Abstract
LMO2, a critical transcription regulator of hematopoiesis, is involved in human T-cell leukemia. The binding site of proline and acidic amino acid–rich protein (PAR) transcription factors in the promoter of the LMO2 gene plays a central role in hematopoietic-specific expression. E2A-HLF fusion derived from t(17;19) in B-precursor acute lymphoblastic leukemia (ALL) has the transactivation domain of E2A and the basic region/leucine zipper domain of HLF, which is a PAR transcription factor, raising the possibility that E2A-HLF aberrantly induces LMO2 expression. We here demonstrate that cell lines and a primary sample of t(17;19)-ALL expressed LMO2 at significantly higher levels than other B-precursor ALLs did. Transfection of E2A-HLF into a non-t(17;19) B-precursor ALL cell line induced LMO2 gene expression that was dependent on the DNA-binding and transactivation activities of E2A-HLF. The PAR site in the LMO2 gene promoter was critical for E2A-HLF-induced LMO2 expression. Gene silencing of LMO2 in a t(17;19)-ALL cell line by short hairpin RNA induced apoptotic cell death. These observations indicated that E2A-HLF promotes cell survival of t(17;19)-ALL cells by aberrantly up-regulating LMO2 expression. LMO2 could be a target for a new therapeutic modality for extremely chemo-resistant t(17;19)-ALL.
Introduction
Transcription factors that regulate normal hematopoiesis are frequently involved in leukemogenesis1,2 through 2 types of chromosomal translocation: one causes in-frame fusion of 2 genes and the resultant chimeric transcription factor acquires novel functions and/or functionally disrupts the normal gene products, and the other causes aberrant activation of a transcription factor gene due to juxtaposition to a strong enhancer of the immunoglobulin or T-cell receptor (TCR) loci. The LMO2 gene, located on the short arm of chromosome 11 at band 13 (11p13), was discovered from a recurrent site of translocations in T-cell acute lymphoblastic leukemia (T-ALL) as a paradigm of the latter type of translocation.3,4 LMO2 is a member of the LIM-only zinc finger protein family and is present in a transcription factor complex5 that also includes E2A, TAL1, GATA1, and LDB1 in erythroid cells.6 Within this complex, LMO2 mediates the protein-protein interactions by recruiting LDB1,7 whereas TAL1, GATA1, and E2A directly bind to the specific DNA target sites.6,8 Homozygous null mutation of Lmo2 showed embryonic lethality due to lack of yolk sac erythropoiesis,9 and chimeric animals produced from homozygous-deficient embryonic stem cells demonstrated a requirement of Lmo2 in adult hematopoiesis10 and angiogenic remodeling of the vasculature.11 Lmo2 is expressed in long-term repopulating hematopoietic stem cells12 and in hematopoietic progenitors,9 and its expression is maintained in erythroid cells during differentiation. In contrast, Lmo2 expression is repressed in terminally differentiated granulocytes, macrophages, T cells, and B cells9 with the exception of germinal center B cells.13-15 During T-cell development, Lmo2 is expressed in immature CD4/CD8 double-negative thymocytes and is down-regulated as maturation progresses,16 and transgenic mice expressing Lmo2 using a thymocyte-specific promoter developed an accumulation of CD4/CD8 double-negative thymocytes and eventually a T-cell lymphoma.17-20 Of note, among X-linked severe combined immunodeficiency patients receiving retroviral IL2Rγc gene therapy, 2 patients developed T-ALL due to aberrant activation of LMO2 via integration of the retroviral vector in the LMO2 gene.16,21,22 These observations suggested that deregulated LMO2 increases susceptibility to T-cell malignancies by blocking differentiation. Although similar down-regulation of LMO2 was suggested during B-cell development,9 the significance of LMO2 expression in B-precursor ALL remains totally unclarified.
The LMO2 gene has 2 transcriptional promoters and comprises 6 exons, of which exons 4, 5, and 6 encode the protein.23 The distal and proximal promoters are located upstream of exon 1 or 3 of the larger transcripts, respectively, and the 2 resultant transcripts encode the same open reading frame. The proximal promoter is active in hematopoietic progenitor and endothelial cells, dependent on activation of 3 conserved Ets sites,24 but transgenic analysis demonstrated that the proximal promoter alone is insufficient for full expression of the Lmo2 gene in hematopoietic cells.24 The distal promoter is involved in hematopoietic-specific LMO2 gene expression that is dependent on activation of the proline and acidic amino acid-rich protein (PAR) site in the LMO2 gene promoter.25 The PAR transcription factors belong to the basic region/leucine zipper (bZIP) factor family and include hepatic leukemic factor (HLF),26,27 albumin gene promoter D-site binding protein (DBP),28 and thyrotroph embryonic factor (TEF).29 Among the PAR transcription factors, it has been demonstrated that TEF shows the highest potential to activate the LMO2 promoter in erythroid cells.25
t(17;19)(q21-q22;p13) is a relatively rare translocation among childhood ALL cases2 and is linked with the B-precursor phenotype. E2A-HLF derived from t(17;19) promotes anchorage-independent growth of murine fibroblasts30,31 and protects cells from apoptosis induced by growth factor deprivation,32-34 and E2A-HLF transgenic mice develop T-cell malignancies.35,36 In E2A-HLF chimera, the transactivation domain of E2A fuses to the bZIP dimerization and DNA-binding domain of HLF, one of the PAR transcription factors.26,27 As a result, E2A-HLF recognizes the consensus sequence of PAR transcription factors as a dimer and transactivates downstream target genes.27,31,37,38 Considering the critical involvement of the PAR site in the distal promoter of the LMO2 gene in the hematopoietic-specific expression of LMO2,25 E2A-HLF might induce aberrant expression of LMO2 through the distal promoter. In the present study, we show aberrantly higher expression of LMO2 in t(17;19)-ALL as one of the direct targets of E2A-HLF. The biologic significance of LMO2 in leukemogenesis of t(17;19)-ALL is also investigated and discussed.
Methods
Leukemia cell lines and patient sample
Four ALL cell lines with 17;19 translocation (UOC-B1, HALO1, YCUB2, Endo-kun) were used in this study. As B-precursor ALL cell lines, 9 MLL-rearranged ALL cell lines (KOPN-1, KOPB-26, KOCL-33, -44, -45, -50, -51, -58, and -69),39 6 Philadelphia chromosome (Ph1)–positive ALL cell lines (KOPN-30bi, -57bi, -66bi, -72bi, YAMN-73, and -91),40 7 t(1;19)-ALL cell lines (697, KOPN-34, -36, -60, -63, YAMN-90, and -92), and 6 other ALL cell lines including 1 with t(12;21) (Reh) and 5 with others (KOPN-35, -61, -62, -79, and -84) were used. Seven T-ALL cell lines (KOPT-K1, -5, -6, -11, YAMT-12, Jurkat, and MOLT4F), 4 Burkitt B-cell lines (KOBK-130, Daudi, Namalwa, and Raji), and 4 Epstein-Barr virus (EBV)–transformed normal B-cell lines (YAMB-1, -3, -4, and -9) were also used. All cell lines were maintained in RPMI1640 medium supplemented with 10% fetal calf serum (FCS) in a humidified atmosphere of 5% CO2 at 37°C. Analysis of a sample from a patient with t(17;19)-ALL was approved by the Ethical Review Board of the University of Yamanashi. Mononuclear cells (blasts > 95%) that had been isolated from bone marrow aspirates of the patient by Ficoll-Hypaque density centrifugation were stored in liquid nitrogen with 15% dimethyl sulfoxide in fetal calf serum (FCS).
Isolation of normal B precursors
The CD34+ population was separated from human cord blood mononuclear cells (MNCs) using MACS MicroBeads (Miltenyi Biotec) and, subsequently, CD34+/CD19− and CD34+/CD19+ populations were sorted by flow cytometry (FACS Vantage; Becton Dickinson) using FITC-Lineage marker (CD3, CD4, CD8, CD11b, CD56, CD235a, CD41a) in combination with PE-CD34 and APC-CD19. CD19+/IgM− and CD19+/IgM+ populations were sorted from human cord blood MNCs by flow cytometry using FITC-Lineage marker in combination with PE-IgM and APC-CD19. The CD19+ population was also directly separated from peripheral blood MNCs with MACS MicroBeads. RNA was extracted from each population using RNeasy mini kit (QIAGEN), and cDNA was synthesized using SuperScript VILO cDNA synthesis kit (Invitrogen).
Western blot analysis
Cells were solubilized in Nonidet P-40 lysis buffer, and total cellular proteins were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) under reducing conditions. After transfer onto nitrocellulose membrane and blocking with 5% nonfat dry milk in 0.05% Tween-20 Tris [tris(hydroxymethyl)aminomethane]–buffered saline (TBS), the membrane was incubated with the primary antibodies in 5% milk in TBS. Goat anti–human LMO2 and mouse anti–human α-Tubulin antibodies were purchased from R&D Systems and Sigma-Aldrich, respectively. Rabbit anti–human antibodies against E2A and HLF(C) were established as previously reported.37 Membranes were incubated with horseradish peroxidase–conjugated rabbit anti–goat, goat anti–mouse, and goat anti–rabbit IgG (1:1000 dilution; MBL) and were then developed using the enhanced chemiluminescence kit (Amersham Pharmacia Biotech).
Real-time PCR analysis
Total RNA was extracted using Trizol reagent (Invitrogen). Reverse transcription (RT) was performed using random hexamer (Amersham Bioscience) by Superscript II reverse transcriptase (Invitrogen), and then the cDNA product was incubated with RNase (Invitrogen). For quantitative real-time polymerase chain reaction (PCR) of LMO2, triplicated samples containing cDNA with Taqman Universal PCR Master Mix (Applied Biosystems) and Gene Expression Product (exons 1/2, HS00951959_m1; exons 4/5, HS00277106_m1; Applied Biosystems) were amplified according to the manufacturer's protocol using KOPT-6 derived from T-ALL with t(11;14) as a control. As an internal control for relative gene expression, quantitative real-time PCR for GAPDH (Hs 99999905_m1, Applied Biosystems) was performed.
Semiquantitative PCR of transcripts derived from distal and proximal promoters
RNA transcripts originating from the distal LMO2 promoter were quantified with forward primer 5′-CAAAGCAGGCAATTAGCCC-3′ and reverse primer 5′-CCTCTCCACTAGCTACTGC-3′, which are situated in exons 1 and 2, respectively. Total LMO2 expression was quantified with forward primer 5′- GAGCTGCGACCTCTGTGG-3′ and reverse primer 5′-CACCCGCATTGTCATCTCAT-3′, which are situated in exons 5 and 6, respectively. Standard curves were created against a single copy of the LMO2 full-length cDNA subcloned into the pGEMT Easy (Promega) backbone. The assay was performed in triplicate, and the mean quantity of proximal promoter-derived transcripts was directly calculated by subtracting the mean quantity of distal promoter-derived transcripts from the mean quantity of total transcripts. To consider the degree of approximation of the calculated mean, the following equation was considered: var(a+b) = var(a)+var(b)+2cov(a,b). The standard deviation of the quantity of proximal promoter-derived transcripts is therefore assumed to be represented as follows: SD prox LMO2 = sqrt [(SD total LMO2)2- (SD distal LMO2)2].
Construction of eukaryotic expression vectors and transfection
Expression plasmids containing wild-type and mutated E2A-HLF cDNA were constructed with the pMT-CB6+ eukaryotic expression vector (a gift from F. Rauscher III, Wistar Institute, Philadelphia, PA),32 which contains the inserted cDNA under control of a sheep metallothionein promoter. ΔAD1/ΔLH mutant and Basic region mutant (BX) were prepared as previously reported.30,33 Transfectants were generated by electroporation followed by selection using neomycin analog G418 as previously reported.33
Electrophoretic mobility shift assay
Nuclear extracts of cells were prepared and binding reactions were performed as previously reported.31,37 Briefly, a 32P-end-labeled oligonucleotide probe containing wild-type HLF consensus sequence (CS; 5′-GCTACATATTACGTAATAAGCGTT-3′) was incubated in 10 μL of binding buffer and 5 μL of nuclear lysates in the presence of 1 μg of shared calf thymus DNA. In the competition inhibition experiments, an approximately 100-fold molar excess of the unlabeled oligonucleotide was added to the reaction mixture. Polyvalent HLF(C) or E2A rabbit antiserum was added to the nuclear lysates before the DNA-binding reaction.
Reporter assays
−512/+428 KpnI-HindIII fragment and −512/+249 KpnI/BglII fragment of the LMO2 gene generated by PCR were cloned into the pGL3 basic vector (Promega). −512/+428 PARM that has the sequence CATCGATCAT instead of ATTACATCAT in the PAR site was generated by PCR mutagenesis. All constructs were subjected to nucleotide sequence analysis to verify the appropriate insertions. pGL3 control vector and pRL-TK vector were used as the positive control and internal control of transfection efficiency, respectively. For transfection, wild type and E2A-HLF-expressing Nalm6 cells were plated at 5 × 105 cells/well in a 24-well plate and a total of 5 μg of luciferase reporter plasmid, 1 μg of pRL-TK and 2 μL of lipofectamine (Invitrogen) in 50 μL of serum-free medium (Opti-MEM I; Invirogen) were added. After 24 hours of culture, 100μM ZnSO4 at final concentration was added to induce E2A-HLF expression. Cells were harvested 48 hours after transfection and lysed in 50 μL of lysis buffer (Promega). Activities of firefly and Renilla luciferases in each lysate were measured sequentially using the Dual-Luciferase reporter assay system from Promega by a luminometer according to the manufacturer's instructions.
Lentivirus shRNA/siRNA expression vectors and infection
pLL3.7 lentiviral vector was engineered to produce short hairpin RNAs (shRNAs) under the control of mouse U6 promoter and co-express green fluorescent protein (GFP) as a reporter gene by cytomegalovirus-derived promoter–GFP expression cassette.41 Short interfering RNA (siRNA) target sequences were designed to be homologous to the wild-type LMO2 cDNA sequence, and oligonucleotides were subcloned into pLL3.7.42 The selected sequences were submitted to BLAST search to assure that only LMO2 was targeted. Among 5 sets of oligonucleotides containing siRNA target sequences, the following set was selected for further analysis due to the specificity and efficiency: 5′-TgacgcatttcggttgagaaTTCAAGAGAttctcaaccgaaatgcgtcTTTTTTC-3′(Blunt-siRNA/sense hairpin-siRNA/antisense-polyA-XhoI/forward);5′-TCGAGAAAAAAgacgcatttcggttgagaa-TCTCTTGAAttctcaaccgaaatgcgtcA-3′(reverse). The control vector contained the following as ineffective set: 5′-TgcaatattacatatacgccTTCAAGAGAggcgtatatgtaatattgcTTTTTTC-3′ (forward); 5′-TCGAGAAA-AAAgcaatattacatatacgccTCTCTTGAAggcgtatatgtaatattgcA-3′(reverse). pLL3.7 shRNA vector or control vector was cotransfected with packaging vector into 293FT cells and the resulting supernatant was collected after 36 hours.42 Lentivirus was recovered after ultracentrifugation and infected to UOC-B1 cells.
Results
Aberrant expression of LMO2 in t(17;19)-ALL
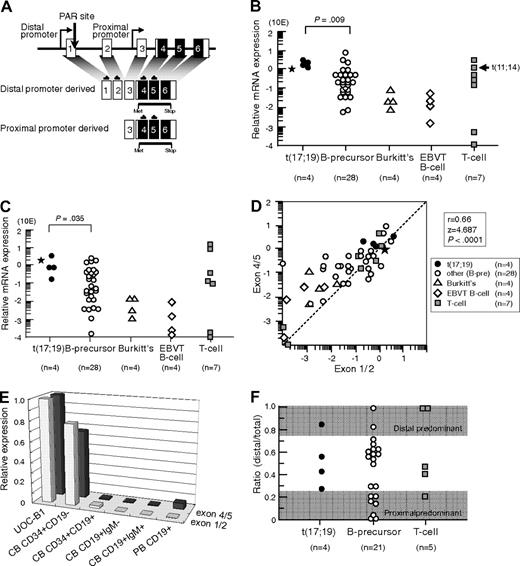
We first analyzed LMO2 gene expression in 4 t(17;19)-ALL cell lines, UOC-B1, HALO1, YCUB2, and Endo-kun, by real-time RT-PCR using 2 different sets of primers (Figure 1A): one set directed toward exons 1 and 2 that is specific for the transcripts derived from the distal promoter, and the other set directed toward exons 4 and 5 that is specific for the transcripts derived from both the distal and proximal promoters. Real-time RT-PCR analysis using the primers specific for exons 4 and 5 demonstrated that all 4 t(17;19)-ALL cell lines expressed LMO2 transcripts at an equivalent level to that in KOPT6, a T-ALL cell line that aberrantly expresses LMO2 as a result of t(11;14) (Figure 1B). The level of LMO2 transcripts in t(17;19)-ALL cell lines was significantly higher than that in 28 other B-precursor ALL cell lines (P = .009, Mann-Whitney test) including Ph1-ALL, t(1;19)-ALL, and MLL+ALL (supplemental Figure 1A, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Real-time RT-PCR analysis using the primers specific for exons 1 and 2 demonstrated that the level of LMO2 transcripts derived from the distal promoter in t(17;19)-ALL cell lines was also significantly higher than that in the other B-precursor ALL cell lines (Figure 1C and supplemental Figure 1B). A strong correlation was observed between the levels of LMO2 transcripts quantified by the 2 sets of primers among the 47 cell lines (r = 0.66, P < .0001; Figure 1D). The primary sample from a t(17;19)-ALL patient also demonstrated high levels of LMO2 transcripts (Figure 1B-C). Consistent with down-regulation of LMO2 gene expression during the progression of normal B-cell development,9 the gene expression level of LMO2 in Burkitt B-cell lines as well as EBV-transformed normal B-cell lines was unanimously low. Thus, the LMO2 gene expression level during B-cell development was analyzed using fractions of cord blood and peripheral blood MNCs (Figure 1E). The LMO2 gene expression level in the CD34+/CD19− population of cord blood MNCs was almost equivalent to that in UOC-B1, and it was markedly down-regulated in the CD34+/CD19+ population. This low expression level was sustained in the CD19+/IgM− and the CD19+/IgM+ populations of cord blood MNCs as well as in the CD19+ population of peripheral blood MNCs.
LMO2 gene expression in t(17;19)-ALL. (A) Schematic representation of 2 LMO2 gene promoters and primers for real-time RT-PCR analysis. Primers directed toward exons 1 and 2 specifically detect transcripts derived from the distal promoter, and those directed toward exons 4 and 5 detect transcripts derived from both the distal and proximal promoters. (B) Relative LMO2 gene expression determined by real-time RT-PCR using the primers for exons 4 and 5. Arrow indicates T-ALL cell line with t(11;14), and asterisk indicates the level of LMO2 transcripts in a primary leukemia sample from the patient with t(17;19)-ALL. The P value determined by Mann-Whitney test is indicated. (C) Relative LMO2 gene expression determined by real-time RT-PCR using the primers for exons 1 and 2. (D) Correlation between the levels of LMO2 transcripts quantified by the primers for exons 4 and 5 (vertical axis) and those for exons 1 and 2 (horizontal axis). (E) LMO2 gene expression in CD34+/CD19−, CD34+/CD19+, CD19+/IgM−, and CD19+/IgM+ populations of cord blood mononuclear cells (MNCs) and CD19+ population of peripheral blood MNCs. Relative LMO2 gene expression was determined by real-time RT-PCR using UOC-B1 as a control with the primers for exons 1 and 2 and the primers for exons 4 and 5. The gene expression level of β-actin was used as an internal control. SE of triplicated samples was always less than 10%. (F) Ratio of LMO2 gene expression derived from the distal promoter to total LMO2 gene expression in ALL cell lines. LMO2 gene expression in the cell lines that expressed a total LMO2 gene level of at least 10 times lower than that in UOC-B1 was semiquantifiedby real-time PCR with the specific primers for LMO2 transcripts sourced at the distal promoter and for total LMO2 transcripts. The dark areas indicate a proximal promoter-predominant pattern (ratio < 0.25) or a distal promoter-predominant pattern (ratio > 0.75).
LMO2 gene expression in t(17;19)-ALL. (A) Schematic representation of 2 LMO2 gene promoters and primers for real-time RT-PCR analysis. Primers directed toward exons 1 and 2 specifically detect transcripts derived from the distal promoter, and those directed toward exons 4 and 5 detect transcripts derived from both the distal and proximal promoters. (B) Relative LMO2 gene expression determined by real-time RT-PCR using the primers for exons 4 and 5. Arrow indicates T-ALL cell line with t(11;14), and asterisk indicates the level of LMO2 transcripts in a primary leukemia sample from the patient with t(17;19)-ALL. The P value determined by Mann-Whitney test is indicated. (C) Relative LMO2 gene expression determined by real-time RT-PCR using the primers for exons 1 and 2. (D) Correlation between the levels of LMO2 transcripts quantified by the primers for exons 4 and 5 (vertical axis) and those for exons 1 and 2 (horizontal axis). (E) LMO2 gene expression in CD34+/CD19−, CD34+/CD19+, CD19+/IgM−, and CD19+/IgM+ populations of cord blood mononuclear cells (MNCs) and CD19+ population of peripheral blood MNCs. Relative LMO2 gene expression was determined by real-time RT-PCR using UOC-B1 as a control with the primers for exons 1 and 2 and the primers for exons 4 and 5. The gene expression level of β-actin was used as an internal control. SE of triplicated samples was always less than 10%. (F) Ratio of LMO2 gene expression derived from the distal promoter to total LMO2 gene expression in ALL cell lines. LMO2 gene expression in the cell lines that expressed a total LMO2 gene level of at least 10 times lower than that in UOC-B1 was semiquantifiedby real-time PCR with the specific primers for LMO2 transcripts sourced at the distal promoter and for total LMO2 transcripts. The dark areas indicate a proximal promoter-predominant pattern (ratio < 0.25) or a distal promoter-predominant pattern (ratio > 0.75).
Next, the contribution of the distal promoter was analyzed in those cell lines that expressed the total LMO2 gene at a level of at least 10 times lower than that in UOC-B1 by real-time PCR using standard curves, which were created against DNA template of the full-length LMO2 cDNA that was subcloned into the vector as a single copy. The ratio of the quantity of LMO2 transcripts sourced at the distal promoter to the quantity of total LMO2 transcripts in the t(17;19)-ALL cell lines was 0.26 to 0.83 (Figure 1F), indicating that both the distal and the proximal promoters contributed to LMO2 gene expression. None of the 4 t(17;19)-ALL cell lines showed a proximal promoter-predominant pattern (ratio < 0.25), while 7 of 21 other B-precursor ALL cell lines and 1 of 5 T-ALL cell lines showed a proximal promoter-predominant pattern.
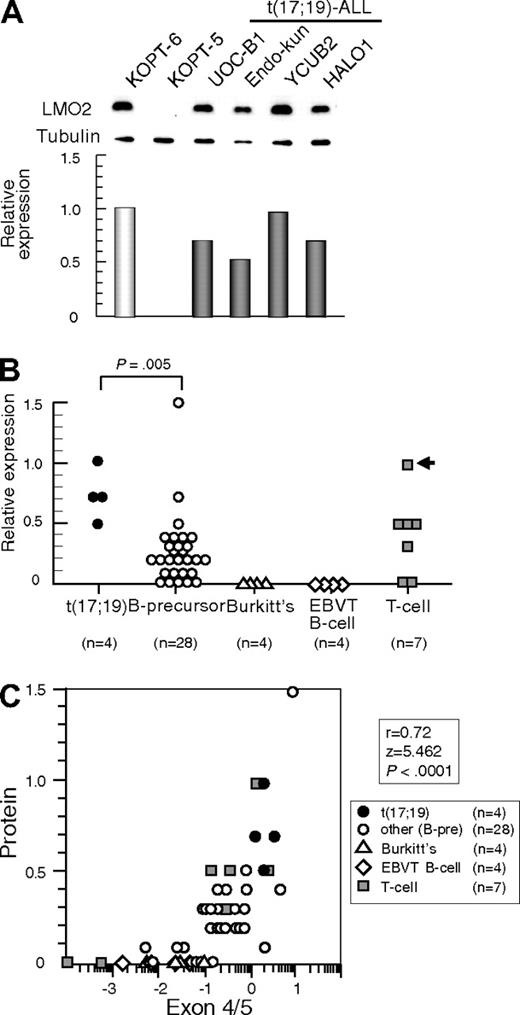
We next analyzed LMO2 protein expression by Western blotting using α-tubulin expression as an internal control. Consistent with the gene expression level, LMO2 protein was aberrantly expressed in all 4 t(17;19)-ALL cell lines at a similarly high level to that in the T-ALL cell line with t(11;14) (Figure 2A). compared with other B-precursor ALL cell lines (Figure 2B), the t(17;19)-ALL cell lines expressed significantly higher levels of LMO2 protein (P = .005, Mann-Whitney test). Consistent with the low gene expression level, protein expression of LMO2 was undetectable in both Burkitt B-cell lines and EBV-transformed normal B-cell lines. Strong correlations were observed between the levels of LMO2 protein and LMO2 transcripts analyzed by the primers specific for exons 4 and 5 (r = 0.72, P < .0001, Figure 2C) and those for exons 1 and 2 (r = 0.42, P = .0089). These observations indicated that t(17;19)-ALL cells aberrantly express LMO2.
LMO2 protein expression in t(17;19)-ALL. (A) Western blot analysis of LMO2. Relative expression of each cell line was determined by quantifying the intensity of each band using KOPT6, a T-ALL cell line with t(11;14), as a positive control and KOPT-5, a T-ALL cell line without LMO2 expression, as a negative control, and normalized by the level of α-tubulin expression as an internal control. (B) Relative level of LMO2 protein expression. The P value determined by Mann-Whitney test is indicated. (C) Correlation between levels of relative protein expression of LMO2 (vertical axis) and gene expression of LMO2 analyzed by real-time RT-PCR using the primers for exons 4 and 5 (horizontal axis).
LMO2 protein expression in t(17;19)-ALL. (A) Western blot analysis of LMO2. Relative expression of each cell line was determined by quantifying the intensity of each band using KOPT6, a T-ALL cell line with t(11;14), as a positive control and KOPT-5, a T-ALL cell line without LMO2 expression, as a negative control, and normalized by the level of α-tubulin expression as an internal control. (B) Relative level of LMO2 protein expression. The P value determined by Mann-Whitney test is indicated. (C) Correlation between levels of relative protein expression of LMO2 (vertical axis) and gene expression of LMO2 analyzed by real-time RT-PCR using the primers for exons 4 and 5 (horizontal axis).
Up-regulation of the LMO2 gene expression by E2A-HLF
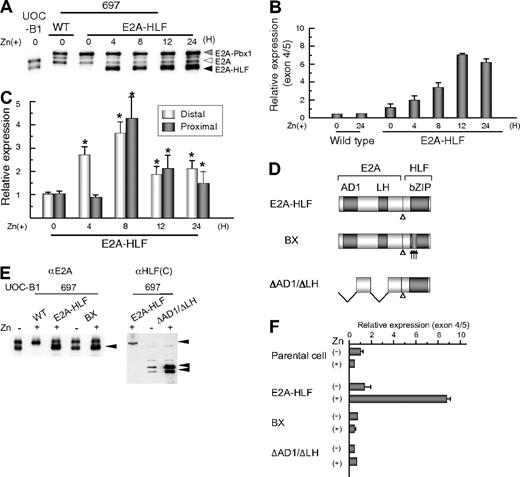
To test the possibility that aberrant expression of LMO2 in t(17;19)-ALL cells is driven by E2A-HLF, we transfected E2A-HLF into B-precursor ALL cell line 697, which has t(1;19) and expresses approximately 100-fold lower level of LMO2 gene than the t(17;19)-ALL cell lines, using a zinc-inducible vector. In E2A-HLF-transfected 697 cells, E2A-HLF was up-regulated to a level equivalent to that in UOC-B1 cells within 4 hours of the addition of zinc to the culture medium (Figure 3A). When analyzed by real-time RT-PCR using primers specific for exons 4 and 5 (Figure 3B), LMO2 gene expression was up-regulated by the addition of zinc in the E2A-HLF-transfected 697 cells but not in the wild-type 697 cells. When the LMO2 gene expression derived from the distal and the proximal promoters was differentially semiquantified by real-time PCR (Figure 3C), significant gene expression derived from the distal promoter was immediately induced within 4 hours after the addition of zinc. Subsequently, significant gene expression derived from the proximal promoter was induced within 8 hours after the addition of zinc. These observations suggest that the distal and the proximal promoters of the LMO2 gene sequentially contribute to E2A-HLF–induced LMO2 gene expression. We further tested 2 types of E2A-HLF mutants in 697 cells. BX contains substitutions of 6 critical basic amino acids in the basic region of HLF to abolish DNA-binding ability, while ΔAD1/ΔLH lacks 2 transactivation domains of E2A to abolish transactivation ability but retain DNA-binding ability (Figure 3D).30,33 Despite almost equivalent levels of expression of each mutant protein to that of E2A-HLF (Figure 3E), the gene expression level of LMO2 remained unchanged after the addition of zinc (Figure 3F), indicating that both the DNA-binding and transactivation abilities of E2A-HLF are required for induction of LMO2 gene expression.
Induction of LMO2 gene expression by E2A-HLF. (A) Induction of E2A-HLF expression. Lysates of wild type (WT) and a clone of E2A-HLF–transfected 697 cells as well as UOC-B1 cells harvested at the indicated time after the addition of zinc were blotted with an anti-E2A serum. Gray, white, and black arrowheads indicate E2A-Pbx1, E2A, and E2A-HLF, respectively. (B) Time course analysis of LOM2 gene expression after induction of E2A-HLF. Levels of LMO2 transcripts were quantified by real-time RT-PCR using the primers for exons 4 and 5, normalized by GAPDH gene expression as an internal control. Changes in fold induction of LMO2 gene expression level to that in wild type 697 cells cultured in the absence of zinc are shown as the mean ± SE of triplicate samples. (C) Time course analysis of LOM2 gene expression derived from the distal and proximal promoters after induction of E2A-HLF. Levels of LMO2 transcripts in E2A-HLF–transfected 697 cells cultured in the presence of zinc were semiquantified by real-time RT-PCR with the specific primers for LMO2 transcripts sourced at the distal promoter and for total LMO2 transcripts. Changes in fold induction of LMO2 gene expression level to that in E2A-HLF–transfected cells cultured in the absence of zinc are shown as the mean ± SE of triplicate samples. Asterisks indicate the significant gene induction determined by t-test. (D) Schematic diagram of mutants of E2A-HLF. (E) Western blot analysis of mutants of E2A-HLF. Lysates of UOC-B1 cells and wild-type (WT) and clones of 697 cells transfected with E2A-HLF, BX, and ΔAD1/ΔLH cultured in the absence or presence of zinc for 24 hours were blotted with E2A (left panel) and HLF(C) (right panel) antisera. (F) LMO2 gene expression in mutant E2A-HLF–transfected 697 cells. Wild-type (WT) and transfectants of 697 cells were cultured in the absence or presence of zinc for 24 hours, and the levels of LMO2 transcripts were quantified by real-time RT-PCR. Changes in fold induction of LMO2 gene expression level to that in wild-type 697 cells cultured in the absence of zinc are shown as the mean ± SE of triplicate samples.
Induction of LMO2 gene expression by E2A-HLF. (A) Induction of E2A-HLF expression. Lysates of wild type (WT) and a clone of E2A-HLF–transfected 697 cells as well as UOC-B1 cells harvested at the indicated time after the addition of zinc were blotted with an anti-E2A serum. Gray, white, and black arrowheads indicate E2A-Pbx1, E2A, and E2A-HLF, respectively. (B) Time course analysis of LOM2 gene expression after induction of E2A-HLF. Levels of LMO2 transcripts were quantified by real-time RT-PCR using the primers for exons 4 and 5, normalized by GAPDH gene expression as an internal control. Changes in fold induction of LMO2 gene expression level to that in wild type 697 cells cultured in the absence of zinc are shown as the mean ± SE of triplicate samples. (C) Time course analysis of LOM2 gene expression derived from the distal and proximal promoters after induction of E2A-HLF. Levels of LMO2 transcripts in E2A-HLF–transfected 697 cells cultured in the presence of zinc were semiquantified by real-time RT-PCR with the specific primers for LMO2 transcripts sourced at the distal promoter and for total LMO2 transcripts. Changes in fold induction of LMO2 gene expression level to that in E2A-HLF–transfected cells cultured in the absence of zinc are shown as the mean ± SE of triplicate samples. Asterisks indicate the significant gene induction determined by t-test. (D) Schematic diagram of mutants of E2A-HLF. (E) Western blot analysis of mutants of E2A-HLF. Lysates of UOC-B1 cells and wild-type (WT) and clones of 697 cells transfected with E2A-HLF, BX, and ΔAD1/ΔLH cultured in the absence or presence of zinc for 24 hours were blotted with E2A (left panel) and HLF(C) (right panel) antisera. (F) LMO2 gene expression in mutant E2A-HLF–transfected 697 cells. Wild-type (WT) and transfectants of 697 cells were cultured in the absence or presence of zinc for 24 hours, and the levels of LMO2 transcripts were quantified by real-time RT-PCR. Changes in fold induction of LMO2 gene expression level to that in wild-type 697 cells cultured in the absence of zinc are shown as the mean ± SE of triplicate samples.
Essential role of the PAR site in the distal promoter of the LMO2 gene for up-regulation of LMO2 expression by E2A-HLF
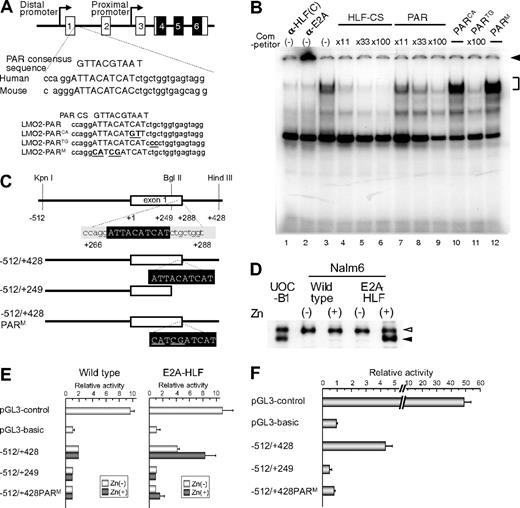
To determine whether E2A-HLF binds to the PAR site in the distal promoter of the LMO2 gene, we performed electrophoretic mobility shift assay using HLF-CS sequence as a probe in the presence of the double-stranded oligomers listed in Figure 4A as competitors.25 The DNA-protein complex of E2A-HLF, which was ablated or supershifted in the presence of anti-HLF(C) (Figure 4B lane 1) or anti-E2A (Figure 4B lane 2) antisera, respectively, was competed by the addition of oligomers centered on the PAR site in the distal promoter region (Figure 4B lanes 7-9), although less effectively than unlabeled HLF-CS probe (Figure 4B lanes 3-6). The oligomers containing 2-bp replacement in the 3′ region immediately outside of the PAR site (PARTG) effectively competed the formation of DNA-protein complex (Figure 4B lane 11), while those containing 2-bp (PARCA) or 4-bp (PARM) replacement within the PAR site (Figure 4B lanes 10,12) did not, indicating that the PAR site in the distal promoter of the LMO2 gene is critical for the binding of E2A-HLF. We also performed electrophoretic mobility shift assay using the sequence of the PAR site in the distal promoter of the LMO2 gene as a probe, but could not find significant binding (data not shown). High levels of sequence conservation among the mammalian sequences were reported across the entire LMO2 genomic region.24 The PAR site sequence in the distal promoter is highly conserved in chimpanzee (Pan troglodytes), cow (Bos Taurus), and mouse (Mus musculus; supplemental Figure 2A), suggesting that the PAR site plays an essential role in the LMO2 gene expression in these mammalian species. Consistent with conservation of the PAR site in the distal promoter of mouse lom2 gene, when E2A-HLF was transfected into FL5.12 cells, an IL-3–dependent mouse pro-B cell line, using zinc inducible vector as reported before,33 lmo2 gene expression derived from the distal promoter was up-regulated by the addition of zinc in the E2A-HLF-transfected FL5.12 cells but not in the empty vector–transfected FL5.12 cells (supplemental Figure 2B).
Involvement of the PAR site in the distal promoter of LMO2 gene in E2A-HLF–induced LMO2 expression. (A) Schematic representation of the proline and acidic amino acid–rich protein (PAR) site in the distal promoter of the LMO2 gene and oligonucleotides used as competitors in electrophoretic mobility shift assay. Mutations in the sequence are underlined. (B) Electrophoretic mobility shift assay performed in nuclear extracts from UOC-B1 cells using an HLF-CS sequence as a probe in the presence of a series of double-stranded oligomers as competitors (lanes 4-12) or anti-HLF(C) (lane 1) and anti-E2A (lane 2) sera. The molar ratio of cold competitor to probe is indicated in each lane. The specific DNA-protein complex is indicated by the bracket and the supershifted complex is indicated by the arrowhead. (C) Schematic representation of 3 reporter constructs for reporter assay. Mutations in the sequence are underlined. (D) Western blot analysis of E2A-HLF–transfected Nalm6 cells. Lysates of UOC-B1 cells and wild type and E2A-HLF transfected-Nalm6 cells cultured in the absence or presence of zinc for 24 hours were blotted with E2A antisera. Open and closed arrowheads indicate E2A and E2A-HLF, respectively. (E) Luciferase assay in E2A-HLF–transfected Nalm6 cells. Assays were performed in wild type and E2A-HLF–transfected Nalm6 cells cultured in the absence or presence of zinc for 24 hours after transient transfection of each reporter plasmid. The values were normalized for transfection efficiencies using a cotransfected Renilla luciferase construct. (F) Luciferase assay in YCUB2 cells. The values were normalized for transfection efficiencies using a cotransfected Renilla luciferase construct.
Involvement of the PAR site in the distal promoter of LMO2 gene in E2A-HLF–induced LMO2 expression. (A) Schematic representation of the proline and acidic amino acid–rich protein (PAR) site in the distal promoter of the LMO2 gene and oligonucleotides used as competitors in electrophoretic mobility shift assay. Mutations in the sequence are underlined. (B) Electrophoretic mobility shift assay performed in nuclear extracts from UOC-B1 cells using an HLF-CS sequence as a probe in the presence of a series of double-stranded oligomers as competitors (lanes 4-12) or anti-HLF(C) (lane 1) and anti-E2A (lane 2) sera. The molar ratio of cold competitor to probe is indicated in each lane. The specific DNA-protein complex is indicated by the bracket and the supershifted complex is indicated by the arrowhead. (C) Schematic representation of 3 reporter constructs for reporter assay. Mutations in the sequence are underlined. (D) Western blot analysis of E2A-HLF–transfected Nalm6 cells. Lysates of UOC-B1 cells and wild type and E2A-HLF transfected-Nalm6 cells cultured in the absence or presence of zinc for 24 hours were blotted with E2A antisera. Open and closed arrowheads indicate E2A and E2A-HLF, respectively. (E) Luciferase assay in E2A-HLF–transfected Nalm6 cells. Assays were performed in wild type and E2A-HLF–transfected Nalm6 cells cultured in the absence or presence of zinc for 24 hours after transient transfection of each reporter plasmid. The values were normalized for transfection efficiencies using a cotransfected Renilla luciferase construct. (F) Luciferase assay in YCUB2 cells. The values were normalized for transfection efficiencies using a cotransfected Renilla luciferase construct.
We next performed luciferase assay of 3 reporter constructs25 (Figure 4C) in Nalm6 cells transfected with E2A-HLF using a zinc-inducible vector. In E2A-HLF-transfected Nalm6 cells, E2A-HLF was faintly expressed in the absence of zinc, and it was up-regulated in the presence of zinc to a level equivalent to that in UOC-B1 cells (Figure 4D). When the −512/+428 reporter construct was transiently transfected, transcriptional activity was up-regulated in E2A-HLF–transfected Nalm6 cells in the presence of zinc whereas it was virtually silent in the wild-type Nalm6 cells (Figure 4E). Transcriptional activity of the distal promoter was completely abolished by deletion or mutation of the PAR site. Exactly the same pattern of promoter activities was verified in YCUB2, one of the t(17;19)-ALL cell lines (Figure 4F), demonstrating that E2A-HLF up-regulates LMO2 gene expression by binding to the PAR site in the distal promoter of the LMO2 gene.
Induction of apoptosis by shRNA for LMO2 in a t(17;19)-ALL cell line
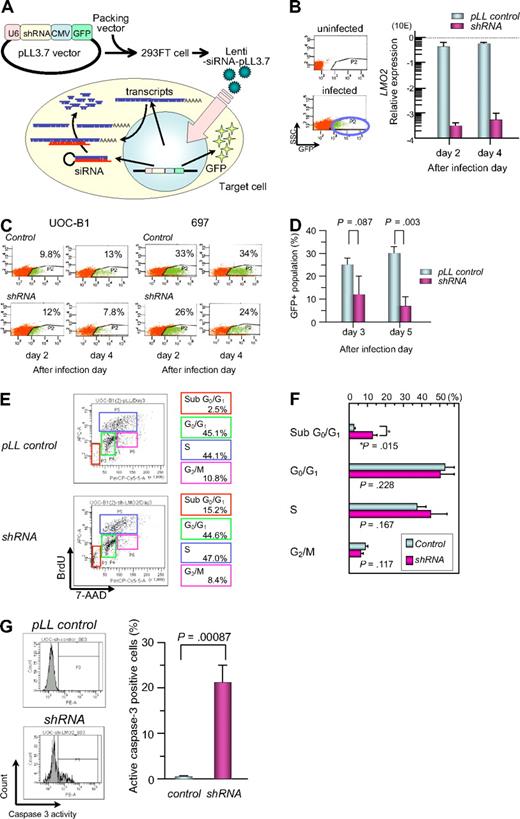
To study the significance of aberrant LMO2 expression in leukemogenesis in t(17;19)-ALL, we introduced shRNA against LMO2 into the t(17;19)-positive UOC-B1 cell line using a lentivirus vector, which contains GFP cDNA for cell sorting (Figure 5A).41 The expression level of the LMO2 gene in the GFP-positive (+) population of shRNA virus–infected UOC-B1 (shRNA/UOC-B1) cells was approximately 1000-fold lower than that in the GFP+ control virus–infected UOC-B1 (control/UOC-B1) cells when analyzed by real-time RT-PCR using the primers for exons 4 and 5 (Figure 5B). Despite infection with the same multiplicity of infection, the percentage of the GFP+ population decreased in the shRNA/UOC-B1 cells, in particular the GFPhigh population that is supposed to express a higher level of shRNA, whereas it was unchanged in the control/UOC-B1 cells (Figure 5C). The percentage of the GFP+ population in shRNA/UOC-B1 cells was significantly lower than that in the control/UOC-B1 cells (Figure 5D, 7.5% vs 30.5% on day 5 after infection; P = .003 by t test). By contrast, when infected into 697 cells, a t(1;19)-ALL cell line used as a control, the percentage of the GFP+ population was stable in both the shRNA-virus-infected and the control virus-infected cells (Figure 5C).
Gene silencing of LMO2 in t(17;19)-ALL cell line by a lentiviral vector. (A) Schematic representation of a short hairpin RNA (shRNA)–expressing lentiviral vector. pLL3.7 lentiviral vector was engineered to co-express green fluorescent protein (GFP) as a reporter gene by cytomegalovirus-derived promoter–GFP expression cassette. pLL3.7 lentiviral vector and packaging vector were cotransfected into 293FT cells and the resulting supernatant was collected after 36 hours. Lentivirus was recovered after ultracentrifugation and infected to UOC-B1 cells. (B) LMO2 expression of GFP-positive population sorted from lentivirus-infected cells. On day 2 or 4 after infection, the GFP-positive population was sorted and processed for real-time RT-PCR analysis using the primer for exons 4 and 5 of LMO2 gene. The gray boxes indicate pLL control vector-infected cells and the purple boxes indicate shRNA-expressing cells. (C) Changes in GFP-positive populations in UOC-B1 and 697 cells on days 2 and 4 after infection. The percentage of the GFP positive population is indicated in each box. (D) Changes in the percentage of GFP-positive populations in UOC-B1 cells infected with shRNA-containing and control lentivirus on day 3 and 5 after infection. The P value in t test is indicated. (E) Flow cytometric analysis of BrdU/7-AAD double staining in the GFP-positive population of shRNA-expressing and control UOC-B1 cells 3 days after infection. Representative data of the percentage of apoptotic cells in the sub G0/G1 phase among the GFP-positive population and the percentage of living cells in the G0/G1, S, and G2/M phases are indicated. (F) Comparison of cell-cycle distribution between control virus-infected cells and shRNA virus-infected cells. The P value in t test is indicated. (G) Flow cytometric analysis of cleaved-caspase3 in the GFP-positive population of shRNA-expressing and control UOC-B1 cells 3 days after infection. Representative data of the percentage of cleaved-caspase3-positive cells among the GFP-positive population are indicated in the left panel. Percentages of cleaved-caspase3-positive cells are compared between control virus-infected cells and shRNA virus-infected cells. The P value in t test is indicated.
Gene silencing of LMO2 in t(17;19)-ALL cell line by a lentiviral vector. (A) Schematic representation of a short hairpin RNA (shRNA)–expressing lentiviral vector. pLL3.7 lentiviral vector was engineered to co-express green fluorescent protein (GFP) as a reporter gene by cytomegalovirus-derived promoter–GFP expression cassette. pLL3.7 lentiviral vector and packaging vector were cotransfected into 293FT cells and the resulting supernatant was collected after 36 hours. Lentivirus was recovered after ultracentrifugation and infected to UOC-B1 cells. (B) LMO2 expression of GFP-positive population sorted from lentivirus-infected cells. On day 2 or 4 after infection, the GFP-positive population was sorted and processed for real-time RT-PCR analysis using the primer for exons 4 and 5 of LMO2 gene. The gray boxes indicate pLL control vector-infected cells and the purple boxes indicate shRNA-expressing cells. (C) Changes in GFP-positive populations in UOC-B1 and 697 cells on days 2 and 4 after infection. The percentage of the GFP positive population is indicated in each box. (D) Changes in the percentage of GFP-positive populations in UOC-B1 cells infected with shRNA-containing and control lentivirus on day 3 and 5 after infection. The P value in t test is indicated. (E) Flow cytometric analysis of BrdU/7-AAD double staining in the GFP-positive population of shRNA-expressing and control UOC-B1 cells 3 days after infection. Representative data of the percentage of apoptotic cells in the sub G0/G1 phase among the GFP-positive population and the percentage of living cells in the G0/G1, S, and G2/M phases are indicated. (F) Comparison of cell-cycle distribution between control virus-infected cells and shRNA virus-infected cells. The P value in t test is indicated. (G) Flow cytometric analysis of cleaved-caspase3 in the GFP-positive population of shRNA-expressing and control UOC-B1 cells 3 days after infection. Representative data of the percentage of cleaved-caspase3-positive cells among the GFP-positive population are indicated in the left panel. Percentages of cleaved-caspase3-positive cells are compared between control virus-infected cells and shRNA virus-infected cells. The P value in t test is indicated.
To determine whether the reduction in the GFP+ cells in shRNA/UOC-B1 cells was due to cell death or cell-cycle arrest, we performed flow cytometric analysis of BrdU/7-AAD double staining on day 3 after infection (Figure 5E). The percentage of sub-G0/G1 apoptotic cells in the GFP+ shRNA/UOC-B1 cells was significantly higher than that in the control/UOC-B1 cells (12.4% vs 3.6%; P = .014 by t test, Figure 5F), while the percentages of cells in the G0/G1, S, and G2/M phases were almost unchanged. Moreover, as shown in Figure 5G, the percentage of caspase-3–activated cells in the GFP+ shRNA/UOC-B1 cells was significantly higher than that in the control/UOC-B1 cells (21.4% vs 0.5%; P = .00087 by t test). These observations demonstrate that induction of apoptotic cell death is responsible for the reduction in the GFP+ population among shRNA/UOC-B1 cells.
Discussion
In T-ALL, 2 mechanisms of aberrant expression of the LMO2 gene have been well characterized: one is translocation of the LMO2 gene, leading to its combination with enhancers or other regulatory elements of TCR genes,1,3,4,43 and the other is cryptic deletion at 11p12-13 resulting in loss of negative regulatory sequences of the LMO2 gene.44 These 2 chromosomal abnormalities directly induce aberrant expression of the LMO2 gene. Here we demonstrated a novel mechanism for aberrant expression of the LMO2 gene as a downstream target of E2A-HLF fusion transcript derived from t(17;19) based on the following observations: all 4 t(17;19)-ALL cell lines studied and a patient's sample expressed a high level of LMO2 gene transcript, and transfection of E2A-HLF into 697 cells induced gene expression of LMO2 that was dependent on the transactivation and DNA-binding activities of E2A-HLF. E2A-HLF specifically bound to the PAR site in the distal promoter of the LMO2 gene at least in vitro and enhanced promoter activity. Moreover, LMO2 transcripts derived from both the proximal and distal promoters were expressed in t(17;19)-ALL cell lines. E2A-HLF rapidly induced LMO2 gene expression originating from the distal promoter, and then induced LMO2 gene expression derived from the proximal promoter, suggesting that E2A-HLF induces LMO2 gene expression not only directly through the PAR site in the distal promoter but also indirectly through the proximal promoter.
It has been reported that LMO2 expression is down-regulated during T-cell development at the transition from immature CD4/CD8 double-negative thymocytes to more mature stages16 and that enforced expression of LMO2 in thymocytes blocks differentiation of CD4/CD8 double-negative thymocytes and induces T-cell malignancies.17-20 In the present study, we confirmed that LMO2 gene expression in both EBV-transformed normal B-cell lines and Burkitt B-cell lines was significantly lower than that in B-precursor ALL cell lines. In particular, LMO2 protein expression was almost undetectable in these normal and leukemic B-cell lines. Moreover, the LMO2 gene expression level was markedly down-regulated during the transition from CD19-negative to CD19-positive population in the cord blood CD34+MNCs. These data are in agreement with the down-regulation of LMO2 gene expression during the early phase in the normal progression of B-cell development.19 Of note, gene silencing of LMO2 by the introduction of shRNA using lentivirus vector induced specific cell death. Interestingly, a recent analysis by Natkunam et al15 demonstrated that the majority of CD10-positive germinal center B cells coexpressed LMO2 while CD79a+ plasma cells lacked LMO2 expression, suggesting the association of LMO2 down-regulation with normal B-cell development. These observations seem to be in agreement with the assumption that aberrant expression of LMO2 is involved in the maturation arrest of t(17;19)-ALL cells at the immature B-precursor stage. Taken together, LMO2 expression is down-regulated during the normal development of both T cells and B cells, and its aberrant expression promotes cell survival of immature lymphocytes, which subsequently contributes to leukemogenesis of both T-ALL and B-precursor ALL. In this context, it should be noted that approximately one-fourth of non-t(17;19) B-precursor ALL cell lines expressed LMO2 at an equivalent level to that in t(17;19)-ALL cell lines, suggesting that LMO2 might also play a role, at least in part, in the leukemogenesis of other types of B-precursor ALL. Finally, considering the dismal outcome of conventional chemotherapy for t(17;19)-ALL cases,45 the development of new therapeutic modalities is urgently needed. Here we demonstrated that gene silencing of LMO2 using shRNA specifically induced cell death of t(17;19)-ALL cells. Thus, there is a possibility that aberrantly expressed LMO2 in t(17;19)-ALL might become a possible target for therapy.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: K.H. performed most of experiments and analyzed the data; T. Inukai designed the project, analyzed the data, and wrote the manuscript; J.K. and Y.F. performed shRNA analysis; T. Ikawa, H. Kawamoto, H. Kurosawa, A.T.L., and H.M. provided tools for analysis; S.H.O., B.G, N.K., Y.M., and H.O. performed gene expression analysis; K.A., X.Z., I.K., H.H., K.K., and K.G. performed analysis of cell line; T. Inaba wrote the manuscript; and K.S. supervised the project and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Dr Takeshi Inukai, Department of Pediatrics, School of Medicine, University of Yamanashi, 1110 Shimokato, Chuo, Yamanashi 409-3898, Japan; e-mail: tinukai@yamanashi.ac.jp.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal