Abstract
Twenty percent to 30% of transient abnormal myelopoiesis (TAM) observed in newborns with Down syndrome (DS) develop myeloid leukemia of DS (ML-DS). Most cases of TAM carry somatic GATA1 mutations resulting in the exclusive expression of a truncated protein (GATA1s). However, there are no reports on the expression levels of GATA1s in TAM blasts, and the risk factors for the progression to ML-DS are unidentified. To test whether the spectrum of transcripts derived from the mutant GATA1 genes affects the expression levels, we classified the mutations according to the types of transcripts, and investigated the modalities of expression by in vitro transfection experiments using GATA1 expression constructs harboring mutations. We show here that the mutations affected the amount of mutant protein. Based on our estimates of GATA1s protein expression, the mutations were classified into GATA1s high and low groups. Phenotypic analyses of 66 TAM patients with GATA1 mutations revealed that GATA1s low mutations were significantly associated with a risk of progression to ML-DS (P < .001) and lower white blood cell counts (P = .004). Our study indicates that quantitative differences in mutant protein levels have significant effects on the phenotype of TAM and warrants further investigation in a prospective study.
Introduction
In children with Down syndrome (DS), the risk of developing acute megakaryocytic leukemia (AMKL) is estimated at 500 times higher than in children without DS. Interestingly, neonates with DS are at a high risk of developing a hematologic disorder referred to as transient abnormal myelopoiesis (TAM). It has been estimated that 5% to 10% of infants with DS exhibit the disorder, and in most cases, it resolves spontaneously within 3 months. However, approximately 20% of the severe cases are still subject to fatal complications and 20% to 30% of patients who escape from early death develop AMKL referred to as myeloid leukemia of DS (ML-DS) within 4 years.1–4
Recent studies found that high white blood cell (WBC) count, failure of spontaneous remission, early gestational age (EGA) and liver fibrosis or liver dysfunction are significantly associated with early death.5–7 Most of the same covariates were found in all of the reports. However, the risk factors for the progression to ML-DS remain elusive.
Blast cells in most patients with TAM and ML-DS have mutations in exon 2 of the gene coding the transcription factor GATA1,8–14 which is essential for normal development of erythroid and megakaryocytic cells.15–18 The mutations lead to exclusive expression of a truncated GATA1 protein (referred to as GATA1s) translated from the second methionine on exon 3. These findings strongly suggest that the qualitative deficit of GATA1 contributes to the genesis of TAM and ML-DS. The analysis of megakaryocyte-specific knockdown of GATA1 in vivo has revealed a critical role for this factor in megakaryocytic development. Reduced expression (or complete absence) of GATA1 in megakaryocytes leads to increased proliferation and deficient maturation as well as a reduced number of circulating platelets.19,20 Mice harboring a heterozygous GATA1 knockdown allele frequently develop erythroblastic leukemia.21 These observations indicate that the expression levels of GATA1 are crucial for the proper development of erythroid and megakaryocytic cells and compromised GATA1 expression is a causal factor in leukemia.22 Nevertheless, the impact of a quantitative deficit of the factor on the pathogenesis of TAM and ML-DS has not been examined.
In this study, we classified the GATA1 mutations observed in TAM patients according to the types of transcripts, and investigated the modalities of gene expression by in vitro transfection assays using GATA1 expression constructs. We report here that the spectrum of the transcripts derived from the mutant genes affects protein expression and the risk of progression from TAM to ML-DS.
Methods
Patients
This study was approved by the Ethics Committee of Hirosaki University Graduate School of Medicine, and all clinical samples were obtained with informed consent from the parents of all patients with TAM, in accordance with the Declaration of Helsinki. The following clinical data were collected: sex, gestational age, birth weight, time of diagnosis, symptom at diagnosis, and clinical presentation. The following laboratory data were obtained: a complete blood cell count at diagnosis including WBC and the percentage of blasts in the peripheral blood, coagulation parameters, liver enzymes (alanine aminotransferase and aspartate aminotransferase), and total bilirubin. The procedure for the detection of GATA1 mutations was described previously.13 Genomic DNA was directly extracted from peripheral blood or bone marrow with the QIAamp blood mini kit (QIAGEN). Total RNA was extracted from white blood cells prepared by removal of erythrocytes by hypotonic buffer treatment of peripheral blood. Clinical features, outcomes, and characteristics of GATA1 mutations are indicated in Table 1.
Clinical features and mutation characteristics in TAM patients with GATA1 mutations
| Patient No. . | Sex . | WBC, ×109/L . | Outcome . | GATA1 mutation* . | Consequence of mutation . | Mutation type . |
|---|---|---|---|---|---|---|
| 113,24 | F | 63.9 | CR | 207 C>G | Tyr69stop | PTC 1-3′ |
| 213 | F | 89.0 | Early death | 199 G>T | Glu67stop | PTC 1-3′ |
| 313 | F | NA | NA | 174 ins 19 bp CAGCCACCGCTGCAGCTGC | Frame shift at codon58, stop at codon 73 | PTC 1-3′ |
| 413 | F | 128.8 | CR | IVS1 to IVS2 del 1415 bp | Splice mutant | Splicing error |
| 513 | F | NA | NA | 49 C>T | Gln17stop | PTC 1-5′ |
| 613 | F | 248.6 | NA | Loss of 2nd exon | Splice mutant | Splicing error |
| 713 | F | 31.2 | CR | Loss of 2nd exon | Splice mutant | Splicing error |
| 813 | M | 199.6 | CR | −11 to +33 del 44 bp | No translation from Met1 | Loss of 1st Met |
| 913 | M | 44.9 | Early death | 45 ins C | Frame shift at codon15, stop at codon 39 | PTC 1-5′ |
| 1013 | M | 50.9 | CR | 37 G>T | Glu13stop | PTC 1-5′ |
| 1113 | F | 103.0 | Early death | 90-91 del AG | Frame shift at codon 30, stop at codon 38 | PTC 1-5′ |
| 1213 | F | 14.6 | Evolved to ML-DS | 116 del A | Frame shift at codon 39, stop at codon 136 | PTC 2 |
| 1313 | M | 423.0 | CR | 185 ins 22 bp GCTGCAGCTGCGGCACTGGCCT | Frame shift at codon 62, stop at codon 74 | PTC 1-3′ |
| 1413 | M | 201.2 | CR | 189 C>A | Tyr63stop | PTC 1-3′ |
| 1513 | M | NA | NA | 1 A>G | No translation from Met1 | Loss of 1st Met |
| 1613 | F | 28.3 | CR | 189 C>A | Tyr63stop | PTC 1-3′ |
| 1713 | M | 203.0 | Evolved to ML-DS | 38-39 del AG | Frame shift at codon 13, stop at codon 38 | PTC 1-5′ |
| 1813 | M | 31.3 | CR | 189 C>A | Tyr63stop | PTC 1-3′ |
| 1913 | M | NA | NA | 90-91 del AG | Frame shift at codon 30, stop at codon 38 | PTC 1-5′ |
| 2013 | F | 114.0 | Early death | 187 ins T | Frame shift at codon 63, stop at codon 67 | PTC 1-3′ |
| 2125 | F | 26.0 | Evolved to ML-DS | 194 ins 20 bp GGCACTGGCCTACTACAGGG | Frame shift at codon 65, stop at codon 143 | PTC 2 |
| 2225 | F | 25.0 | Evolved to ML-DS | 194 ins 20 bp GGCACTGGCCTACTACAGGG | Frame shift at codon 65, stop at codon 143 | PTC 2 |
| 23 | F | 49.9 | CR | 3 G>T | No translation from Met1 | Loss of 1st Met |
| 24 | F | 46.2 | NA | IVS1 3′ boundary AG>AA | Splice mutant | Splicing error |
| 25 | F | 10.5 | CR | 194 ins 19 bp GCACTGGCCTACTACAGGG | Frame shift at codon 65, stop at codon 73 | PTC 1-3′ |
| 2624 | F | 244.0 | Evolved to ML-DS | 1 A>G | No translation from Met1 | Loss of 1st Met |
| 27 | F | 38.3 | CR | Loss of 2nd Exon | Splice mutant | Splicing error |
| 2824 | F | 34.6 | CR | IVS1 to exon2 del 148 bp | Splice mutant | Splicing error |
| 29 | M | 25.9 | Evolved to ML-DS | 160 ins TC | Frame shift at codon 54, stop at codon 137 | PTC 2 |
| 30 | F | 52.3 | Evolved to ML-DS | 187 ins CCTAC | Frame shift at codon 63, stop at codon 138 | PTC 2 |
| 3124 | F | 221.0 | CR | 183-193 del 11 bp CTACTACAGGG | Frame shift at codon 62, stop at codon 63 | PTC 1-3′ |
| 32 | M | 149.7 | CR | 2 T>G | No translation from Met1 | Loss of 1st Met |
| 3324 | M | 132.3 | Evolved to ML-DS | 101-108 del 8 bp TCCCCTCT | Frame shift at codon 34, stop at codon 36 | PTC 1-5′ |
| 3424 | F | 220.0 | Early death | 90-91 del AG | Frame shift at codon 30, stop at codon 38 | PTC 1-5′ |
| 3524 | M | 166.0 | Early death | IVS2 5′ boundary GT>CT | Splice mutant | Splicing error |
| 3624 | M | 57.6 | Early death | 193-199 GACGCTG>TAGTAGT | Asp65stop | PTC 1-3′ |
| 3724 | M | 247.6 | Early death | Exon2 to IVS2 del 218 bp | Splice mutant | Splicing error |
| 3824 | M | 93.3 | Early death | IVS1 3′ boundary AG>AA | Splice mutant | Splicing error |
| 3924 | M | 290.8 | Early death | 186 ins 12 bp GGCACTGGCCTA | Tyr62stop | PTC 1-3′ |
| 40 | F | 7.8 | CR | 2 T>C | No translation from Met1 | Loss of 1st Met |
| 4124 | M | 136.6 | Early death | IVS2 5′ boundary GT>GC | Splice mutant | Splicing error |
| 42 | M | 33.1 | Early death | 187 ins 8 bp TGGCCTAC | Frame shift at codon 63, stop at codon 139 | PTC 2 |
| 43 | M | 9.0 | CR | 22 ins G | Frame shift at codon 8, stop at codon 39 | PTC 1-5′ |
| 44 | M | 24.1 | Evolved to ML-DS | 149 ins 20 bp AGCAGCTTCCTCCACTGCCC | Frame shift at codon 50, stop at codon 143 | PTC 2 |
| 4524 | F | 53.3 | CR | 173 C>TGCTGCAGTGTAGTA | Frame shift at codon 58, stop at codon 141 | PTC 2 |
| 46 | F | 119.0 | CR | 1 A>C | No translation from Met1 | Loss of 1st Met |
| 47 | M | 33.0 | CR | 189 C>A | Tyr63stop | PTC 1-3′ |
| 48 | M | 178.2 | Early death | 188 ins 22 bp GCAGCTGCGGCACTGGCCTACT | Frame shift at codon 63, stop at codon 74 | PTC 1-3′ |
| 49 | F | 73.6 | CR | 3 G>A | No translation from Met1 | Loss of 1st Met |
| 50 | F | 12.9 | CR | 158 ins 7 bp AGCACAG | Frame shift at codon 53, stop at codon 69 | PTC 1-5′ |
| 51 | M | 13.0 | CR | 154-161 del 8 bp ACAGCCAC | Frame shift at codon 52, stop at codon 64 | PTC 1-5′ |
| 52 | M | 105.5 | Early death | 4 G>T | Glu2stop | PTC 1-5′ |
| 53 | F | 98.3 | CR | 4 G>T | Glu2stop | PTC 1-5′ |
| 54 | F | 356.9 | CR | 219 A>C | Splice mutant | Splicing error |
| 55 | F | 25.8 | Evolved to ML-DS | 157 ins CA | Frame shift at codon 53, stop at codon 137 | PTC 2 |
| 56 | M | 97.4 | Evolved to ML-DS | 185-188 del 4 bp ACTA | Frame shift at codon 62, stop at codon 135 | PTC 2 |
| 57 | F | 97.3 | Early death | 3 G>A | No translation from Met1 | Loss of 1st Met |
| 58 | M | NA | CR | 3 G>A | No translation from Met1 | Loss of 1st Met |
| 59 | M | 20.2 | CR | 150 ins 5 bp TGGCT | Frame shift at codon 50, stop at codon 52 | PTC 1-5′ |
| 60 | M | 133.4 | CR | 174 ins 19 bp CAAAGCAGCTGCAGCGGTG | Frame shift at codon 58, stop at codon 73 | PTC 1-3′ |
| 61 | M | NA | CR | 220 G>T | Splice mutant | Splicing error |
| 62 | M | 120.2 | CR | 220 G>A | Splice mutant | Splicing error |
| 63 | F | 39.0 | CR | 97-139 del 43 bp | Frame shift at codon 33, stop at codon 122 | PTC 2 |
| 64 | F | NA | NA | 156 ins C | Frame shift at codon 52, stop at codon 67 | PTC 1-5′ |
| 65 | F | 32.4 | CR | 174 ins 7 bp CTGCAGC | Frame shift at codon 58, stop at codon 69 | PTC 1-3′ |
| 66 | M | 69.4 | Early death | 174-177 GGCA>TGCGGTGG | Frame shift at codon 58, stop at codon 68 | PTC 1-3′ |
| Patient No. . | Sex . | WBC, ×109/L . | Outcome . | GATA1 mutation* . | Consequence of mutation . | Mutation type . |
|---|---|---|---|---|---|---|
| 113,24 | F | 63.9 | CR | 207 C>G | Tyr69stop | PTC 1-3′ |
| 213 | F | 89.0 | Early death | 199 G>T | Glu67stop | PTC 1-3′ |
| 313 | F | NA | NA | 174 ins 19 bp CAGCCACCGCTGCAGCTGC | Frame shift at codon58, stop at codon 73 | PTC 1-3′ |
| 413 | F | 128.8 | CR | IVS1 to IVS2 del 1415 bp | Splice mutant | Splicing error |
| 513 | F | NA | NA | 49 C>T | Gln17stop | PTC 1-5′ |
| 613 | F | 248.6 | NA | Loss of 2nd exon | Splice mutant | Splicing error |
| 713 | F | 31.2 | CR | Loss of 2nd exon | Splice mutant | Splicing error |
| 813 | M | 199.6 | CR | −11 to +33 del 44 bp | No translation from Met1 | Loss of 1st Met |
| 913 | M | 44.9 | Early death | 45 ins C | Frame shift at codon15, stop at codon 39 | PTC 1-5′ |
| 1013 | M | 50.9 | CR | 37 G>T | Glu13stop | PTC 1-5′ |
| 1113 | F | 103.0 | Early death | 90-91 del AG | Frame shift at codon 30, stop at codon 38 | PTC 1-5′ |
| 1213 | F | 14.6 | Evolved to ML-DS | 116 del A | Frame shift at codon 39, stop at codon 136 | PTC 2 |
| 1313 | M | 423.0 | CR | 185 ins 22 bp GCTGCAGCTGCGGCACTGGCCT | Frame shift at codon 62, stop at codon 74 | PTC 1-3′ |
| 1413 | M | 201.2 | CR | 189 C>A | Tyr63stop | PTC 1-3′ |
| 1513 | M | NA | NA | 1 A>G | No translation from Met1 | Loss of 1st Met |
| 1613 | F | 28.3 | CR | 189 C>A | Tyr63stop | PTC 1-3′ |
| 1713 | M | 203.0 | Evolved to ML-DS | 38-39 del AG | Frame shift at codon 13, stop at codon 38 | PTC 1-5′ |
| 1813 | M | 31.3 | CR | 189 C>A | Tyr63stop | PTC 1-3′ |
| 1913 | M | NA | NA | 90-91 del AG | Frame shift at codon 30, stop at codon 38 | PTC 1-5′ |
| 2013 | F | 114.0 | Early death | 187 ins T | Frame shift at codon 63, stop at codon 67 | PTC 1-3′ |
| 2125 | F | 26.0 | Evolved to ML-DS | 194 ins 20 bp GGCACTGGCCTACTACAGGG | Frame shift at codon 65, stop at codon 143 | PTC 2 |
| 2225 | F | 25.0 | Evolved to ML-DS | 194 ins 20 bp GGCACTGGCCTACTACAGGG | Frame shift at codon 65, stop at codon 143 | PTC 2 |
| 23 | F | 49.9 | CR | 3 G>T | No translation from Met1 | Loss of 1st Met |
| 24 | F | 46.2 | NA | IVS1 3′ boundary AG>AA | Splice mutant | Splicing error |
| 25 | F | 10.5 | CR | 194 ins 19 bp GCACTGGCCTACTACAGGG | Frame shift at codon 65, stop at codon 73 | PTC 1-3′ |
| 2624 | F | 244.0 | Evolved to ML-DS | 1 A>G | No translation from Met1 | Loss of 1st Met |
| 27 | F | 38.3 | CR | Loss of 2nd Exon | Splice mutant | Splicing error |
| 2824 | F | 34.6 | CR | IVS1 to exon2 del 148 bp | Splice mutant | Splicing error |
| 29 | M | 25.9 | Evolved to ML-DS | 160 ins TC | Frame shift at codon 54, stop at codon 137 | PTC 2 |
| 30 | F | 52.3 | Evolved to ML-DS | 187 ins CCTAC | Frame shift at codon 63, stop at codon 138 | PTC 2 |
| 3124 | F | 221.0 | CR | 183-193 del 11 bp CTACTACAGGG | Frame shift at codon 62, stop at codon 63 | PTC 1-3′ |
| 32 | M | 149.7 | CR | 2 T>G | No translation from Met1 | Loss of 1st Met |
| 3324 | M | 132.3 | Evolved to ML-DS | 101-108 del 8 bp TCCCCTCT | Frame shift at codon 34, stop at codon 36 | PTC 1-5′ |
| 3424 | F | 220.0 | Early death | 90-91 del AG | Frame shift at codon 30, stop at codon 38 | PTC 1-5′ |
| 3524 | M | 166.0 | Early death | IVS2 5′ boundary GT>CT | Splice mutant | Splicing error |
| 3624 | M | 57.6 | Early death | 193-199 GACGCTG>TAGTAGT | Asp65stop | PTC 1-3′ |
| 3724 | M | 247.6 | Early death | Exon2 to IVS2 del 218 bp | Splice mutant | Splicing error |
| 3824 | M | 93.3 | Early death | IVS1 3′ boundary AG>AA | Splice mutant | Splicing error |
| 3924 | M | 290.8 | Early death | 186 ins 12 bp GGCACTGGCCTA | Tyr62stop | PTC 1-3′ |
| 40 | F | 7.8 | CR | 2 T>C | No translation from Met1 | Loss of 1st Met |
| 4124 | M | 136.6 | Early death | IVS2 5′ boundary GT>GC | Splice mutant | Splicing error |
| 42 | M | 33.1 | Early death | 187 ins 8 bp TGGCCTAC | Frame shift at codon 63, stop at codon 139 | PTC 2 |
| 43 | M | 9.0 | CR | 22 ins G | Frame shift at codon 8, stop at codon 39 | PTC 1-5′ |
| 44 | M | 24.1 | Evolved to ML-DS | 149 ins 20 bp AGCAGCTTCCTCCACTGCCC | Frame shift at codon 50, stop at codon 143 | PTC 2 |
| 4524 | F | 53.3 | CR | 173 C>TGCTGCAGTGTAGTA | Frame shift at codon 58, stop at codon 141 | PTC 2 |
| 46 | F | 119.0 | CR | 1 A>C | No translation from Met1 | Loss of 1st Met |
| 47 | M | 33.0 | CR | 189 C>A | Tyr63stop | PTC 1-3′ |
| 48 | M | 178.2 | Early death | 188 ins 22 bp GCAGCTGCGGCACTGGCCTACT | Frame shift at codon 63, stop at codon 74 | PTC 1-3′ |
| 49 | F | 73.6 | CR | 3 G>A | No translation from Met1 | Loss of 1st Met |
| 50 | F | 12.9 | CR | 158 ins 7 bp AGCACAG | Frame shift at codon 53, stop at codon 69 | PTC 1-5′ |
| 51 | M | 13.0 | CR | 154-161 del 8 bp ACAGCCAC | Frame shift at codon 52, stop at codon 64 | PTC 1-5′ |
| 52 | M | 105.5 | Early death | 4 G>T | Glu2stop | PTC 1-5′ |
| 53 | F | 98.3 | CR | 4 G>T | Glu2stop | PTC 1-5′ |
| 54 | F | 356.9 | CR | 219 A>C | Splice mutant | Splicing error |
| 55 | F | 25.8 | Evolved to ML-DS | 157 ins CA | Frame shift at codon 53, stop at codon 137 | PTC 2 |
| 56 | M | 97.4 | Evolved to ML-DS | 185-188 del 4 bp ACTA | Frame shift at codon 62, stop at codon 135 | PTC 2 |
| 57 | F | 97.3 | Early death | 3 G>A | No translation from Met1 | Loss of 1st Met |
| 58 | M | NA | CR | 3 G>A | No translation from Met1 | Loss of 1st Met |
| 59 | M | 20.2 | CR | 150 ins 5 bp TGGCT | Frame shift at codon 50, stop at codon 52 | PTC 1-5′ |
| 60 | M | 133.4 | CR | 174 ins 19 bp CAAAGCAGCTGCAGCGGTG | Frame shift at codon 58, stop at codon 73 | PTC 1-3′ |
| 61 | M | NA | CR | 220 G>T | Splice mutant | Splicing error |
| 62 | M | 120.2 | CR | 220 G>A | Splice mutant | Splicing error |
| 63 | F | 39.0 | CR | 97-139 del 43 bp | Frame shift at codon 33, stop at codon 122 | PTC 2 |
| 64 | F | NA | NA | 156 ins C | Frame shift at codon 52, stop at codon 67 | PTC 1-5′ |
| 65 | F | 32.4 | CR | 174 ins 7 bp CTGCAGC | Frame shift at codon 58, stop at codon 69 | PTC 1-3′ |
| 66 | M | 69.4 | Early death | 174-177 GGCA>TGCGGTGG | Frame shift at codon 58, stop at codon 68 | PTC 1-3′ |
We previously reported the GATA1 mutations of the indicated patients.
F indicates female; M, male; CR, complete remission; NA, not available; and IVS, intervening sequence.
For cDNA nucleotide numbering, nucleotide number 1 corresponds to the A of the ATG translation initiation codon in the reference sequence.
Construction of GATA1 expression vectors
To construct GATA1 minigene expression vectors, fragments of the normal human GATA1 gene from a part of intron 1 to the stop codon located on exon 6 were amplified by polymerase chain reaction (PCR; Prime STAR HS: Takara Bio) and subcloned to mammalian expression vector pcDNA3.1 (+)/Neo (Invitrogen). To introduce mutations identical to those observed in TAM patients into the expression vector, the regions containing mutations were amplified by PCR from patient samples and inserted into the expression plasmid. To construct expression vectors carrying cDNA, we performed PCR using cDNA derived from baby hamster kidney 21 (BHK-21) cells transfected with GATA1 minigene vectors. The PCR products were subcloned to pcDNA3.1(+)/Neo. Details of the sequence of each expression construct are described in Table 2.
GATA1 expression vectors used in this study
| Name . | Patient no. . | GATA1 mutation* . | Last normal GATA1 amino acid . | PTC . | Mutation type . |
|---|---|---|---|---|---|
| WG | – | – | Ser413 | – | Normal |
| SP1 | 24, 38 | intron1 3′ boundary AG>AA | Ser413 | – | Splicing error |
| SP2 | 41 | intron2 5′ boundary GT>GC | Ser413 | – | Splicing error |
| L | 46 | 1 A>C | (Met1 is replaced by Val1) | – | Loss of 1st Met |
| P1-1 | 11, 19, 34 | 90, 91 del AG | Gly31 | 38 | PTC 1-5′ |
| P1-2 | 14, 16, 18, 47 | 189 C>A | Tyr62 | 63 | PTC 1-3′ |
| P1-3 | 25 | 194 ins 19 bp | Arg64 | 73 | PTC 1-3′ |
| P1-4 | 17 | 38, 39 del AG | Ser12 | 38 | PTC 1-5′ |
| P1-5 | 33 | 101-108 del 8 bp | Phe33 | 36 | PTC 1-5′ |
| P1-6 | 50 | 158 ins 7 bp | Tyr52 | 69 | PTC 1-5′ |
| P1-7 | 3 | 174 ins 19 bp | Ala58 | 73 | PTC 1-3′ |
| P1-8 | 48 | 188 ins 22 bp | Try62 | 74 | PTC 1-3′ |
| P2-1 | 21, 22 | 194 ins 20 bp | Arg64 | 143 | PTC 2 |
| P2-2 | 44 | 149 ins 20 bp | Ala49 | 143 | PTC 2 |
| P2-3 | 29 | 160 ins TC | Ala53 | 137 | PTC 2 |
| Name . | Patient no. . | GATA1 mutation* . | Last normal GATA1 amino acid . | PTC . | Mutation type . |
|---|---|---|---|---|---|
| WG | – | – | Ser413 | – | Normal |
| SP1 | 24, 38 | intron1 3′ boundary AG>AA | Ser413 | – | Splicing error |
| SP2 | 41 | intron2 5′ boundary GT>GC | Ser413 | – | Splicing error |
| L | 46 | 1 A>C | (Met1 is replaced by Val1) | – | Loss of 1st Met |
| P1-1 | 11, 19, 34 | 90, 91 del AG | Gly31 | 38 | PTC 1-5′ |
| P1-2 | 14, 16, 18, 47 | 189 C>A | Tyr62 | 63 | PTC 1-3′ |
| P1-3 | 25 | 194 ins 19 bp | Arg64 | 73 | PTC 1-3′ |
| P1-4 | 17 | 38, 39 del AG | Ser12 | 38 | PTC 1-5′ |
| P1-5 | 33 | 101-108 del 8 bp | Phe33 | 36 | PTC 1-5′ |
| P1-6 | 50 | 158 ins 7 bp | Tyr52 | 69 | PTC 1-5′ |
| P1-7 | 3 | 174 ins 19 bp | Ala58 | 73 | PTC 1-3′ |
| P1-8 | 48 | 188 ins 22 bp | Try62 | 74 | PTC 1-3′ |
| P2-1 | 21, 22 | 194 ins 20 bp | Arg64 | 143 | PTC 2 |
| P2-2 | 44 | 149 ins 20 bp | Ala49 | 143 | PTC 2 |
| P2-3 | 29 | 160 ins TC | Ala53 | 137 | PTC 2 |
– indicates not applicable.
For cDNA nucleotide numbering, nucleotide number 1 corresponds to the A of the ATG translation initiation codon in the reference sequence.
Transfection
BHK-21, a baby hamster kidney fibroblast cell line, was cultured with Dulbecco modified Eagle medium supplemented 10% fetal bovine serum. GATA1 expression vectors were transfected into BHK-21 cells using FuGENE HD transfection reagent (Roche Diagnostics) according to the manufacturer's methods. After 24 hours, protein and total RNA were extracted.
Western blot analysis
Lysates of transfected BHK-21 cells were transferred to Hybond-P (GE Healthcare) and processed for reaction with anti-GATA1 antibody M-20 (Santa Cruz Biotechnology) or anti-neomycin phosphotransferase II (NeoR) antibody (Millipore) as described previously.23
Northern blot analysis
Two micrograms of total RNA were transferred to Hybond-N+ (GE Healthcare) and hybridized with GATA1 or NeoR DNA probe. Hybridization and detection were performed with the Gene Images AlkPhos Direct Labeling and Detection System (GE Healthcare) according to the manufacturer's instructions.
RT-PCR
To detect alternatively spliced transcripts derived from GATA1 minigene constructs or from patients' peripheral blood mononuclear cells (obtained by Ficoll-Hypaque fractionation), we performed reverse transcription (RT)–PCR using primers T7: 5′ AATACGACTCACTATAG 3′ and GATA1 AS1, and GATA1 S1 and GATA1 AS1, respectively.13 Densitometric analyses were performed by the Quantity-One software (Version 4.5.2; Bio-Rad Laboratories).
Statistical analysis
The cumulative incidence of the progression to ML-DS was analyzed with the Gray test. Differences in the distribution of individual parameters among patient subsets were analyzed using the Pearson χ2 test or Fisher exact test for categorized variables and the Mann-Whitney U test for continuous variables. The univariate Cox proportional hazards model was used to obtain the estimates and the 95% confidence interval of the relative risk for prognostic factors.
Results
Patient characteristics and outcomes
From 2003 to 2008, we screened GATA1 mutations in clinical samples obtained from 78 patients with TAM upon request from referring hospitals. Acquired GATA1 mutations were detected in a total of 72 (92.3%) patients among them. Of the 72 patients, 6 harbored multiple GATA1 mutant clones and were excluded from this study because we could not determine a dominant clone in these patients. Those 6 have not progressed to ML-DS. For the remaining 66 patients (32 male and 34 female), the clinical characteristics and laboratory data at diagnosis are described in Table 1 and summarized in Table 3. Early death within the first 6 months of life occurred in 16 patients (24.2%). The covariates correlated with early death were as follows: EGA, low birth weight, high WBC count at diagnosis, high percentage of peripheral blast cells, complication of effusions, and bleeding diatheses. These prognostic factors were identified in previous studies.5–7 Eleven (16.7%) cases subsequently developed ML-DS. The median age at diagnosis of ML-DS was 396 days (range 221-747 days). Univariate analysis revealed no covariates correlated with progression to ML-DS except the low total bilirubin level at diagnosis (P = .023).
Findings at diagnosis and during the course of TAM were significantly associated with early death and the progression to leukemia (univariate analysis)
| Variable . | Total (n = 66) . | Early death (n = 16) . | P . | Progressed to ML-DS (n = 11) . | P . |
|---|---|---|---|---|---|
| Sex | |||||
| Male, n (%) | 32 (48.5) | 11 (68.8) | 5 (45.5) | ||
| Female, n (%) | 34 (51.5) | 5 (31.3) | .088 | 6 (54.5) | .947 |
| Median gestational age, wk (range) | 37.35 (30.0-40.6) | 34.6 (30.0-38.4) | 38.1 (32.6-40.6) | ||
| Term versus preterm | |||||
| Term (≥ 37 weeks), n (%) | 27 (58.7) | 4 (30.8) | 5 (71.4) | ||
| Preterm (< 37 weeks), n (%) | 19 (41.3) | 9 (69.2) | .021 | 2 (28.6) | .465 |
| Median birth weight, kg (range) | 2.5 (1.4-3.5) | 2.2 (1.6-2.7) | 2.5 (1.6-3.5) | ||
| Not LBW versus LBW | |||||
| Not LBW (≥ 2.5 kg), n (%) | 24 (52.2) | 3 (23.1) | 3 (42.9) | ||
| LBW (< 2.5 kg), n (%) | 22 (47.8) | 10 (76.9) | .025 | 4 (57.1) | .184 |
| Median WBC, ×109/L (range) | 69.4 (7.8-423.0) | 104.3 (33.1-290.8) | 26 (14.6-244.0) | ||
| WBC < 70 × 109/L vs WBC > 70 × 109/L | |||||
| WBC < 70 × 109/L, n (%) | 30 (50.8) | 4 (25.0) | 7 (63.6) | ||
| WBC > 70 × 109/L, n (%) | 29 (49.2) | 12 (75.0) | .020 | 4 (36.4) | .755 |
| Median peripheral blasts, % (range) | 56.0 (4.0-94.0) | 78.0 (8.0-93.0) | .031 | 49.5 (6.0-66.0) | .752 |
| Median AST, IU/L (range) | 61 (16-4341) | 79 (41-3866) | .620 | 51 (16-153) | .553 |
| Median ALT, IU/L (range) | 39 (4-653) | 41 (7-473) | .455 | 12 (4-96) | .615 |
| Median T-Bil mg/dL (range) | 6.3 (0.6-46.0) | 6.06 (2.4-16.5) | .922 | 3.01 (1.82-6.50) | .023 |
| Effusions, n (%) | 16 of 44 (36.4) | 8 of 11 (72.7) | .007 | 1 of 7 (14.3) | .912 |
| Bleeding diatheses, n (%) | 13 of 45 (28.9) | 8 of 12 (66.7) | .001 | 1 of 7 (14.3) | .123 |
| Variable . | Total (n = 66) . | Early death (n = 16) . | P . | Progressed to ML-DS (n = 11) . | P . |
|---|---|---|---|---|---|
| Sex | |||||
| Male, n (%) | 32 (48.5) | 11 (68.8) | 5 (45.5) | ||
| Female, n (%) | 34 (51.5) | 5 (31.3) | .088 | 6 (54.5) | .947 |
| Median gestational age, wk (range) | 37.35 (30.0-40.6) | 34.6 (30.0-38.4) | 38.1 (32.6-40.6) | ||
| Term versus preterm | |||||
| Term (≥ 37 weeks), n (%) | 27 (58.7) | 4 (30.8) | 5 (71.4) | ||
| Preterm (< 37 weeks), n (%) | 19 (41.3) | 9 (69.2) | .021 | 2 (28.6) | .465 |
| Median birth weight, kg (range) | 2.5 (1.4-3.5) | 2.2 (1.6-2.7) | 2.5 (1.6-3.5) | ||
| Not LBW versus LBW | |||||
| Not LBW (≥ 2.5 kg), n (%) | 24 (52.2) | 3 (23.1) | 3 (42.9) | ||
| LBW (< 2.5 kg), n (%) | 22 (47.8) | 10 (76.9) | .025 | 4 (57.1) | .184 |
| Median WBC, ×109/L (range) | 69.4 (7.8-423.0) | 104.3 (33.1-290.8) | 26 (14.6-244.0) | ||
| WBC < 70 × 109/L vs WBC > 70 × 109/L | |||||
| WBC < 70 × 109/L, n (%) | 30 (50.8) | 4 (25.0) | 7 (63.6) | ||
| WBC > 70 × 109/L, n (%) | 29 (49.2) | 12 (75.0) | .020 | 4 (36.4) | .755 |
| Median peripheral blasts, % (range) | 56.0 (4.0-94.0) | 78.0 (8.0-93.0) | .031 | 49.5 (6.0-66.0) | .752 |
| Median AST, IU/L (range) | 61 (16-4341) | 79 (41-3866) | .620 | 51 (16-153) | .553 |
| Median ALT, IU/L (range) | 39 (4-653) | 41 (7-473) | .455 | 12 (4-96) | .615 |
| Median T-Bil mg/dL (range) | 6.3 (0.6-46.0) | 6.06 (2.4-16.5) | .922 | 3.01 (1.82-6.50) | .023 |
| Effusions, n (%) | 16 of 44 (36.4) | 8 of 11 (72.7) | .007 | 1 of 7 (14.3) | .912 |
| Bleeding diatheses, n (%) | 13 of 45 (28.9) | 8 of 12 (66.7) | .001 | 1 of 7 (14.3) | .123 |
Some clinical data were not available. We defined the number of patients for whom clinical data was available as (n).
LBW indicates low birth weight; AST, aspartate transaminase; ALT, alanine transaminase; and T-Bil, total bilirubin.
GATA1 mutations affect expression levels of GATA1s protein
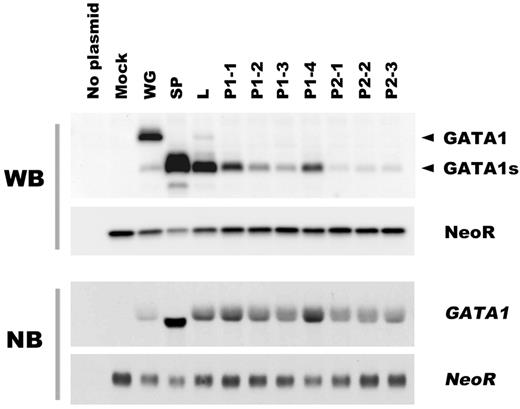
We first asked whether the spectrum of transcripts derived from the mutant GATA1 genes affected the expression levels of the translation products. The transcripts coding GATA1s protein were categorized into 3 groups as follows: loss of the first methionine, splicing errors, and premature termination codon (PTC). Furthermore, the PTC group was divided into 2 subcategories by the location of introduced PTC. In this report, we refer to the mutation that causes PTC before the second methionine at codon 84 as PTC type 1, and after codon 84 as PTC type 2. We constructed cDNA expression vectors for each class of mutations observed in TAM patients, and transfected these constructs into BHK-21 cells (Figure 1). The details of the GATA1 mutations are described in Table 2. Western blot analysis revealed that GATA1s proteins were most abundantly expressed in mutants with splicing errors. The transcripts from mutants that had lost the first methionine were also efficiently translated. In contrast, in the cells expressing PTC type 1 or type 2 constructs, GATA1s expression levels were uniformly low. Note that the translation efficiency of the PTC type 2 transcripts was lowest among them.
Effects of mutant transcripts of GATA1 on the expression level of the truncated protein. The GATA1 mutations observed in TAM patients are classified according to the types of transcripts. The translational efficiency of each transcript was assessed by Western blot analysis in BHK-21 cells transfected with GATA1 cDNA expression vectors (top part of the panel) and Northern blot analysis (bottom part of the panel), respectively. WG indicates wild type GATA1; SP, splicing error mutation (Δexon 2); L, loss of first methionine mutation; P1, PTC type 1 mutation; P2, PTC type 2 mutation. The details of the GATA1 mutations are summarized in Table 1. NeoR indicates Neomycin phosphotransferase II.
Effects of mutant transcripts of GATA1 on the expression level of the truncated protein. The GATA1 mutations observed in TAM patients are classified according to the types of transcripts. The translational efficiency of each transcript was assessed by Western blot analysis in BHK-21 cells transfected with GATA1 cDNA expression vectors (top part of the panel) and Northern blot analysis (bottom part of the panel), respectively. WG indicates wild type GATA1; SP, splicing error mutation (Δexon 2); L, loss of first methionine mutation; P1, PTC type 1 mutation; P2, PTC type 2 mutation. The details of the GATA1 mutations are summarized in Table 1. NeoR indicates Neomycin phosphotransferase II.
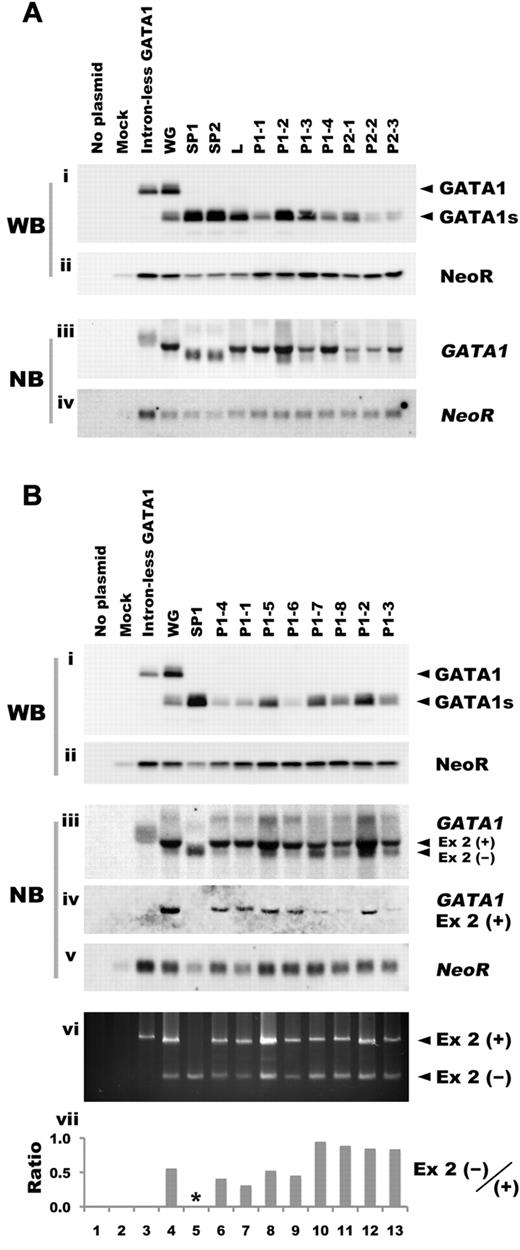
To test the possibility that mutations in GATA1 have an effect on the quantity of the transcripts, we next prepared human GATA1 minigene expression vectors, and assessed the expression levels. Consistent with the results using cDNA expression vectors, Western blot analysis showed that the expression levels of GATA1s were lower in cells expressing PTC type 2 mutations, whereas the expression levels of the proteins from PTC type 1 mutations were not uniformly low (Figure 2Ai). Northern blot analysis revealed that the lowest expression levels of GATA1 mRNAs were observed in cells transfected with PTC type 2 constructs, whereas the mRNA levels in mutants that had lost the first methionine and PTC type 1 mutants were almost comparable to those of control minigene constructs harboring wild type GATA1 gene (Figure 2Aiii). Thus, abundant proteins were produced from GATA1 mRNAs in mutants with splicing errors and those that lost the first methionine. Conversely, relatively low levels of protein were produced by PTC type 2 mutants because of inefficient translation and reduced levels of message (Figure 2Ai,iii). However, in the case of PTC type 1 mutations, especially P1-1 and P1-4, we could find no correlation between the amount of transcripts or translation efficiency and the expression levels of GATA1s proteins (Figure 2Ai,iii).
GATA1 mutations affect the expression level of the truncated protein. (A) The expression levels of GATA1s protein and mRNA were assessed in BHK-21 cells transfected with human GATA1 minigene expression vectors carrying mutations observed in TAM patients. Western blot analysis was performed with anti-GATA1 (i) or anti-NeoR antibody (ii). Northern blot analysis was carried out with GATA1 exon 3-6 fragment (iii) or NeoR cDNA (iv) as probe. (B) The expression levels of GATA1s protein and mRNA in BHK-21 cells transfected with human GATA1 minigene expression vectors with PTC type 1 mutation. Levels were assessed by Western blot analysis with anti-GATA1 antibody (i), anti-NeoR antibody (ii). Northern blot analysis was performed with GATA1 exon 3-6 (iii), exon 2 (iv), or NeoR cDNA (v). To detect the transcripts derived from the human GATA1 minigene expression construct, RT-PCR analysis was carried out using primers described in “RT-PCR” (vi). Ex 2(+) and Ex 2(−) indicate PCR products or transcripts with or without exon 2, respectively. Ratio of Ex 2(−)/(+) was calculated from the results of a densitometric analysis of the RT-PCR. The asterisk denotes unavailable data (vii).
GATA1 mutations affect the expression level of the truncated protein. (A) The expression levels of GATA1s protein and mRNA were assessed in BHK-21 cells transfected with human GATA1 minigene expression vectors carrying mutations observed in TAM patients. Western blot analysis was performed with anti-GATA1 (i) or anti-NeoR antibody (ii). Northern blot analysis was carried out with GATA1 exon 3-6 fragment (iii) or NeoR cDNA (iv) as probe. (B) The expression levels of GATA1s protein and mRNA in BHK-21 cells transfected with human GATA1 minigene expression vectors with PTC type 1 mutation. Levels were assessed by Western blot analysis with anti-GATA1 antibody (i), anti-NeoR antibody (ii). Northern blot analysis was performed with GATA1 exon 3-6 (iii), exon 2 (iv), or NeoR cDNA (v). To detect the transcripts derived from the human GATA1 minigene expression construct, RT-PCR analysis was carried out using primers described in “RT-PCR” (vi). Ex 2(+) and Ex 2(−) indicate PCR products or transcripts with or without exon 2, respectively. Ratio of Ex 2(−)/(+) was calculated from the results of a densitometric analysis of the RT-PCR. The asterisk denotes unavailable data (vii).
GATA1s expression levels largely depend on the amount of the alternative splicing form
To investigate the precise relationship between PTC type 1 mutations and GATA1s protein levels, we examined more type 1 mutations using the minigene constructs. Western blot analysis showed relatively higher expression of the proteins in samples expressing P1-5, P1-7, P1-8, P1-2, and P1-3 than the other constructs (Figure 2Bi). Each mutation in the mutant minigene construct is described in Table 2. Interestingly, all samples that expressed higher levels of GATA1s protein exhibited intense signals at lower molecular weights than the dominant GATA1 signal (Figure 2Biii). Because the size of the lower molecular weight band was identical to that observed in the splicing error mutant (Figure 2Biii), we speculated that the signal might be derived from a transcript lacking exon 2 (Δexon 2) by alternative splicing. To examine that possibility, we attempted Northern blot analysis using the GATA1 exon 2 fragment as a probe, and as expected, only the longer transcript was detected (Figure 2Biv). To confirm the correlation between the amount of Δexon 2 transcript and GATA1s protein, we performed a quantitative assessment by densitometric analysis. The results showed a strong correlation between Δexon 2 transcript and GATA1s protein levels (r = 0.892, P = .003), but not with the long transcript containing exon2 nor total GATA1 mRNA (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Next, we performed RT-PCR using primers recognizing both transcripts, and calculated the ratio of Δexon 2 to the long transcript (Figure 2Bvi-vii). The intensive short transcript was detected in all samples with higher expression of GATA1s (P1-5, P1-7, P1-8, P1-2, and P1-3; Figure 2Bvii). Interestingly, most of these mutations were clustered in the 3′ region of exon 2 (Table 2, Figure 2Bvii). These results suggest that the location of the mutation predicts the efficiency of alternative splicing and GATA1s expression levels.
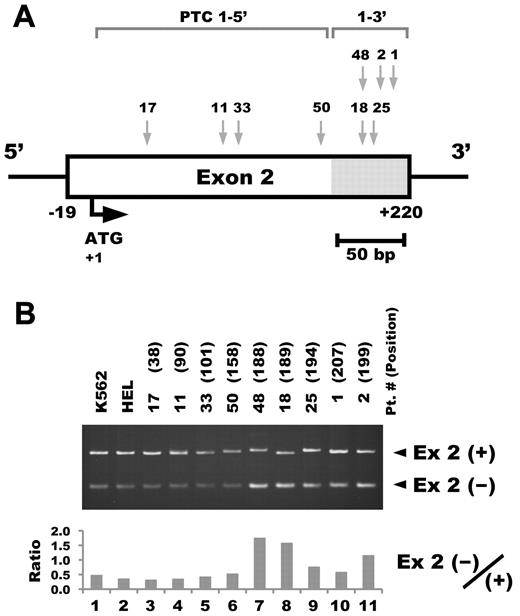
To examine whether differential splicing efficiency could also be observed in TAM blasts with PTC type 1 mutations, RT-PCR analysis was performed using patients' clinical samples. Intense transcription of the short form was observed in the samples from the patients who had GATA1 mutations located on the 3′ side of exon 2 (+169 to +218 in mRNA from the ATG translation initiation codon; Figure 3A-B). We refer to them as PTC type 1-3′ and the mutations located on the 5′ side of exon 2 as PTC type 1-5′.
The location of the PTC type 1 mutation affects the efficiency of alternative splicing in TAM blast cells. (A) The location of the GATA1 mutation in each TAM patient. Details of the mutation in each sample are described in Table 1. (B) RT-PCR analysis of GATA1 in TAM blast cells harboring PTC type 1 mutations. RT-PCR was performed using primers recognizing both the long transcript including exon 2 and Δexon 2 (top). All of the patient samples consisted of mononuclear cells from peripheral blood. The numbers in parentheses indicate the number of nucleotides in mRNA from the translation initiation codon. Ex 2(+) and Ex 2(−) indicate PCR products with or without exon 2, respectively (middle). Ratio of Ex 2(−)/(+) was calculated from the results of a densitometric analysis of the RT-PCR (bottom). Note that the intense bands of the short form were observed in the samples from the patients who have GATA1 mutations located on the 3′ side of exon 2 (lanes 7-11).
The location of the PTC type 1 mutation affects the efficiency of alternative splicing in TAM blast cells. (A) The location of the GATA1 mutation in each TAM patient. Details of the mutation in each sample are described in Table 1. (B) RT-PCR analysis of GATA1 in TAM blast cells harboring PTC type 1 mutations. RT-PCR was performed using primers recognizing both the long transcript including exon 2 and Δexon 2 (top). All of the patient samples consisted of mononuclear cells from peripheral blood. The numbers in parentheses indicate the number of nucleotides in mRNA from the translation initiation codon. Ex 2(+) and Ex 2(−) indicate PCR products with or without exon 2, respectively (middle). Ratio of Ex 2(−)/(+) was calculated from the results of a densitometric analysis of the RT-PCR (bottom). Note that the intense bands of the short form were observed in the samples from the patients who have GATA1 mutations located on the 3′ side of exon 2 (lanes 7-11).
Correlation of the phenotype and GATA1 mutations in TAM patients
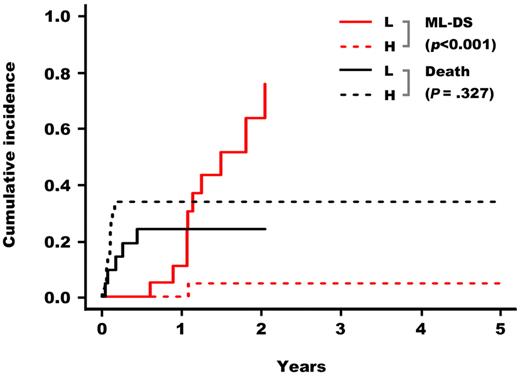
Based on these results, GATA1 mutations were classified into 2 groups: a high GATA1s expression group (GATA1s high group) including the loss of first methionine type, the splicing error type, and PTC type 1-3′, and a low GATA1s expression group (GATA1s low group) including PTC type 1-5′ and PTC type 2. We classified TAM patients into these 2 groups in accordance with the GATA1s expression levels estimated from the mutations and compared their clinical data. High counts of WBC and blast cells were significantly associated with the GATA1s high group (P = .004 and P = .008, respectively; Table 4). Although high WBC count was correlated with early death, there were no significant differences in the cumulative incidence of early death between the 2 groups (Figure 4). Importantly, TAM patients in the GATA1s low group had a significantly higher risk for the development of leukemia (P < .001; Figure 4). Of 11 TAM patients who progressed to ML-DS, 10 belonged to the GATA1s low group. Notably, 8 patients among them had PTC type 2 mutations (Tables 1, 5).
Correlations between patient covariates and GATA1 expression levels
| . | GATA1s expression group . | P . | |
|---|---|---|---|
| High (n = 40) . | Low (n = 26) . | ||
| Sex: male/female, n | 19/21 | 13/13 | .843* |
| Gestational age, wk | 37.3 (30.0-40.0) | 37.9 (32.6-40.6) | .487 |
| Birth weight, kg | 2.5 (1.6-3.3) | 2.5 (1.4-3.5) | .698 |
| WBC, ×109/L | 105.65 (7.8-423.0) | 39.0 (9.0-220.0) | .004 |
| Number of blasts, ×109/L | 72.1 (0.42-301.6) | 13.4 (0.45-189.2) | .008 |
| AST, IU/L | 68.5 (23-501) | 46.5 (16-4341) | .113 |
| ALT, IU/L | 41.0 (5-407) | 12.5 (4-653) | .075 |
| T-Bil mg/dL | 6.7 (0.6-15.3) | 4.65 (1.82-46.0) | .270 |
| Effusions, n (%) | 11 of 27 (40.7) | 5 of 17 (29.4) | .447† |
| Bleeding diatheses, n (%) | 8 of 29 (27.6) | 5 of 16 (31.3) | .528† |
| . | GATA1s expression group . | P . | |
|---|---|---|---|
| High (n = 40) . | Low (n = 26) . | ||
| Sex: male/female, n | 19/21 | 13/13 | .843* |
| Gestational age, wk | 37.3 (30.0-40.0) | 37.9 (32.6-40.6) | .487 |
| Birth weight, kg | 2.5 (1.6-3.3) | 2.5 (1.4-3.5) | .698 |
| WBC, ×109/L | 105.65 (7.8-423.0) | 39.0 (9.0-220.0) | .004 |
| Number of blasts, ×109/L | 72.1 (0.42-301.6) | 13.4 (0.45-189.2) | .008 |
| AST, IU/L | 68.5 (23-501) | 46.5 (16-4341) | .113 |
| ALT, IU/L | 41.0 (5-407) | 12.5 (4-653) | .075 |
| T-Bil mg/dL | 6.7 (0.6-15.3) | 4.65 (1.82-46.0) | .270 |
| Effusions, n (%) | 11 of 27 (40.7) | 5 of 17 (29.4) | .447† |
| Bleeding diatheses, n (%) | 8 of 29 (27.6) | 5 of 16 (31.3) | .528† |
Values are given as the median (range). P values estimated by Mann-Whitney U test.
Pearson χ2 test.
Fisher exact test.
Cumulative incidence of early death and of ML-DS in children with TAM. Based on the estimated GATA1s expression levels, patients were classified in 2 groups: GATA1s high and low groups. TAM patients in the GATA1s low group had a significantly higher risk for the development of leukemia (P (gray) < .001).
Cumulative incidence of early death and of ML-DS in children with TAM. Based on the estimated GATA1s expression levels, patients were classified in 2 groups: GATA1s high and low groups. TAM patients in the GATA1s low group had a significantly higher risk for the development of leukemia (P (gray) < .001).
Summary of outcomes and GATA1 mutation types in TAM patients
| Mutation type . | Outcome of TAM . | TAM . | ML-DS . | |||||
|---|---|---|---|---|---|---|---|---|
| CR . | Early death . | Evolved to ML-DS . | NA . | Total (n = 66) . | Total (n = 40) . | |||
| High group | ||||||||
| Loss of 1st Met, n (%) | 7 | 1 | 1 | 1 | 10 (15.2) | 3 (7.5) | ||
| Splicing error, n (%) | 7 | 4 | 0 | 2 | 13 (19.7) | 40 (15.2) | 6 (15.0) | 16 (40.0) |
| PTC 1-3′, n (%) | 10 | 6 | 0 | 1 | 17 (25.8) | 7 (17.5) | ||
| Low group | 26 (39.4) | 24 (60.0) | ||||||
| SPTC 1-5′, n (%) | 6 | 4 | 2 | 3 | 15 (22.7) | 14 (35.0) | ||
| PTC 2, n (%) | 2 | 1 | 8 | 0 | 11 (16.7) | 10 (25.0) | ||
| Mutation type . | Outcome of TAM . | TAM . | ML-DS . | |||||
|---|---|---|---|---|---|---|---|---|
| CR . | Early death . | Evolved to ML-DS . | NA . | Total (n = 66) . | Total (n = 40) . | |||
| High group | ||||||||
| Loss of 1st Met, n (%) | 7 | 1 | 1 | 1 | 10 (15.2) | 3 (7.5) | ||
| Splicing error, n (%) | 7 | 4 | 0 | 2 | 13 (19.7) | 40 (15.2) | 6 (15.0) | 16 (40.0) |
| PTC 1-3′, n (%) | 10 | 6 | 0 | 1 | 17 (25.8) | 7 (17.5) | ||
| Low group | 26 (39.4) | 24 (60.0) | ||||||
| SPTC 1-5′, n (%) | 6 | 4 | 2 | 3 | 15 (22.7) | 14 (35.0) | ||
| PTC 2, n (%) | 2 | 1 | 8 | 0 | 11 (16.7) | 10 (25.0) | ||
To validate this observation, we examined the proportion of mutation types in 40 ML-DS patients observed in the same period of time as this surveillance. The results showed a significantly higher incidence of GATA1s low type mutations in ML-DS than in TAM (P = .039; Table 5). These results further support the present findings that quantitative differences in the mutant protein have a significant effect on the risk of progression to ML-DS.
Discussion
In TAM, GATA1 mutations lead to the expression of proteins lacking the N-terminal transactivation domain. In addition to this qualitative change, we showed here that the mutations affect the expression level of the truncated protein. The mutations were classified into 2 groups according to the estimated GATA1s expression level. Comparison of the clinical features between the 2 groups revealed that GATA1s low mutations were significantly associated with a high risk of progression to ML-DS and lower counts of both WBC and blast cells. These results suggest that quantitative differences in protein expression caused by GATA1 mutations have significant effects on the phenotype of TAM.
GATA1s was shown previously to be produced from wild-type GATA1 through 2 mechanisms: use of the alternative translation initiation site at codon 84 of the full-length transcript and alternative splicing of exon 2.12,26 However, the translation efficiencies of GATA1s from the full-length of mRNA and short transcripts have not been investigated. Our results clearly showed that the Δexon 2 transcript produced GATA1s much more abundantly than did the full-length transcript. The translation efficiencies of GATA1s from full-length transcripts containing PTC were also lower than the alternative spliced form. These results support our contention that GATA1s expression levels largely depend on the amount of the Δexon 2 transcript. Thus, one cannot predict the expression level of GATA1s protein from the total amount of the transcript.
The differences in the quantities of GATA1s proteins expressed by PTC type 1-5′ and -3′ mutations revealed the importance of the location of the mutation for splicing efficiency and protein expression. The splicing efficiency is regulated by cis-elements located in exons and introns (referred to as exonic and intronic splicing enhancers or silencers), and transacting factors recognizing these elements.27,28 The PTC type 1-3′ mutations induced efficient skipping of exon 2 (Figures 2Bvi-vii, 3A-B). These mutations might affect exonic splicing enhancers or silencers located in exon 2. To predict the splicing pattern from the mutations more accurately, the elucidation of cis-elements and transacting splicing factors, which regulate the splicing of exon 2 of GATA1, will be very important.
The nonsense mediated RNA decay pathway (NMD), a cellular mechanism for detection of PTC and prevention of translation from aberrant transcripts,29,30 might regulate the expression of GATA1s protein derived from PTC type 2 mutations, which contained PTCs after the second methionine at codon 84. We consistently detected low amounts of transcripts of GATA1 in samples expressing PTC type 2 mutations, whereas the expression levels of GATA1 mRNA from PTC type 1 mutations were comparable with that from wild-type GATA1 (Figure 2Aiii). These results suggest that the location of PTC relative to alternative translation initiation sites is important for effective NMD surveillance.
Available evidence indicates that acute leukemia arises from cooperation between one class of mutations that interferes with differentiation (class II mutations) and another class that confers a proliferative advantage to cells (class I mutations).31 Recent reports showed that introducing high levels of exogenous GATA1 lacking the N-terminus did not reduce the aberrant growth of GATA1-null megakaryocytes, but instead induced differentiation.32,33 This observation suggested that abundant GATA1s protein functions like a class I mutation in TAM blasts. In contrast, reducing GATA1 expression leads to differentiation arrest and aberrant growth of megakaryocytic cells.19,20 The present data suggest that GATA1s is expressed at very low levels in TAM blasts with GATA1s low mutations. These levels may not be sufficient to provoke normal maturation. Together, these findings suggest that the low expression of GATA1s might function like class II mutations in TAM blasts. Additional class I mutations or epigenetic alterations might be more effective in the development of leukemia in blast cells expressing GATA1s at low levels.
In the present study, we identified a subgroup of TAM patients with a higher risk of developing ML-DS. Of 66 children, 11 (16.7%) with TAM subsequently developed ML-DS and 10 of them belonged to the GATA1s low group harboring the PTC type 2 or PTC type 1-5′ mutations. Surprisingly, 8 of 11 patients (73%) with the PTC type 2 mutations developed ML-DS (Tables 1, 5), whereas 2 of 15 patients (13.3%) with PTC type 1-5′ mutations developed leukemia. The estimated expression levels of GATA1s from PTC type 2 mutations were lower than those from PTC type 1-5′ mutations (Figures 1, 2Ai). These results suggest that the type 2 mutations may be a more significant risk factor for developing ML-DS (supplemental Figure 2). However, our classification of GATA1 mutations mainly rested on extrapolation from in vitro transfection experiments (Figures 1–2) and RT-PCR analyses of a small number of patient samples (Figure 3). The stability of the transcripts and the splicing efficiency of the second exon of GATA1 will be regulated through complex mechanisms. To confirm our findings, precise mapping of the mutations that affect the expression levels of GATA1s and a prospective study with a large series of TAM patients are necessary.
Finally, we proposed the hypothesis that the quantitative differences in GATA1s protein expression caused by mutations have a significant effect on the phenotype of TAM. The observations described here provide valuable information about the roles of GATA1 mutations on multistep leukemogenesis in DS patients. Moreover, the results might have implications for management of leukemia observed in DS infants and children. Because the blast cells in both TAM and subsequent ML-DS appear highly sensitive to cytarabine,34–39 the preleukemic clone could be treated with low-dose cytarabine without severe side effects, and elimination of the preleukemic clone might prevent progression to leukemia.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Dr Tetsuo Mitsui (Yamagata University School of Medicine), Shingo Morinaga (National Hospital Organization Kumamoto Medical Center), Takahide Nakano (Kansai Medical University), Masahiro Migita (Japan Red Cross Kumamoto Hospital), Hiroshi Kanda (Kurume University School of Medicine), Koji Kato (The First Nagoya Red Cross Hospital), and Takahiro Uehara (Kameda Medical Center) for providing patient samples. We thank Dr Eiki Tsushima, Ko Kudo (Hirosaki University Graduate School of Medicine), and Ms Hitomi Iwabuchi for statistical analysis, helpful discussions, and technical assistance, respectively.
This work was supported by grants-in-aid from the Ministry of Education, Culture, Sports, Science, and Technology of Japan and Health and Labor Sciences Research Grants (research on intractable diseases) the Ministry of Health, Labor, and Welfare of Japan.
Authorship
Contribution: R.K. and T. Toki designed, organized, and performed research, analyzed data, and wrote the paper; K.T. designed research and collected and analyzed clinical data; G.X. and R.W. performed mutation screening; A.S., H.K., K. Kawakami, M.E., D.H., K. Kogawa, S.A., Y.I., S.I., T. Taga, Y.K., and Y.H. provided clinical samples and data; A.H. and S.K. performed mutation screening and provided clinical samples and data; and E.I. designed and organized research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Etsuro Ito, Department of Pediatrics, Hirosaki University Graduate School of Medicine, 5 Zaifu-cho, Hirosaki, Aomori, 036-8563, Japan; e-mail: eturou@cc.hirosaki-u.ac.jp.
References
Author notes
R.K. and T.T. contributed equally to this work.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal