Abstract
The most frequent translocation t(8;21) in acute myeloid leukemia (AML) generates the chimeric AML1/ETO protein, which blocks differentiation and induces self-renewal in hematopoietic progenitor cells. The underlying mechanisms mediating AML1/ETO-induced self-renewal are largely unknown. Using expression microarray analysis, we identified the Groucho-related amino-terminal enhancer of split (AES) as a consistently up-regulated AML1/ETO target. Elevated levels of AES mRNA and protein were confirmed in AML1/ETO-expressing leukemia cells, as well as in other AML specimens. High expression of AES mRNA or protein was associated with improved survival of AML patients, even in the absence of t(8;21). On a functional level, knockdown of AES by RNAi in AML1/ETO-expressing cell lines inhibited colony formation. Similarly, self-renewal induced by AML1/ETO in primary murine progenitors was inhibited when AES was decreased or absent. High levels of AES expression enhanced formation of immature colonies, serial replating capacity of primary cells, and colony formation in colony-forming unit-spleen assays. These findings establish AES as a novel AML1/ETO-induced target gene that plays an important role in the self-renewal phenotype of t(8;21)-positive AML.
Introduction
The acquisition of unlimited growth and proliferation capabilities is an essential component of cancerogenesis.1 In acute myeloid leukemia (AML), enhanced self-renewal of hematopoietic progenitor cells is thought to be a crucial event for leukemogenesis. This hypothesis is supported by the fact that full-length AML1/ETO, one of the most frequently detected oncogenes in AML, not only blocks differentiation of hematopoietic progenitor cells but also induces self-renewal in hematopoietic progenitor cells.2-4 Expression of AML1/ETO is not sufficient on its own to induce a leukemia-like disease in mouse models.5,6 These features of AML1/ETO suggest that it is an appropriate candidate to study the mechanisms of oncogene-induced self-renewal in leukemia.
Unfortunately, the mechanisms of AML1/ETO-induced self-renewal remain largely unknown. A critical role of Wnt signaling in the regulation of hematopoietic stem and progenitor cell self-renewal and proliferation has been described.7 A couple of findings indicate that translocation products, including AML1/ETO, may mediate the enhanced self-renewal phenotype by influencing Wnt signaling. The products of several balanced translocations, including AML1/ETO, activate the Wnt signaling pathway in hematopoietic cells,8 and we and others have shown the involvement of Wnt signaling in leukemogenesis.8-14 Downstream effectors of the Wnt signaling pathway (eg, lymphoid enhancer-binding factor 1 [LEF-1]) have also been shown to be important in granulopoiesis and forced overexpression of LEF-1 induces leukemia in mice.15
LEF-1 and other proteins of the TCF (T-cell specific transcription factor)/LEF family of transcription factors are regulated by members of the Groucho protein family. Groucho was first identified in Drosophila and mapped to enhancer of split complex. In mammals, there are at least 5 known genes encoding Groucho homologs, termed TLE (transducin-like enhancer of split) in humans and Grg (Groucho-related genes) in mice.16 Groucho proteins specifically function as corepressors together with a variety of DNA-binding proteins. GRG1-4 and TLE1-4 share 2 highly conserved N-terminal glutamine-rich (Q) domains, a variable glycine/proline-rich region (GP domain), a CcN domain containing phosphorylation sites in close proximity of nuclear import signal sequence, and a serine/proline-rich region (SP domain) followed by a C-terminal WD40 repeat. The product of Grg5 in mice and its homolog AES (amino-terminal enhancer of split) in humans possess a shorter form of the highly conserved N-terminal domain and lack the repressor domain. As a consequence, they counteract the repressive functions of other GRG/TLE proteins and induce transcription of target genes.16
In this study, we show that AML1/ETO induces the Groucho-inhibitory AES protein. Moreover, AES expression is associated with a favorable prognosis in AML. On a functional level, AES enhances self-renewal of hematopoietic progenitor cells. Suppression of AES by RNAi abolishes the self-renewal inducing properties of AML1/ETO.
Methods
Cells, culture conditions, additional stimulants, and FACS
Adherent HeLa cell line was cultured in DMEM with l-glutamine and 10% fetal calf serum (FCS). Human U937, Kasumi-1, K562, KCL-22, NB4, HL60 cells and murine 32D cells were cultured in RPMI with l-glutamine and 10% FCS; 32D cultures were supplemented with murine IL-3 obtained from supernatant of WEHI-3B cells. Stably transfected U937 cell lines expressing in a Zn2+-inducible fashion AML1/ETO or empty vector as control have been previously described.8 For induction of the fusion genes in these cells, 0.1mM ZnSO4 was added for 12 hours. Trichostatin A (1μM) was added for 2 hours. All-trans retinoic acid (10μM; Sigma-Aldrich) was added as differentiation agent for NB4 cells. Cells were harvested at the indicated time points for analysis. Cells were washed with phosphate-buffered saline (PBS), resuspended in 100 μL PBS, and incubated with PE-labeled anti-CD11b (clone M1/7015.1, Cymbus Biotechnology) and FITC-labeled anti-CD14 antibody for 10 minutes on ice for staining of granulocytes and macrophages, respectively. Cells were analyzed by flow cytometry (FACSCalibur, BD Biosciences).
The Grg5−/− mice in a C57BL6 background were kindly provided by Dr Tom Gridley (The Jackson Laboratory).17 Animal experiments were approved and performed in accordance with all regulatory guidelines. For all clinical patient samples, informed consent was obtained in accordance with the Declaration of Helsinki, and the study was approved by the Ethics committee.
RNA isolation, cDNA preparation, and quantitative PCR
RNA was isolated using RNAeasy kit (QIAGEN) according to the manufacturer's instructions. RNA (1 μg) was reverse transcribed using oligo-d(T) primer and random hexamers. cDNA was generated with M-MLV reverse transcriptase (Promega) for 1 hour at 42°C. The quantitative reverse-transcribed polymerase chain reaction (RT-PCR) for gene expression was performed with TaqMan technology (Applied Biosystems). Reaction volume was 12 μL containing mastermix (Eurogentec) with TaqMan probe or SYBR Green. The primer sequence for AES was: TGTCCTACGGATTGAACATCGA (forward), ATGCACAAACAGGCGGAAATTGTGAAG (probe) and GGCGCAAATCCCATTCAG (reverse). Values were normalized to GAPDH expression levels.
Antibodies, protein extraction, and Western blotting
Cell pellets were resuspended in lysis puffer (150mM NaCl, 1% NP-40, 0.5% deoxycholate [DOC], 0.1% SDS, 50mM Tris-HCl, pH 8.0), supplemented with protease inhibitor complete (Roche Diagnostics), and incubated for 10 minutes on ice. After centrifugation at 14 000g for 5 minutes at 4°C, the supernatants were collected as whole cell lysates. Protein concentration was determined with BCA assay (Pierce Chemical). Western blotting was performed with equal amounts of total protein separated by SDS-PAGE and transferred to polyvinylidene difluoride (PVDF) membrane. After blocking, the membrane was incubated with primary antibody for 1 hour, washed, and incubated with HRP-conjugated secondary antibody. Immunoreactive stainings were developed using ECL (Invitrogen) or ECL plus (GE Healthcare). An autoradiograph on film was obtained with an appropriate exposure time. Polyclonal antibody anti-AES were raised against the epitopes aa183-197 and aa2-16 (Eurogentech) and tested for specificity. Polyclonal anti-AES and anti-AML1/ETO antibodies were obtained from Imgenex and Merck, respectively. As internal loading control, anti–β-actin (Sigma-Aldrich) was used for lysates on immunoblot procedures.
TMA
Tissue microarray (TMA) construction of formalin-fixed, paraffin-embedded bone marrow (BM) biopsies of patients diagnosed with primary, untreated AML was performed as duplicate cores, previously described in detail.8,18 A diagnostic Giemsa-stained section served as control to enable the definition of areas with the highest amount of blast cells. Cell lines and primary CD34+ cells were mounted in agarose, formalin-fixed, paraffin-embedded. Tissue sections from the TMA block (3 μm thickness) were mounted on SuperFrost Plus slides and dewaxed in xylene followed by demasking with steam of 10mM citrate buffer, pH 6.0, for 30 minutes. Primary antibody anti-AES (1:250) was incubated 25 minutes in autostainer (Dako North America). Secondary antibody was conjugated with Dako REAL Link biotinylated streptavidin alkaline phosphatase. The reaction was visualized by a RED chromogen (Fast RED, Dako North America) and contrasted with hematoxylin (Meyer) to ensure a proper staining. Light microscopy was performed with an Axioplan microscope (Zeiss) using a 25× Plan-Neofluar 0.80 or 100× Plan-Neofluar 1.30 oil lens. Images were captured using Axiovision (Zeiss) and Photoshop Version 5.5 (Adobe Systems).
Sequences and generation of stable RNAi expressing cells
For 32D cells, the vector pRNAT (Genescript) was used to express GRG5/AES or nonfunctional control scrambled (scr) shRNA constructs, which were introduced into the SalI/BglII sites. The GRG5/AES target sequence that gave the best suppression and used in this study was ATCTCGATGTTCAATCCGTAG. The shRNA scrambled control target sequence was AGATCCGTATAGTGTACCTTA. 32D cells were electroporated with 10 μg of vector DNA coexpressing green fluorescent protein (GFP). Subsequently isolated cell batches were analyzed for GFP and positively sorted. Bulk subclones of cultures were selected with neomycin (G418, 800 μg/mL) and tested for shRNA knockdown in quantitative RT-PCR and Western blotting.
For stable transduction of human cell lines, LKO.1 vectors containing shRNA sequence against human AES (MISSION TRCN#20031 and #20032, Sigma-Aldrich) were used for infections. For stable transduction of AML1/ETO-transformed primary murine BM cells, LKO.1-puro vectors expressing shRNA sequence against murine GRG (MISSION TRCN#97716 and #97717, Sigma-Aldrich) were used.
Retroviral transduction of primary BM cells
All retroviral vector pseudotypes were generated by transient transfection of Phoenix cells (kindly provided by Gary Nolan, Stanford University) with plasmids expressing appropriate viral packaging proteins, as described previously.19 The γ-retroviral vector SF91iGFP or its derivative SF91iYFP6 were used for expression of AML1/ETO and AES, respectively. For colony-forming unit-spleen (CFU-S) assays, BM cells were transduced with pMY-AES-IRES-EGFP or control retroviruses. For GRG5-knockout BM, MIG-AML1/ETO vectors were used for transduction.20 The lentiviral vector LKO.1-puro (Sigma-Aldrich) or its derivative LKO.1-YFP were used to express AES shRNA. For retroviral transduction of primary BM cells, C57Bl/6 mice or Balb/C mice were given 5-fluorouracil (5-FU) intraperitoneally (150 μg/g body weight) and BM cells were isolated after 2 days from humera and femura. Alternatively, GRG5/AES knockout or wild-type BM cells were isolated from untreated mice and progenitors were enriched on the basis of the Sca1 stem cell marker and/or the absence of lineage differentiation markers (Sca1+/lin−) by immunomagnetic beads using “MACS” cell separation columns (Miltenyi Biotec). BM cells were cultured with 20% FCS, 1% penicillin/streptomycin, and a growth factor cocktail of murine recombinant IL-3 (10 ng/mL), huIL-6 (5 ng/mL), and murine stem cell factor (muSCF; 50 ng/mL). On the following 2 days, 3 rounds of transduction were performed. The cells were seeded at a concentration of 1 × 106/mL on freshly thawed, cell-free supernatant containing retroviral particles in the presence of the aforementioned cytokine cocktail and protamine sulfate (4 mg/mL). For each round of transduction, the cells were centrifuged at 400g for 30 minutes and incubated at 37°C for 2 hours. The cells were incubated overnight in the last viral supernatant at 37°C. Sorting of GFP+ cells was performed with FACSAriaII (BD Biosciences), using FACSDiva software Version 6.1.3.
CFU-S assays
An enriched population of HSCs of the surface phenotype Lin−Sca1+ (LS) from C57Bl6 mice was isolated using the lineage depletion kit from Miltenyi Biotec (catalog no. 130-090-858) and the mouse Sca1 selection cocktail from StemCell Technologies (no. 18756) according to the manufacturer's protocols. Freshly isolated LS cells were infected on 2 consecutive days with AES-expressing and control retroviruses, coexpressing enhanced GFP, by spinoculation at 1200g, 32°C, 90 minutes. Transduced cells were cultured in MethoCult M3234 methylcellulose medium (StemCell Technologies) at a final concentration in Iscove modified Dulbecco medium (IMDM) containing: 10% FBS, IL-3, IL-6, both at 10 ng/mL, and SCF at 100 ng/mL (all recombinant cytokines from PeproTech). Seven days later, cells were washed out of methylcellulose media and incubated overnight in IMDM supplemented with cytokines before FACS sorting. Enhanced GFP+ cells were rested for an additional 24 hours in IMDM with cytokines. A total of 8 × 103 cells was administered intravenously into lethally irradiated (11 Gy) recipient C57Bl6 mice 10 to 12 weeks of age. Spleens were harvested at 12 days after transplantation, fixed in Tellesnizky solution (70% [vol/vol] ethanol, 1% [vol/vol] formaldehyde, 10% [vol/vol] acetic acid), and macroscopic colonies were documented. In a second CFU-S assay, the spleens were embedded in paraffin for microscopic detection of colonies; 5 μm thick paraffin sections were deparaffinized before staining twice for 10 minutes in xylene, then washed twice from xylene with 96% ethanol, rehydrated by incubation for 5 minutes in serial ethanol dilutions (2 times in every solution) 96%, 80%, 70%, 50%, and finally in PBS buffer. Then the sections were washed several times with distilled water and stained for 6 minutes in Mayer hematoxylin solution. After 10 minutes under running tap water, sections were stained in 1% eosin G solution for 2 minutes, washed again, and finally mounted using Mowiol solution. Microscopic CFU-Ss of mice transplanted with AES (n = 8) or empty vector (n = 8) being transduced into lineage-depleted primary BM cells were analyzed. CFU-Ss were counted on 2 different sections of each spleen by 2 independent investigators; the numbers of colonies were compared using t test.
Colony growth and apoptosis assays
Fluorescein isothiocyanate–labeled annexin A5 (Beckman Coulter) staining was used to detect apoptosis in starved 32D stable cell lines were subjected to annexin A5 as described before. Positive cells were analyzed with CellQuest software (BD Biosciences). For terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling (TUNEL) assay, 32D cells fixed onto fibronectin-coated slides were washed and 4% paraformaldehyde/PBS fixed. After blocking and permeabilization, cells were labeled for in situ cell death detection kit TMR red (Roche Diagnostics) according to the manufacturer's instructions. Terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling-positive apoptotic cells (red) were subjected to fluorescence microscopy.
For colony growth assays, BM cells from Balb/C mice infected with AES-expressing vectors (SF91-iYFP; infection frequency, 70%-80%) were seeded at a density of 103 cells per dish (diameter, 35 mm) in triplicates containing IMDM, 1% methylcelluose (Invitrogen), 20% FCS, recombinant murine IL-3 (10 ng/mL), and recombinant human IL-6 (5 ng/mL). After incubation at 37°C, colonies with more than 50 agglomerated cells were counted on day 7. Determination of BM colony-forming units was carried out as colony-forming units-granulocyte, erythrocyte, monocyte/macrophage, megakaryocyte (CFU-GEMM), colony-forming units-granulocyte-macrophage (CFU-GM), and colony-forming units-megakaryocyte (CFU-M) indications.
BM cells from GRG5/AES knockout mice were lineage depleted before retroviral transduction with MIG and sorted for GFP+ cells. Colonies were counted and serially replated after 7 days in culture. Three independent experiments were performed in triplicates each, and the number of colonies was compared using paired t test.
For BM cells infected with AML1/ETO-expressing vectors (SF91-GFP), cells were seeded at a density of 103 cells per dish in methylcellulose cultures (M3434, StemCell Technologies). Cells were serially replated at 1-week intervals, leading to the selection of AML1/ETO-expressing cells after 4 rounds. Cells were harvested, subjected to infection with LKO.1 lentiviral vectors expressing shRNA against AES (see “Retroviral transduction of primary BM cells”), selected for 4 days in the presence of puromycin (1 μg/mL), and then plated again in methylcellulose. Two independent experiments were performed.
For BM cells infected with AES-expressing vectors (SF91-GFP), cells were seeded at a density of 103 cells per dish in methylcellulose cultures (M3434, StemCell Technologies). Cells were serially replated at 10-day intervals. Two independent experiments in triplicates were performed.
Chromatin immunoprecipitation
CD34+ progenitor cells derived from G-CSF–mobilized donors without leukemia were transfected using Nucleofection (Amaxa) with either an AES expression construct or with empty pcDNA3 control vector. Cells were subjected to ChIP analysis essentially as described before8 with the exception that preimmune serum (before producing polyclonal Abs anti-AES serum) and polyclonal Abs anti-AES (Eurogentech) were used. PCR reactions were performed as described previously8 and visualized on a 1.5% agarose gel stained with ethidium bromide.
Confocal microscopy
Adherent HeLa and suspension HL-60 cells were seeded on coverslips coated with 50 μg/mL fibronectin. Cells were fixed for 10 minutes using 4% paraformaldehyde/PBS, permeabilized with 0.2% Triton X-100/PBS for 2 minutes at room temperature and stained with anti-AES Ab (Imgenex) and the appropriate secondary Ab goat anti–rabbit IgG coupled with Alexa 488 diluted in 2% BSA. Afterward, actin cytoskeleton was detected with Alexa Fluor 568 phalloidin (Invitrogen). Washed and Mowiol-mounted cells were analyzed by confocal fluorescence microscopy (LSM510, Carl Zeiss).
RNA extraction and hybridization
Total RNA was extracted from cells using TRIzol reagent (Invitrogen), and 10 μg of total RNA was reverse transcribed into cDNA using an oligo-d(T)-T7-primer. Subsequently, T7 polymerase was used for in vitro transcription. During transcription, cRNA was labeled using biotinylated oligonucleotides. The resulting labeled cRNAs were fragmented and hybridized to HuGeneFL oligonucleotide microarrays, containing probes for approximately 6800 independent transcripts (Affymetrix). Arrays were scanned after staining with streptavidin-phycoerythrin, signal amplification with biotinylated anti-streptavidin antibodies, and subsequent staining with streptavidin-phycoerythrin. Raw data were normalized and scaled to an average level of 2500.
Microarray data analyses and statistics
Microarray analyses to identify AML1/ETO target genes were described previously. To identify AES target genes, CD34+ progenitor cells derived from granulocyte colony-stimulating factor mobilized donors without leukemia were transfected using Nucleofection (Amaxa) with either an AES expression construct (n = 2) or with empty pcDNA3 control vector (n = 2). Nontransfected CD34+ cells (n = 4) served as another control. mRNA was isolated 24 hours after transfection and hybridized onto ABI 1700 whole genome arrays. Microarray data were quantile normalized. Genes that were altered in AES-expressing CD34+ cells at least 3-fold compared with control vector-transfected cells and at least 3-fold compared with nontransfected cells were considered to be altered on AES expression. The resulting gene lists (induced and repressed) were then analyzed for their association with biologic pathways and functions using the Panther database at www.pantherdb.org.
Differences between 2 groups were compared using Student t test if not indicated otherwise. Survival analyses were performed using SPSS (Statistical Package for the Social Sciences Version 7.0). Overall, 236 patients were included into the multivariate analysis using a stepwise (forward) proportional Cox regression hazard model. The following parameters were included as categorical variables into the analysis: cytogenetics (3 groups: high-risk, low-risk, and intermediate/unknown), FLT3-TKD mutations, NRAS mutations, KRAS mutations, C/EBPα mutations, FLT3-ITD mutations, and AES expression levels as low/high according to the median expression. Information on patient age and NPM mutation status was not available. In all statistical tests, a P less than .05 was regarded as statistically significant.
Results
Identification of AES as AML1/ETO target gene
In microarray analyses aimed to define target genes of common translocations in AML,8 we found the mRNA of AES consistently induced by AML1/ETO. These data were confirmed by real-time RT-PCR and Western blot analysis in U937 cells transfected with an inducible AML1/ETO vector (Figure 1A-B). Trichostatin A, a histone deacetylase inhibitor, prevented AML1/ETO-enhanced AES expression (Figure 1A). This suggests that AES is induced by an indirect mechanism that involves repression of genomic target genes by AML1/ETO. In a published microarray dataset (GDS1074, Gene Expression Omnibus; National Institutes of Health, National Center for Biotechnology Information), AES up-regulation by AML1/ETO was confirmed in retrovirally AML1/ETO-transduced CD34+ progenitor cells (Figure 1C). Although statistically not significant (only 6 samples were analyzed), it still suggested a trend toward higher AES levels on AML1/ETO transduction.
AML1/ETO induces AES mRNA and protein expression. (A) AES expression on the mRNA level (quantitative RT-PCR) in U937 cells transfected with an inducible AML1/ETO vector, compared with control cells. The effect was reverted to normal levels of AES expression by histone deacetylase inhibition with trichostatin A (TSA) exposure. (B) U937 cells that express AML1/ETO show high levels of AES protein. Western blot analysis was performed with anti-AES and anti-actin antibody as loading control. (C) AES induction by AML1/ETO (mRNA) in retrovirally transduced CD34+ progenitor cells (published data; GDS1074, Gene Expression Omnibus, National Institutes of Health, National Center for Biotechnology Information). (D) AES cDNA expression patterns in hematopoiesis. Total RNA was isolated from sorted murine BM cells; cells were sorted according to indicated surface markers by fluorescence-activated cell sorter. (E) Confocal microcopy pictures show subcellular nuclear localization of AES in HeLa cells and HL-60 cells as indicated.
AML1/ETO induces AES mRNA and protein expression. (A) AES expression on the mRNA level (quantitative RT-PCR) in U937 cells transfected with an inducible AML1/ETO vector, compared with control cells. The effect was reverted to normal levels of AES expression by histone deacetylase inhibition with trichostatin A (TSA) exposure. (B) U937 cells that express AML1/ETO show high levels of AES protein. Western blot analysis was performed with anti-AES and anti-actin antibody as loading control. (C) AES induction by AML1/ETO (mRNA) in retrovirally transduced CD34+ progenitor cells (published data; GDS1074, Gene Expression Omnibus, National Institutes of Health, National Center for Biotechnology Information). (D) AES cDNA expression patterns in hematopoiesis. Total RNA was isolated from sorted murine BM cells; cells were sorted according to indicated surface markers by fluorescence-activated cell sorter. (E) Confocal microcopy pictures show subcellular nuclear localization of AES in HeLa cells and HL-60 cells as indicated.
GRG5/AES expression patterns in normal hematopoiesis and AML
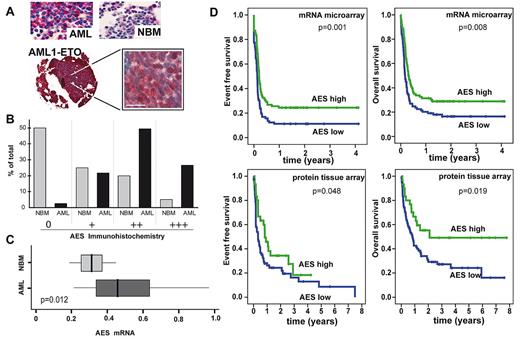
In murine BM, the murine AES homolog GRG5 is mainly expressed in myeloid cells and in lin−/ckit+ progenitor cells (Figure 1D). Immunofluorescence staining with an anti-AES antibody in HeLa and HL-60 cells was predominantly nuclear (Figure 1E). AES immunostaining in primary AML patient samples was nuclear and cytoplasmic in most cases (Figure 2A) with a predominant cytoplasmic staining observed in some AML cases (eg, small AML photo). Interestingly, AES expression was repressed on all-trans retinoic acid-induced differentiation of NB4 leukemia cells (supplemental Figure 2A-B).
AES expression in AML patients and its association with patient prognosis. (A) Representative micrographs of TMA from NBM and AML patients stained with anti-AES antibody and Fast-Red secondary antibody contrasted with hematoxylin and eosin. Overview and 63× magnification of one sample with the t(8; 21) (AML1/ETO) chromosomal translocation. Bar represents 20 μm. (B) Quantitative TMA analysis of AES expression using categories of staining intensity (0 indicates nothing; + low; ++ moderate; and +++ strong) shows comparison between NBM specimens (N = 40; AML, N = 163) in percentage of total number of samples. Statistical significance using χ2 test was P less than .05. (C) RT-PCR analysis of AML patients (n = 50) versus NBM (n = 8) of AES mRNA level demonstrates higher AES expression levels in AML (P = .012). (D) Kaplan-Meier plots of event-free survival and overall survival in AML patients with low or high levels of AES expression. For mRNA analyses, a published microarray dataset was used, and patients were regarded as high or low on AES expression based on the median expression. Immunohistochemistry was performed by tissue array of an independent patient population. In both groups, patients with a favorable karyotype (t(15;17), t(8;21), inv(16)) or C/EBPα mutations were excluded. Statistical significance was evaluated using the log-rank test (Mantel-Cox).
AES expression in AML patients and its association with patient prognosis. (A) Representative micrographs of TMA from NBM and AML patients stained with anti-AES antibody and Fast-Red secondary antibody contrasted with hematoxylin and eosin. Overview and 63× magnification of one sample with the t(8; 21) (AML1/ETO) chromosomal translocation. Bar represents 20 μm. (B) Quantitative TMA analysis of AES expression using categories of staining intensity (0 indicates nothing; + low; ++ moderate; and +++ strong) shows comparison between NBM specimens (N = 40; AML, N = 163) in percentage of total number of samples. Statistical significance using χ2 test was P less than .05. (C) RT-PCR analysis of AML patients (n = 50) versus NBM (n = 8) of AES mRNA level demonstrates higher AES expression levels in AML (P = .012). (D) Kaplan-Meier plots of event-free survival and overall survival in AML patients with low or high levels of AES expression. For mRNA analyses, a published microarray dataset was used, and patients were regarded as high or low on AES expression based on the median expression. Immunohistochemistry was performed by tissue array of an independent patient population. In both groups, patients with a favorable karyotype (t(15;17), t(8;21), inv(16)) or C/EBPα mutations were excluded. Statistical significance was evaluated using the log-rank test (Mantel-Cox).
We next analyzed AES mRNA levels based on a large microarray expression dataset.21 AES mRNA was expressed in all French-American-British (FAB) subtypes (supplemental Figure 2C). Patients with C/EBPα mutations (n = 16) showed significantly lower levels of AES than other (n = 230) patients (P < .001, Mann-Whitney U test). For survival analyses, we included all samples from AML patients who did not harbor t(8;21), inv(16), t(15;17), or C/EBPα mutations because these leukemias are all associated with a favorable prognosis. The median AES expression value of all patients was used to define low (n = 89) and high (n = 86) expressers. Event-free survival was significantly longer in patients with high AES mRNA expression (median, 59.8 months) compared with low AES-expressing patients (median, 30.4 months) (P = .001). In addition, overall survival was prolonged in patients with high (median, 72.3 months) AES expression compared with those with low (median, 45.6 months) levels of AES expression (P = .008). In addition, we performed multivariate analyses of all AML patients with available data in the cohort using cytogenetics (3 groups), low/high levels of AES, and the following mutations: FLT3-ITD, FLT3-TKD, NRAS, KRAS, and C/EBPα. In a stepwise Cox regression analysis of 236 AML patients, cytogenetics, C/EBPα mutations, FLT3-ITD and AES expression were significantly associated with event-free survival and overall survival (Table 1).
Multivariate survival analysis
| Parameter . | Overall survival . | Event-free survival . | ||||
|---|---|---|---|---|---|---|
| Hazard ratio . | 95% CI . | P . | Hazard ratio . | 95% CI . | P . | |
| Cytogenetics | < .001 | < .001 | ||||
| High risk | 3.7 | 2.1-6.6 | < .001 | 2.7 | 1.6-4.6 | < .001 |
| FLT3-ITD mutation | 1.5 | 1.1-2.2. | .017 | 1.4 | 1.0-2.0 | .044 |
| CEBPα mutation | 0.3 | 0.1-0.7 | .005 | 0.3 | 0.1-0.7 | .003 |
| AES expression | 0.7 | 0.5-1.0 | .047 | 0.7 | 0.5-1.0 | .046 |
| Parameter . | Overall survival . | Event-free survival . | ||||
|---|---|---|---|---|---|---|
| Hazard ratio . | 95% CI . | P . | Hazard ratio . | 95% CI . | P . | |
| Cytogenetics | < .001 | < .001 | ||||
| High risk | 3.7 | 2.1-6.6 | < .001 | 2.7 | 1.6-4.6 | < .001 |
| FLT3-ITD mutation | 1.5 | 1.1-2.2. | .017 | 1.4 | 1.0-2.0 | .044 |
| CEBPα mutation | 0.3 | 0.1-0.7 | .005 | 0.3 | 0.1-0.7 | .003 |
| AES expression | 0.7 | 0.5-1.0 | .047 | 0.7 | 0.5-1.0 | .046 |
The mRNA expression values for AES were recorded as low or high with regard to median expression levels of all AML patients in the microarray study. Overall, 236 patients with available data were included in the multivariate analysis using a stepwise (forward) proportional Cox regression hazard model. The following parameters were included as categorical variables into the analysis: cytogenetics (3 groups: high risk, low risk, or intermediate/unknown), FLT3-TKD mutations, NRAS mutations, KRAS mutations, C/EBPα mutations, FLT3-ITD mutations, and AES expression levels as low/high according to the median expression.
Complementary to the data from the microarray dataset, we also analyzed AES mRNA levels by real-time RT-PCR and found that AES expression was significantly increased in AML patient blasts compared with normal BM (NBM; Figure 2C).
We used a TMA of AML and NBM biopsies to corroborate these findings on the protein level. Expression of AES was confirmed in more than 95% of AML specimens (Figure 2). In contrast, significant expression was observed in less than 50% of the specimens in NBM. AES levels were much higher in AML specimens than in NBM (Figure 2B). Interestingly, AML1/ETO+ AML samples showed high levels of AES expression (Figure 2A). We then analyzed the association between AES levels and clinical features. Higher amounts of AES protein were associated with significantly improved long-term survival in univariate analysis (Figure 2D). Multivariate analyses were not performed because cytogenetics and mutation status were available for a minority of patients, only. Taken together, these findings indicated that AES is highly expressed on the mRNA and protein level in AML and that high levels of AES mRNA and protein are associated with a favorable prognosis.
AES is required for AML1/ETO-induced self-renewal
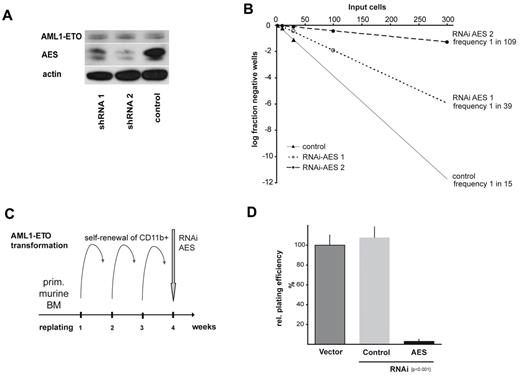
These findings prompted us to analyze the functional importance of AES for AML1/ETO function. The t(8;21)-positive human Kasumi-1 cells show high levels of endogenous AES expression, which was reduced by more than 70% on lentiviral infection of different shRNA against AES (Figure 3A). Using limiting dilution analyses of these cells, we quantified the fraction of colony-forming cells transduced with either shRNA against AES (2 sequences) or a nonfunctional scrambled control. Colonies were counted after 10 days, and the relative cloning efficiency was calculated according to Poisson distribution. As shown in Figure 3B, shRNA directed against AES strongly increased the frequency of wells that did not contain Kasumi-1 colonies, demonstrating the importance of AES for the clonal growth of AML1/ETO-expressing cells. Whereas the frequency of colony-forming cells was estimated to be 1:15 for cells transfected with a scrambled control shRNA, the frequency of colony-forming cells after AES suppression was reduced to 1:39 and 1:109, respectively (Figure 3B). This reduction of colony-forming cells after AES suppression was not found in any other tested cell line (Table 2). These observations were a first indication that AES is important for self-renewal in AML1/ETO+ cells. Interestingly, neither apoptosis nor cell proliferation was affected by AES suppression in either Kasumi-1 or any other of the human leukemia cell lines (supplemental Figure 1A-B, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). In 32D cells, a hematopoietic progenitor cell line GRG5/AES suppressed by siRNA reduced colony growth and increased apoptosis to approximately 20% (supplemental Figure 1C-F).
AES expression is required for AML1/ETO-induced proliferation. (A) Two different shRNAs against AES delivered by lentiviral vectors were shown to down-regulate AES expression in Kasumi-1 cells, as demonstrated in Western blot analyses stained with anti-AES antibody. Anti-actin serves as loading control. (B) Limited dilution analyses of cloning capacity of the stably transduced and sorted Kasumi-1 cells showed a significantly reduced frequency of self-renewal capacity. Relative cloning efficiency was calculated as the frequency of growing colonies in correlation to seeded cell number. The log fraction of negative wells on input cells per plate is indicated (Poisson distribution). (C) Scheme of experimental setup to analyze self-renewal capacity in primary murine BM. The transforming capacity of AML1/ETO in CD11b+ BM cells was used to select for cells with self-renewal capacity after several rounds of replating. Immortalized cells were subsequently infected with LKO.1-puro vectors expressing shRNA against AES, selected and replated in colony assays. (D) The ability to form colonies on replating is shown after AES knockdown compared with nontransduced or scrambled transduced (control) cells. Data are mean plus or minus SD of 2 independent experiments each performed in triplicate (total 6 dishes). Differences between AES-shRNA and scrambled control transduced colony numbers were statistically significant (P < .001).
AES expression is required for AML1/ETO-induced proliferation. (A) Two different shRNAs against AES delivered by lentiviral vectors were shown to down-regulate AES expression in Kasumi-1 cells, as demonstrated in Western blot analyses stained with anti-AES antibody. Anti-actin serves as loading control. (B) Limited dilution analyses of cloning capacity of the stably transduced and sorted Kasumi-1 cells showed a significantly reduced frequency of self-renewal capacity. Relative cloning efficiency was calculated as the frequency of growing colonies in correlation to seeded cell number. The log fraction of negative wells on input cells per plate is indicated (Poisson distribution). (C) Scheme of experimental setup to analyze self-renewal capacity in primary murine BM. The transforming capacity of AML1/ETO in CD11b+ BM cells was used to select for cells with self-renewal capacity after several rounds of replating. Immortalized cells were subsequently infected with LKO.1-puro vectors expressing shRNA against AES, selected and replated in colony assays. (D) The ability to form colonies on replating is shown after AES knockdown compared with nontransduced or scrambled transduced (control) cells. Data are mean plus or minus SD of 2 independent experiments each performed in triplicate (total 6 dishes). Differences between AES-shRNA and scrambled control transduced colony numbers were statistically significant (P < .001).
Cloning efficiency of leukemic cell lines on lentiviral transduction with shRNA against AES
| . | Scrambled control . | shRNA-AES-#1 . | shRNA-AES#2 . |
|---|---|---|---|
| Kasumi-1 cloning | 1 in 15 | 1 in 39 | 1 in 109 |
| 95% CI | 12-17 | 33-46 | 92-129 |
| P | .0001 | .0001 | |
| KCL-22 cloning | 1 in 82 | 1 in 72 | 1 in 79 |
| 95% CI | 65-104 | 57-91 | 63-100 |
| P | .42 | .82 | |
| K562 cloning | 1 in 28 | 1 in 20 | 1 in 43 |
| 95% CI | 22-35 | 16-25 | 34-55 |
| P | .05 | .01 | |
| U937 cloning | 1 in 2 | 1 in 3 | 1 in 3 |
| 95% CI | 2-3 | 2-3 | 3-4 |
| P | .29 | .04 |
| . | Scrambled control . | shRNA-AES-#1 . | shRNA-AES#2 . |
|---|---|---|---|
| Kasumi-1 cloning | 1 in 15 | 1 in 39 | 1 in 109 |
| 95% CI | 12-17 | 33-46 | 92-129 |
| P | .0001 | .0001 | |
| KCL-22 cloning | 1 in 82 | 1 in 72 | 1 in 79 |
| 95% CI | 65-104 | 57-91 | 63-100 |
| P | .42 | .82 | |
| K562 cloning | 1 in 28 | 1 in 20 | 1 in 43 |
| 95% CI | 22-35 | 16-25 | 34-55 |
| P | .05 | .01 | |
| U937 cloning | 1 in 2 | 1 in 3 | 1 in 3 |
| 95% CI | 2-3 | 2-3 | 3-4 |
| P | .29 | .04 |
Cloning efficiency was calculated by plating 1, 3, 10, 100, and 300 transduced cells (GFP+) each into 48-well colony assays (300 cells only for Kasumi-1 and KCL-22). The number of wells with colony growth was counted. Poisson distributions were calculated using L-Calc limiting dilution analysis software Version 1.1.1 (StemSoft Company). The calculated cloning efficiency as well as the 95% confidence intervals (CI) are indicated. Two-sided P values were calculated for each cell line and each shRNA construct with regard to the cloning efficiency in the scrambled control. The data indicate the mean of 2 independent experiments. Strong effects of shRNA against AES were found only in AML1/ETO+ Kasumi-1 cells with 2.6-fold and 7.3-fold reduction in cloning efficiency. Minor effects (< 2-fold reduction) were observed in K562 cells and in U937 cells.
Next, we transduced primary murine BM with an AML1/ETO-expressing vector to induce self-renewal. AML1/ETO-expressing cells that had gained self-renewal potential were selected in methylcellulose replating assays, giving rise to significant colony numbers after 3 rounds of replating in AML1/ETO-induced cells. At this time, cells were transduced with shRNA against either AES or a scrambled sequence. Cells were selected with puromycin and then replated (Figure 3C). AML1/ETO-transduced cells with scrambled shRNA still produced further colonies. In contrast, suppression of AES by RNAi almost completely abolished colony formation (Figure 3D; P < .001). These data suggested that AES is required for AML1/ETO mediated continuous colony formation potential in GRG5/AES-expressing hematopoietic progenitor cells.
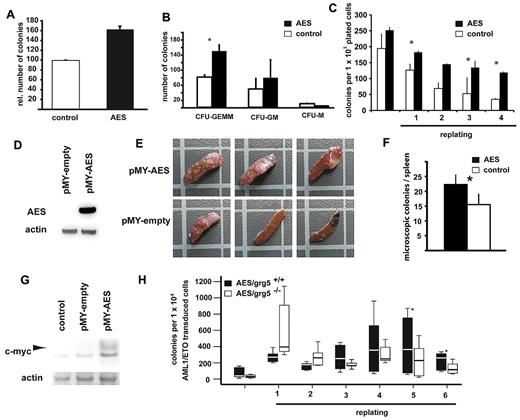
Next, we analyzed whether AES overexpression could enhance proliferation and/or self-renewal. For this purpose, wild-type murine BM cells were transduced with AES or empty control vector. AES expression consistently increased colony formation (P = .001, Figure 4A). Interestingly, a large percentage of the colonies formed by AES-expressing cells remained immature as evidenced by the higher numbers of colony-forming unit-granulocyte, erythrocyte, monocyte/macrophage, megakaryocyte colonies (P = .017, paired t test; Figure 4B). In further experiments, we determined the induction of self-renewal in AES-expressing BM cells. Overexpression of AES-induced enhanced self-renewal in vitro as shown by the ability to replate colonies over several rounds (fourth replating: P = .002; Figure 4C). To obtain information from in vivo experiments, we performed day 12 CFU-S assays to test the short-term self-renewal capacity of progenitor cells overexpressing AES (Figure 4D). Spleens of mice transplanted with AES-overexpressing cells showed a strong increase in macroscopic (3 of 5 spleens) and microscopic (P = .001, t test) CFU-S numbers compared with control transduced cells (Figure 4E-F). Within the colony-forming tissue of the spleens, we found increased c-myc protein levels (Figure 4G). These results indicated that AES induced increased self-renewal and up-regulation of c-myc in vivo.
AES enhances proliferation and self-renewal in hematopoietic progenitor cells. Colony formation of murine primary BM cells retrovirally transduced with AES or empty control vector. (A) Higher numbers of relative colonies after AES overexpression. Bars represent relative colony numbers of 3 experiments. (B) The increase in the colony count was associated with an increased fraction of immature colonies (results and SE are shown). *Statistical significance at P < .05. White bars represent empty vector control; and black bars AES overexpression. (C) AES-transduced murine BM cells were serially plated twice. The results of 4 rounds of replating with SE are shown. *Statistical significance at P < .05. White bars represent empty vector control; and black bars AES overexpression. (D) AES-transduced murine BM cells were analyzed for AES expression. (E) CFU-S d12 assays were performed with AES-overexpressing, transduced BM cells and control cells. Three of 5 spleens of mice transplanted with AES-overexpressing cells showed a strong increase in macroscopic CFU-S numbers. These spleens and representative controls are shown. (F) A second set of CFU-S d12 assays with AES-overexpressing, transduced BM cells and control cells was performed, and microscopic colonies were counted. Data are mean plus or minus SD. The difference was statistically significant (P = .001). (G) Protein lysates of spleen tissue with colonies were analyzed for c-myc expression by Western blot analysis. (H) Primary BM cells from GRG5 KO-mice and wt-mice were transduced with AML1/ETO and serially transplated. The results of 6 rounds of replating are shown. *Statistical significance at P < .01 (paired t test).
AES enhances proliferation and self-renewal in hematopoietic progenitor cells. Colony formation of murine primary BM cells retrovirally transduced with AES or empty control vector. (A) Higher numbers of relative colonies after AES overexpression. Bars represent relative colony numbers of 3 experiments. (B) The increase in the colony count was associated with an increased fraction of immature colonies (results and SE are shown). *Statistical significance at P < .05. White bars represent empty vector control; and black bars AES overexpression. (C) AES-transduced murine BM cells were serially plated twice. The results of 4 rounds of replating with SE are shown. *Statistical significance at P < .05. White bars represent empty vector control; and black bars AES overexpression. (D) AES-transduced murine BM cells were analyzed for AES expression. (E) CFU-S d12 assays were performed with AES-overexpressing, transduced BM cells and control cells. Three of 5 spleens of mice transplanted with AES-overexpressing cells showed a strong increase in macroscopic CFU-S numbers. These spleens and representative controls are shown. (F) A second set of CFU-S d12 assays with AES-overexpressing, transduced BM cells and control cells was performed, and microscopic colonies were counted. Data are mean plus or minus SD. The difference was statistically significant (P = .001). (G) Protein lysates of spleen tissue with colonies were analyzed for c-myc expression by Western blot analysis. (H) Primary BM cells from GRG5 KO-mice and wt-mice were transduced with AML1/ETO and serially transplated. The results of 6 rounds of replating are shown. *Statistical significance at P < .01 (paired t test).
We also analyzed the potential of AML1/ETO to immortalize progenitor cells and to sustain proliferation in a mouse model with constitutive genetic ablation of Grg5/AES. Targeted deletion of Grg5/AES has been performed in mice, and no overt abnormalities in hematopoiesis have been reported.17 AML1/ETO was transduced into wild-type and Grg5/AES−/− lineage-negative hematopoietic progenitor cells. AML1/ETO-transduced Grg5−/− BM cells showed a trend toward lower numbers of colonies on plating in methylcellulose (P = .053, paired t test). When the formed colonies were replated, the AML1/ETO-transduced Grg5/AES−/− cells formed more colonies in the first (P = .043, paired t test) and second (P = .043, paired t test) replating than their wild-type counterparts. However, Grg5/AES−/− cells constantly formed fewer colonies starting with the third replating, and colony formation efficiency of these cells further decreased. Significantly lower numbers of colonies were formed by AML1/ETO-transduced Grg5/AES−/− cells after the fifth (P = .009, paired t test) and sixth replating (P = .002, paired t test) (Figure 4H). These analyses indicated that the absence of GRG5 reduced AML1/ETO-induced self-renewal in serial replating assays. It also suggested that compensatory mechanisms exist in Grg5/AES−/− hematopoietic progenitor cells that allow AML1/ETO-induced proliferation in the absence of GRG5/AES. We performed microarray expression analyses between LSK (lin−Sca1+kit+) cells derived from Grg5/AES−/− mice and from wild-type siblings. A Panther protein classification analysis of the results indicated that nucleic acid-binding proteins, RNA-binding proteins, and ribosomal proteins (all P < .01) were expressed at higher levels in Grg5/AES−/− LSK cells (data not shown). Colony formation frequency of primary murine BM cells from wild-type and Grg5/AES−/− mice did not differ (data not shown). In addition, no differences in serial replating potential were observed on retroviral transduction of an empty vector (data not shown). These findings, in combination with the altered gene expression profile in LSK cells, indicate that compensatory mechanisms exist on constitutive deletion of Grg5/AES in mice.
Identification of AES targets of repression
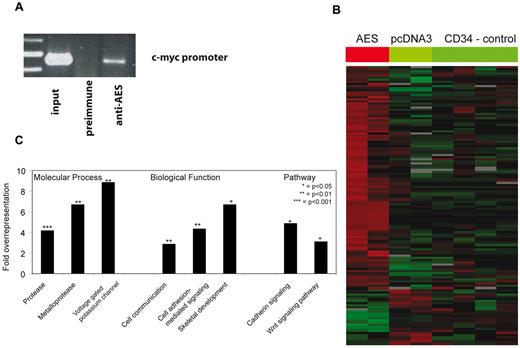
AES is thought to inhibit Groucho repressor functions by binding to specific promoters. We therefore expressed AES in normal human CD34+ progenitor cells and verified its binding to the c-myc promoter in vivo (Figure 5A). We performed whole genome microarray analyses of AES-expressing CD34+ cells and control cells. Overall, 130 transcripts were significantly altered on up-regulation of AES (Figure 5B; supplemental Table 1). Whereas 104 genes were induced on AES expression, 26 genes were repressed. A Panther classification revealed that cadherin signaling and the Wnt signaling pathway were significantly overrepresented (Figure 5C). In addition, among the biological functions, “skeletal development,” “cellular signaling,” and “communication” were enriched. These findings are in line with the skeletal deformations observed in GRG5 knockout mice17 and the proposed functions of AES as an enhancer of Wnt signaling. The enriched molecular processes (proteases and potassium channels) are less well explained at the moment but may offer a starting point to elucidate further functions of AES.
Identification of AES targets of repression. (A) AES-overexpressing CD34+ progenitor cells were analyzed by chromatin immunoprecipitation assay. Preimmune serum and serum against AES were used. The binding of AES to the known target promoter of c-myc was verified by PCR in vivo. (B) Whole genome microarray analyses of AES-overexpressing CD34+ cells and control cells. Gene expression results are shown as a heatmap of target genes. (C) Results of a Panther classification of the microarray analyses. Bars represent the overexpression of genes in different molecular processes, biological functions, and pathways after AES overexpression in CD34+ progenitor cells.
Identification of AES targets of repression. (A) AES-overexpressing CD34+ progenitor cells were analyzed by chromatin immunoprecipitation assay. Preimmune serum and serum against AES were used. The binding of AES to the known target promoter of c-myc was verified by PCR in vivo. (B) Whole genome microarray analyses of AES-overexpressing CD34+ cells and control cells. Gene expression results are shown as a heatmap of target genes. (C) Results of a Panther classification of the microarray analyses. Bars represent the overexpression of genes in different molecular processes, biological functions, and pathways after AES overexpression in CD34+ progenitor cells.
Discussion
Self-renewal is an intrinsic property of stem cells. Acquisition or preservation of self-renewal capability constitutes an important step in leukemogenesis. The ability to confer enhanced self-renewal capacity has been ascribed to several oncogenes in leukemia, but the underlying mechanisms have remained elusive. Here, we provide evidence that AML1/ETO leads to enhanced expression of AES, which counteracts repressive Groucho/TLE proteins. Up-regulation of AES at sufficient levels is necessary for AML1/ETO effects on long-term self-renewal and immortalization of hematopoietic cells. Experiments with siRNA against GRG5/AES show a strict dependency of self-renewal in AML1/ETO-transduced BM, whereas the effect in AML1/ETO-transduced GRG5/AES knockout BM is much milder. Gene expression analyses indicated altered gene expression patterns in Grg5/AES−/− LSK cells. It is probable that compensatory gene expression changes occur when Grg5/AES is already lacking during development.
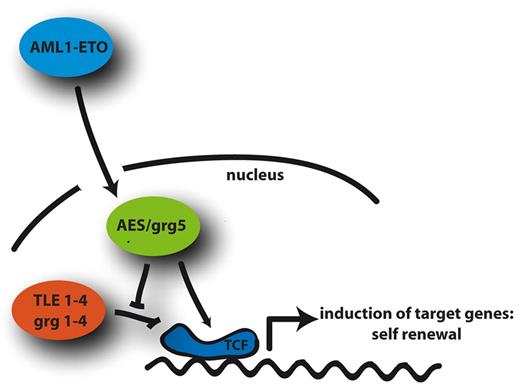
The proliferation of AML1/ETO-transduced Grg5/AES−/− BM is not impaired, whereas immortalization is reduced in the absence of GRG5/AES. Importantly, overexpression of AES enhanced self-renewal in hematopoietic progenitors but did not induce leukemia. Apparently, colonies arising on AES overexpression were more immature than those from control cells. Colony-forming activity of short-term repopulating hematopoietic stem cells in spleens is increased by overexpressing AES. These findings suggest that AES overexpression is required for the establishment of a self-renewing cell that permits progression to leukemia. Recently, it was shown that loss of TLE1 and TLE4 cooperates with AML1/ETO in induction of proliferation and survival of myeloid cells.9 In line with this, we have found that TLE1 to TLE4 are generally down-regulated in AML and in t(8;21)-positive AML (data not shown). It appears that the critical repressor (TLE1-TLE4) activity, regulated in part by AES expression, mediates important signals that are crucial for hematopoietic self-renewal. TLEs and AES regulate the activity of TCF/LEF transcription factors, which are leukemogenic when overexpressed. In addition, TCF/LEF factors are the mediators of β-catenin and γ-catenin/plakoglobin transcriptional activity. We have previously demonstrated that AML1/ETO and other translocation products in AML activate the Wnt signaling pathway via γ-catenin up-regulation.8 Thus, induction of γ-catenin and AES might act synergistically in enhancing Wnt signaling activity (Figure 6).
Model of the functional consequence of AML1/ETO-induced AES expression. AES expression is induced by AML1/ETO, which counteracts the transcriptional repression of Wnt target genes by TLE1-4 and thereby induces self-renewal via TCF in hematopoietic progenitor cells.
Model of the functional consequence of AML1/ETO-induced AES expression. AES expression is induced by AML1/ETO, which counteracts the transcriptional repression of Wnt target genes by TLE1-4 and thereby induces self-renewal via TCF in hematopoietic progenitor cells.
The probable importance of this pathway is further evidenced by its frequent activation in AML. Previously, we showed that FLT3 mutations can increase β-catenin protein levels and activity.14 In the current study, we found that AES expression is very widespread in AML, indicating that mechanisms besides AML1/ETO exist that can up-regulate AES expression. Sekkai et al have shown that STAT3-induced AES in murine embryonic stem cells.22 AES has also been demonstrated to interact with other transcription factors. For example, Wang et al17 have demonstrated a positive regulation of RUNX function by a dominant negative variant of GRG5, whereas GRG3 inhibits the RUNX2 function required for murine skeletal development and postnatal growth.17 Thus, regulating the inhibitor of TLE-repressors might be a fine-tuned process that is altered in AML.
Not only does the biologic function of AES resemble that of AML1/ETO, we also found that AES expression was associated with a favorable prognosis in patients. Remarkably, the favorable prognosis was evident by examining either AES mRNA levels or protein levels in independent patient cohorts in univariate analysis. The effect persisted even when patients with t(8;21) were excluded from the analyses. In multivariate analyses, AES mRNA expression emerged as an independent predictor of event-free and overall survival. But further analyses are necessary to establish AES expression as an independent biomarker given that several known prognostic factors, such as age and nucleophosmin mutation status, were not available for the analyzed patients. Nonetheless, the combined analyses indicate that AES expression is generally associated with a good prognosis. The prognostic role of both AES-overexpressing and AML1/ETO-expressing leukemias might be the result of higher susceptibility of these leukemic cells to chemotherapy-induced apoptosis or of the mechanisms that induce self-renewal. In AML, balanced translocations as well as some other alterations (eg, C/EBPα mutations) are pathogenetically relevant and, at the same time, associated with a favorable prognosis.23,24 It is possible that the good response to chemotherapy in these cases might rely on the specific mechanisms of leukemogenesis.
Taken together, AES is a target gene of AML1/ETO that is important for AML1/ETO-induced self-renewal. AES and TLE factors might constitute novel targets for therapy approaches that specifically aim at inhibiting self-renewal.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Sandra Gelsing, Britta Körner, Frank Berkenfeld, Beate Surmann, Ramona Gölzer, and Silvia Münch for excellent technical assistance; Dr Tom Gridley for providing Grg5/AES−/− mice; and Drs Peter Valk, Roel Verhaak, and Ruud Delwel for providing microarray expression data.
This work was supported by the Deutsche Forschungsgemeinschaft, the German José-Carreras Leukemia Foundation, the Wilhelm Sander-Stiftung, the Deutsche Krebshilfe (Oncogene Networks), the Interdisziplinäres Zentrum für Klinische Forschung Münster, the LeukemiaNet of the National Genome Network-Plus, and the Innovative Medizinische Forschung at the University of Münster.
Authorship
Contribution: B.S., C.S., M.K., H.S., and C.M.-T. designed and performed research, analyzed data, and wrote the paper; U.B., P.T., M.-P.H., E.B., S.H., M.R., O.V., and M.S. performed research; W.E.B. and S.K. wrote the paper; M.D. and N.B. analyzed data; and G.K. and C.B. designed research.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Carsten Müller-Tidow, Department of Hematology and Oncology, University of Münster, Domagkstrasse 3, 48129 Münster, Germany; e-mail: muellerc@uni-muenster.de.
References
Author notes
On behalf of the Study Alliance Leukemias.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal