A tumor watch was established by crossing both mCG-PRWT and mCG-PR2VR heterozygotes with WT B6 × 129 F1 mice. Moribund animals were killed and analyzed for the presence of acute leukemia.
Progenitor assays and flow cytometry
Hematopoietic progenitor assays were performed as described (MethoCult 3534; StemCell Technologies).15 Flow cytometry of spleen and marrow cells were performed using antibodies to Gr-1, CD11b, CD34, and CD117 with isotype controls (BD Biosciences). Data were acquired on a modified FACScan flow cytometer (Cytek Development), and analyzed using FlowJo Version 7.2 (TreeStar).
qRT-PCR
PR gene expression was measured by 1-step reverse transcription-amplification polymerase chain reaction (PCR) of mRNA isolated from murine bone marrow or NB4 cell lines using published primer and probe sequences with a 5′-6-FAM, 3′-BHQ1 dual-labeled Taqman probe.16 PR transcripts were quantified by the comparative CΤ method relative to a glyceraldehyde 3-phosphate dehydrogenase control. To exclude genomic DNA amplification, controls were performed concurrently without reverse transcriptase.
Detection of PR protein
Bone marrow and spleen samples from PR mice were incubated with diisopropyl fluorophosphate (2mM) for 15 minutes on ice to inactivate intracellular proteases, and were then lysed using a 1% nonyl phenoxypolyethoxylethanol buffer containing a protease inhibitor cocktail (Sigma-Aldrich). Proteins were resolved using 8% sodium dodecyl sulfate–polyacrylamide gel electrophoresis, transferred to a polyvinylidene fluoride membrane and blotted as described.17
Results
In vitro cleavage of PML-RARα
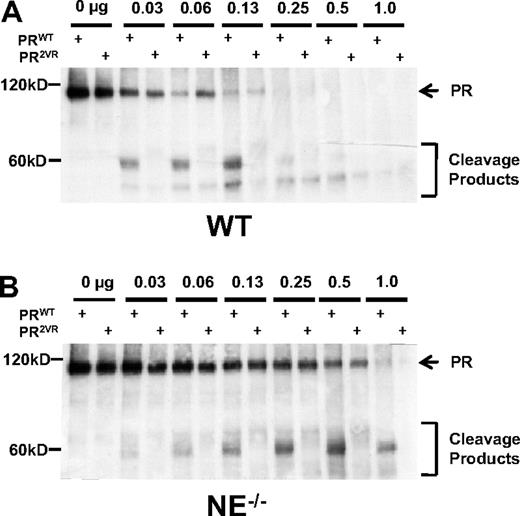
To determine the relative efficiency of cleavage of PRWT versus PR2VR by murine bone marrow proteases, we subjected in vitro translated protein to cleavage using unfractionated murine bone marrow extracts. In concordance with our previously published data, WT bone marrow extracts efficiently cleave PRWT, generating dominant approximately 58-kDa cleavage products, while mutations at V420 and V432 reduce cleavage at these sites by approximately one-half (56.3% ± 11.6%, n = 3, Figure 1A). Bone marrow extracts derived from NE−/− mice also cleave PRWT, but less efficiently than WT bone marrow extracts (23.2% ± 10.5%, n = 3), indicating that NE is the dominant PR-cleaving protease in marrow-derived cells (Figure 1B). PR2VR was relatively resistant to cleavage by both WT and NE deficient marrow extracts and also displayed a different cleavage pattern from PRWT. While these data reinforce the concept that NE is the primary protease that cleaves PR at V420 and V432, it also demonstrates that other marrow-derived proteases can also cleave PR at alternative sites in vitro.
In vitro cleavage of PR. Representative Western blots of in vitro translated PRWT or PR2VR incubated with serial dilutions of bone marrow extracts from either (A) WT or (B) NE−/− mice using an antibody against RARα.
In vitro cleavage of PR. Representative Western blots of in vitro translated PRWT or PR2VR incubated with serial dilutions of bone marrow extracts from either (A) WT or (B) NE−/− mice using an antibody against RARα.
PR2VR does not have novel gain-of-function properties
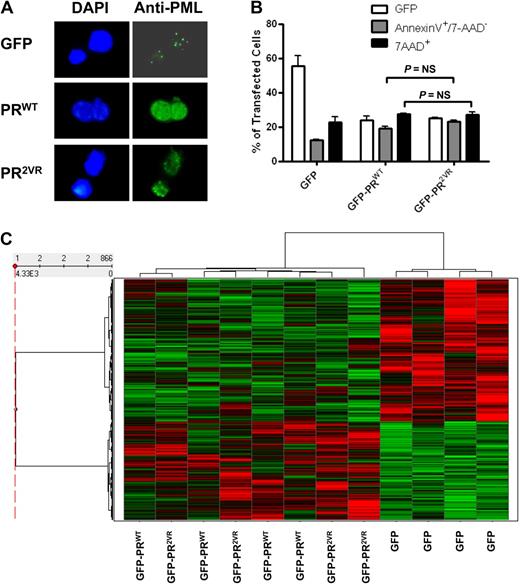
To determine whether PR2VR has properties that are independent of cleavage resistance, we expressed GFP-PR fusions in U937 cells. Similar to PRWT, PR2VR localized to the nucleus and retained the ability to induce a microspeckled distribution of PML (Figure 2A). Furthermore, like PRWT, PR2VR induces cell death when transiently transfected in U937 cells (Figure 2B). To detect differences in the proliferation or differentiation of myeloid progenitors between the different mouse strains, we used a granulocyte colony-stimulating factor-dependent myeloid differentiation system developed by McLemore et al18 in which hematopoietic progenitors from preleukemic bone marrow are treated for 7 days with granulocyte colony-stimulating factor and SCF, which induces a coordinate wave of myeloid maturation. Using this assay, we did not detect any differences between PRWT and PR2VR in the proliferation, cell cycle status or differentiation of myeloid progenitors. (supplemental Figures 1-2, available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Expression of PR2VR in myeloid cell lines. (A) Staining for PML oncogenic domains in K562 cell lines transfected with GFP-PRWT or GFP-PR2VR (B) Analysis of apoptosis by annexin V staining in U937 cells at 24 hours after transfection with GFP-PRWT or GFP-PR2VR (C) Heatmap of hierarchical clustering of z-score normalized microarray data of U937 cells transfected with GFP control versus GFP-PRWT or GFP-PR2VR.
Expression of PR2VR in myeloid cell lines. (A) Staining for PML oncogenic domains in K562 cell lines transfected with GFP-PRWT or GFP-PR2VR (B) Analysis of apoptosis by annexin V staining in U937 cells at 24 hours after transfection with GFP-PRWT or GFP-PR2VR (C) Heatmap of hierarchical clustering of z-score normalized microarray data of U937 cells transfected with GFP control versus GFP-PRWT or GFP-PR2VR.
To assess for differences in transcriptional effects of the 2 PR proteins, we performed gene expression arrays of U937 cells 6 to 9 hours after transfection. A heatmap of hierarchical clustering of z-score normalized microarray data demonstrates clear separation of significantly up- and down-regulated genes in GFP-PRWT or GFP-PR2VR transfected cells compared with cells transfected with a GFP control plasmid (Figure 2C). Only a small subset of probesets demonstrated differences in expression between GFP-PRWT and GFP-PR2VR; of these 866 probesets, only 5 had a greater than 2-fold change when comparing GFP-PRWT and GFP-PR2VR transfected cells (supplemental Table 1). In addition, we observed that p21/Cip, a well-characterized transcriptional target of PR,19 is down-regulated to a similar degree in cells transfected with either GFP-PRWT or GFP-PR2VR. These results suggest that the transcriptional activities of PRWT are retained in the PR2VR mutant.
Generation and analysis of mCG-PR2VR mice
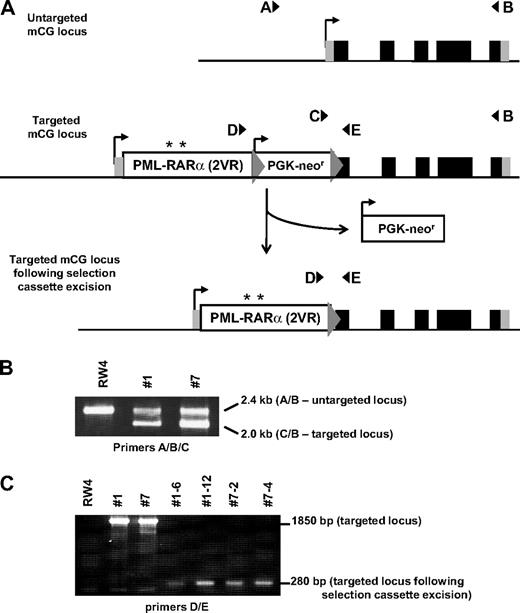
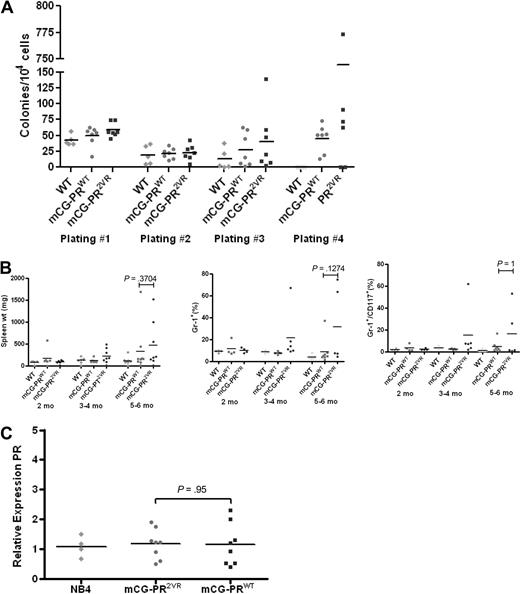
We generated knock-in mice with the PR2VR mutation targeted to the 5′ untranslated region of the early myeloid protease cathepsin G, exactly as described previously for mCG-PRWT knock-in mice14 (Figure 3). Analysis of 6- to 8-week-old mCG-PR2VR mice revealed no difference in PR mRNA abundance in the bone marrow, compared with mCG-PRWT mice (Figure 4C). No difference was observed between the numbers of progenitors in WT versus mCG-PR2VR versus mCG-PRWT bone marrow cells using methylcellulose-based colony assays. However, serial replating of colonies revealed that both PRWT and PR2VR derived progenitors have increased replating efficiency (Figure 4A). Similar to PRWT mice, PR2VR mice developed a myeloproliferative disorder characterized by splenomegaly, with an increase in the percentage of Gr-1+ and Gr-1+/CD117+ cells in the spleen (Figure 4B). No alterations were observed in the peripheral blood counts of nonleukemic PRWT and PR2VR mice (Figure 5A).
Targeting of PR2VR to mCG locus. (A) A targeting vector containing the PR cDNA with the V420 and V432R mutations (PR2VR, with position of mutations designated by asterisk) and a PGK-neo selectable marker cassette flanked by targeting arms from the mCG locus was transfected into an embryonic stem cell line (RW4; 129/SvJ). The 5 exons of the endogenous mCG gene are represented by black (coding regions) and gray (5′ and 3′ untranslated regions) rectangles. The PGK-neor cassette was flanked by loxP sites (gray triangles), which allowed its removal by transient transfection of ES cells with a Cre recombinase expression vector. (B) Genotyping of parental untargeted RW4 ES cells and 2 independently targeted mCG-PR2VR clones (#1 and #7) is shown. mCG locus targeting was identified by PCR using a 3-primer multiplex cocktail (A/B/C). The untargeted mCG allele is identified by a 2.4-kb product of the forward “A” primer and a common reverse “B” primer (external to the targeting construct). The correctly targeted mCG-PR2VR alleles are recognized as a 2.0-kb product of the forward “C” primer (within the PGK-neo cassette) and the reverse “B” primer. (C) Removal of the PGK-neo cassette was indicated by disappearance of a 1850-bp band and appearance of a 280-bp band identified by PCR (using primers “D” and “E”). 2 independent targeted mCG-PR2VR parental clones (#1 and #7), and 4 subclones (##1-6, 1-12, 7-2, and 7-4) that have undergone successful excision of the PGK-neor cassette are shown. Mice generated from 2 ES clones containing independent Cre excision events (7-2 and 7-4) were used for all subsequent experiments.
Targeting of PR2VR to mCG locus. (A) A targeting vector containing the PR cDNA with the V420 and V432R mutations (PR2VR, with position of mutations designated by asterisk) and a PGK-neo selectable marker cassette flanked by targeting arms from the mCG locus was transfected into an embryonic stem cell line (RW4; 129/SvJ). The 5 exons of the endogenous mCG gene are represented by black (coding regions) and gray (5′ and 3′ untranslated regions) rectangles. The PGK-neor cassette was flanked by loxP sites (gray triangles), which allowed its removal by transient transfection of ES cells with a Cre recombinase expression vector. (B) Genotyping of parental untargeted RW4 ES cells and 2 independently targeted mCG-PR2VR clones (#1 and #7) is shown. mCG locus targeting was identified by PCR using a 3-primer multiplex cocktail (A/B/C). The untargeted mCG allele is identified by a 2.4-kb product of the forward “A” primer and a common reverse “B” primer (external to the targeting construct). The correctly targeted mCG-PR2VR alleles are recognized as a 2.0-kb product of the forward “C” primer (within the PGK-neo cassette) and the reverse “B” primer. (C) Removal of the PGK-neo cassette was indicated by disappearance of a 1850-bp band and appearance of a 280-bp band identified by PCR (using primers “D” and “E”). 2 independent targeted mCG-PR2VR parental clones (#1 and #7), and 4 subclones (##1-6, 1-12, 7-2, and 7-4) that have undergone successful excision of the PGK-neor cassette are shown. Mice generated from 2 ES clones containing independent Cre excision events (7-2 and 7-4) were used for all subsequent experiments.
Analysis of mCG-PR2VR. (A) Colony-forming units from serial replating of bone marrow progenitors (n = 5-6/group) in 6- to 12-week-old mice. (B) Analysis of spleens from animals 2 to 6 months old for weight and immunophenotypes by flow cytometry (n = 4-9 mice/group). (C) RT-PCR for PR expression relative to GAPDH in bone marrow of 6- to 12-week-old animals (n = 4/group, performed in duplicate).
Analysis of mCG-PR2VR. (A) Colony-forming units from serial replating of bone marrow progenitors (n = 5-6/group) in 6- to 12-week-old mice. (B) Analysis of spleens from animals 2 to 6 months old for weight and immunophenotypes by flow cytometry (n = 4-9 mice/group). (C) RT-PCR for PR expression relative to GAPDH in bone marrow of 6- to 12-week-old animals (n = 4/group, performed in duplicate).
Analysis of APL arising from mCG-PR2VR mice. (A) Analysis of leukemic mCG-PRWT (n = 11) and mCG-PR2VR (n = 29) mice for spleen weights, complete blood counts, and immunophenotyping by flow cytometry for Gr-1+/CD117+ blasts. (B) Immunophenotyping and (C) Wright-Giemsa staining of spleen cells from representative leukemic mCG-PRWT and mCG-PR2VR mice. (D-E) Western blotting of cell lysates from (D) bone marrow of preleukemic mice (E) spleen from leukemic mice. * represents a nonspecific band observed in the bone marrow extracts. Controls are a U937-PR9 cell line that contains the PR cDNA under the control of a Zinc (Zn2+) inducible promoter treated with 100μM ZnSO4 for 1 hour.
Analysis of APL arising from mCG-PR2VR mice. (A) Analysis of leukemic mCG-PRWT (n = 11) and mCG-PR2VR (n = 29) mice for spleen weights, complete blood counts, and immunophenotyping by flow cytometry for Gr-1+/CD117+ blasts. (B) Immunophenotyping and (C) Wright-Giemsa staining of spleen cells from representative leukemic mCG-PRWT and mCG-PR2VR mice. (D-E) Western blotting of cell lysates from (D) bone marrow of preleukemic mice (E) spleen from leukemic mice. * represents a nonspecific band observed in the bone marrow extracts. Controls are a U937-PR9 cell line that contains the PR cDNA under the control of a Zinc (Zn2+) inducible promoter treated with 100μM ZnSO4 for 1 hour.
APL arising in mCG-PR2VR mice
A tumor watch was established with mCG-PR2VR mice generated from 2 independent ES subclones (designated 7-2 and 7-4), and APL development was compared with WT littermates and a concurrent cohort of strain-matched mCG-PRWT mice. Most mCG-PR2VR mice developed an acute myeloid leukemia with promyelocytic features, characterized by a marked peripheral leukocytosis and infiltration of the spleen and marrow with tumor cells that coexpress CD117 and Gr-1 (Figure 5A-C). Like mCG-PRWT, tumors derived from mCG-PR2VR respond to the differentiating effects of all-trans retinoic acid (supplemental Figure 3). In preleukemic mice, PR protein could be detected in neither mCG-PRWT nor mCG-PR2VR bone marrow extracts (Figure 5D). In leukemic mice, full-length PR protein was detectable in tumors derived from both strains, but PR2VR mice displayed an altered cleavage pattern in vivo compared with PRWT mice (Figure 5E), as expected.
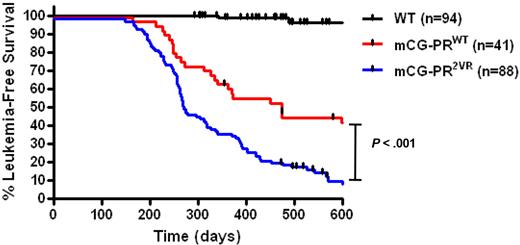
No difference in leukemia-free survival was observed in PR2VR mice generated from the 7-2 (n = 38) versus 7-4 (n = 50) founder strains (Kaplan-Meier estimate of median survival 290 vs 269 days, P = not significant [NS]). However, mice bearing the PR2VR mutation had a median leukemia-free survival of 274 days, compared with 473 days for PRWT mice maintained in the same F1 strain background (P < .001, Figure 6). These cohorts were generated and analyzed simultaneously in our mouse facility.
Discussion
In this report, we describe the phenotype of mice carrying a form of PR that is relatively resistant to proteolytic cleavage by both NE and other marrow-derived proteases. We demonstrated that expression of PR2VR in early myeloid cells is associated with the development of a myeloproliferative disorder, followed by progression to AML with promyelocytic features. Although these leukemias are morphologically and phenotypically identical to that of mCG-PRWT mice, mCG-PR2VR unexpectedly displayed a significant decrease in leukemia latency, a result that is the opposite of what we had originally predicted.
Although we were unable to detect differences in PR abundance in fully leukemic animals, our current results suggest that PR2VR may be a more potent oncogene because it is relatively resistant to proteolysis (resulting in a higher “dose” of full-length PR in early myeloid cells). Increased PR levels may confer a subtle competitive advantage at a critical point in hematopoiesis, when a stem (or progenitor) cell is capable of being fully transformed by PR. These data are consistent with our previous results demonstrating that a 2-fold increase in PR gene dosage in mCG-PR mice accelerates leukemia development by approximately 6 weeks.14 While we cannot completely exclude the possibility that the PR2VR mutation confers a novel gain of function activity that is unrelated to its protease resistance, we doubt this possibility. We have shown that PRWT and PR2VR have equal abilities to induce apoptosis and disrupt PML oncogenic domains. Furthermore, RNA expression profiles induced by PRWT and PR2VR expression in myeloid cells are essentially equivalent. Ultimately, the most important function of PR was preserved in mCG-PR2VR animals; the mutant protein initiates an acute leukemia with promyelocytic features that is phenotypically identical to that of mCG-PRWT mice.
Importantly, these results strongly suggest that the reduced penetrance of leukemia in NE deficient mice is not caused by the lack of cleavage of PR per se. Although the relationship between NE and PR was initially identified based on biochemical purification of a protease that cleaves PR in vitro, the data presented here make it extremely unlikely that PR cleavage by NE is relevant for the pathogenesis of APL. A number of recent studies suggest that alternative substrates and/or activities of NE may be relevant. NE deficiency produces no discernable hematopoietic phenotype in mice,10 or in patients with dipeptidyl peptidase I deficiency (an enzyme required for the processing of NE), which results in functional deficiency of NE.20 Although NE has primarily been implicated in host defense mechanisms, heterozygous mutations in ELA2, the gene encoding NE, are found in patients with severe congenital neutropenia who possess a high risk of transformation to acute leukemia.21 NE also has other substrates, both intra- and extracellular, which may be important for the development of cancer. Tavor et al demonstrated that neutrophil elastase can cleave CXCL12/SDF1, and that release of cell-surface elastase regulated the proliferation and migration of AML cells in vitro.22 More recently, NE deficiency in an inducible K-ras mouse model of lung adenocarcinoma was found to reduce tumor burden and protect these mice from death.23 In this study, NE was found to be actively transported to a subcellular location where it cleaves the insulin receptor substrate-1 (IRS-1), leading to PI3-kinase pathway activation and tumor cell proliferation. In the absence of NE, IRS-1 protein was more abundant, and tumor cell growth was impaired. Although IRS-1 protein levels are unaltered in APL tumors (data not shown), alternative NE substrates may be present in APL cells that perform similar functions and future studies will be designed to identify these substrates. Collectively, these results suggest that NE may play a role in governing cellular proliferation in inflammatory states, and that NE deficiency may alter APL pathogenesis via indirect mechanisms that are not yet fully understood. Further studies of the role of NE in the pathogenesis of AML as well as the potential role of neutrophil elastase inhibitors as therapy for AML will be pursued.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Mieke Hoock for mouse colony management, Nancy Reidelberger for editorial assistance, and McGarry Houghton II and Steve Shapiro for measuring IRS-1 in APL tumors.
This work was supported by National Institutes of Health grants T32 HL007088 and K23 CA140707 to G.L.U. and CA083962 to T.J.L and by the Barnes-Jewish Hospital Foundation to T.J.L. G.L.U is an American Society of Hematology Scholar in Clinical/Translational Research. J.S.W. is a Leukemia & Lymphoma Society Fellow. The Siteman Cancer Center and High Speed Cell Sorter Core is supported in part by a National Cancer Institute Cancer Center Support Grant P30 CA91842.
National Institutes of Health
Authorship
Contribution: G.L.U. designed and performed research, analyzed and interpreted data, and wrote the manuscript; J.S.W., J.E.P., N.R.G., and A.A.L. designed and performed research, analyzed and interpreted data; and T.J.L. designed research, analyzed and interpreted data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Timothy J. Ley, Washington University School of Medicine, 660 S Euclid Ave, Campus Box 8007, St Louis, MO 63110; e-mail: tley@dom.wustl.edu.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal