Abstract
Evaluating hematopoietic stem cell (HSC) function in vivo requires a long-term transplantation assay. Although zebrafish are a powerful model for discovering the genetics of hematopoiesis, hematopoietic transplantation approaches have been underdeveloped. Here we established a long-term reconstitution assay in adult zebrafish. Primary and secondary recipients showed multilineage engraftment at 3 months after transplantation. Limiting dilution data suggest that at least 1 in 65 000 zebrafish marrow cells contain repopulating activity, consistent with mammalian HSC frequencies. We defined zebrafish haplotypes at the proposed major histocompatibility complex locus on chromosome 19 and tested functional significance through hematopoietic transplantation. Matching donors and recipients dramatically increased engraftment and percentage donor chimerism compared with unmatched fish. These data constitute the first functional test of zebrafish histocompatibility genes, enabling the development of matched hematopoietic transplantations. This lays the foundation for competitive transplantation experiments with mutant zebrafish HSCs and chemicals to test for effects on engraftment, thereby providing a model for human hematopoietic diseases and treatments not previously available.
Introduction
Positioned at the base of the hematopoietic hierarchy, the long-term hematopoietic stem cell (HSC) is capable of both self-renewal and lineage-specific differentiation to provide all the blood cell lineages for the lifetime of an animal, a phenotype that is functionally evaluated using long-term repopulation (LTR) assays.1,2 Murine hematopoietic stem and progenitor cells are prospectively isolated by flow cytometry using surface markers and transplanted into recipient animals to reconstitute various differentiated cell fates (reviewed by Challen et al3 ). Although the murine system has been invaluable for delineating the developmental relationship of hematopoietic cell fates, the molecular mechanisms governing HSC self-renewal versus differentiation decisions are largely unknown.
Zebrafish have emerged as a prominent model system for studying the developmental genetics of hematopoiesis based largely on high fecundity, transparency during embryonic and larval stages, and the ability to be manipulated for forward screening.4,5 Large-scale mutagenesis and small-molecule screens have identified novel pathways regulating expansion of definitive HSCs during development and differentiation of various hematopoietic lineages.6-8 These studies suggest that the zebrafish system can uncover novel programs essential for vertebrate HSC induction and self-renewal decisions, yet functional LTR assays have been lacking.
The first successful short-term hematopoietic cell transplants between adult zebrafish were reported only a few years ago.9 As transplantation technology continues to develop in the zebrafish system, 2 major areas for advancement have emerged: development of quantitative long-term repopulating assays and generation of histocompatible zebrafish lines. In human bone marrow transplantation, matching the donor and recipient major histocompatibility complex (MHC) haplotypes is critical to achieve a high rate of donor cell engraftment and recipient survival.10,11 The MHC locus on human chromosome 6 includes both class I and class II human leukocyte antigen (HLA) genes, as well as a number of genes important for antigen processing. Class I MHC genes encode transmembrane receptors that heterodimerize with β2-microglobulin to form an antigen-presenting complex expressed on virtually all nucleated cells. In humans, these include the HLA-A, HLA-B, and HLA-C genes. Class II MHC molecules are heterodimeric complexes consisting of HLA-DP, -DQ, and -DR genes, expressed by antigen-presenting cells, including macrophages, monocytes, and some B cells. In humans, the class I and II MHC genes are inherited as a parental haplotype with rare recombination events. Matching of the MHC genes of donors and transplantation recipients decreases immunologic rejection of donor tissue, and in the case of HSC transplantation also decreases the risk of graft-versus-host disease (GVHD). In mice, LTR and competitive transplantation assays are performed using inbred, congenic strains such that immune rejection and GVHD are not confounding factors. Attempts to develop inbred congenic zebrafish strains have not been successful because of lack of fecundity in highly inbred fish lines (Stephen Johnson, Washington University, oral communication, L.I.Z.). Therefore, defining the functional MHC locus (or loci) in the zebrafish would be useful to prospectively select suitable donors and recipients for transplantation purposes.
Unlike human MHC genes, zebrafish MHC class I (HLA-A, -B, and -C equivalent) and MHC class II (HLA-DR and -DQ equivalent) genes are not localized to a single chromosome. MHC genes and their paralogs are scattered throughout the zebrafish genome, having been identified in a genome-wide study using sequenced bacterial artificial chromosomes from the zebrafish genome sequencing project at the Sanger Center.12 A proposed zebrafish MHC core region located on chromosome 19 contains classic class I MHC genes, including mhc1uaa, mhc1uba, mhc1uca, mhc1uda, mhc1uea, and mhc1ufa.12,13 This chromosome 19 locus also encodes tightly linked antigen processing genes, making it syntenic with the human MHC locus. A second proposed MHC class I locus is found on chromosome 1.12,14 Additional nonclassic MHC class I genes are positioned at 3 different loci on chromosomes 3, 8, and 25 and have no apparent orthologs outside teleost fish.15
Using the zebrafish model as a discovery mechanism for regulators of HSC homeostasis and engraftment has already been successful.6,16 However, the fish model remains limited by the lack of immune-matched LTR assays that could be used for discovery and functional analysis of genes regulating HSCs. Knowledge of MHC function is lacking in zebrafish, as is the contribution of particular MHC genes to transplantation. As reported here, our initial quantitative HSC transplant experiments between unrelated persons resulted in high mortality and a variable level of engraftment. We hypothesized that our transplant results might be affected by mismatched MHC genes between donor and recipient fish. In the setting of MHC mismatch, donor HSCs could be rejected by recipient lymphocytes in a sublethally irradiated host, leading to graft failure, and ultimately death. Alternatively, engrafted but mismatched donor cells could attack the recipient resulting in GVDH, which could also be fatal.
To attempt to resolve this issue, we first defined haplotypes of MHC class I genes at the core locus on chromosomes 19 and then performed the first biologic studies of their function using hematopoietic transplantation in zebrafish. By matching the donors to recipients at this one locus, we report a significant increase in donor cell engraftment and percentage donor chimerism. These studies provide the foundation for functionally assessing and quantifying stem cell activity between genetic strains. Our assay significantly advances hematopoietic transplantation technology in the zebrafish, in that multipotent progenitors can now be functionally distinguished from HSCs. In addition, we identify chromosome 19 as a functionally significant locus of MHC genes in zebrafish transplant biology.
Methods
Fish care and strains
Cell harvesting
Labeled whole kidney marrow (WKM) from Tg(β-actin:GFP) adults was dissected from killed animals and prepared in 0.9 times phosphate-buffered saline (PBS) with 5% heat-inactivated fetal calf serum (FCS), as previously reported.9 For carrier cells, the skin and heart of deeply anesthetized adult zebrafish were punctured with a p20 micropipette under the pectoral fin, and the fish was subsequently killed. Peripheral blood (∼ 5 μL) was then drawn into a heparin-coated tip and expelled into a fluorescence-activated cell sorter (FACS) tube containing 2 mL of the same PBS/FCS buffer. Peripheral blood was pooled from several wild-type donor animals, filtered over a 40μM mesh, and cell counts were performed on a hemocytometer or cellometer (Nexcelom Bioscience). Cells were pelleted and resuspended in PBS/FCS buffer in the desired volume for injections.
Adult transplantation procedure
Wild-type adult zebrafish were subjected to γ-irradiation using a Cesium-137 irradiator as previously described at doses ranging from 20 to 40 Gy delivered as a single dose and transplanted 2 days later as previously described.19 Each recipient fish was anesthetized in a 0.02% tricaine solution and placed on its back on a small sponge. While lightly squeezing either side of the animal near the pectoral fins to push the heart forward, a 5-μL cell suspension of WKM with 105 peripheral blood carrier cells was delivered via direct cardiac puncture with a Hamilton syringe (33/1 in./30DEG model, 80300 catalog number).20 Later experiments were performed using a retro-orbital injection method as this resulted in significantly decreased procedure-related mortality.21 Injected fish were placed in a recovery tank (∼ 8-10 fish per tank) and returned to the aquatic system later that day. Fish were monitored daily, and dead animals were removed promptly. After 13 to 16 weeks, transplant recipients were killed for analysis of WKM by flow cytometry as previously described using a FACSVantage flow cytometer (BD Biosciences).9 Propidium iodide (1 mg/mL; Sigma-Aldrich) was added to exclude dead cells. The myeloid, lymphoid, and precursor gates were set as previously described.9 Data were analyzed using FloJo software Version 7.6.3 for Mac.
FACS analysis
Standard gates for GFP+ cells were determined based on the baseline GFP+ autofluorescence in AB wild-type fish serving as negative controls.
Tail clip and DNA preparation
For genotyping, DNA was prepared from individual tail-clip samples. The HotSHOT DNA preparation protocol was used.22 A 1:20 dilution of the original DNA stock was used in each 10 μL polymerase chain reaction (PCR).
PCR conditions
Standard PCR reactions contained the following: 2 μL of DNA, 7.3 μL reaction buffer (10mM Tris-HCl, pH 8.3, 50mM KCl, 1.5mM MgCl2, 0.01% [weight/volume] gelatin, 0.01% NP-40, and 0.01% Tween 20), 2.6 μL of water, 0.1 μL of a primer mix containing 20μM of each primer, and 0.05 μL of Taq polymerase (5 units/μL). The cycling conditions involved an initial enzyme activation step at 94°C for 5 minutes, followed by 35 cycles consisting of denaturing at 94°C for 30 seconds, annealing at 52°C for 30 seconds, and polymerization at 72°C for 1 minute, followed by 10 cycles consisting of denaturing at 94°C for 30 seconds, annealing at 58°C for 30 seconds, polymerization at 72°C for 1 minute, and a final extension for 10 minutes at 72°C. The PCR was performed using GeneAmp 9700 systems thermocycler (PE Applied Biosystems).
Sequencing (standard) and sequence analysis
PCR products were purified using the Agencourt AMPure magnetic bead nucleotide purification system (Beckman Coulter Genomics) and sequenced using the BigDye Terminator Version 3.1 Cycle Sequencing Kit (Applied Biosystems). Sequencing reaction products were resolved and read using an ABI 3730xl DNA Analyzer (PE Applied Biosystems). Obtained sequences were analyzed using Mutation Surveyor Version 3.01 (SoftGenetics LLC; www.softgenetics.com/mutationSurveyor.html).
Results
Quantification of percentage donor chimerism and detection of multilineage engraftment in a long-term repopulating assay
Prior experiments documented engraftment of WKM cells into embryonic and adult zebrafish recipients.9,19 Experiments showed short-term (30-day) multilineage engraftment but did not provide reproducible methods for regulating the number of cells delivered to the host or for quantifying percentage donor chimerism of engrafted animals. To address these basic issues, we standardized an intracardiac method for delivering a predetermined volume (5 μL) of WKM donor cells into recipient hosts using a Hamilton syringe, allowing precise delivery of the donor cell dose. To determine donor chimerism, we used fluorescently labeled donor cells from Tg(β-actin:GFP) fish.23 Recipient WKM was analyzed by forward and side scatter profile as published previously9 and the percentage of GFP+ cells determined for each cell-specific gate (Figure 1A). Although the erythroid gate was devoid of fluorescence (data not shown), the average percentage of GFP+ cells was 37.8% ± 20.6% in the lymphoid gate, 49.6% ± 16.7% in the precursor gate, and 82.9% ± 6.7% in the myeloid gate (n = 6; Figure 1A-B). The same analysis was applied to WKM transplant recipients 12 to 15 weeks after transplantation. To detect multilineage engraftment, we examined GFP fluorescence in the myeloid, lymphoid, and precursor marrow populations of recipient fish. We found multilineage engraftment of recipient fish at 12 to 15 weeks after transplantation comparable with the level of GFP expression seen in the Tg(β-actin:GFP) donors (Figure 1C-D).
Transplantation of GFP-labeled transgenic whole kidney marrow shows long-term engraftment in adult zebrafish. (A) FACS analysis of control Tg(β-actin:GFP) WKM cells showing the forward scatter vs side scatter profile from a representative donor animal. The erythroid gate is marked in red, the myeloid gate is green, the precursor gate is black, and the lymphoid gate is blue. (B) Histograms for GFP expression of cells within the myeloid, lymphoid, and precursor lineage gates for a representative Tg(β-actin:GFP) donor fish. The percentage of GFP+ cells in each lineage gate is shown. (C) Forward scatter (FSC) vs side scatter (SSC) profile of marrow from an animal 3 months after transplantation with 500 × 103 marrow cells showing full reconstitution with donor cells. (D) Histograms for GFP expression of cells within the myeloid, lymphoid, and precursor gates for a representative transplant recipient fish analyzed 3 months after transplantation showing multilineage engraftment with GFP+ donor cells. (E) Kaplan-Meier survival curves of adult zebrafish transplanted with 500 × 103 whole kidney marrow cells after graded doses of total body irradiation. (F) Percentage GFP+ cells in the myeloid and lymphoid populations of control Tg(β-actin:GFP) animals (left), in myeloid cells of transplant recipients (middle), and in lymphoid cells of transplant recipients (right). Each diamond represents an individual animal. Each host was transplanted with 500 × 103 WKM cells after exposure to 20, 25, or 30 Gy of total body irradiation. Percentage of GFP+ cells in the myeloid gate (middle) and lymphoid gate (right) at 90 days after transplantation is shown. Percentages plotted correspond to raw data numbers. Red lines indicate the lower threshold for successful myeloid (> 4%) and lymphoid (> 0.6%) engraftment as determined by negative control animals. (G) Percentage GFP+ cells in the myeloid and lymphoid populations of WKM from secondary transplant recipients. Each unique symbol represents an individual animal.
Transplantation of GFP-labeled transgenic whole kidney marrow shows long-term engraftment in adult zebrafish. (A) FACS analysis of control Tg(β-actin:GFP) WKM cells showing the forward scatter vs side scatter profile from a representative donor animal. The erythroid gate is marked in red, the myeloid gate is green, the precursor gate is black, and the lymphoid gate is blue. (B) Histograms for GFP expression of cells within the myeloid, lymphoid, and precursor lineage gates for a representative Tg(β-actin:GFP) donor fish. The percentage of GFP+ cells in each lineage gate is shown. (C) Forward scatter (FSC) vs side scatter (SSC) profile of marrow from an animal 3 months after transplantation with 500 × 103 marrow cells showing full reconstitution with donor cells. (D) Histograms for GFP expression of cells within the myeloid, lymphoid, and precursor gates for a representative transplant recipient fish analyzed 3 months after transplantation showing multilineage engraftment with GFP+ donor cells. (E) Kaplan-Meier survival curves of adult zebrafish transplanted with 500 × 103 whole kidney marrow cells after graded doses of total body irradiation. (F) Percentage GFP+ cells in the myeloid and lymphoid populations of control Tg(β-actin:GFP) animals (left), in myeloid cells of transplant recipients (middle), and in lymphoid cells of transplant recipients (right). Each diamond represents an individual animal. Each host was transplanted with 500 × 103 WKM cells after exposure to 20, 25, or 30 Gy of total body irradiation. Percentage of GFP+ cells in the myeloid gate (middle) and lymphoid gate (right) at 90 days after transplantation is shown. Percentages plotted correspond to raw data numbers. Red lines indicate the lower threshold for successful myeloid (> 4%) and lymphoid (> 0.6%) engraftment as determined by negative control animals. (G) Percentage GFP+ cells in the myeloid and lymphoid populations of WKM from secondary transplant recipients. Each unique symbol represents an individual animal.
Engraftment thresholds were determined by analyzing the mean percentage plus or minus SD of GFP+ cells in WT fish lacking the β-actin:GFP transgene (n = 33). To determine multilineage engraftment, we focused on the lymphoid and myeloid gates. The precursor gate was not included in our analysis because this gate contains a mixture of both erythroid and myeloid precursors. To avoid mis-scores, we decided to set the engraftment threshold at least 3 SDs above the mean for each cell type. In the myeloid fraction, the mean background level of GFP fluorescence was 1.23% ± 0.81%, with a maximum value of 3.96%. The mean GFP fluorescence in the lymphoid population was 0.16% ± 0.14%, with a maximum of 0.53%. Therefore, animals showing > 4% and > 0.6% GFP positivity in the myeloid and lymphoid gates, respectively, were considered engrafted with donor cells.
Optimal recipient survival and engraftment with 25 Gy conditioning radiation dose
To determine the optimal conditioning dose of radiation for transplant recipients, we injected 500 000 WKM cells from Tg(β-actin:GFP) animals into wild-type hosts treated with various doses of γ-irradiation. Previously published work has established 40 Gy as a lethal radiation dose for the adult zebrafish, whereas 20 Gy is sublethal, as more than 85% of animals survive.19 We elected to irradiate recipients with various doses of γ-irradiation at 5 Gy increments between 20 and 40 Gy and monitored survival daily (Figure 1E). Engraftment was analyzed after 90 days. WKM transplantation was unable to rescue animals receiving a 40 Gy dose (n = 21; Figure 1E), suggesting that tissues other than the hematopoietic compartment are damaged. The highest dose capable of being radioprotected by WKM was 30 Gy. Whereas 100% of the survivors showed multilineage engraftment (n = 8 of 8) in the marrow, only 12.5% (n = 8 of 63) of the animals survived 3 months after transplantation (Figure 1E-F). In contrast, animals conditioned with 20 Gy irradiation had excellent survival at 90 days (85%) but lower levels of donor chimerism, and only 6 of 17 (29%) of the survivors were engrafted. At 25 Gy, however, we found improved engraftment (n = 46 of 50, 92%) and increased survival (n = 50 of 115, 44%) compared with the 20 and 30 Gy doses, respectively. The fact that 4 of these animals survived and recovered their own blood system without showing significant donor cell contribution suggests that the 25 Gy dose is still within the sublethal range. The 25 Gy radiation dose was used for pretransplantation conditioning for all subsequent experiments.
Previous studies showed that approximately 88% of animals exposed to 25 Gy survive 4 weeks.19 We repeated this experiment and assayed survival of nontransplanted (n = 20) or mock-transplanted (n = 30; injected with PBS/FCS) animals for 90 days and found that, in either case, only 15% to 17% survived this extended period of time, whereas animals receiving transplanted marrow cells had 46% survival (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). We concluded that the 25 Gy dose is largely lethal to adult zebrafish by 3 months after irradiation, and a portion of this lethality can be rescued by hematopoietic transplantation. In summary, these data define a radiation dose that optimizes donor cell engraftment and animal survival, thus creating a reproducible assay that can be used as the foundation for long-term reconstitution experiments in adult zebrafish.
Multilineage engraftment of secondary transplant recipients after serial transplantation of whole kidney marrow
To ensure that our assay measured long-term reconstituting cells, we serially transplanted 50 000 or 100 000 chimeric marrow cells isolated from transplant recipients at 3 months after transplantation into irradiated secondary recipients. Multilineage reconstitution was detected at 12 weeks after transplantation in 5 of 12 recipients, whereas engraftment of myeloid cells alone was observed in one recipient, indicating engraftment of a committed myeloid progenitor and not a true HSC (Figure 1G). These results indicate that HSCs can be detected using this transplantation assay by their ability to reconstitute multilineage hematopoiesis in both primary and secondary recipients.
Limiting dilution analyses
Like those performed in the mouse, limiting dilution analyses provide the ideal system for comparing HSC function between 2 experimental groups. Here, we aimed to generate a similar assay in the zebrafish that would allow quantification of repopulating cell number per marrow volume. As the number of transplanted WKM cells decreases, the quantity of HSCs capable of long-term reconstitution declines. We hypothesized that this decrease in HSC number would be directly related to animal mortality. Although we could not control for death from radiation effects, infection, or graft rejection because of immune causes, we speculated that the frequency of these events would be similar at every limiting dilution dose. Thus, we assumed that any change in the rate of survival was the result of engraftment of donor HSCs.
We found maximal survival (50.5%) and engraftment (37.9%) of compromised recipients when 50 000 WKM cells were transplanted (Table 1; Figure 2). As the WKM dose was lowered, we found a gradual decline in both survival and repopulation, suggesting that fewer HSCs were being donated to irradiated hosts. Surprisingly, at the highest WKM cell doses tested (75 000 and 100 000), we also observed a decline in survival and engraftment. This result suggests that immune rejection may be stronger when a higher number of donor cells, and hence an increased number of mature lymphocytes, are transplanted into non–MHC-matched animals. We devised the following statistical model to interpret these limiting dilution data in a manner that takes all of the parameters described into consideration.
Dose-dependent effects on engraftment success
| Cells injected per recipient, C . | Recipients, N(C) . | Survivors, S(C) . | Engraftment successes, G(C) . | ||
|---|---|---|---|---|---|
| No. . | % of recipients . | No. . | % of recipients . | ||
| 100 000 | 86 | 25 | 29.1 | 25 | 29.0 |
| 75 000 | 48 | 22 | 45.8 | 16 | 33.3 |
| 50 000 | 95 | 48 | 50.5 | 36 | 37.8 |
| 25 000 | 46 | 11 | 23.9 | 7 | 15.2 |
| 15 000 | 40 | 17 | 42.5 | 10 | 25.0 |
| 10 000 | 82 | 24 | 29.3 | 16 | 19.5 |
| 5000 | 46 | 15 | 32.6 | 6 | 13.0 |
| 1000 | 45 | 6 | 13.3 | 1 | 2.2 |
| 500 | 37 | 3 | 8.1 | 1 | 2.7 |
| 100 | 48 | 4 | 8.3 | 1 | 2.1 |
| Cells injected per recipient, C . | Recipients, N(C) . | Survivors, S(C) . | Engraftment successes, G(C) . | ||
|---|---|---|---|---|---|
| No. . | % of recipients . | No. . | % of recipients . | ||
| 100 000 | 86 | 25 | 29.1 | 25 | 29.0 |
| 75 000 | 48 | 22 | 45.8 | 16 | 33.3 |
| 50 000 | 95 | 48 | 50.5 | 36 | 37.8 |
| 25 000 | 46 | 11 | 23.9 | 7 | 15.2 |
| 15 000 | 40 | 17 | 42.5 | 10 | 25.0 |
| 10 000 | 82 | 24 | 29.3 | 16 | 19.5 |
| 5000 | 46 | 15 | 32.6 | 6 | 13.0 |
| 1000 | 45 | 6 | 13.3 | 1 | 2.2 |
| 500 | 37 | 3 | 8.1 | 1 | 2.7 |
| 100 | 48 | 4 | 8.3 | 1 | 2.1 |
These are the raw data used to calculate the limiting dilution assay graphed in Figure 2.
Limiting dilution analysis reveals that survival and engraftment are tightly linked. Data from Table 1 were graphed for each cell dose in an HSC limiting dilution transplantation experiment (bars) and the SE calculated and shown. Recipients were injected with increasing numbers of marrow cells, and a constant number of peripheral blood carrier cells (105 per recipient). After 3 months, recipients were killed and the marrow was dissected for FACS analysis. GFP+ expression in the myeloid and lymphoid gates was used to determine donor engraftment. The percentage of animals surviving 90 days (A) and the percentage of animals engrafted (B) are shown. Transplanted marrow cell dose is depicted on the x-axis. The solid black line depicts the statistical fitted model.
Limiting dilution analysis reveals that survival and engraftment are tightly linked. Data from Table 1 were graphed for each cell dose in an HSC limiting dilution transplantation experiment (bars) and the SE calculated and shown. Recipients were injected with increasing numbers of marrow cells, and a constant number of peripheral blood carrier cells (105 per recipient). After 3 months, recipients were killed and the marrow was dissected for FACS analysis. GFP+ expression in the myeloid and lymphoid gates was used to determine donor engraftment. The percentage of animals surviving 90 days (A) and the percentage of animals engrafted (B) are shown. Transplanted marrow cell dose is depicted on the x-axis. The solid black line depicts the statistical fitted model.
The equation we present accounts for the number of injected fish surviving to 12 weeks and the number that, having survived, showed evidence of engraftment. Following Smith et al,24 we let p denote the fraction of HSC among the injected cells. It follows that a recipient of C cells has probability 1 − (1 − p)C of including at least one HSC and thus successfully engrafting. In addition, we postulate that engraftment enhances the chance of host survival, increasing the probability of survival from s0 when engraftment fails to a larger probability s1 when engraftment is successful. Counterbalancing the benefit of the graft, we hypothesize that any given injected cell has a probability a of provoking an immune response, which would lead to death of the host. Conversely, the chance of averting such a response is (1 − a)C. Taking all these factors into account, the chance of survival with engraftment for a given fish is PG = s1(1 − a)C(1 − (1 − p)C), and the total probability of survival (with or without donor cell engraftment) is PS = PG + s0 (1 − a)C(1 − p)C.
In place of the least-squares method described by Smith et al,24 we determined the optimal estimates for these parameters by maximizing the likelihood of the data,
where, among the N(C) fish injected with C cells, S(C) survived and G(C) showed engraftment. Our trinomial model, which is a generalization of the Poisson statistics applied to mammalian limiting dilution data, was devised to describe the more complex outcomes generated by the fish system. In the degenerate case, where the transplant does not evoke a lethal immune response (a = 0) and engraftment of the host animal can be assessed (s0 = s1 = 1), the trinomial model reduces to a simple Poisson formula (PG = 1 − (1 − p)C). From the data in Table 1, we obtained p = 1.53 ± 0.22 × 10−5, or 1 HSC per 65.5 ± 9.3 × 103 WKM cells (estimate ± SE). Without engraftment, the estimated probability of survival based on this model was s0 = 16.8% ± 2.2%. Validating our statistical analyses, we found in a completely separate experiment that when no transplantation was administered to adults irradiated at 25 Gy, 15% (3 of 20) survived 90 days (data not shown). With successful donor engraftment, the probability s1 was indistinguishable from 100%. The probability of any given injected cell provoking a lethal immune response was estimated at a = 8.0 ± 1.4 × 10−4%. In summary, our statistical analyses predict the probability that any cell in the marrow contains long-term repopulating potential by fitting the data to a model. These data provide the ability to compare HSC frequencies in the WKM of different wild-type or mutant zebrafish strains.
Identification and typing of MHC genes
Having designed an assay to evaluate the function of HSCs in zebrafish, the unpredictable contribution of the immune system in our assay posed a severe limitation to interpreting any experiments comparing different zebrafish strains. We hypothesized that matching the MHC genes in the core MHC region on chromosome 19 might be sufficient to improve the survival and engraftment rates in an HSC transplantation experiment designed between siblings. From previous studies, 2 publicly known haplotypes of the core MHC region had been identified, each containing different MHC genes.12,13 The Zv8 chromosome AB haplotype contained 4 MHC class I U genes, whereas the Zv8 chromosome 19 haplotype contained 2 U genes (Figure 3A). Using the sequences from these 6 MHC class I U genes, 5 additional U genes with different nucleotide sequences were retrieved from EST and cDNA sequence databases at National Center for Biotechnology Information using BLAST searches.
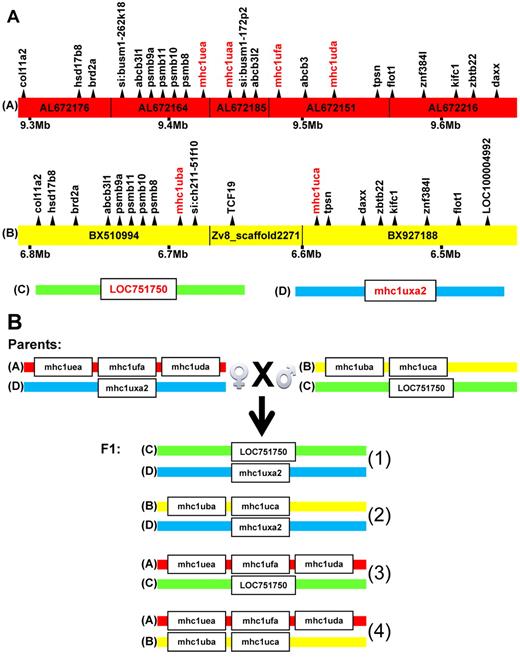
Four haplotypes identified at the MHC locus on chromosome 19. (A) Haplotype A is found on Zv8 chromosome AB and is formed by alignment of 5 bacterial artificial chromosomes: AL672176, AL672164, AL672185, AL672151, and AL672216. Haplotype B is found on Zv8 chromosome 19 and is formed by alignment of 2 bacterial artificial chromosomes (BX927188 and BX510994) and 6 pieces of shotgun sequences from Zv8_scaffold2271. MHC class I U genes are in red, and flanking genes are in black. Haplotype A contains 4 MHC class I U genes and 18 flanking genes, whereas haplotype B contains 2 MHC class I U genes and 17 flanking genes. Both haplotypes are defined by the same flanking genes, such as col11a2, daxx, and flot1, in a region 350 kb in length. The megabase pair (Mb) positions under the haplotypes are from Ensembl Zebrafish genome browser. Haplotypes C and D are identified by direct sequencing of PCR amplified DNA sequences and are defined by the 2 MHC class I U genes (LOC751750 and mhc1uxa2) that independently segregated in the family of F1 siblings. (B) MHC genotypes for the wild-type AB male and Tg(β-actin:GFP) female parents are shown, as well as the 4 genotypes identified in their F1 progeny.
Four haplotypes identified at the MHC locus on chromosome 19. (A) Haplotype A is found on Zv8 chromosome AB and is formed by alignment of 5 bacterial artificial chromosomes: AL672176, AL672164, AL672185, AL672151, and AL672216. Haplotype B is found on Zv8 chromosome 19 and is formed by alignment of 2 bacterial artificial chromosomes (BX927188 and BX510994) and 6 pieces of shotgun sequences from Zv8_scaffold2271. MHC class I U genes are in red, and flanking genes are in black. Haplotype A contains 4 MHC class I U genes and 18 flanking genes, whereas haplotype B contains 2 MHC class I U genes and 17 flanking genes. Both haplotypes are defined by the same flanking genes, such as col11a2, daxx, and flot1, in a region 350 kb in length. The megabase pair (Mb) positions under the haplotypes are from Ensembl Zebrafish genome browser. Haplotypes C and D are identified by direct sequencing of PCR amplified DNA sequences and are defined by the 2 MHC class I U genes (LOC751750 and mhc1uxa2) that independently segregated in the family of F1 siblings. (B) MHC genotypes for the wild-type AB male and Tg(β-actin:GFP) female parents are shown, as well as the 4 genotypes identified in their F1 progeny.
An F1 generation of siblings was then created by crossing a Tg(β-actin:GFP) female fish and a male AB wild-type fish (Figure 3B). Sequence specific PCR primers designed against the unique MHC I U gene sequences (Table 2) were used for specific amplification and sequencing of these U genes in both parents and 214 of their F1 progeny. Existence of specific U genes in an individual genome was evaluated by genomic PCR reactions with carefully designed positive and negative controls. Single nucleotide polymorphisms in U genes were genotyped by direct sequencing of amplified fragments of U genes. Both Mendelian segregation of linked single nucleotide polymorphisms and the presence of specific U gene(s) were used to define haplotypes at the core MHC locus. Within this family, both of the publicly available haplotypes were identified, as well as 2 additional haplotypes of the core MHC region defined by 2 single U genes that each segregated independently (Figure 3B).
Primers used for genotyping of MHC class I U genes
| Gene . | Exon . | Primer pair . | Nucleotide sequence . | bp . |
|---|---|---|---|---|
| mhc1uxa2 | 2 | 1 | AGGCATACACTCTCTGAAATACTTCTTC | 264 |
| CCCTGTGACTGGTTAAACCTCTCC | ||||
| mhc1uea | 2 | 2 | TACACACTCCCTGAGGTACTTC | 204 |
| TGTGCGCCAGTAAAGATCTGAG | ||||
| mhc1uda | 2 | 1 | AGGCATACACTCTCTGAAATACTTCTTC | 264 |
| CCCTGTGACTGGTTAAACCTCTCC | ||||
| mhc1uda | 3 | 3 | GAAATGTACGGCTGTGAGTG | 252 |
| CAGGCTGCTCTTTCCATAC | ||||
| mhc1ufa | 2 | 4 | GAAAGCATTCTCACACTGTTATCT | 258 |
| GTGTTTGGTTAAAACGTTCCATTG | ||||
| mhc1ufa | 3 | 5 | CAGTTCATGGTCGGCTGTGA | 250 |
| AAGAATCTTTGCCGTAACCC | ||||
| LOC751750 | 2 | 6 | CACACCTGGAATGCCTATTACA | 245 |
| TTGAAGCGCTCTATTGCAACTT | ||||
| LOC751750 | 3 | 3 | GAAATGTACGGCTGTGAGTG | 252 |
| CAGGCTGCTCTTTCCATAC | ||||
| mhc1uba | 2 | 7 | CCACAGCTGGAAAGCCTATT | 266 |
| ATTTTTTACCTTGTGTTTGG | ||||
| mhc1uba | 3 | 8 | GTGTTCACACCTTCCAGTTC | 268 |
| TCCAAAGTGTCCTTCCCATA | ||||
| mhc1uca | 2 | 6 | CACACCTGGAATGCCTATTACA | 245 |
| TTGAAGCGCTCTATTGCAACTT | ||||
| mhc1uca | 3 | 9 | ACACATTACAGGAAATTTACGG | 260 |
| GCTGCTCTTTCCATACTCCAAG |
| Gene . | Exon . | Primer pair . | Nucleotide sequence . | bp . |
|---|---|---|---|---|
| mhc1uxa2 | 2 | 1 | AGGCATACACTCTCTGAAATACTTCTTC | 264 |
| CCCTGTGACTGGTTAAACCTCTCC | ||||
| mhc1uea | 2 | 2 | TACACACTCCCTGAGGTACTTC | 204 |
| TGTGCGCCAGTAAAGATCTGAG | ||||
| mhc1uda | 2 | 1 | AGGCATACACTCTCTGAAATACTTCTTC | 264 |
| CCCTGTGACTGGTTAAACCTCTCC | ||||
| mhc1uda | 3 | 3 | GAAATGTACGGCTGTGAGTG | 252 |
| CAGGCTGCTCTTTCCATAC | ||||
| mhc1ufa | 2 | 4 | GAAAGCATTCTCACACTGTTATCT | 258 |
| GTGTTTGGTTAAAACGTTCCATTG | ||||
| mhc1ufa | 3 | 5 | CAGTTCATGGTCGGCTGTGA | 250 |
| AAGAATCTTTGCCGTAACCC | ||||
| LOC751750 | 2 | 6 | CACACCTGGAATGCCTATTACA | 245 |
| TTGAAGCGCTCTATTGCAACTT | ||||
| LOC751750 | 3 | 3 | GAAATGTACGGCTGTGAGTG | 252 |
| CAGGCTGCTCTTTCCATAC | ||||
| mhc1uba | 2 | 7 | CCACAGCTGGAAAGCCTATT | 266 |
| ATTTTTTACCTTGTGTTTGG | ||||
| mhc1uba | 3 | 8 | GTGTTCACACCTTCCAGTTC | 268 |
| TCCAAAGTGTCCTTCCCATA | ||||
| mhc1uca | 2 | 6 | CACACCTGGAATGCCTATTACA | 245 |
| TTGAAGCGCTCTATTGCAACTT | ||||
| mhc1uca | 3 | 9 | ACACATTACAGGAAATTTACGG | 260 |
| GCTGCTCTTTCCATACTCCAAG |
The same pairs of primers were used for amplifying and sequencing.
Transplantation of MHC-matched donor cells yields increased donor chimerism compared with unmatched recipients
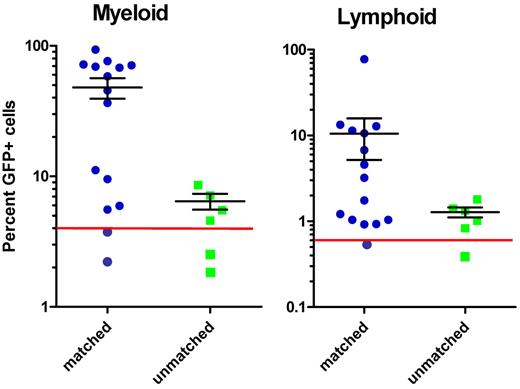
Based on the genotyping results at the chromosome 19 MHC locus, the sibling progeny were categorized into 4 groups, as expected by Mendelian segregation of the parental haplotypes (Figure 3B). WKM was isolated from GFP+ donors and transplanted into irradiated GFP− matched sibling recipients or unrelated recipients with unknown MHC typing. Fourteen to 16 weeks after transplantation, recipient fish were killed and WKM was analyzed by flow cytometry to assess for engraftment of GFP+ donor cells. Multilineage (lymphoid and myeloid) engraftment was observed in 13 of 15 (87%) matched recipients, with a single recipient displaying only lymphoid engraftment (n = 1 of 15; Figure 4). The mean percentage donor chimerism for the matched engrafted recipients was 47.86% ± 30.9% for myeloid cells and 10.51% ± 19.88% (mean ± SD) for lymphoid cells. In contrast, unmatched recipients showed multilineage engraftment in 4 of 6 surviving fish (67%), but with much lower levels of donor chimerism. One additional unmatched recipient was engrafted in the lymphoid lineage only. The mean percentage of donor chimerism for engrafted unmatched recipients was 6.45% ± 1.77% and 1.28% ± 0.38%, for the myeloid and lymphoid cells, respectively (Figure 4). The levels of donor chimerism in the matched recipients were significantly higher than those of the unmatched recipients (P = .0002 and P = .05 for the myeloid and lymphoid gates, respectively). These data indicate that matching the MHC genes at the chromosome 19 locus is important for engraftment of zebrafish HSCs. This experiment represents the first functional evaluation of zebrafish MHC molecules in a transplantation setting.
Increased percentage donor chimerism in MHC-matched transplant recipients. The percentage of GFP+ cells within the myeloid and lymphoid gates was plotted for each individual transplant recipient. Each recipient animal received 50 to 75 × 103 WKM cells in addition to 2 × 105 peripheral blood carrier cells. Each unique symbol represents a single transplant recipient. Sixteen recipients were evaluated 16 weeks after transplantation, including all the unmatched recipients. Five of the matched recipients were evaluated at 14 weeks. The red lines indicate the thresholds for myeloid (> 4%) and lymphoid (> 0.6%) engraftment. Mean percentage of GFP+ cells ± SEM is shown for the engrafted animals in each group. The level of donor chimerism in the engrafted matched recipients was significantly higher than for the engrafted unmatched recipients (P = .0002 and P = .05 for the myeloid and lymphoid gates, respectively).
Increased percentage donor chimerism in MHC-matched transplant recipients. The percentage of GFP+ cells within the myeloid and lymphoid gates was plotted for each individual transplant recipient. Each recipient animal received 50 to 75 × 103 WKM cells in addition to 2 × 105 peripheral blood carrier cells. Each unique symbol represents a single transplant recipient. Sixteen recipients were evaluated 16 weeks after transplantation, including all the unmatched recipients. Five of the matched recipients were evaluated at 14 weeks. The red lines indicate the thresholds for myeloid (> 4%) and lymphoid (> 0.6%) engraftment. Mean percentage of GFP+ cells ± SEM is shown for the engrafted animals in each group. The level of donor chimerism in the engrafted matched recipients was significantly higher than for the engrafted unmatched recipients (P = .0002 and P = .05 for the myeloid and lymphoid gates, respectively).
Discussion
As the zebrafish continues to emerge as a prominent model for exploring the genetics of stem cell biology, methodologies to assess the function of HSCs at the cellular level must be developed. Contributing to this effort, assays in the fish have progressed significantly over the past 5 years with the ability to separate the major blood lineages by flow cytometry, the establishment of the minimal sublethal and lethal irradiation doses, and the achievement of short-term hematopoietic donor cell engraftment.9,19 When genetic screens uncover mutants that display phenotypic stem cell defects or advantages, quantitative assays are critical to compare HSC function of these mutants with wild-type HSCs. Because transplantation is the “gold standard” for assessing stem and progenitor cell activity, we conclude that these approaches need to be applied to the zebrafish to more accurately define mutant hematopoietic phenotypes.
To use this assay for quantitative transplantation experiments, significant modifications were made to the previously published method. Previously, the number of cells injected into host zebrafish animals was not amenable to quantification based on the method of delivery with a microinjection needle that is difficult to calibrate.9,19 To circumvent this limitation, we created a procedure to deliver a precise number of cells to the circulatory system. Early experiments used intracardiac injection, whereas later experiments adopted a retro-orbital injection method that decreased procedure-related mortality. Here we report experiments analyzed at 13 to 16 weeks after transplant. For future experiments, we recommend analysis at 4 to 6 months to ensure long-term engraftment.
Another major change to the transplant protocol was the addition of carrier cells to the WKM cell preparation. In the mouse, the standard method for limiting dilution assays uses a fixed number of wild-type marrow cells mixed with decreasing numbers of test marrow cells. The recipient animal receives sufficient wild-type HSCs to be radioprotected, and engraftment efficiency of the test marrow is determined at 3 to 6 months after transplantation. This method ensures the survival of the recipient. Because of the significant percentage of lymphocytes in WKM, and immunologic concerns in unmatched zebrafish transplants, we opted to use peripheral blood as the source of carrier cells, as > 99% of the cells are mature erythrocytes, and < 0.5% are lymphocytes (data not shown). This allowed the serial dilution of donor marrow cells so that as few as one single WKM cell could be accurately delivered to a recipient fish in the context of 100 000 carrier cells. Injection of carrier cells alone did not lead to engraftment, indicating that if HSCs are present in the peripheral blood of zebrafish the frequency is less than 1 per 100 000 cells. Using peripheral blood carrier cells prolonged the survival of recipients in the first 25 days after transplantation leading to enhanced survival at 3 months for recipients receiving carrier cells and WKM compared with WKM alone (supplemental Figure 2). The disadvantage of using peripheral blood carrier cells is that recipients not receiving an adequate number of donor stem cells will die because of lack of engraftment, and the data for those animals are lost. For future experiments, the use of unfractionated marrow as carrier cells would be preferable so that all recipients are radioprotected, allowing more animals to survive until the end of the experiment. The use of compromised marrow cells is common in murine limiting dilution experiments (reviewed by Purton and Scadden).2 Immunologic matching would be critically important for these WKM carrier cells, as is true in murine experiments. Approximately 10%-25% of the WKM preparation contains lymphocytes, which if immunologically mismatched would be capable of rejecting the test marrow, or engrafting and causing GVHD in the host. In either case, the recipient fish would not likely survive, thereby defeating the purpose of using WKM to radioprotect the recipient.
Defining a radiation dose that allows for efficient donor cell engraftment and recipient survival has been a major challenge. In these studies, we tested single exposures at 5 Gy increments between the reported minimal sublethal and lethal dose and revealed that hosts conditioned with 25 Gy radiation show the best engraftment and survival long term. However, this dose is still within the sublethal range as ∼17% of animals survive for > 3 months with no transplant. Moreover, the mortality of transplant recipients can be high even with a “radioprotective” dose of marrow cells. For example, our data show that radiation doses > 30 Gy are uniformly fatal, despite injection of a radioprotective dose of marrow cells (Figure 1E), suggesting that nonhematopoietic tissues, such as the intestine or gills may be damaged. To circumvent this problem, split radiation doses could be tested, as are commonly used in murine HSC transplantation, to ameliorate deleterious effects on other organs while still ablating the hematopoietic compartment.
Highly variable mortality of marrow transplant recipients has been another problem. Many factors contribute to post-transplant mortality, including age and health of recipients, post-transplant care, and the genetic background and radiation sensitivity of a given strain. As these factors can be difficult to control or predict from one experiment to the next, we suggest it is impossible to compare mortality rates between independent transplant experiments. Despite this, the basic engraftment biology remains constant, such that comparisons can be made between 2 groups, as long as enough animals survive to the end of the experiment. Future experiments must include negative controls receiving radiation alone to establish the baseline mortality under given conditions, and sufficient transplanted animals to have adequate survivors for analysis.
An example of this is the poor survival observed in the limit dilution experiments for the 25 000 recipients compared with the 15 000 and 50 000 recipients (Figure 2). To overcome this variability, our statistical model takes the survival variation into account by reporting the SE to indicate the level of uncertainly for each of the fitted parameters. For our data, the fitted line in Figure 2 is minimally influenced by the 25 000 data, as the results are very similar, even when the analysis is repeated with these data omitted (supplemental Table 1). Therefore, despite the 25 000 data, we think our results accurately represent the HSCs in the WKM, with the limitation of the SE as reported.
As transplantation technology continues to develop in the zebrafish, the major area for advancement is the generation of histocompatible lines. The use of congenic zebrafish strains for transplant experiments would be optimal, and parthenogenesis may facilitate development of these animals over the next few years.25 This will help normalize the variable mortality in the assay. In the meantime, knowledge of functional MHC genes in the zebrafish is invaluable. Our data show that the MHC class I genes on chromosome 19 are functionally important for engraftment and that immune function clearly plays a role in engraftment and donor chimerism. MHC genes at other chromosomal loci may also prove to be important for transplantation. Likely candidates include additional class I genes on chromosome 1 and MHC class II genes on chromosome 8.
As immunologic issues are abolished, competitive transplants to determine relative engraftment efficiency between genetically different marrow populations will become possible. The results presented here not only set the foundation on which immune-matched transplantation in zebrafish can develop, but also provide the first functional assay in fish to distinguish hematopoietic stem and progenitor cell fates and to quantify HSC numbers. As zebrafish mutants and chemical treatments continue to be identified with a postulated effect on HSCs and engraftment, understanding all the functional MHC genes in the zebrafish will allow rapid identification of immune-matched recipients for competitive transplantation experiments. Such experiments will advance the use of the zebrafish for modeling humans diseases of HSCs including bone marrow failure syndromes and hematopoietic malignancies.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Teresa Bowman for critical comments, A. McCollum, C. Lawrence, and B. Barut for animal and laboratory management, and A. Flint for flow cytometry assistance.
This work was supported by the National Institute of Diabetes and Digestive and Kidney Diseases (Career Development Award 1K01DK067179, C.E.B.; and Career Development Award 5K08DK074595, J.L.O.d.J.). H.A.F. was supported by the Harvard Clinical and Translational Science Center (Catalyst; National Institutes of Health UL1 RR-025758). L.I.Z. is supported by Howard Hughes Medical Institute and the National Institutes of Health (National Heart, Lung, and Blood Institute grant 5R01HL48801-13).
National Institutes of Health
Authorship
Contribution: J.L.O.d.J., C.E.B., A.T.C., E.P., E.A.M., and A.C.H.S. performed experiments; J.L.O.d.J., C.E.B., E.A.M., A.T.C., Y.Z., and L.I.Z. designed the research and analyzed the data; H.A.F. was responsible for the limiting dilution model; and J.L.O.d.J., C.E.B., and L.I.Z. wrote the manuscript.
Conflict-of-interest disclosure: L.I.Z. is a founder and stockholder of Fate Inc and a scientific advisor for Stemgent. The remaining authors declare no competing financial interests.
The current affiliation of J.L.O.d.J. is University of Chicago, Department of Pediatrics, Section of Hematology-Oncology and Stem Cell Transplant, Chicago, IL.
Correspondence: Leonard I. Zon, Children's Hospital Boston, Harvard Medical School, 1 Blackfan Cir, Karp Bldg, 7th Fl, Boston, MA 02115; e-mail: zon@enders.tch.harvard.edu.
References
Author notes
J.L.O.d.J. and C.E.B. contributed equally to this study.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal