Abstract
Natural killer (NK) cells are innate lymphocytes capable of immediate effector functions including cytokine production and cytotoxicity. Compared with B and T cells, the factors that control the peripheral maturation of NK cells are poorly understood. We show that Blimp1, a transcriptional repressor required for the differentiation of plasma cells and short-lived effector T cells, is expressed by NK cells throughout their development. Interleukin 15 (IL-15) is required for the early induction of Blimp1 in NK cells, with expression increasing in the most mature subsets of mouse and human NK cells. We show that Blimp1 is required for NK-cell maturation and homeostasis and for regulating their proliferative potential. It is also essential for high granzyme B expression, but not for most cytokine production and cytotoxicity. Surprisingly, interferon regulatory factor 4 (IRF4) and B-cell lymphoma 6 (Bcl6), 2 transcription factors crucial for the regulation of Blimp1 in B and T cells, are largely dispensable for Blimp1 expression in NK cells. T-bet deficiency, however, leads to attenuated Blimp1 expression. We have identified NK cells as the first hematopoietic cell type in which the IRF4-Blimp1-Bcl6 regulatory axis is not in operation, highlighting the distinct nature of the NK-cell gene-regulatory network.
Introduction
Natural killer (NK) cells are innate lymphocytes specialized in cytokine production and cytotoxicity toward tumors and virus-infected cells. NK cells develop in the bone marrow from hematopoietic stem cells (HSCs) via lymphoid precursors.1 The earliest committed NK-cell precursors can be isolated from lineage marker–negative fetal thymus or adult bone marrow and express high levels of the interleukin 15 (IL-15)/interleukin 2 receptorβ (IL-2Rβ) chain (CD1222 ). NK-cell precursors then undergo maturation and functional diversification in the bone marrow, thymus, and a variety of peripheral organs including the liver, spleen, and lymph nodes.1 In the mouse, peripheral maturation in the spleen is characterized by the up-regulation of the markers CD43, Mac-1 (CD11b), CD94, and KLRG1, with concomitant down-regulation of CD27 and ckit.1,3-6 Mature Mac-1highKLRG1+CD27low NK cells are the dominant population in nonlymphoid organs (with the exception of the liver).3,7
In contrast to B and T cells, knowledge about the transcriptional circuitries controlling NK-cell development and maturation remains limited. Several factors, including E4BP4,8,9 Tox,10 and Id2,11 are required for the development of NK cells from early progenitors, whereas GATA3 is essential for thymic NK cells12 and modulates the function of mature NK cells.13 A second group of transcription factors, including Ets1,14 E74-like factor 4,15 interferon regulatory factor 2 (IRF2),16 and T-bet,17,18 is more specifically required for the later stages of NK-cell differentiation and function. Interestingly, a number of these factors, including Id2 and T-bet, also have important functions in CD8+ T-cell differentiation.19,20
Blimp1 (also known as Prdm1) is a pleiotropic transcription factor that plays crucial roles in the differentiation of plasma cells and CD8+ effector T cells.21-25 In both processes, Blimp1 is required for terminal differentiation and Blimp1 deficiency leads to loss of functional antibody-secreting cells and short-lived cytotoxic T cells. Moreover, in CD8+ T cells, Blimp1 is required for the optimal expression of cytotoxic molecules such as granzyme B and perforin and for the regulation of chemokine receptors such as CC-chemokine receptor 7 (CCR7), CCR5, and CCR6, but is dispensable for most cytokine production.24,25 Blimp1 is also broadly required for the maintenance of T-cell homeostasis, because mice that lack Blimp1 in all T cells or mice reconstituted with Blimp1-deficient fetal liver cells show an accumulation of activated T cells and develop immunopathology.22,26 In CD4+ T cells, Blimp1 has been suggested to attenuate Th1 differentiation by repressing the genes encoding T-bet and interferonγ (IFNγ).27 Blimp1 expression within B and T cells is controlled by 2 transcription factors, IRF4 and Bcl6. While Bcl6 and Blimp1 mutually repress the expression of each other in activated B and CD4+ T cells,28 IRF4 directly activates Blimp1 expression.29
We show that Blimp1 is expressed by NK cells and is vital for their peripheral maturation and tissue distribution, as well as for restricting their proliferative potential. In striking contrast to B and T lymphocytes, Blimp1 is induced in immature NK cells in an IL-15–dependent manner and maintained throughout the lineage. Blimp1 expression is increased in the most mature subsets of mouse and human NK cells and is further up-regulated by cytokines such as IL-12 and IL-21. Although Blimp1 is dispensable for most cytokine production and cytotoxicity, it is required for maximal granzyme B expression. Blimp1 expression is independent of IRF4 and Bcl6, despite their presence in NK cells, but relies on T-bet, suggesting fundamental differences in the transcriptional network of NK cells compared with other lymphocytes.
Methods
Mice
Blimp1/GFP reporter (Blimp1gfp/+) mice were generated and maintained on a C57BL/6 (Ly5.2+) background, as described previously.30 To generate Blimp1gfp/gfp or Rag2−/−Blimp1gfp/gfp mice, fetal liver cells were isolated from E14.5 Blimp1gfp/gfp or Rag2−/−Blimp1gfp/gfp embryos and intravenously injected into lethally irradiated Rag1−/−Ly5.1 or Rag2−/−γc−/− recipients, respectively (1-3 × 106 fetal liver cells per recipient). Irf4−/− mice,31 Bcl6−/− mice,32 and Tbx21−/− mice33 have been described previously. Blimp1gfp/gfp: Ly5.1 mixed bone-marrow chimeras were generated by reconstituting lethally irradiated Ly5.1 recipients with a mixture of bone-marrow cells isolated from Blimp1gfp/gfp and wild-type Ly5.1 mice. Chimeric mice were analyzed after a minimum of 6 weeks after reconstitution. To allow for variation in the reconstitution frequency, data were normalized for the NK-cell compartment in the spleen. All mice were maintained on a C57BL/6 background at The Walter and Eliza Hall Institute. Animal experiments were undertaken according to the institution's animal ethics committee guidelines and approval.
Antibodies and flow cytometry
The following anti–mouse monoclonal antibodies (mAbs) were used for flow cytometric analysis: NK1.1 (PK136), Ly5.2 (104), Ly5.1 (A20), CD27 (LG.7F9/LG.3A10), KLRG1 (2FY), CD49b (HMα2), CD4 (RM4-5), CD122 (TM-β1), CD43 (S7), Mac-1 (M1/70), ckit (2B8), T-cell receptorβ (TCRβ; H57-597), CD8 (53-6-7), CD44 (IM7), CD62L (Mel-14) IFNγ (XMG1.2), Ly49A (YE132), Ly49C/I (SW5E6), Ly49D (4E5/E1), Ly49F (BHF-719), Ly49H (3D10), and CD94 (18d3). Biotinylated mAbs were revealed with streptavidin Cy5 (Southern Biotech) or streptavidin PerCPCy5.5. Anti–human CD3 (SK7), CD56 (B159), NKp46 (9E2), and CD16 (3G8) were also used. Mouse anti–T-bet antibody (4B10) was obtained from Santa Cruz Biotechnology and revealed with anti–immunoglobulin 1 (anti–IgG1) (eBioscience, ×56). Staining for Eomes (Dan11mag) and T-bet was performed using the Foxp3-staining kit (eBioscience). Anti–granzyme B (GB12) was purchased from Invitrogen and the remaining reagents were from BD Biosciences. Viable cells were identified by propidium iodide or Cytox blue exclusion. Cells were analyzed on LSRII, FACSCalibur, or FACSCanto cytometers (Beckman Dickinson), and cell sorting was performed on MoFlo (Becton Coulter) or Aria cytometers (Becton Dickinson). Data were processed using FlowJo Version 9.0 (TreeStar) and Weasel Version 2.7 software (Walter and Eliza Hall Institute).
Cell isolation and culture
Unless stated otherwise, splenic NK cells were enriched using DX5 magnetic beads (Miltenyi Biotec) and cultured in IL-15 (50 ng/mL; R&D Systems) for 5 days. The cells were then used or recultured in the presence of IL-12 (5 ng/mL), IL-18 (20 ng/mL), or IL-21 (100 ng/mL), all from R&D Systems. For some experiments, IL-15–expanded NK cells were cultured in the presence of 20 μg/mL of anti–NK1.1, anti–NKG2D, or anti–CD16 (24G.2) or various cytokine concentrations for 2 days, as described previously.34 The differentiation of NK cells from purified HSCs was carried out using OP9 and OP9-DL1 stromal cells as described previously.35 Lung lymphocytes were purified from single cell suspensions by centrifugation on Histopaque (1.077 g/mL; Sigma-Aldrich) for 20 minutes at 400g at room temperature.
Intracellular cytokine stains
DX5-enriched NK cells were stained for relevant surface molecules, fixed, permeabilized using the Cytofix/Cytoperm reagent (BD Biosciences), and costained for intracellular cytokines. Intracellular granzyme B content and IFNγ were measured after stimulation of the cells with 5 ng/mL of IL-12, 50 ng/mL of IL-18 or 50 ng/mL of phorbol myristate acetate (PMA), and 500 ng/mL of ionomoycin for 5 hours in the presence of GolgiPlug, followed by subsequent surface-marker staining and permeabilization using the Cytofix/Cytoperm reagent (BD Biosciences).
Cytokine bead array
NK cells were expanded in IL-15 for 5 days before being transferred into IL-15, IL-12/IL-18, or IL-15/IL-21. After another 2 days in culture, cell numbers were determined and equal numbers of cells seeded in the same conditions. Supernatants were collected after an additional 16 hours and assayed for cytokines using the Bio-Rad Bioplex cytokine bead assay (Mouse 23-Plex Panel).
PCR analysis
Total RNA was prepared from flow cytometry–purified human NK cells using an RNeasy kit (QIAGEN). cDNA was synthesized from total RNA with random hexamers and SuperScript III reverse transcriptase (Invitrogen). Primer sequences were as follows: BLIMP1: 5′-GATGCGGATATGACTCTGTGG-3′; 5′-CTCGGTTGCTTTAGACTGCT-3′; HPRT: 5′-GTGATGAAGGAGATGGGAGG-3′; and 5′-ACTGCCTGACCAAGGAAAG-3′.
Cytotoxicity assays
The cytotoxicity of NK cells was assessed using RMAS lymphoma cells transduced with either the control murine stem cell leukemia virus vector (RMAS-MSCV) or the MSCV vector expressing Rae1β (RMAS-Rae1b), as we have described previously.36
In vivo tumor model
Groups of 4-7 Rag2−/−Blimp1+/+ or Rag2−/−Blimp1gfp/gfp mice were inoculated subcutaneously with RMA-S tumor cells at the indicated doses, as described previously.37 Tumor growth was examined every second day. Recorded data are the means ± SEM of mice in each group.
Western blotting
Proteins were extracted with radioimmunoprecipitation assay buffer (2 × 107 cells/mL) and subjected to Western blotting according to standard techniques. The monoclonal antibody to Blimp1 (5E7) was described previously.30 Rat anti–mouse IRF4 (3E4) and Bcl6 (7D1) monoclonal antibodies were raised against a glutathione S-transferase fusion protein containing the carboxy-terminal 65-amino acid sequence of IRF4 and amino acids #261-386 of Bcl6, respectively, and were a gift from Lynn Corcoran (The Walter and Eliza Hall Institute of Medical Research, Parkville, Australia). Anti–β-actin (Santa Cruz Biotechnology) was used as a loading control.
Proliferation assay
Sorted TCRβ−NK1.1+CD49b+ NK cells were cultured for 2 days in a range of IL-15 concentrations before being pulsed with 1μCi/well of 3H-thymidine for the last 8 hours of culture. Cells were harvested and 3H-thymidine incorporation determined by scintillation counting.
In vivo proliferation assay
Purified wild-type (Ly5.1+) or Blimp1gfp/gfp (Ly5.2+) TCRβ−NK1.1+CD49b+ NK cells were cultured in IL-15 for 7 days before being mixed at a 1:1 ratio (total 2 × 106 cells), injected into Rag2−/−γc−/− hosts, and analyzed after 6 days by flow cytometry.
Statistics
Data were analyzed using a 2-tailed Student t test (paired or unpaired as appropriate). A P value less than .05 was considered significant.
Results
Blimp1 is constitutively expressed in NK cells and is up-regulated during maturation
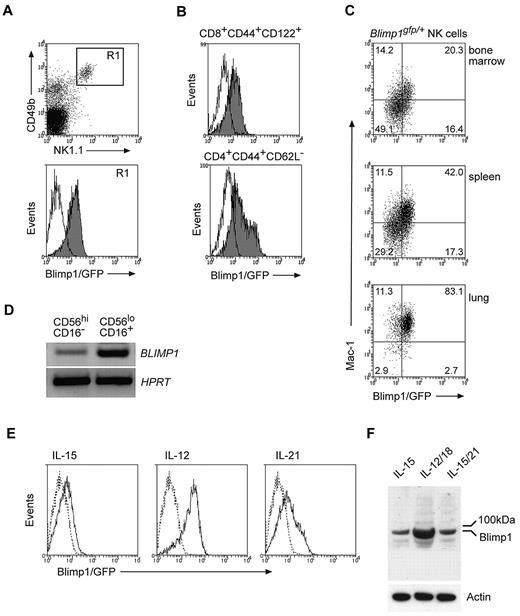
To determine whether Blimp1 is expressed by NK cells, we made use of Blimp1/GFP reporter mice (Blimp1gfp/+).30 Flow cytometric analysis of NK cells revealed GFP fluorescence that was similar to that observed for the corresponding effector T-cell populations of the same mouse, indicating that Blimp1 was expressed in NK cells (Figure 1A-B). Blimp1/GFP was detected in all NK cells independent of their localization, including NK cells from the bone marrow, lymph nodes, spleen, blood, liver, and lung (Figure 1 and data not shown). In B and T cells, Blimp1 expression was closely linked to terminal differentiation of effector cells. NK cells undergo significant phenotypic and functional changes during their maturation in the periphery, and we speculated that Blimp1 might be up-regulated during peripheral NK-cell differentiation. Consistent with this idea, Blimp1/GFP was highest in the Mac-1high population, the most mature NK-cell subset,4 independent of their anatomical location (Figure 1C). In agreement with this finding and with data recently published,38 mature human CD16+ NK cells exhibited distinctly higher BLIMP1 expression than their more immature CD16− counterparts, suggesting that BLIMP1 is also up-regulated during maturation of human NK cells (Figure 1D).
Blimp1 is constitutively expressed in NK cells and is up-regulated during their maturation. (A-B) Blimp1gfp/+ splenocytes were stained with antibodies as indicated and gated on either (A) NK1.1+CD49b+TCRβ− NK cells (R1) or (B) TCRβ+CD8+CD44+CD122+ or TCRβ+CD4+CD62L− effector/memory phenotype T cells. (C) NK cells from indicated organs of Blimp1gfp/+ mice were identified as in (A) and analyzed for GFP and Mac-1 expression. Numbers in the plots show the proportion of NK cells in each quadrant. (D) Peripheral blood NK cells were isolated from healthy human donors and sorted into CD16+ and CD16− subpopulations. After RNA isolation and cDNA synthesis, transcripts were measured for BLIMP1 and HPRT by semiquantitative PCR. (E-F) DX5+ NK cells isolated from Blimp1gfp/+ (solid line) and wild-type mice (dotted line) were cultured in IL-15 for 5 days before being transferred into IL-15 plus cytokines as indicated. GFP expression was measured after 2 days (E). Wild-type NK cells were also subjected to Western blotting using antibodies as indicated (F). The results are representative of 3-6 independent experiments.
Blimp1 is constitutively expressed in NK cells and is up-regulated during their maturation. (A-B) Blimp1gfp/+ splenocytes were stained with antibodies as indicated and gated on either (A) NK1.1+CD49b+TCRβ− NK cells (R1) or (B) TCRβ+CD8+CD44+CD122+ or TCRβ+CD4+CD62L− effector/memory phenotype T cells. (C) NK cells from indicated organs of Blimp1gfp/+ mice were identified as in (A) and analyzed for GFP and Mac-1 expression. Numbers in the plots show the proportion of NK cells in each quadrant. (D) Peripheral blood NK cells were isolated from healthy human donors and sorted into CD16+ and CD16− subpopulations. After RNA isolation and cDNA synthesis, transcripts were measured for BLIMP1 and HPRT by semiquantitative PCR. (E-F) DX5+ NK cells isolated from Blimp1gfp/+ (solid line) and wild-type mice (dotted line) were cultured in IL-15 for 5 days before being transferred into IL-15 plus cytokines as indicated. GFP expression was measured after 2 days (E). Wild-type NK cells were also subjected to Western blotting using antibodies as indicated (F). The results are representative of 3-6 independent experiments.
Cytokines have been shown to be important modulators of NK-cell activation, function, and maturation.1 In agreement with this notion, we found that IL-12, a combination of IL-12 and IL-18, and IL-21, cytokines known to promote NK-cell differentiation, were potent in inducing Blimp1 expression in NK cells higher than the constitutive level maintained by IL-15 alone (Figure 1E-F). Whereas these cytokines induced equivalent Blimp1/GFP over a broad range of concentrations, cross-linking of NK-cell–activating receptors did not result in any substantial up-regulation of Blimp1/GFP (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Interestingly, murine NK cells predominantly expressed the full-length Blimp1 protein (Figure 1F), unlike human NK cells, which were recently shown to express 3 distinct isoforms of BLIMP1.38
Blimp1 expression in NK cells is IL-15–dependent
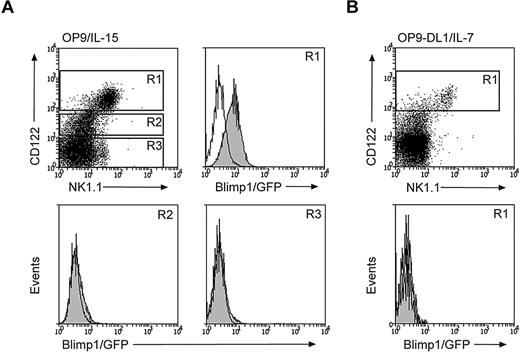
Our data so far indicate that NK cells constitutively express Blimp1, which can be further up-regulated during maturation. This is strikingly different from the situation in B and T cells, in which the Blimp1 protein is restricted to terminally differentiating effector cells.23 To determine at what stage Blimp1 expression is initiated in the NK-cell lineage, we sorted HSC-enriched populations from Blimp1gfp/+ and control mice and cultured them on OP9 cells in the presence of IL-15 to induce NK-cell development, or on OP9-DL1 cells in the presence of IL-7 to induce T-cell development.35 HSCs did not express Blimp1 (data not shown); however, when cultured under NK-cell–inducing conditions (OP9/IL-15), they gave rise to Blimp1/GFP+NK1.1+CD122+ cells (Figure 2A R1). In contrast, the NK1.1−CD122+ NK-cell progenitors (R2) were Blimp1/GFP negative in the same culture, as were NK1.1−CD122− cells (R3). Transient NK-cell differentiation is also observed when HSCs are cultured in conditions favorable for T-cell differentiation (OP9-DL1/IL-7).35 The NK1.1+CD122+ NK cells induced in these conditions were Blimp1/GFP negative (Figure 2B R1 and data not shown), suggesting that IL-15 signals are required for the induction of Blimp1 expression.
IL-15 is required for Blimp1 induction in developing NK cells. (A-B) Lineage marker–negative Sca1+ckit+ HSC-enriched progenitors were isolated from the bone marrow of Blimp1gfp/+ or wild-type mice and cultured on OP9 cells in the presence of IL-15 (A) or on OP9-DL1 cells in the presence of IL-7 (B). GFP expression of the indicated populations was measured at day 9 of culture (filled histograms, Blimp1gfp/+; open histograms, wild-type). Results are representative of 2 independent experiments.
IL-15 is required for Blimp1 induction in developing NK cells. (A-B) Lineage marker–negative Sca1+ckit+ HSC-enriched progenitors were isolated from the bone marrow of Blimp1gfp/+ or wild-type mice and cultured on OP9 cells in the presence of IL-15 (A) or on OP9-DL1 cells in the presence of IL-7 (B). GFP expression of the indicated populations was measured at day 9 of culture (filled histograms, Blimp1gfp/+; open histograms, wild-type). Results are representative of 2 independent experiments.
Blimp1 is required for NK-cell maturation and homeostasis
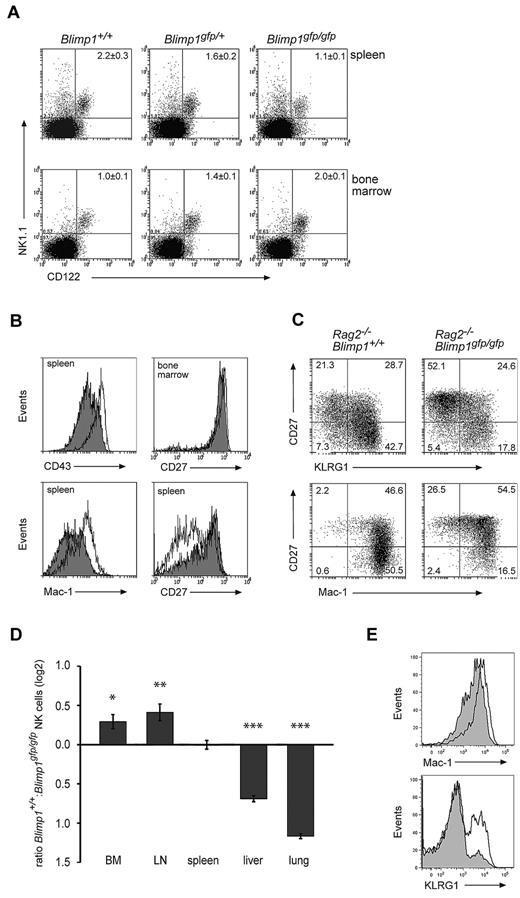
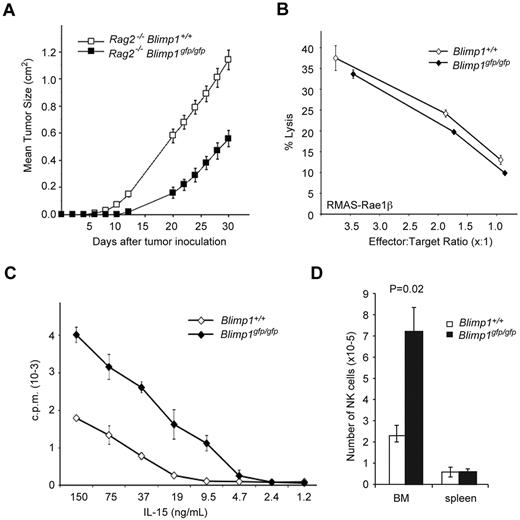
To determine the functional relevance of our findings, we generated chimeric mice using lethally irradiated Rag2−/−γc−/− mice reconstituted with Blimp1gfp/gfp fetal liver cells (termed Blimp1gfp/gfp or Blimp1-deficient mice thereafter) or with control cells. As described previously, this approach results in mice lacking functional Blimp1 in all hematopoietic cells including NK cells.30 This strategy was necessary because Blimp1 deficiency is embryo lethal30 and a reliable Cre-lox system for the conditional ablation of genes in NK cells is lacking. Examination of Blimp1gfp/gfp and control mice revealed that while the proportion of Blimp1-deficient NK cells was increased in the bone marrow and reduced in the spleen compared with heterozygous or wild-type controls (Figure 3A), the absolute numbers of splenic NK cells were largely unchanged (1.78 ± 0.3 × 106 cells in +/+, 1.72 ± 0.6 × 106 in Blimp1gfp/gfp). Therefore, Blimp1 was not required for the generation of normal numbers of NK cells.
Loss of Blimp1 alters NK-cell maturation and homeostasis. (A) Rag1−/−Ly5.1 mice were lethally irradiated and reconstituted with E14.5 fetal liver cells of the indicated genotypes. Donor derived (Ly5.2+) cells from the spleen and bone marrow of these mice were analyzed 6-8 weeks after reconstitution for the indicated markers. Numbers are the mean ± SEM for 4-6 mice. (B-C) Lethally irradiated Rag2−/−γc−/− mice were reconstituted with E14.5 Rag2−/−Blimp1+/+ or Rag2−/−Blimp1gfp/gfp fetal liver cells. After reconstitution, NK cells (NK1.1+CD49b+) in the spleen or bone-marrow cells were analyzed for the markers as indicated. Open histograms, Rag2−/−Blimp1+/+; filled histograms, Rag2−/−Blimp1gfp/gfp. Numbers in the plots show the proportion of NK cells in each quadrant. Results are representative of at least 3 experiments each containing 3-4 mice. (D) Bone marrow chimeras were generated by reconstitution of lethally irradiated C57BL/6 Ly5.1+ mice with a 1:1 mixture of Ly5.1+ and Blimp1gfp/gfp (Ly5.2+) bone marrow and analyzed after 10-12 weeks. The ratio of Ly5.1+ NK cells and Ly5.2+Blimp1gfp/gfp NK cells in different organs is shown as the mean ± SEM (n = 6-9). P value compares the indicated values to zero: *P = .03; **P = .007; ***P < .00001 (E) Marker expression on Ly5.2+Blimp1gfp/gfp (filled histogram) or Ly5.1+ wild-type NK cells (open histogram) in mixed bone-marrow chimeras.
Loss of Blimp1 alters NK-cell maturation and homeostasis. (A) Rag1−/−Ly5.1 mice were lethally irradiated and reconstituted with E14.5 fetal liver cells of the indicated genotypes. Donor derived (Ly5.2+) cells from the spleen and bone marrow of these mice were analyzed 6-8 weeks after reconstitution for the indicated markers. Numbers are the mean ± SEM for 4-6 mice. (B-C) Lethally irradiated Rag2−/−γc−/− mice were reconstituted with E14.5 Rag2−/−Blimp1+/+ or Rag2−/−Blimp1gfp/gfp fetal liver cells. After reconstitution, NK cells (NK1.1+CD49b+) in the spleen or bone-marrow cells were analyzed for the markers as indicated. Open histograms, Rag2−/−Blimp1+/+; filled histograms, Rag2−/−Blimp1gfp/gfp. Numbers in the plots show the proportion of NK cells in each quadrant. Results are representative of at least 3 experiments each containing 3-4 mice. (D) Bone marrow chimeras were generated by reconstitution of lethally irradiated C57BL/6 Ly5.1+ mice with a 1:1 mixture of Ly5.1+ and Blimp1gfp/gfp (Ly5.2+) bone marrow and analyzed after 10-12 weeks. The ratio of Ly5.1+ NK cells and Ly5.2+Blimp1gfp/gfp NK cells in different organs is shown as the mean ± SEM (n = 6-9). P value compares the indicated values to zero: *P = .03; **P = .007; ***P < .00001 (E) Marker expression on Ly5.2+Blimp1gfp/gfp (filled histogram) or Ly5.1+ wild-type NK cells (open histogram) in mixed bone-marrow chimeras.
Because a deficiency of Blimp1 in the hematopoietic system leads to immunopathology associated with an accumulation of activated T cells and an altered cytokine environment,22,26 we wished to examine Blimp1-deficient NK cells in the absence of other lymphocytes. To do this, we generated chimeric mice from Rag2−/−Blimp1gfp/gfp fetal liver cells, which, due to the lack of T and B cells, remained healthy and showed no immunopathology (data not shown). High cell-surface expression of Mac-1 and CD43 are commonly used to identify the most mature mouse NK cells4 and, interestingly, the expression of both molecules was markedly reduced on splenic NK cells in the absence of Blimp1 (Figure 3B left). More recently, CD27 in conjunction with KLRG1 and Mac-1 was used to describe NK-cell maturation. Spleen CD27+ NK cells first up-regulate Mac-1 before differentiating through Mac-1highCD27+KLRG1− and Mac-1highCD27−KLRG1+ stages.3,6,7 CD27 expression was uniformly high in the bone marrow of Blimp1-deficient and control mice, whereas Blimp1-deficient spleens showed a reduction in the frequency of the Mac-1highCD27−KLRG1+ mature NK cells (Figure 3B-C), with the proportion of KLRG1+ splenic NK cells decreasing from 57.2 ± 4.7% in Blimp1+/+ to 28.7 ± 3.8% in Blimp1gfp/gfp mice (P = .002, each n = 5), indicating that Blimp1 was required for the peripheral maturation of NK cells.
To rigorously test if the maturation phenotype and the altered homeostasis of Blimp1-deficient NK cells was indeed intrinsic to NK cells or was a result of an altered cellular composition or cytokine environment in Blimp1-deficient mice, we generated chimeric mice by reconstituting lethally irradiated Ly5.1 mice with a mixture of congenically marked bone marrow from wild-type (Ly5.1+) and Blimp1gfp/gfp (Ly5.2+) mice. We have shown before that this strategy significantly delays the onset of pathology caused by Blimp1-deficient T cells,26 thereby allowing a comparison of the homeostasis and tissue distribution of Blimp1-deficient and control NK cells under the same conditions. Examination of NK cells in various organs of these mice revealed that Blimp1-deficient NK cells were present at significantly increased frequencies in bone marrow and lymph nodes but were dramatically reduced in livers and lungs compared with their wild-type competitors (Figure 3D). Blimp1-deficient NK cells showed a normal repertoire of NK-cell receptors (supplemental Figure 2) while maintaining their immature KLRG1− phenotype (Figure 3E), indicating that the requirement of Blimp1 for the correct tissue localization and peripheral maturation of NK cells was cell intrinsic.
Blimp1 is dispensable for most NK-cell cytokine production and cytotoxicity
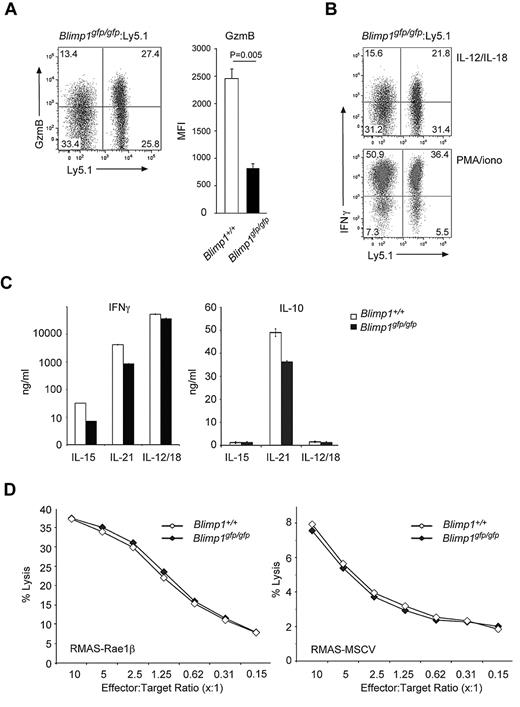
To test the functional properties of Blimp1-deficient NK cells, we first examined granzyme B and IFNγ levels in NK cells from mixed bone-marrow chimeric mice (Blimp1gfp/gfp:Ly5.1). Similar to our findings in CD8+ effector T cells,24 granzyme B expression was significantly decreased in Blimp1-deficient NK cells (Figure 4A). In contrast, the level of IFNγ produced by Blimp1gfp/gfp NK cells was only mildly reduced compared with control cells after activation with IL-12 and IL-18, and was unchanged after strong ex vivo stimulation using PMA and ionomycin (Figure 4B). This was surprising and in contrast to data obtained from mouse CD4+ T cells and human NK cells, in which Blimp1 was found to directly repress Ifnγ27 ; however, these data agree with our findings in CD8+ effector T cells, which in the absence of Blimp1 display unaltered IFNγ secretion.24
Blimp1 is dispensable for most effector functions of NK cells. (A-B) Gated TCRβ−NK1.1+CD49b+ NK cells from mixed bone-marrow chimeras, generated as described in Figure 3D, were examined for (A) granzyme B and (B) IFNγ expression by intracellular staining and flow cytometry. Numbers in the plots show the proportion of NK cells in each quadrant. Before analysis, the cells were stimulated with either (A-B) IL-12 and IL-18 or (B) PMA and ionomycin for 5 hours as indicated. Graph in (A) shows the mean fluorescence index (MFI) of granzyme B in Blimp1+/+ and Blimp1gfp/gfp NK cells. Data are the means ± SEM of 6 mice of each genotype. P value compares the indicated genotypes. (C-D) DX5+ NK cells from wild-type and Blimp1gfp/gfp mice were expanded for 5 days in IL-15, transferred into conditions as indicated for another 2 days, and the quantity of secreted IFNγ and IL-10 (C) and cytotoxic activity against RMAS-Rae1β and RMAS-MSCV (D) was determined. Data in panels C-D are the means of triplicate measurements ± SEM and are representative of 3-5 experiments.
Blimp1 is dispensable for most effector functions of NK cells. (A-B) Gated TCRβ−NK1.1+CD49b+ NK cells from mixed bone-marrow chimeras, generated as described in Figure 3D, were examined for (A) granzyme B and (B) IFNγ expression by intracellular staining and flow cytometry. Numbers in the plots show the proportion of NK cells in each quadrant. Before analysis, the cells were stimulated with either (A-B) IL-12 and IL-18 or (B) PMA and ionomycin for 5 hours as indicated. Graph in (A) shows the mean fluorescence index (MFI) of granzyme B in Blimp1+/+ and Blimp1gfp/gfp NK cells. Data are the means ± SEM of 6 mice of each genotype. P value compares the indicated genotypes. (C-D) DX5+ NK cells from wild-type and Blimp1gfp/gfp mice were expanded for 5 days in IL-15, transferred into conditions as indicated for another 2 days, and the quantity of secreted IFNγ and IL-10 (C) and cytotoxic activity against RMAS-Rae1β and RMAS-MSCV (D) was determined. Data in panels C-D are the means of triplicate measurements ± SEM and are representative of 3-5 experiments.
Because IL-12 and IL-21 have been shown to induce the functional maturation of NK cells3,36 and also to up-regulate Blimp1, we cultured Blimp1gfp/gfp and wild-type NK cells with these factors and measured NK-cell functions such as cytokine secretion and cytotoxicity. Surprisingly, in all conditions tested, Blimp1-deficient NK cells secreted similar amounts of IFNγ and IL-10 (Figure 4C and supplemental Figure 3) and showed a similar ability to lyse target cells (Figure 4D) compared with the wild-type controls, suggesting that Blimp1 is not required for most NK-cell effector functions.
Blimp1 controls the proliferative potential of NK cells
Given the crucial roles of Blimp1 in late B- and T-cell differentiation, the finding that NK cells were broadly functional in the absence of Blimp1 was intriguing and led us to test NK-cell functions in vivo. To this end, we injected RMAS lymphoma cells subcutaneously into Rag2−/−Blimp1gfp/gfp and control Rag2−/− mice and measured tumor burden. Interestingly, Blimp1-deficient NK cells were superior to their wild-type counterparts in their ability to control the growth of the tumor cells in vivo (Figure 5A). Because the cytotoxic function of freshly isolated NK cells per se did not seem to be altered (Figure 5B), this result suggested that NK cells in the absence of Blimp1 showed enhanced proliferation, survival, or recruitment to the site of tumor growth. To determine the proliferative potential of NK cells in the absence of Blimp1, we cultured Blimp1gfp/gfp and wild-type NK cells in a wide range of IL-15 concentrations. Blimp1-deficient NK cells exhibited enhanced proliferation in the presence of low concentrations of IL-15 compared with wild-type controls (Figure 5C). Increased numbers of Blimp1-deficient NK cells were also observed in an in vivo transfer model in which Blimp1gfp/gfp and wild-type NK cells were injected together into Rag2−/−γc−/− mice that lack all lymphocytes including NK cells. Examination of these mice after 6 days revealed increased numbers of Blimp1-deficient NK cells in the bone marrow compared with wild-type cells, which is consistent with enhanced proliferative potential and developmental immaturity of Blimp1-deficient NK cells (Figure 5D). These results indicate that Blimp1 regulates NK-cell homeostasis and function in vivo by promoting their differentiation and restricting their proliferation.
Blimp1 controls the proliferative potential of NK cells. (A) Rag2−/−Blimp1+/+ or Rag2−/−Blimp1gfp/gfp mice, generated as described in Figure 3C, were injected subcutaneously with 5 × 105 RMAS cells. Tumor growth was measured and is presented as mean tumor size ± SEM. Results are representative of 3 independent experiments, each with 4-7 mice per group. (B-C) Wild-type (Ly5.1+) or Blimp1gfp/gfp (Ly5.2+) TCRβ−NK1.1+CD49b+ NK cells were sorted from the spleens of mixed bone-marrow chimeric mice, generated as described in Figure 3D, and either subjected to an ex vivo killing assay using RMAS-Rae1β target cells (B) or cultured in titrated amounts of IL-15 as indicated (C). Proliferation was measured 48 hours later. Data in (B) and (C) are the means of triplicate measurements ± SEM. (D) Purified wild-type (Ly5.1+) or Blimp1gfp/gfp (Ly5.2+) TCRβ−NK1.1+CD49b+ NK cells were cultured in IL-15 for 7 days before being mixed at a 1:1 ratio (total 2 × 106 cells), injected into Rag2−/−γc−/− hosts, and analyzed after 6 days by flow cytometry. Total numbers of the NK-cell population in spleen and total bone marrow (BM) are shown as the means ± SEM. P values compare the indicated genotypes. Results are representative of 2-3 individual experiments
Blimp1 controls the proliferative potential of NK cells. (A) Rag2−/−Blimp1+/+ or Rag2−/−Blimp1gfp/gfp mice, generated as described in Figure 3C, were injected subcutaneously with 5 × 105 RMAS cells. Tumor growth was measured and is presented as mean tumor size ± SEM. Results are representative of 3 independent experiments, each with 4-7 mice per group. (B-C) Wild-type (Ly5.1+) or Blimp1gfp/gfp (Ly5.2+) TCRβ−NK1.1+CD49b+ NK cells were sorted from the spleens of mixed bone-marrow chimeric mice, generated as described in Figure 3D, and either subjected to an ex vivo killing assay using RMAS-Rae1β target cells (B) or cultured in titrated amounts of IL-15 as indicated (C). Proliferation was measured 48 hours later. Data in (B) and (C) are the means of triplicate measurements ± SEM. (D) Purified wild-type (Ly5.1+) or Blimp1gfp/gfp (Ly5.2+) TCRβ−NK1.1+CD49b+ NK cells were cultured in IL-15 for 7 days before being mixed at a 1:1 ratio (total 2 × 106 cells), injected into Rag2−/−γc−/− hosts, and analyzed after 6 days by flow cytometry. Total numbers of the NK-cell population in spleen and total bone marrow (BM) are shown as the means ± SEM. P values compare the indicated genotypes. Results are representative of 2-3 individual experiments
Blimp1 expression in NK cells is independent of IRF4 and Bcl6
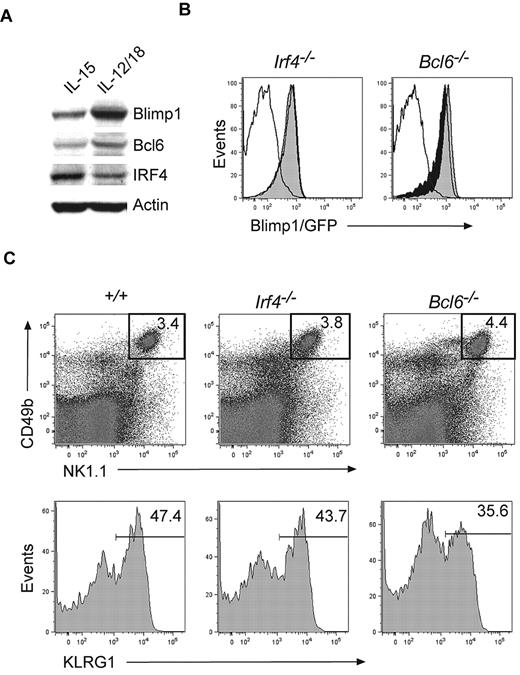
Two transcription factors that directly regulate Blimp1 expression in B and T cells, IRF429 and Bcl6, have been identified.28 While IRF4 induces Blimp1 expression in B cells and CD4+ T cells, Bcl6 and Blimp1 are part of a negative regulatory circuit active in B cells, T cells, and osteoclasts.28,39 Because IRF4 and Bcl6 were present in NK cells (Figure 6A), we reasoned that they might control Blimp1 expression in this lineage. To test this hypothesis, we generated Irf4−/− and Bcl6−/− mice on a Blimp1gfp/+ background and monitored Blimp1/GFP expression in the NK-cell compartment. Surprisingly, NK cells expressed normal amounts of Blimp1/GFP in the absence of either IRF4 or Bcl6 (Figure 6B), indicating that both factors were dispensable for Blimp1 expression in NK cells. Neither IRF4- nor Bcl6-deficient NK cells showed significant changes in the expression of markers associated with NK-cell maturation (Figure 6C), suggesting that neither factor plays a major developmental role in NK cells.
Blimp1 expression in NK cells is independent of IRF4 and Bcl6. (A) DX5+ NK cells were expanded in IL-15 for 5 days and then transferred into cytokines for 2 days as indicated. Protein extracts were subjected to Western blotting with antibodies to the indicated proteins. (B) Blimp1gfp/+ mice were bred on Irf4−/− or Bcl6−/− backgrounds and TCRβ−NK1.1+CD49b+ NK cells were analyzed for Blimp1/GFP expression (empty histogram, wild-type control; filled black histogram, Blimp1gfp/+; filled gray histogram, Blimp1gfp/+ and deficient for the indicated transcription factor). (C) Flow cytometric analysis of splenocytes isolated from Irf4−/−, Bcl6−/−, or control mice (top panel). Gated TCRβ−NK1.1+CD49b+ NK cells are shown in the lower panel. Numbers indicate the proportion of cells of the indicated phenotype. The results shown are representative of 3-6 independent experiments.
Blimp1 expression in NK cells is independent of IRF4 and Bcl6. (A) DX5+ NK cells were expanded in IL-15 for 5 days and then transferred into cytokines for 2 days as indicated. Protein extracts were subjected to Western blotting with antibodies to the indicated proteins. (B) Blimp1gfp/+ mice were bred on Irf4−/− or Bcl6−/− backgrounds and TCRβ−NK1.1+CD49b+ NK cells were analyzed for Blimp1/GFP expression (empty histogram, wild-type control; filled black histogram, Blimp1gfp/+; filled gray histogram, Blimp1gfp/+ and deficient for the indicated transcription factor). (C) Flow cytometric analysis of splenocytes isolated from Irf4−/−, Bcl6−/−, or control mice (top panel). Gated TCRβ−NK1.1+CD49b+ NK cells are shown in the lower panel. Numbers indicate the proportion of cells of the indicated phenotype. The results shown are representative of 3-6 independent experiments.
Blimp1 expression in NK cells requires T-bet
The maturation defect we have found in Blimp1-deficient NK cells resembles that previously reported for mice deficient in T-bet (encoded by Tbx21).17,18 Moreover, Blimp1 has been shown to directly repress Tbx21 in CD4+ T cells27 and to modulate the expression of a related T-box transcription factor, Eomesodermin (Eomes), in CD8+ T cells.24 Thus, it was possible that the defective NK-cell maturation in the absence of Blimp1 was due to altered Tbx21 or Eomes expression. However, expression of both factors in Blimp1-deficient and control NK cells was similar, with only a relatively minor increase in Eomes (mean fluorescence index [MFI] 7781 ± 585 in +/+, 9096 ± 610 in Blimp1gfp/gfp; P = .02) and no difference in T-bet (MFI 2756 ± 116 in +/+, 2840 ± 137 in Blimp1gfp/gfp; P > .05), suggesting that Blimp1 was not essential for the regulation of Tbx21 or Eomes in NK cells (Figure 7A). Interestingly, T-bet–deficient NK cells showed a substantial up-regulation of Eomes (Figure 7A; MFI 7602 ± 536 in +/+, 14 364 ± 1138 in Tbx21−/−; P = .008), suggesting that Eomes may compensate for some T-bet functions.20
T-bet–dependent Blimp1 expression in NK cells. Flow cytometric analysis of gated splenic TCRβ−NK1.1+CD49b+ NK cells from mixed bone-marrow chimeric mice that were generated as described in Figure 3D. (A) Splenic NK cells from mixed bone-marrow chimeric mice of the genotypes indicated were examined for T-bet and Eomes levels by intracellular staining and flow cytometry. Shown is the expression in TCRβ−NK1.1+CD49b+ NK cells of the indicated genotype (Blimp1gfp/gfp [Ly5.2+], filled gray histograms) and wild-type (Ly5.1+, empty histograms). Tbx21−/− NK cells are shown as a control for the T-bet staining; naive CD4+ T cells are shown as a control for the Eomes stain (dashed lines). (B) Blimp1/GFP expression in Tbx21−/−Blimp1gfp/+ (filled gray histogram) and control (empty histogram, wild-type; filled black histogram, Blimp1gfp/+) NK cells. (C-D) Gated splenic TCRβ−NK1.1+CD49b+ NK cells from mixed bone-marrow chimeric mice were analyzed by flow cytometry for the indicated markers. Data in panel C show analysis of Ly5.2+ NK cells of the indicated genotypes. Data in panel D show total NK cells. Numbers in the plots show the proportion of NK cells in each quadrant. The results are representative of 2 experiments each with 3 mice.
T-bet–dependent Blimp1 expression in NK cells. Flow cytometric analysis of gated splenic TCRβ−NK1.1+CD49b+ NK cells from mixed bone-marrow chimeric mice that were generated as described in Figure 3D. (A) Splenic NK cells from mixed bone-marrow chimeric mice of the genotypes indicated were examined for T-bet and Eomes levels by intracellular staining and flow cytometry. Shown is the expression in TCRβ−NK1.1+CD49b+ NK cells of the indicated genotype (Blimp1gfp/gfp [Ly5.2+], filled gray histograms) and wild-type (Ly5.1+, empty histograms). Tbx21−/− NK cells are shown as a control for the T-bet staining; naive CD4+ T cells are shown as a control for the Eomes stain (dashed lines). (B) Blimp1/GFP expression in Tbx21−/−Blimp1gfp/+ (filled gray histogram) and control (empty histogram, wild-type; filled black histogram, Blimp1gfp/+) NK cells. (C-D) Gated splenic TCRβ−NK1.1+CD49b+ NK cells from mixed bone-marrow chimeric mice were analyzed by flow cytometry for the indicated markers. Data in panel C show analysis of Ly5.2+ NK cells of the indicated genotypes. Data in panel D show total NK cells. Numbers in the plots show the proportion of NK cells in each quadrant. The results are representative of 2 experiments each with 3 mice.
We also determined whether Blimp1 expression in NK cells depends on T-bet. Intriguingly, Tbx21−/−Blimp1gfp/+ NK cells expressed a markedly reduced amount of Blimp1/GFP (Figure 7B), suggesting that T-bet acts upstream of Blimp1 in NK cells. In agreement with earlier reports,17 T-bet–deficient NK cells showed a distinct block in peripheral maturation at the CD27+KLRG1− stage that was similar to the defects observed in Blimp1-deficient NK cells (Figure 7C). The notion that in NK cells, T-bet but not Bcl6 acts in the same transcriptional network as Blimp1 was further supported by the finding that both T-bet– and Blimp1-deficient NK cells expressed high amounts of ckit, a marker of immature NK cells,4 and showed a similar reduction of granzyme B expression, whereas Bcl6-deficient NK cells were indistinguishable from wild-type cells (Figure 7D). Interestingly, neither T-bet nor IRF4 was required for the IL-12– or IL-21–induced up-regulation of Blimp1 in cultured NK cells, further indicating that the expression of Blimp1 in NK cells differs fundamentally from that in T or B cells (supplemental Figure 4).
Discussion
In this study, we identify Blimp1 as an important player in the transcriptional network governing NK-cell differentiation. Our data demonstrate that NK cells express Blimp1 constitutively from a very early point in their development. We further show that Blimp1 levels increase during functional maturation of NK cells and that Blimp1 controls the differentiation and homeostasis of NK cells. Surprisingly, Blimp1 expression in NK cells is independent of IRF4 and Bcl6, 2 well-characterized regulators of Blimp1 in other hematopoietic lineages.
Constitutive expression of Blimp1 is a unique feature of NK cells among the hematopoietic lineages and is in sharp contrast to B cells and CD8+ T cells, in which Blimp1 expression marks terminally differentiated effector cells, including plasma cells and short-lived cytotoxic effector T cells.40 The unique expression pattern of Blimp1 may be reflective of the innate nature of NK cells, which show effector functions such as cytotoxicity and cytokine production throughout their development without the need for extensive proliferation and discrete peripheral differentiation stimuli.
NK cells crucially depend on IL-15, which controls their proliferation and survival in the periphery.41,42 Our results indicate that IL-15 signals are also required for the induction of Blimp1 in early NK-cell development. This again contrasts with findings in CD8+ T cells, in which IL-15 down-regulated Blimp1 expression.43 Interestingly, however, the constitutive Blimp1 expression in NK cells is paralleled in memory CD8+ T cells,24 and both NK cells and CD8 memory T cells require IL-15 for their survival and turnover.42 Thus, IL-15 may act as a modulator of Blimp1 expression in NK cells and memory CD8+ T cells that balances Blimp1 effects on cytotoxicity with those that limit the proliferation and survival of effector cells. The baseline level of Blimp1 was also increased by IL-12, IL-18, and IL-21, cytokines that induce NK-cell maturation. This is again analogous to other lymphocytes in which several cytokines, including IL-21, promote Blimp1 expression.29 Interestingly, activation of NK cells through antibody-mediated cross-linking of activating receptors did not significantly enhance Blimp1 expression, suggesting that these receptors function in a manner similar to that of the B- and T-cell antigen receptors, which on their own do not transduce signals to induce Blimp1 expression.43
Peripheral maturation of NK cells is known to be a multistep process.3,6,7 The most immature NK cells in the mouse spleen express the marker combination Mac-1lowCD27+ckit+. During maturation, these cells first lose ckit expression before up-regulating KLRG1 and silencing CD27 to undergo their final maturation into Mac-1highCD27−KLRG1+ cells. Blimp1-deficient NK cells had altered expression of several of these maturation markers, including a lower amount of Mac-1, and were impaired in the transition from the CD27+KLRG1− to the CD27−KLRG1+ stage. This was in agreement with the hyperresponsiveness of Blimp1-deficient NK cells to IL-15 and their enhanced tumor clearance capacity, because it is known that the CD27+KLRG1− subset of wild-type NK cells has the greatest proliferative potential and superior effector potential.3,7 In addition to its role in peripheral NK-cell maturation, Blimp1 was also required to maintain the appropriate homeostatic numbers of NK cells in nonlymphoid organs such as the lung and liver. Failure to populate the lung may be a consequence of the splenic maturation defect, because CD27−KLRG1+ NK cells predominate in the lungs of wild-type mice3 ; however, Blimp1 is also known the regulate the expression of several chemokine receptors in CD8+ T cells, including CCR5, CCR6, and CCR7, the modulation of which is important for tissue homing.24,25 Further analysis is required to investigate the Blimp1-dependent factors that control NK-cell homing to nonlymphoid tissues.
Several transcription factors are known to control the peripheral maturation of NK cells, including GATA-3,13 Irf2,16,44 and T-bet.17,18 Of these, the T-bet–deficient phenotype most resembles that of Blimp1 mutant NK cells in that they are blocked at a CD27+ckit+ state while having relatively normal expression of NK-cell receptors such as those in the Ly49 family (17 and Figure 7). In keeping with the similarities of the phenotypes, Blimp1 expression was markedly reduced in the absence of T-bet, suggesting that in contrast to CD4+ T cells, in which Tbx21 may be directly repressed by Blimp1,27 T-bet was required for Blimp1 expression in NK cells. Interestingly, T-bet and Blimp1 are both necessary for normal granzyme B production by both NK cells and CD8+ T cells.24,25,45 The functions of these transcription factors, however, are not identical, because a deficiency in T-bet leads to a more severe block in NK-cell maturation and plays a modest role in NK-cell cytotoxicity and IFNγ production,18 and we found no role for Blimp1 in these effector functions. This was the case despite the reduced expression of granzyme B, which contributes to target killing but is not essential for this process.46
During the revision of this manuscript, a study describing BLIMP1 expression and function human NK cells was published.38 This study showed that, in contrast to the mouse homolog, human BLIMP1 is present in 3 isoforms in NK cells. In agreement with our data, Smith et al also found that human BLIMP1 is induced by cytokines and is present in the CD16+ NK-cell subset in the peripheral blood.38 However, in contrast to our data, Smith et al did not observe constitutive expression of BLIMP1 in human NK cells. Whether this was due to a lower sensitivity in the detection method or represents a real biologic difference is presently unclear and requires further study. Smith et al also found a role for BLIMP1 in limiting the production of effector cytokines such as IFNγ and tumor necrosis factorα in human NK cells through direct binding to both loci.38 This finding agrees with the proposed role of Blimp1 in repressing Ifng in mouse CD4+ T cells, but contrasts with our finding of normal IFNγ expression in mouse NK cells in the absence of Blimp1. Independence of IFNγ expression from Blimp1 was also found in CD8+ T cells after viral infection,24,25,45 suggesting that the regulation of Ifng by Blimp1 displays cell-type and species specificity, potentially at the level of the chromatin structure of the Ifng locus. It is also important to note that in human NK cells38 and mouse CD4+ T cells,27 Blimp1 only modulates expression of Ifng, indicating that other factors are also involved in controlling the effector cytokine response.
Our analysis of transcription factors that are intimately linked to the regulation of Blimp1 expression in B and T cells revealed that the transcriptional control of Blimp1 in NK cells is distinct from other lymphocyte lineages. IRF4, which binds at multiple places in the Blimp1 locus and is essential for Blimp1 induction in activated T cells and plasma cells,29,47 was dispensable for Blimp1 expression in NK cells. Another transcription factor implicated in Blimp1 regulation, Bcl6, was coexpressed with Blimp1 in NK cells, and deficiency of Bcl6 led only to a subtle increase in the Blimp1 and granzyme B levels. The coexpression of these 2 factors is surprising, because Bcl6 and Blimp1 have been shown to antagonize each other's expression in all hematopoietic cell types yet examined, including germinal center B cells, follicular helper T cells,28 and, most recently, osteoclasts.39 Therefore, our data indicate that the regulation of Blimp1 expression is fundamentally different in NK cells compared with B and T lymphocytes. While neither IRF4 nor Bcl6 was required for Blimp1 expression, loss of T-bet led to attenuated Blimp1 expression and abrogated NK-cell maturation. This also highlights the unique nature of the genetic network controlling the NK-cell lineage, because Blimp1 and T-bet are independently regulated in differentiating CD8+ effector and memory T cells,24,48 while in CD4+ T cells Blimp1 is proposed to act upstream of T-bet.27 Remarkably, neither T-bet nor IRF4 was required for cytokine-mediated up-regulation of Blimp-1. T-bet is known to up-regulate the Il12rb gene and could mediate the IL-12 induction of Blimp1 expression,49 whereas IRF4 is essential for the IL-21–induced Blimp1 expression in B and T cells; however, our loss-of-function experiments showed that these factors were dispensable for the up-regulation of Blimp1 in NK cells mediated by IL-12 and IL-21, respectively. One explanation for the T-bet–independent expression of Blimp1 in the presence of IL-12 is that Eomes levels were increased in Tbx21−/− NK cells and thus potentially compensated for the loss of T-bet. Further studies are now required to investigate how Blimp1 is regulated in NK cells. In particular, it will be important to determine how the multiple signals that mature NK cells integrate, induce, and fine-tune Blimp1 expression, ultimately promoting NK-cell maturation and tissue homeostasis.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Y. Hayakawa, R. Thong, S. Sterle, J. Brady, and K. Elder for technical assistance; T. Mak for providing the Irf4−/− mice; L. Corcoran for antibodies; and C. Vinuessa for the Bcl6−/− mice.
This work was supported by grants from the National Health and Medical Research Council (NHMRC) of Australia (to A.K., S.C., D.M.T., M.J.S., and S.L.N.). A.K. was supported by an Australian Research Council (ARC) Future Fellowship and a Leukemia & Lymphoma Society Fellowship; S.C. by an ARC Discovery Fellowship; N.D.H. by a Human Frontiers Science Program Long-term Fellowship; M.J.S. by an NHMRC Australia Fellowship; D.M.T. by NHMRC fellowships; and S.L.N. by a Pfizer Australia Research Fellowship.
Authorship
Contribution: A.K., S.C., N.D.H., N.J.B., and M.J.S. designed and performed the research; A.K., M.J.S., D.M.T., and S.L.N. analyzed the data; and A.K. and S.L.N. wrote the paper.
Conflict-of-interest declaration: The authors declare no competing financial interests.
Correspondence: Stephen L. Nutt or Axel Kallies, The Walter and Eliza Hall Institute of Medical Research, 1G Royal Parade, Parkville, Victoria, 3050, Australia; e-mail nutt@wehi.edu.au or kallies@wehi.edu.au.

![Figure 7. T-bet–dependent Blimp1 expression in NK cells. Flow cytometric analysis of gated splenic TCRβ−NK1.1+CD49b+ NK cells from mixed bone-marrow chimeric mice that were generated as described in Figure 3D. (A) Splenic NK cells from mixed bone-marrow chimeric mice of the genotypes indicated were examined for T-bet and Eomes levels by intracellular staining and flow cytometry. Shown is the expression in TCRβ−NK1.1+CD49b+ NK cells of the indicated genotype (Blimp1gfp/gfp [Ly5.2+], filled gray histograms) and wild-type (Ly5.1+, empty histograms). Tbx21−/− NK cells are shown as a control for the T-bet staining; naive CD4+ T cells are shown as a control for the Eomes stain (dashed lines). (B) Blimp1/GFP expression in Tbx21−/−Blimp1gfp/+ (filled gray histogram) and control (empty histogram, wild-type; filled black histogram, Blimp1gfp/+) NK cells. (C-D) Gated splenic TCRβ−NK1.1+CD49b+ NK cells from mixed bone-marrow chimeric mice were analyzed by flow cytometry for the indicated markers. Data in panel C show analysis of Ly5.2+ NK cells of the indicated genotypes. Data in panel D show total NK cells. Numbers in the plots show the proportion of NK cells in each quadrant. The results are representative of 2 experiments each with 3 mice.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/117/6/10.1182_blood-2010-08-303123/4/m_zh89991066040007.jpeg?Expires=1769098544&Signature=kA5sjBRX-TAKHVbhF5coZ7AzJ9uyX0z2z6H-psWimkilE4B0l8XP8sIN090~y9vX5xea4nsdvhcTzmXF3YjAC2Aegt22oyTLIqigkx2ID-wPiqJOWG04jyPBLXXGYZxdMTbE134sUg5wvA98alqKUeJxh94tfV7Xnm0Cwd5XiBWhvQc6XY3pxQsHvxiXUJVDS-PMj3WcxIIF971fTo4H0e3qqa3gkipLafhfvTxGPUOjn-7diQ02NZGU6rAE8eH723l3okNf5t-WD0u8D2~lxIagpm-b4oZE2NJav6C03wpARxL2qBXvDh4bb4Cr~g6NriLzEyEDQMJQm2Ohc4O32A__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal