Abstract
The genome of mantle cell lymphoma (MCL) is, in addition to the translocation t(11;14), characterized by a high number of secondary chromosomal gains and losses that probably account for the various survival times of MCL patients. We investigated 77 primary MCL tumors with available clinical information using high-resolution RNA expression and genomic profiling and applied our recently developed gene expression and dosage integrator algorithm to identify novel genes and pathways that may be of relevance for the pathobiology of MCL. We show that copy number neutral loss of heterozygosity is common in MCL and targets regions that are frequently affected by deletions. The molecular consequences of genomic copy number changes appear complex, even in genomic loci with identified tumor suppressors, such as the region 9p21 containing the CDKN2A locus. Moreover, the deregulation of novel genes, such as CUL4A, ING1, and MCPH1, may affect the 2 crucial pathogenetic mechanisms in MCL, the disturbance of the proliferation, and DNA damage response pathways. Deregulation of the Hippo pathway may have a pathogenetic role in MCL because decreased expression of its members MOBKL2A, MOBKL2B, and LATS2 was associated with inferior outcome, including an independent validation series of 32 MCLs.
Introduction
Mantle cell lymphoma (MCL) is an aggressive subtype of B-cell lymphomas that accounts for approximately 6% of all lymphoid neoplasms.1 The median survival time for patients with MCL is approximately 4 to 5 years and appears to be on the rise because of improved treatment regimens and an expanded number of novel therapeutic options. Nevertheless, survival times in MCL range between a few months only and more than 10 years,2-5 and it is widely assumed that genetic alterations that develop secondary to the hallmark translocation t(11;14) involving cyclin D1 account, at least in part, for this clinical variability.2 Consequently, several studies have investigated chromosomal alterations in MCL, for example, by conventional comparative genomic hybridization (CGH),6,7 by array CGH,8-11 and, very recently, by single nucleotide polymorphism (SNP) arrays.12-14 Taken together, these studies identified the most frequently altered chromosomal regions in MCL, including gains of chromosomal material in 3q, 7q, and 8q and losses in 1p, 6q, 8p, 9p, 9q, 11q, 13q, and 17p. In addition, these studies have narrowed down some of the minimal regions affected by genomic copy number alterations (CNAs) in MCL and have led to the suggestion of putative target genes of these chromosomal abnormalities, with particular emphasis on genes involved in cell cycle regulation and DNA damage response pathways. Furthermore, recent SNP array studies of MCL cell lines and small series of primary MCL samples revealed recurrent regions of copy number neutral loss of heterozygosity (CNN-LOH; also referred to as acquired/partial uniparental disomy [UPD]) that may represent an alternative mechanism to an allelic deletion in the process of a tumor suppressor gene inactivation because the CNN-LOH regions appear to cluster in chromosomal locations that are also frequently affected by deletions.12-15
Previous copy number profiling studies in MCL6,8-15 were performed on relatively small numbers of primary tumor specimens, used low resolution techniques, or did not include accompanying gene expression profiling experiments or survival data. In contrast, we here present high-resolution gene expression and 500K SNP array data (mean intermarker spacing, 5.8 kb) from77 primary MCL samples, including 72 cyclin D1–positive and 5 cyclin D1–negative MCLs with available survival data representing the largest MCL series studied to date. Using an integrated analysis approach, gene expression and dosage integrator (GEDI), which was recently developed by us,16 we here refine the minimal regions of recurrent CNAs and CNN-LOH in MCL and identify novel putative target genes and pathways that might be pathogenetically relevant and show an association with the clinical outcome. Several genes of the Hippo signaling pathway exhibit altered expression in MCL and may therefore deserve more detailed future studies.
Methods
Patient samples
Seventy-seven primary MCL specimens from previously untreated patients were investigated in this study by high-resolution Affymetrix 500K SNP arrays (median/mean intermarker spacing, 2.5 kb/5.8 kb), including 72 cyclin D1–positive and 5 cyclin D1–negative cases from a cohort previously studied by the Leukemia and Lymphoma Molecular Profiling Project using Lymphochip DNA microarrays.3,17 A subset of 64 samples was investigated with Affymetrix HG U133 plus 2.0 gene expression arrays. DNA and RNA were extracted from fresh frozen lymph node specimens. Tumor histology was reviewed by a panel of expert hematopathologists, and the diagnosis of MCL was established according to current criteria of the World Health Organization classification.1 All cyclin D1–negative cases showed classic morphology, a B-cell immunophenotype, and expression of CD5, but no cyclin D1 expression. Their gene expression profile was similar to cyclin D1–positive MCL.17 An overview of the clinical characteristics of the MCL cases is provided in Table 1. Clinical data were obtained from all patients according to a protocol approved by the National Cancer Institute Institutional Review Board. In addition, the expression of selected genes was investigated in an independent group of 32 untreated MCL patients from the Hospital Clinic of Barcelona, Spain. Ethics approval for the use of these materials was obtained in accordance with the Declaration of Helsinki.
Clinical data of the MCL patients investigated with 500K SNP arrays
| . | All cases, no. (%) . | Cyclin D1+, no. (%) . | Cyclin D1−, no. (%) . |
|---|---|---|---|
| No. of patients | 77 | 72 (94) | 5 (6) |
| Median age at diagnosis, y (range) | 61 (38-93) | 63 (38-93) | 60 (54-77) |
| Sex | |||
| Male | 58 (75) | 54 (75) | 4 (80) |
| Female | 19 (25) | 18 (25) | 1 (20) |
| Morphology | |||
| Classic | 67 (87) | 62 (86) | 5 (100) |
| Blastoid | 10 (13) | 10 (14) | 0 |
| B symptoms | 26 (34) | 25 (35) | 1 (20) |
| Ann Arbor stage | |||
| I/II | 9 (12) | 9 (13) | 0 |
| III/IV | 68 (88) | 63 (87) | 5 (100) |
| BM involvement | 46 (60) | 41 (57) | 5 (100) |
| LDH level more than normal | 26 (34) | 25 (35) | 1 (20) |
| Therapy* | |||
| Combined chemotherapy | 63 (82) | 58 (81) | 5 (100) |
| Combined chemotherapy plus rituximab | 8 (10) | 8 (11) | 0 |
| No chemotherapy | 4 (5) | 4 (6) | 0 |
| Response to therapy (CR + PR) | 58 of 74 (78) | 53 of 69 (77) | 5 of 5 (100) |
| Median follow-up, months | 33 | 32 | 38 |
| Median survival (range), months† | 37 (24-50) | 35 (21-48) | NR |
| . | All cases, no. (%) . | Cyclin D1+, no. (%) . | Cyclin D1−, no. (%) . |
|---|---|---|---|
| No. of patients | 77 | 72 (94) | 5 (6) |
| Median age at diagnosis, y (range) | 61 (38-93) | 63 (38-93) | 60 (54-77) |
| Sex | |||
| Male | 58 (75) | 54 (75) | 4 (80) |
| Female | 19 (25) | 18 (25) | 1 (20) |
| Morphology | |||
| Classic | 67 (87) | 62 (86) | 5 (100) |
| Blastoid | 10 (13) | 10 (14) | 0 |
| B symptoms | 26 (34) | 25 (35) | 1 (20) |
| Ann Arbor stage | |||
| I/II | 9 (12) | 9 (13) | 0 |
| III/IV | 68 (88) | 63 (87) | 5 (100) |
| BM involvement | 46 (60) | 41 (57) | 5 (100) |
| LDH level more than normal | 26 (34) | 25 (35) | 1 (20) |
| Therapy* | |||
| Combined chemotherapy | 63 (82) | 58 (81) | 5 (100) |
| Combined chemotherapy plus rituximab | 8 (10) | 8 (11) | 0 |
| No chemotherapy | 4 (5) | 4 (6) | 0 |
| Response to therapy (CR + PR) | 58 of 74 (78) | 53 of 69 (77) | 5 of 5 (100) |
| Median follow-up, months | 33 | 32 | 38 |
| Median survival (range), months† | 37 (24-50) | 35 (21-48) | NR |
BM indicates bone marrow; LDH, lactate dehydrogenase; CR, complete remission; PR, partial remission; and NR, not reached.
No information available for 2 patients.
Estimated median (95% confidence interval).
SNP array studies
SNP arrays were performed using 500K SNP arrays (250K NSP and STY arrays, Affymetrix) according to the manufacturer's instructions (www.Affymetrix.com). Data files were generated using the Affymetrix gene chip operating and gene chip genotyping analysis (GTYPE) software suites. GTYPE Version 4.1 was used for the analysis of signal intensity and to determine genotype calls. Because no matched normal DNA was available, unpaired analysis using an independent reference set was performed. The complete dataset was analyzed as combined subarrays (500K) by visual inspection using CNAT 4.0 (Affymetrix) and the copy number analyzer for gene chip (CNAG), Version 3.0 (available at http://www.genome.umin.jp/) and by an algorithm described by Lenz et al16 (supplemental Data, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). In brief, this algorithm divides SNP array data generated with CNAG into segments of statistically similar copy number, classified as amplification, gain, loss, double loss, and normal. Subsequently, minimal common regions (MCRs) are defined by assembling segments of abnormal copy number of a specific type that affect the same chromosomal region in different samples. Specifically, 4 classes of recurrent abnormalities are defined, namely, (1) abnormal chromosome arm (> 60% of the chromosomal arm is covered with segments of the specified type of abnormality), (2) abnormal whole chromosome (both arms of a chromosome are scored as being > 60% abnormal), (3) short recurrent abnormalities (based on segments < 20 Mb in length), and (4) long recurrent abnormalities (based on segments > 15 Mb in length, but which are not part of an “abnormal chromosome arm” MCR).
Regions of CNN-LOH/UPD were determined by visual inspection using the CNAG and CNAT tools. To focus on UPDs that were most probably acquired during tumorigenesis, only UPDs more than 10 Mb in size were considered because random stretches of homozygosity of this size occur rarely in normal samples.18
Gene expression arrays and integrated analysis of copy number and gene expression data
Gene expression profiling using the HG U133 plus 2.0 gene expression arrays (Affymetrix) was performed in 64 MCL samples based on the availability of RNA and following the Affymetrix expression analysis technical manual (www.Affymetrix.com). The average probe array signals were scaled to a target signal of 500 using the GeneChip operating software. All original microarray data can be found on the Gene Expression Omnibus public database under accession number GSE21452.
Datasets from the SNP array and gene expression studies were merged using the GEDI algorithm previously described by Lenz et al.16 After the division of the SNP array data into segments of statistically similar copy numbers and the definition of MCRs (“SNP array studies”), the GEDI algorithm determines the degree of expression alteration of each gene within each MCR and computes a statistic that summarizes these correlations across all genes in the MCR.16
Quantitative real-time PCR
Quantitative real-time reverse transcription polymerase chain reaction (RT-PCR) was used to determine relative mRNA expression levels of selected genes using TaqMan gene expression assays (Applied Biosystems), including MOBKL2A (Hs00370031_m1), MOBKL2B (Hs00257351_m1), LATS1 (Hs01125524_m1), and LATS2 (Hs00324396_m1). Reverse transcription was performed using 1 μg of total RNA with the SuperScript First-Strand kit (Invitrogen). PCR reactions were performed using the ABI Prism 7900 HT Sequence Detection System (Applied Biosystems) according to standard protocols as recommended by the manufacturer. Quantitative RT-PCR data were analyzed applying the comparative threshold cycle (Ct) method (2−ΔΔCt method) using GUSB as endogenous reference.
Other statistical methods
Unless otherwise specified, data were analyzed using SPSS software (Version 15.0). Survival was estimated with the Kaplan-Meier method and compared by log-rank tests. P values less than .05 were considered statistically significant.
Results
CNAs and CNN-LOH
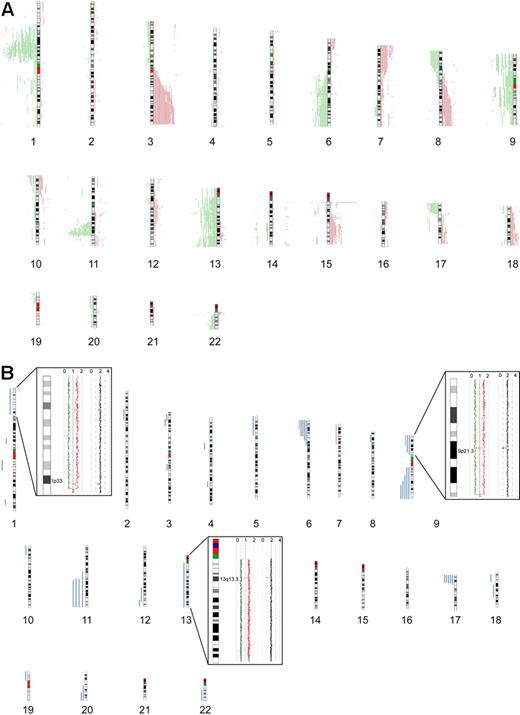
Seventy-seven primary MCL samples, including 5 cyclin D1–negative cases, were successfully analyzed by 500K SNP arrays. Chromosomal alterations were analyzed by visual inspection and by an alternative algorithm described in “Methods,”16 with highly similar results. CNAs were identified in all of the investigated cases, with a median number of 15 CNAs (10 losses, 5 gains) per case, determined by visual inspection. The most frequently identified CNAs were distributed in a pattern typical of MCL. Figure 1A provides a genome-wide overview of gains and losses detected in our series, including regions of putative copy number variation (CNVs), such as the regions 15q11.2 and 17q21.31. CNVs in this study were identified by comparison with a public database (http://projects.tcag.ca/variation/).
Chromosomal alterations in 77 MCL cases determined by 500K SNP arrays. Chromosomal alterations in 72 cyclin D1–positive and 5 cyclin D1–negative primary MCL cases determined by 500K SNP arrays. (A) Copy number alterations (including putative copy number variations) according to the CNAG software. Gains are displayed in red on the right side of the ideograms, and losses are shown in green on the left side of the ideograms. (B) Regions of CNN-LOH, also termed UPD; more than 10 Mb determined by the CNAG/AsCNAR and CNAT software tools. Regions of CNN-LOH/UPD are shown in blue, and regions of uniparental trisomy are shown in purple. (Insets) Regions of partial CNN-LOH/UPD in chromosomes 1, 9, and 13 that include small regions of homozygous deletions in 1p32.3/33 (contains the genomic loci of CDKN2C and FAF1), 9p21.3 (CDKN2A locus), and 13q13.3 (containing C13orf36, RFXAP, and SMAD9), indicating that the CNN-LOH/UPD arose after the loss of one copy of the respective genomic locus. The black line indicates total gene dosage; the diploid state is indicated by 2. The red and green lines indicate allele-specific gene dosage levels, with loss of one parental copy and gain of the other in the region of partial CNN-LOH/UPD.
Chromosomal alterations in 77 MCL cases determined by 500K SNP arrays. Chromosomal alterations in 72 cyclin D1–positive and 5 cyclin D1–negative primary MCL cases determined by 500K SNP arrays. (A) Copy number alterations (including putative copy number variations) according to the CNAG software. Gains are displayed in red on the right side of the ideograms, and losses are shown in green on the left side of the ideograms. (B) Regions of CNN-LOH, also termed UPD; more than 10 Mb determined by the CNAG/AsCNAR and CNAT software tools. Regions of CNN-LOH/UPD are shown in blue, and regions of uniparental trisomy are shown in purple. (Insets) Regions of partial CNN-LOH/UPD in chromosomes 1, 9, and 13 that include small regions of homozygous deletions in 1p32.3/33 (contains the genomic loci of CDKN2C and FAF1), 9p21.3 (CDKN2A locus), and 13q13.3 (containing C13orf36, RFXAP, and SMAD9), indicating that the CNN-LOH/UPD arose after the loss of one copy of the respective genomic locus. The black line indicates total gene dosage; the diploid state is indicated by 2. The red and green lines indicate allele-specific gene dosage levels, with loss of one parental copy and gain of the other in the region of partial CNN-LOH/UPD.
The statistical algorithm developed by us16 revealed a total of 650 MCRs, including 78 amplifications, 191 gains, 294 losses, and 87 double losses (supplemental Table 1). To focus on CNAs with potential biologic relevance and to limit the number of CNVs in the dataset, we put particular emphasis on CNAs that were significantly associated with corresponding changes in gene expression determined by the GEDI algorithm.16 Of 650 MCRs, 193 were found to be significantly associated with altered gene expression. These included 18 amplifications, 44 gains, 101 losses, and 30 double losses (supplemental Table 1). Table 2 summarizes the most frequently detected MCRs (present in > 10% of the cases) associated with changes in gene expression. Overall, the most frequently detected alterations in our series of MCL cases that are not considered CNVs were losses of chromosomal material in 1p (47%), 6q (21%), 8p (21%), 9p (27%), 11q (50%), 13q (27%), and 17p (23%), and gains in 3q (31%), 7p (13%), and 8q (13%). Losses of genomic material outnumbered gains, a finding typical of MCL. Notably, deletions in the ATM region (11q) correlated with the genomic complexity (CNAs + CNN-LOH) of the tumor cell genome (P = .02).
Most frequently detected CNAs associated with gene expression changes (> 8 of 77 samples, 10%; determined by the GEDI algorithm)
| MCR ID . | Chromosomal location (band; MCR core) . | Type, class of alteration . | Start core . | End core . | MCR peak . | Cyclin D1+, no. . | Cyclin D1−, no. . | Total, no. . |
|---|---|---|---|---|---|---|---|---|
| 280 | 1p31.1-p13.3 | Loss/double loss, long | 78.845412 | 108.161907 | 93.046159 | 34 | 2 | 36 |
| 519 | 17q21.31* | Loss/double loss, short | 41.560151 | 41.707907 | 41.703504 | 33 | 2 | 35 |
| 108 | 3q | Gain/amp, q-arm | 95.031169 | 199.322068 | NA | 21 | 3 | 24 |
| 238 | 17q21.31* | Gain/amp, short | 41.522422 | 41.719832 | 41.590303 | 23 | 0 | 23 |
| 479 | 13q | Loss/double loss, chromosome | 17.960319 | 114.09298 | NA | 20 | 1 | 21 |
| 421 | 9p | Loss/double loss, p-arm | 0.03091 | 44.108554 | NA | 19 | 2 | 21 |
| 451 | 11q21-q23.2 | Loss/double loss, long | 92.710683 | 114.244057 | 99.00843 | 16 | 4 | 20 |
| 452 | 11q22.3-q23.2 | Loss/double loss, short | 107.419503 | 113.384238 | 110.616957 | 18 | 1 | 19 |
| 525 | 17p | Loss/double loss, p-arm | 0.006888 | 22.069633 | NA | 18 | 0 | 18 |
| 649 | 22q11.22† | Double loss, short | 21.388199 | 21.576155 | 21.463635 | 18 | 0 | 18 |
| 122 | 5p15.33* | Gain/amp, short | 0.75219 | 0.87267 | 0.797863 | 16 | 1 | 17 |
| 379 | 6q | Loss/double loss, q-arm | 62.030184 | 170.823609 | NA | 16 | 0 | 16 |
| 408 | 8p | Loss/double loss, p-arm | 0.180568 | 43.820269 | NA | 14 | 2 | 16 |
| 637 | 17q21.31* | Double loss, short | 41.62507 | 41.707907 | 41.63558 | 13 | 2 | 15 |
| 550 | 21q11.2‡ | Loss/double loss, short | 9.887804 | 10.189118 | 9.887804 | 12 | 3 | 15 |
| 607 | 9p21.3 | Double loss, short | 21.81811 | 22.232054 | 21.976218 | 13 | 1 | 14 |
| 422 | 9q | Loss/double loss, q-arm | 64.197638 | 138.363203 | 12 | 1 | 13 | |
| 430 | 10q11.22* | Loss/double loss, short | 46.494897 | 47.030118 | 46.500913 | 12 | 1 | 13 |
| 281 | 1p21.2‡ | Loss/double loss, short | 100.510057 | 101.760343 | 101.292504 | 11 | 2 | 13 |
| 478 | 13q34 | Loss/double loss, short | 110.898938 | 114.09398 | 113.944993 | 10 | 2 | 12 |
| 416 | 9q21.11-q31.1 | Loss/double loss, long | 69.575157 | 102.187565 | 84.13048 | 10 | 0 | 10 |
| 147 | 7p | Gain/amp, p-arm | 0.141322 | 57.730637 | NA | 10 | 0 | 10 |
| 163 | 8q | Gain/amp, q-arm | 47.043376 | 146.264218 | NA | 9 | 1 | 10 |
| 283 | 1p11.2-q12 | Loss/double loss, long | 120.903122 | 142.129196 | 120.903122 | 9 | 1 | 10 |
| 424 | 10p15.2–10p15.3 | Loss/double loss, short | 0.101955 | 3.663599 | 1.913396 | 9 | 1 | 10 |
| 227 | 16p11.2* | Gain/amp, short | 34.328205 | 34.618467 | 34.454744 | 9 | 1 | 10 |
| 279 | 1p22 | Loss/double loss, short | 87.4228 | 87.424276 | 87.4228 | 8 | 2 | 10 |
| 616 | 11p15.1* | Double loss, short | 18.905796 | 18.918254 | 18.911384 | 9 | 0 | 9 |
| 420 | 9 complete | Loss/double loss, chromosome | 0.03091 | 138.363203 | NA | 8 | 1 | 9 |
| 391 | 8p23.2‡ | Loss/double loss, short | 3.469284 | 3.479449 | 3.469284 | 8 | 0 | 8 |
| 537 | 19p13.3 | Loss/double loss, short | 3.232586 | 4.439925 | 3.931367 | 8 | 0 | 8 |
| 526 | 18p11.32* | Loss/double loss, short | 1.903316 | 1.976262 | 1.960916 | 8 | 0 | 8 |
| 57 | 15q11.2* | Amp, short | 19.111608 | 19.876833 | 19.407629 | 7 | 1 | 8 |
| 646 | 21q11.2‡ | Double loss, short | 9.887804 | 10.189118 | 9.887804 | 6 | 2 | 8 |
| 220 | 15q22.2-q26.1 | Gain/amp, long | 57.914389 | 86.625684 | 65.011755 | 6 | 2 | 8 |
| MCR ID . | Chromosomal location (band; MCR core) . | Type, class of alteration . | Start core . | End core . | MCR peak . | Cyclin D1+, no. . | Cyclin D1−, no. . | Total, no. . |
|---|---|---|---|---|---|---|---|---|
| 280 | 1p31.1-p13.3 | Loss/double loss, long | 78.845412 | 108.161907 | 93.046159 | 34 | 2 | 36 |
| 519 | 17q21.31* | Loss/double loss, short | 41.560151 | 41.707907 | 41.703504 | 33 | 2 | 35 |
| 108 | 3q | Gain/amp, q-arm | 95.031169 | 199.322068 | NA | 21 | 3 | 24 |
| 238 | 17q21.31* | Gain/amp, short | 41.522422 | 41.719832 | 41.590303 | 23 | 0 | 23 |
| 479 | 13q | Loss/double loss, chromosome | 17.960319 | 114.09298 | NA | 20 | 1 | 21 |
| 421 | 9p | Loss/double loss, p-arm | 0.03091 | 44.108554 | NA | 19 | 2 | 21 |
| 451 | 11q21-q23.2 | Loss/double loss, long | 92.710683 | 114.244057 | 99.00843 | 16 | 4 | 20 |
| 452 | 11q22.3-q23.2 | Loss/double loss, short | 107.419503 | 113.384238 | 110.616957 | 18 | 1 | 19 |
| 525 | 17p | Loss/double loss, p-arm | 0.006888 | 22.069633 | NA | 18 | 0 | 18 |
| 649 | 22q11.22† | Double loss, short | 21.388199 | 21.576155 | 21.463635 | 18 | 0 | 18 |
| 122 | 5p15.33* | Gain/amp, short | 0.75219 | 0.87267 | 0.797863 | 16 | 1 | 17 |
| 379 | 6q | Loss/double loss, q-arm | 62.030184 | 170.823609 | NA | 16 | 0 | 16 |
| 408 | 8p | Loss/double loss, p-arm | 0.180568 | 43.820269 | NA | 14 | 2 | 16 |
| 637 | 17q21.31* | Double loss, short | 41.62507 | 41.707907 | 41.63558 | 13 | 2 | 15 |
| 550 | 21q11.2‡ | Loss/double loss, short | 9.887804 | 10.189118 | 9.887804 | 12 | 3 | 15 |
| 607 | 9p21.3 | Double loss, short | 21.81811 | 22.232054 | 21.976218 | 13 | 1 | 14 |
| 422 | 9q | Loss/double loss, q-arm | 64.197638 | 138.363203 | 12 | 1 | 13 | |
| 430 | 10q11.22* | Loss/double loss, short | 46.494897 | 47.030118 | 46.500913 | 12 | 1 | 13 |
| 281 | 1p21.2‡ | Loss/double loss, short | 100.510057 | 101.760343 | 101.292504 | 11 | 2 | 13 |
| 478 | 13q34 | Loss/double loss, short | 110.898938 | 114.09398 | 113.944993 | 10 | 2 | 12 |
| 416 | 9q21.11-q31.1 | Loss/double loss, long | 69.575157 | 102.187565 | 84.13048 | 10 | 0 | 10 |
| 147 | 7p | Gain/amp, p-arm | 0.141322 | 57.730637 | NA | 10 | 0 | 10 |
| 163 | 8q | Gain/amp, q-arm | 47.043376 | 146.264218 | NA | 9 | 1 | 10 |
| 283 | 1p11.2-q12 | Loss/double loss, long | 120.903122 | 142.129196 | 120.903122 | 9 | 1 | 10 |
| 424 | 10p15.2–10p15.3 | Loss/double loss, short | 0.101955 | 3.663599 | 1.913396 | 9 | 1 | 10 |
| 227 | 16p11.2* | Gain/amp, short | 34.328205 | 34.618467 | 34.454744 | 9 | 1 | 10 |
| 279 | 1p22 | Loss/double loss, short | 87.4228 | 87.424276 | 87.4228 | 8 | 2 | 10 |
| 616 | 11p15.1* | Double loss, short | 18.905796 | 18.918254 | 18.911384 | 9 | 0 | 9 |
| 420 | 9 complete | Loss/double loss, chromosome | 0.03091 | 138.363203 | NA | 8 | 1 | 9 |
| 391 | 8p23.2‡ | Loss/double loss, short | 3.469284 | 3.479449 | 3.469284 | 8 | 0 | 8 |
| 537 | 19p13.3 | Loss/double loss, short | 3.232586 | 4.439925 | 3.931367 | 8 | 0 | 8 |
| 526 | 18p11.32* | Loss/double loss, short | 1.903316 | 1.976262 | 1.960916 | 8 | 0 | 8 |
| 57 | 15q11.2* | Amp, short | 19.111608 | 19.876833 | 19.407629 | 7 | 1 | 8 |
| 646 | 21q11.2‡ | Double loss, short | 9.887804 | 10.189118 | 9.887804 | 6 | 2 | 8 |
| 220 | 15q22.2-q26.1 | Gain/amp, long | 57.914389 | 86.625684 | 65.011755 | 6 | 2 | 8 |
NA indicates not applicable.
Putative CNVs.
Immunoglobulin gene locus.
Unconfirmed CNV.
The 5 cyclin D1–negative MCLs in our series showed similar genomic copy number abnormalities compared with their cyclin D1–positive counterparts using this high-resolution approach (Table 2). Among the MCRs that were significantly associated with gene expression changes, no MCR was detected that occurred solely within cyclin D1–negative MCL. Of note, 4 of 5 cyclin D1–negative MCLs showed a deletion in 11q, including the ATM locus, and the remaining case displayed a large UPD in this region. These results once again support the notion that cyclin D1–negative MCLs represent a bona fide subset of the entity “mantle cell lymphoma” because nearly 50% of cyclin D1–positive MCLs also showed this alteration.
In addition to changes in the genomic copy number, we identified large regions (> 10 Mb) of CNN-LOH as a frequent phenomenon in our series of MCLs. An example of a large CNN-LOH in chromosome 17 is provided in supplemental Figure 1. Furthermore, we also identified several smaller regions of CNN-LOH, ranging from 2 to 9 Mb in size. Given that their significance is unclear in the setting of an unpaired analysis approach, these were excluded from further analysis. In total, 46 large regions of CNN-LOH (> 10 Mb) with a median size of 25 Mb were detected in 27 of 77 MCLs (35%, Figure 1B). They were nonrandomly distributed with recurrently affected regions in chromosome 6p, 9p, 9q, 11q, 17p, and 20q, most of which are also frequently affected by genomic losses. These results suggest that CNN-LOH in MCLs may target identical chromosomal regions that are otherwise hit by genomic losses. Cyclin D1–negative MCLs showed 3 CNN-LOH localized in 9p, 11q13.2-qter, and 22q12-qter. Overall, the short arm of chromosome 17 was among the regions that were most frequently affected by large CNN-LOH (6%) as well as by deletions (23%; supplemental Table 2). Concomitant mutations of TP53 that had been studied previously19 were detected in 4 of 19 cases with deletion of 17p (21%), whereas 3 of 5 MCL (60%) with a CNN-LOH in 17p carried TP53 mutations. Moreover, one additional MCL with a CNN-LOH in 17p without a detectable TP53 mutation had very low expression of TP53 determined by gene expression analysis. Overall, TP53 mutations were detected in 12 of 67 MCL samples investigated (18%), and these mutations were associated with an unfavorable clinical course (P = .001).
Regions of homozygous deletions and amplifications
The most frequently detected regions of homozygous deletions were located in 9p21.3 (MCR 607), which includes the genomic loci of CDKN2A, CDKN2B, and methylthioadenosine phosphorylase (MTAP), and in 22q11.22 (MCR 649), which harbors the Igλ locus. Using our statistical approach,16 30 regions of homozygous deletions (of 87, supplemental Table 2) were found to be significantly associated with gene expression changes. Not unexpectedly, the most strongly down-regulated gene in MCR 607 was CDKN2A (Figure 2B); likewise, probe sets annotated to the Igλ locus were expressed at very low levels in the MCR 649, demonstrating the power of our approach to detect biologically meaningful associations between genomic alterations and consequences in gene expression in the affected regions. The most frequently detected amplified region was in 3q (MCRs 12 and 15), leading to up-regulated expression of several genes, including APOD, MFI2, DVL3, DGKG, and EVI1 (data not shown). Altogether, 18 amplified regions with associated changes in gene expression were detected (supplemental Table 2).
Chromosomal alterations associated with an unfavorable prognosis and their putative target genes. (A) Loss of 9p. (B) Double loss of 9p21.3. (C) Loss of 1q32. (D) Double loss of 1p32.3/33. (E) Amplification of 12q14. (F) Double loss of 2q13. Kaplan-Meier curves visualize survival differences between MCL patients with and without the respective CNA. (Right panels) Genes within the MCR that had the greatest difference in fold change in cases with the alteration compared with the cases without the alteration. In the gene expression panels, only genes within the core region of the respective MCR are shown. Genes with a P value more than .05 are shown in gray.
Chromosomal alterations associated with an unfavorable prognosis and their putative target genes. (A) Loss of 9p. (B) Double loss of 9p21.3. (C) Loss of 1q32. (D) Double loss of 1p32.3/33. (E) Amplification of 12q14. (F) Double loss of 2q13. Kaplan-Meier curves visualize survival differences between MCL patients with and without the respective CNA. (Right panels) Genes within the MCR that had the greatest difference in fold change in cases with the alteration compared with the cases without the alteration. In the gene expression panels, only genes within the core region of the respective MCR are shown. Genes with a P value more than .05 are shown in gray.
CNAs of prognostic significance
The potential to integrate high-resolution genomic data with gene expression profiling results and clinical data in a large series of MCL is unique to our dataset and allows us to define survival-associated genetic alterations and to search for altered expression of single genes or pathways that may be of pathogenetic relevance. We identified 8 MCRs associated with gene expression changes that were strongly associated with overall survival (P < .002), 3 of which affected chromosomal regions in chromosome 9 (whole chromosome 9, 9p, and 9p21.3). The remaining 5 MCRs were located in 1q32, 1p33, 1q42, 12q14, and 2q13 (Figure 2; supplemental Table 3). Even though some of these MCRs were present in only a few MCLs, the effect on overall survival was dramatic and statistically highly significant, suggesting that these genetic alterations have major pathogenetic consequences and result in a poor clinical course (Figure 2). Next, we took advantage of a combined analysis of copy number and gene expression alterations to identify genes in these MCRs that were most strongly altered in their expression levels, as these genes may point to the involvement of relevant oncogenic pathways (Figure 2). Not surprisingly, in the most commonly altered region in our series, in 9p, CDKN2A and MTAP had reduced expression in MCL with homozygous deletions in 9p21.3 (Figure 2B). However, more frequently, only one allele in this region is deleted in MCLs, often of a larger size spanning many megabases, and this genetic event may indeed affect additional genes or pathways exemplified by the strong down-regulation of PLAA or MOBKL2B (Figure 2A). In MCLs with a double loss in 1p33, CDKN2C had been suggested earlier as a potential target, but Fas-associated factor 1 (FAF1) actually showed a much stronger down-regulation in our series (Figure 2D). In MCLs with a loss in 1q32, reduced expression of PROX1 may be of importance (Figure 2C) and in MCLs with amplifications in 12q14 CDK4 expression is not as strongly affected as several other genes in this region, including CENTG1, which showed an impressive 16-fold up-regulation in MCLs with this genetic alteration (Figure 2E).
Of note, 6 of these 8 survival-associated MCRs were also significantly associated with an increased proliferation signature3 in MCLs, suggesting that molecular events in these regions have a direct impact on the tumor cell proliferation that is the most important biologic feature of MCL determining outcome (supplemental Table 3).
Putative target genes of frequent, not survival-associated, CNAs in MCL
Although some of the most frequently occurring MCRs in our series were not directly associated with altered survival times of the MCL patients, these alterations are nevertheless probably of pathogenetic relevance. To reveal putative target genes in these CNAs, we used again the integrated analysis approach of the gene expression and SNP array data to identify genes that were most strongly altered in their expression levels. Selected regions and potential target genes are shown in Figure 3.
Putative target genes of frequent CNAs in MCL identified by combined SNP and gene expression profiling. (A) Gain of 7p. (B) Loss of 8p. (C) Loss of 13q34. (D) Double loss of 13q33-q34. (Left panels) Genes within each MCR that had the greatest difference in fold change in cases with the alteration compared with the cases without the alteration. (Right panels) Gene expression levels of the candidate genes in each case; red bars represent the cases with the respective CNA.
Putative target genes of frequent CNAs in MCL identified by combined SNP and gene expression profiling. (A) Gain of 7p. (B) Loss of 8p. (C) Loss of 13q34. (D) Double loss of 13q33-q34. (Left panels) Genes within each MCR that had the greatest difference in fold change in cases with the alteration compared with the cases without the alteration. (Right panels) Gene expression levels of the candidate genes in each case; red bars represent the cases with the respective CNA.
Gain of 7p (MCR 147) was one of the most frequently detected gains in our series (13% of MCLs); moreover, this genetic event was associated with an increased proliferation signature of the tumor cells.3 Several genes were affected in their gene expression levels, but IGF2BP3 stands out with a more than 8-fold up-regulation in MCLs showing 7p gains (Figure 3A). Losses in 8p were also frequent and also associated with an increased proliferation signature. The most strongly down-regulated gene in MCLs carrying this alteration was MCPH1 (Figure 3B).
Chromosome 13 was among the chromosomes most frequently affected by CNAs in this series. Most often, a single copy loss of the complete chromosome 13 occurred (21 of 77 MCLs; Table 2). Among the most strongly down-regulated genes in this MCR was CUL4A located in 13q34 (not shown). The second most frequent alteration in chromosome 13 was a monoallelic or biallelic deletion of 13q33-q34, leading to reduced expression of CUL4A and ING1 (Figure 3C-D). Although chromosome 13 was mostly affected by deletions, there was also a region with recurrent gains/amplifications located in 13q31.3 (MCR 51). The most significant up-regulation in this region was observed for MIHG1, encoding the mir17-92 micro-RNA cluster. For 2 selected genes, namely, FZR1 (located in 19p) and IGF2BP3 (located in 7p), quantitative RT-PCR experiments were performed in a subset of 13 cases confirming the results obtained by microarray analysis (data not shown).
Potential relevance of the Hippo signaling pathway in MCL
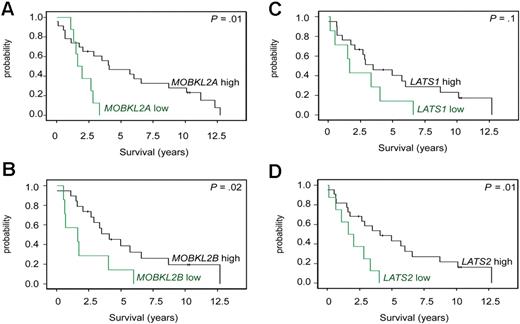
We noticed that, in 3 deleted regions, members of the Hippo signaling pathway had statistically reduced expression levels. Specifically, MOBKL2B was down-regulated in 9p21.2, as were MOBKL2A in 19p13.3 and LATS1 in 6q24/25. The Hippo pathway is supposed to be involved in cancer development by the regulation of proliferation and mitotic checkpoints and therefore of considerable interest in MCLs. Notably, as already shown in Figure 2A, loss of 9p, including MOBKL2B as one of the most affected genes in this region, correlated with an unfavorable clinical course. In this dataset, reduced gene expression levels (cut-off ≤ 25th percentile) of MOBKL2A and MOBKL2B were associated with inferior survival of MCL patients (Figure 4) with P values of .01 and .02, respectively, whereas no significant association between reduced mRNA expression and survival was evident for LATS1. Importantly, the genetic losses in 9p, 19p, and 6q24/25 harboring MOBKL2B, MOBKL2A, and LATS1 were mutually exclusive and affected altogether 29 of 77 MCL cases (38%). Moreover, cases with genetic losses in any one of these regions showed a higher proliferation (P = .01), a higher genetic complexity (CNAs and CNN-LOH, P = .025), and a shorter survival (P = .0001) compared with cases without alterations in any of these genes.
Candidate tumor suppressor genes involved in the Hippo signaling pathway in MCL identified by combined SNP and gene expression profiling. (A) Loss of 9p. (B) Loss of 19p13.3. Kaplan-Meier curves visualizing survival groups according to the mRNA expression determined by gene expression array analysis of MOBKL2B (PS 226844 on HG U133 plus 2.0) located in 9p21.2 and MOBKL2A (PS 235163) located in 19p13.3. For MCR537/loss19p13.3 genes located in the core and extended region are displayed.
Candidate tumor suppressor genes involved in the Hippo signaling pathway in MCL identified by combined SNP and gene expression profiling. (A) Loss of 9p. (B) Loss of 19p13.3. Kaplan-Meier curves visualizing survival groups according to the mRNA expression determined by gene expression array analysis of MOBKL2B (PS 226844 on HG U133 plus 2.0) located in 9p21.2 and MOBKL2A (PS 235163) located in 19p13.3. For MCR537/loss19p13.3 genes located in the core and extended region are displayed.
To validate the potential relevance of the Hippo signaling pathway in MCLs, we studied the mRNA expression levels of MOBKL2A, MOBKL2B, LATS1, and LATS2 in an independent set of 32 MCL cases by quantitative RT-PCR. LATS2 was chosen to be additionally studied because of its location in chromosome 13, which is frequently altered in MCLs. As shown in Figure 5, an association between inferior survival and reduced expression of MOBKL2A, MOBKL2B, and LATS2 could also be demonstrated in this series, thus providing a validation in an independent set of MCL cases. Reduced expression of LATS1 showed a trend toward inferior survival but did not reach statistical significance, perhaps because of the small sample size of the validation group.
Kaplan-Meier curves visualizing MCL survival groups according to the mRNA expression of Hippo pathway associated genes in a validation series. mRNA expression of (A) MOBKL2A, (B) MOBKL2B, (C) LATS1, and (D) LATS2 (cutoff ≤ 25th percentile) in an independent group of 32 untreated MCL patients. Expression was determined by quantitative RT-PCR and the comparative Ct method.
Kaplan-Meier curves visualizing MCL survival groups according to the mRNA expression of Hippo pathway associated genes in a validation series. mRNA expression of (A) MOBKL2A, (B) MOBKL2B, (C) LATS1, and (D) LATS2 (cutoff ≤ 25th percentile) in an independent group of 32 untreated MCL patients. Expression was determined by quantitative RT-PCR and the comparative Ct method.
Discussion
Compared with other lymphoma subtypes, MCL is characterized by a particularly high number of chromosomal gains and losses that are thought to occur secondarily to the underlying hallmark translocation t(11;14) involving cyclin D1. Given the wide range of survival times of MCL patients, it seems probable that the secondary genetic alterations and their molecular consequences are of major importance in determining the aggressiveness of the tumor and therefore the clinical course.2 Although the common chromosomal gains and losses in the MCL genomes have been well established over the last decade,6-14 many of the molecular downstream effects of these alterations remain unresolved. We here present the largest high-resolution gene expression and genomic profiling study available to date that includes 77 primary MCL cases. Moreover, we applied our recently developed algorithm GEDI16 to identify novel putative target genes and pathways.
In line with previous results,12,13 the MCL tumors in our series had highly aberrant karyotypes, and the overall pattern of genomic imbalances is in good concordance with published data from conventional CGH, CGH array, and 100K SNP array analyses.6-8,11,12,14,20 We can also confirm that CNN-LOH is a frequent and nonrandomly distributed event in MCL.12,13 Recurrent large regions of CNN-LOH were detected in chromosomes 6p, 9p, 9q, 11q, and 17p, which are regions that are also frequently affected by deletions in MCL. Interestingly, 3 of 5 MCLs with a CNN-LOH in 17p showed an inactivating mutation of TP53, demonstrating a pathogenetic role of CNN-LOH for the inactivation of this tumor suppressor gene and, potentially, also for tumor suppressor genes in other regions.
The view that cyclin D1–negative MCL exists is now widely established. In this context, it is noteworthy that, also with a high-resolution copy number profiling approach such as ours, no differences in the spectrum of genomic gains and losses could be detected between cyclin D1–positive and cyclin D1–negative MCLs. Intriguingly, the ATM locus in 11q23 that is affected by deletions and/or mutations in approximately 50% of conventional MCLs10,21-23 was found to be altered in all 5 cyclin D1–negative MCLs (4 MCLs with deletions and 1 CNN-LOH), suggesting a common pathogenesis of these 2 MCL subtypes.
The availability of survival data in this study allowed us to specifically search for chromosomal gains and losses and associated gene expression changes that impact on survival of MCL patients. Eight such CNAs could be identified, all of which were associated with inferior survival. Interestingly, 6 of these 8 CNAs were correlated with an increased proliferation signature3 of the tumor cells, demonstrating the central role of this biologic feature for the aggressiveness of the tumor and for the clinical course. In some of these CNAs, putative tumor suppressor genes or potential oncogenes have been described, including CDKN2A in 9p21, CDK4 in 12q14, and BCL2L11 in 2q13. However, our integrated analysis of copy number and gene expression alterations suggests that the situation may be more complex and pinpoints genes or pathways that may have been neglected thus far. For example, reduced or even lost expression of CDKN2A is widely viewed as the most important consequence of genomic losses in 9p21. However, our analysis also demonstrated strong down-regulation of MTAP, in agreement with a previous report, in which reduced or absent MTAP expression was associated with poor survival in MCL.24 Moreover, a member of the Hippo signaling pathway, MOBKL2B, also showed decreased expression in MCL cases with CNAs in 9p21 (later in “Discussion”). In 1q32, PROX1 might be an interesting candidate because it was the only significantly down-regulated gene in the core region and because it has been suggested as a tumor suppressor gene in solid tumors25,26 as well as in hematologic malignancies.27 Homozygous deletions in 1p33 are infrequent in MCLs (in our series, 3 of 77 MCLs), but this genetic event has been repeatedly described previously in MCL cell lines12,13,28 and in primary MCL samples.29 An obvious target gene in this region could be CDKN2C, but a more detailed investigation of this gene by Mestre-Escorihuela et al28 in MCL has been inconclusive. These authors found a decreased expression of CDKN2C in a proportion of MCL cases compared with normal B cells by immunohistochemistry but failed to demonstrate causative mutations or promoter methylation in the majority of investigated cases.28 Our data might suggest that the dramatic loss of expression of FAF1 that is involved in apoptotic and nuclear factor-κB signaling could be the dominant molecular event in MCL cases with single or doubles losses in 1p33.
The comprehensive gene expression and high-resolution analysis of chromosome 13, one of the most frequently altered genomic regions in MCL,6-8,10,20,30-32 may add a few novel candidates that are involved in the 2 main deregulated pathways in MCL, namely, the proliferation and DNA damage response pathways. The most frequent CNA of chromosome 13 according to our algorithm was the deletion of the entire chromosome (MCR 479), followed by monoallelic or biallelic deletions of a minimal region in 13q33-q34. Among the most strongly down-regulated genes in MCL with monoallelic or biallelic deletions in 13q33-q34 were CUL4A and ING1. CUL4A is an E3 ubiquitin ligase that has been shown to be involved in the DNA damage response.33 Very recently, it has been demonstrated that CUL4A binds to the CDKN2A locus, and it has been suggested that the DDB1-CUL4 and MLL1 complexes constitute a novel pathway that mediates p16INK4A activation during an oncogenic checkpoint response.34 ING1 encodes a tumor suppressor protein that is a component of the p53 signaling pathway35 and is involved in the restriction of cell growth and proliferation, apoptosis, maintenance of genomic stability, and modulation of cell cycle checkpoints.36 Down-regulation of CUL4A and ING1 is frequent in MCLs; and because of their involvement in 2 central genetic pathways, the investigation of their potential role in the pathogenesis of MCL appears attractive.
Among the most frequently observed regions with gains/amplifications detected in our MCL series were gains/amplifications of 7p, and the most strongly up-regulated gene in MCL carrying this aberration was IGF2BP3 located in 7p15.3. IGF2BP3 belongs to the VICKZ family of RNA-binding proteins, which are highly conserved in vertebrates and encode for proteins involved in the posttranscriptional regulation processes of RNA and have been implicated in a variety of cellular functions, including cell polarity, migration, and proliferation.37 Increased expression of IGF2BP3 was found to be a marker of unfavorable prognosis, progression, or metastasis in solid tumors.38,39 It is noteworthy that IGF2BP3 is one of the genes in the MCL gene expression signature3 and was found to be predictive of outcome in MCL in an in silico analysis.40 Providing further support for a pathobiologic relevance of IGF2BP3 in MCL, gains of 7p in our series were significantly associated with an increased proliferation of MCL tumors.
Among the most frequently detected CNAs associated with gene expression changes was chromosomal loss of 8p (MCR 408). Loss of 8p has been previously described as a common chromosomal imbalance in MCL10,31 and has been suggested to harbor a putative tumor suppressor locus.41 In our integrated analysis, the most strongly down-regulated gene in MCR408 was MCPH1 located in 8p23. MCPH1 has been suggested to function as a tumor suppressor, as decreased levels of MCPH1 have been associated with genomic instability and metastasis in human cancer.42 MCPH1 interacts with E2F1, and MCPH1 depletion significantly inhibits E2F1-induced apoptosis.43 In MCL, this might be of relevance: given the constitutive expression of cyclin D1 as a result of the translocation t(11;14), RB is nontransiently hyperphosphorylated triggering continuous E2F1 release.
A key finding of our study is the frequent down-regulation of genes associated with the Hippo signaling pathway. This pathway appears to be important for cancer development by the regulation of proliferation and mitotic checkpoints.44,45 Two prominent members of this pathway are MOBKL2B, located in 9p21.2 and MOBKL2A, located in 19p13.3, and low expression of these genes was associated with poor survival in our MCL series. Intriguingly, MOBKL2B has been recently found to be homozygously deleted in the MCL cell line MAVER-1, and monoallelic deletions of this gene are also present in other MCL cell lines.13 MOBKL2A and MOBKL2B are homologs of the MOB1 gene that has potential tumor suppressor properties based on a positive interaction with LATS.44 Two reasons argue for a potential relevance of altered Hippo signaling in MCL: (1) decreased expression of the Hippo members MOBKL2A, MOBKL2B, and LATS2 was associated with inferior survival in an independent validation series of 32 MCL cases; and (2) genomic losses in 9p, 19p, and 6q24/25 harboring MOBKL2B, MOBKL2A, and LATS1 can be observed in 38% of MCLs and, most importantly, are mutually exclusive in our series, suggesting that these alterations might represent alternative mechanisms to affect the Hippo pathway. Importantly, cases with genetic losses in any one of these regions showed a higher proliferation, a higher genetic complexity, and a shorter survival compared with cases without alterations in any of these genes.
In conclusion, our integrated high-resolution analysis of copy number and genome-wide gene expression changes in the largest series of MCL published so far points to novel genes and pathways that affect the 2 crucial pathogenetic mechanisms in MCLs: the disturbed proliferation and DNA damage response pathways. Therefore, this study sets the stage for a more detailed, functional investigation of these genes and pathways in the future, and investigation of the Hippo signaling pathway and its potential pathogenetic relevance in MCL may have a high priority in these efforts.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank all members of the Leukemia and Lymphoma Molecular Profiling Project who are not coauthors of this study, Dr Heike Horn and Dr Ellen Leich for helpful discussions, and Irina Eichelbrönner for expert technical assistance.
This work was supported by the Instituto de Salud Carlos III, Fondo Investigaciones Sanitarias (FIS06/0150 and PI08/0077; S.B.), Comisión Interministerial de Ciencia y Tecnología Española (SAF08/3630; E.C.), Instituto de Salud Carlos III Red Temática de Investigación Cooperativa de Cancer (2006RET2039; E.C.), National Institutes of Health (SPECS grant 5 U01 CA114778-03; E.C., A.R.), and the Lymphoma Research Foundation (LRFMCLI-05-023; E.C., A.R.). A.R. and E.M.H. were supported by the Interdisciplinary Center for Clinical Research, University of Würzburg, Würzburg, Germany. G.O. was supported by the Robert-Bosch-Stiftung GmbH.
National Institutes of Health
Authorship
Contribution: E.M.H. designed and performed research, collected and analyzed the data, and wrote the manuscript; G.W. and W.X. performed statistical analysis; P.J., G.L., and I.S. performed research and analyzed the data; R.M.B., L.M.R., W.-C.C, D.D.W., J.D., E.S.J., R.D.G., S.S.D., H.-K.M.-H., L.M.S., G.O., E.C., and A.R. provided and classified patient samples and collected and analyzed the data; and E.C., S.B., G.O., and A.R. designed research, supervised the study, and wrote the manuscript. All authors critically reviewed the final version of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Andreas Rosenwald, Institute of Pathology, University of Würzburg, Josef-Schneider-Str 2, 97080 Würzburg, Germany; e-mail: Rosenwald@mail.uni-wuerzburg.de.
References
Author notes
G.O., S.B., and A.R. are equal senior authors of this study.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal