Abstract
Among acute myeloid leukemia (AML) patients with a normal karyotype (CN-AML), NPM1 and CEBPA mutations define World Health Organization 2008 provisional entities accounting for approximately 60% of patients, but the remaining 40% are molecularly poorly characterized. Using whole-exome sequencing of one CN-AML patient lacking mutations in NPM1, CEBPA, FLT3-ITD, IDH1, and MLL-PTD, we newly identified a clonal somatic mutation in BCOR (BCL6 corepressor), a gene located on chromosome Xp11.4. Further analyses of 553 AML patients showed that BCOR mutations occurred in 3.8% of unselected CN-AML patients and represented a substantial fraction (17.1%) of CN-AML patients showing the same genotype as the AML index patient subjected to whole-exome sequencing. BCOR somatic mutations were: (1) disruptive events similar to the germline BCOR mutations causing the oculo-facio-cardio-dental genetic syndrome; (2) associated with decreased BCOR mRNA levels, absence of full-length BCOR, and absent or low expression of a truncated BCOR protein; (3) virtually mutually exclusive with NPM1 mutations; and (4) frequently associated with DNMT3A mutations, suggesting cooperativity among these genetic alterations. Finally, BCOR mutations tended to be associated with an inferior outcome in a cohort of 422 CN-AML patients (25.6% vs 56.7% overall survival at 2 years; P = .032). Our results for the first time implicate BCOR in CN-AML pathogenesis.
Introduction
Acute myeloid leukemia (AML) is a molecularly and clinically heterogeneous disease.1 Approximately 30% of patients carry recurrent chromosomal translocations and are grouped into a distinct AML category of the 2008 World Health Organization (WHO) classification of myeloid neoplasms.2
During the past decade, there have been important advances in the molecular characterization of a large group of AML patients who, at cytogenetical resolution, demonstrate a normal karyotype (CN-AML). Several genetic lesions have been found to be closely associated with CN-AML, including mutations in NPM1, CEBPA, FLT3-ITD, and MLL-PTD.1-3 Among these, NPM1 mutations and biallelic CEBPA mutations are likely to be primary genetic events, as shown by their specificity for AML, high stability at relapse, mutual exclusion of other recurrent cytogenetic abnormalities, expression in leukemic stem cells, unique gene expression signatures, and distinct microRNA profiles.4-7 Therefore, NPM1-mutated AML and CEBPA-mutated AML have already been included as provisional entities in the 2008 WHO classification.2 Increased knowledge about molecular lesions underlying CN-AML has greatly improved our capability to stratify CN-AML patients into prognostic risk groups8,9 and to monitor minimal residual disease,10 thus influencing therapeutic decisions.
Recently, next-generation sequencing studies of CN-AML led to the identification of new mutations in the IDH1, IDH2, and DNMT3A genes.11-13 Whereas IDH1 and DNMT3A mutations mainly cluster with NPM1-mutated AML, the subgroup of CN-AML patients that are devoid of NPM1 and CEBPA mutations remain molecularly poorly characterized. To address this issue, we used in-solution exome capture followed by massively parallel sequencing. The study was designed to compare the sequences of the leukemic cells at diagnosis and of the paired normal hematopoietic cells at the time of complete remission from a patient with CN-AML without mutations of NPM1, CEBPA, or FLT3 (both internal tandem duplication and tyrosine kinase domain mutations) and negative for MLL-PTD. The absence of IDH1, IDH2, and DNMT3A mutations was not considered as an inclusion criterion for analysis, because these genetic alterations had not yet been discovered when the index patient was selected for whole-exome sequencing (WES). Our final goal was to identify CN-AML–associated somatic mutations in protein-coding genes and to assess their recurrence in a cohort of 553 AML patients to improve our knowledge of this poorly characterized subgroup.
Methods
Leukemic and normal samples
The characteristics of the AML index patient subjected to WES are described in “Results.” WES was approved by the ethics committee at the University of Perugia, Italy. Validation of WES findings in the AML index patient was performed in an independent cohort of 200 adult AML patients (160 CN-AML and 40 AML patients with recurrent translocations) who were sent to the MLL Munich Leukemia Laboratory for diagnostic assessment between October 2005 and August 2010. The 160 CN-AML patients were characterized extensively for known gene mutations associated with the normal karyotype.
We also analyzed a total of 353 AML patients from Italy, including 192 patients (157 CN-AML patients and 35 patients with recurrent translocations) from the Northern Italy Leukemia Group and 161 patients (105 CN-AML patients and 56 patients with cytogenetic abnormalities, including 21 with recurrent translocations) from various Italian hematologic centers. The Italian CN-AML series was characterized for mutations in NPM1, FLT3, and DNMT3A. Combining the Italian and German series, among the 411 CN-AML patients for whom the NPM1 mutation status was known, 197 patients (48%) were NPM1 mutated. In 184 of 197 patients, FLT3-ITD status was available and was mutated in 67 patients (35%). The study design adhered to the tenets of the Declaration of Helsinki. Written informed consent for analysis of leukemic samples was obtained at each participating center in accordance with the Declaration of Helsinki.
Diagnostic assessment included cytochemistry with myeloperoxidase, nonspecific esterase, and iron staining. Criteria for dysplasia were those established by the 2008 WHO classification.2 Immunophenotyping was performed applying multiparameter flow cytometry and 5-color combinations of mAbs selected for the identification of aberrant immunophenotypes.14 Chromosome banding analysis and FISH were performed as described previously.15
WES and bioinformatic analyses
WES and bioinformatic analyses were carried out as described previously.16 Paired-end massively parallel sequencing was performed on the Illumina Genome Analyzer GAIIx for 2 × 108 cycles using the Chrysalis sequencing kit Version 4.0. The base-calling was performed with GAPipeline Version 1.5.1 and produced approximately 44.5 and 42.2 million pass filter reads (in the leukemic and normal samples, respectively) of 108 bases per library (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article), which represents 4.8-4.6 Gb per library. Mapping on the human genome assembly hg18/NCBI36.1 was performed using MAQ Version 0.7.1 software (http://maq.sourceforge.net/maq-man.shtml), accepting up to 2 mismatches in the first 24 bases (seed) and considering a maximum insert size of 600 bp. Approximately 66% of all mapped reads mapped to an exon target ± 100 bases, documenting the successful technical outcome of this experiment.
Low-quality reads (average Phred score < 5) and duplicate reads were removed (supplemental Table 1). Sequence variants—differences from the reference human genome sequence—were identified in each sample separately, and variants present in the paired normal DNA or likely representing sequencing errors were removed using the SAVI algorithm (Statistical Algorithm for Variant Identification) developed at Columbia University16 (supplemental Figure 1 and supplemental Table 2). Algorithm sensitivity (proportion of true variants identified) was inferred by considering the variants called by the algorithm at known single nucleotide polymorphism database (dbSNP) sites using an error P value cutoff of at most 1 × 10−6 for a variant in both the tumor and normal sample, which produced 13 385 known SNPs. The total number of variants identified in the tumor (16 105), including the ones not previously reported (2720) and the known SNPs (reported in dbSNP 130), was similar to other estimates using different capture and sequencing platforms, including the one used in this study17 (supplemental Table 3), testifying to the sensitivity (proportion of true variants identified) of the SAVI algorithm.16
Verification of WES findings by direct Sanger sequencing in the AML index patient
Candidate somatic variants (present in the tumor but not in its paired normal DNA) that were nonsynonymous or affected the consensus splice site (ie, the 4 nucleotides surrounding the exon-intron boundary) were verified by PCR amplification and direct DNA sequencing of the corresponding regions in the index patient (primers available on request). Because the sensitivity of Sanger sequencing allows the detection of heterozygous mutations only when present in a major clonal population, sequence variants reported in < 25% of the reads were not included in this validation phase.
Screening of mutated genes in independent patient cohorts
The genes found mutated by WES of the index patient were further screened in 2 independent cohorts of 200 and 353 adult AML patients at first diagnosis, respectively. Cohort 1 was from the MLL (n = 200) and was analyzed using a next-generation amplicon deep-sequencing assay. Cohort 2 was from Italy (n = 353) and was studied using conventional direct Sanger sequencing.
In cohort 1, isolation of blast cells, DNA extraction (n = 171), or mRNA extraction (n = 29) and random-primed cDNA synthesis was performed as described previously.18 We applied amplicon deep-sequencing to investigate the complete coding region of 4 candidate genes that had been identified by WES analysis: DNMT3A, BCOR, SSRP1, and YY2. All amplicons were generated from cDNA or genomic DNA specimens. Corresponding accession numbers, primer sequences and PCR amplification protocols are listed in supplemental Tables 4 through 9. Deep sequencing was performed using the 454 GS FLX Titanium amplicon chemistry (Roche Applied Science).19,20
Mapping results and detected variants were exported to R/Bioconductor for further analysis.21 For variant detection, filters were set to display sequence alterations occurring in > 5% of bidirectional reads per amplicon in at least one patient. At least 150 reads were generated for each amplicon. In addition, variant comparisons were performed against dbSNP (http://www.ncbi.nlm.nih.gov/projects/SNP), and the occurrence of small indels was investigated using SEQUENCE Pilot Version 3.4 software (JSI Medical Systems).
In the cohort of 353 AMLs from Italy, genomic DNA extraction, PCR amplification, and bidirectional direct Sanger sequencing of all BCOR coding exons (total of 21 amplicons) were done according to standard methods (Genewiz) using the primers listed in supplemental Table 10. All samples analyzed by direct Sanger sequencing contained at least 12.5% leukemic cells in the case of male patients or at least 25% leukemic cells in the case of female patients, which were used as thresholds for this method to unequivocally detect the presence of a hemizygous or heterozygous mutation in BCOR (located on chromosome X), in a major tumor clone. Analysis of the chromatograms was performed with Mutation Surveyor Version 3.98 software (SoftGenetics).
Although in a few AML patients we observed missense BCOR variants, we decided not to discuss them in this paper as they often turned out to be germline polymorphisms when non-leukemic DNA was available for analysis.
Detection of BCOR mRNA and protein
Total RNA extraction from the AML samples, followed by randomly primed reverse transcription, was performed according to standard methods. BCOR mRNA levels were quantified in duplicate wells by real-time PCR using the TaqMan Gene Expression assays; Hs00372378_m1* for BCOR and Hs99999907_m1 for B2M (housekeeping gene) in an AB7900HT Fast Real-Time PCR System (Applied Biosystems). As a calibrator, we used the average ΔCt of primary leukemic cell samples from 14 BCOR wild-type AML patients. Quantification of BCOR transcript levels in 5 primary AML samples with BCOR mutations that introduced a premature stop codon (before the 2nd last exon) and in the 14 BCOR-unmutated AML samples was performed according to the ΔCt method relative to the calibrator, which was set as 100%.
Expression of the BCOR protein was explored by Western blot in 5 AML patients carrying BCOR disruptive mutations using standard procedures. Briefly, lysates of 1 × 106 leukemic cells were run on 4%-15% Mini-Protean TGX gradient gel (Bio-Rad), transferred to nitrocellulose membrane, and probed with an anti–BCOR mouse polyclonal Ab (ab88112; Abcam), an anti–β-tubulin (clone TUB 2.1; Sigma-Aldrich), or an anti–NPM1 mouse mAb (produced in B.F.'s laboratory) as loading control. CD34+ hematopoietic cells from 2 normal donors and primary AML samples from 19 patients, all lacking BCOR mutations, were also analyzed for comparison. All patient samples had ≥ 70% leukemic cells. The OCI-AML3 cells not carrying BCOR mutations were used as a positive control for full-length BCOR protein expression.
Mutational analyses
Screening of NPM1, FLT3-ITD, CEBPA, IDH1/2, and RUNX1 mutations and analysis for MLL-PTD was performed as described previously.18,22-26 Further details on statistical analyses and gene expression profiling are provided online. All microarray data are available in the Gene Expression Omnibus (GEO) under accession number GSE30442.
Results
Features of the AML index patient subjected to WES
A 67-year-old female patient presented with pancytopenia (WBC, 2540/μL; hemoglobin, 7.4 g/dL; and platelets, 78 000/μL). The BM was massively infiltrated by myeloid blasts (83%) that were positive for myeloperoxidase (30%) and negative for nonspecific esterase. No Auer rods were detectable. The patient was diagnosed with AML with maturation according to WHO 2008 criteria.2 Multiparameter flow cytometric studies were consistent with AML. Cytogenetic analysis of leukemic cells revealed a normal karyotype (46,XX) in 20 of 20 metaphases analyzed. Molecular screening showed no mutations of NPM1, CEBPA, FLT3-ITD, or MLL-PTD. No studies on IDH1 and IDH2 could be performed because mutations in these genes had not yet been discovered at the time the patient was selected for WES. Subsequent analysis of this patient revealed an absence of IDH1 mutations but a mutation in IDH2 (R172L).
Therapy was administered with 2 induction cycles of ICE (idarubicin, cytarabine, etoposide), followed by 2 consolidation cycles with high-dose cytarabine. The patient achieved complete remission but relapsed after 15 months. Salvage treatment was performed with the s-HAM regimen (sequential high-dose Ara-C, mitoxantrone, plus GM-CSF), followed by allogeneic stem cell transplantation with fludarabine and treosulfan as the conditioning regimen. The patient died with no signs of leukemia from septic shock after sigmoid diverticulitis that required sigmoid resection 24 months after her initial diagnosis.
Identification of candidate somatic mutations in the AML index patient
The whole-exome capture and sequencing approach produced in total approximately 86 million mapped reads (approximately 43.9 million from the tumor DNA and approximately 41.7 million from the normal DNA) of 108 nucleotides (supplemental Table 1). After removal of low-quality and clonal reads, the mean depth of the covered exome was 69-fold (tumor) and 66-fold (normal; median, 51-fold and 49-fold, respectively), with 99% of the target exome being covered by at least 1 read and 87%-88% by at least 10 reads (supplemental Table 1).
The SAVI algorithm16 (supplemental Figure 1) identified 13 unique nonsynonymous variants (12 missense and 1 out-of-frame deletion) that were present specifically in the tumor DNA (frequencies ranging from 21%-64% of the reads). These variants were validated by PCR amplification and direct Sanger sequencing of the same leukemic and nonleukemic genomic DNA, and corresponded to 11 distinct genes, including SPATA16, DSP, DNMT3A, SFRS11, CDH12, SSRP1, BCOR, YY2, TCEB3B, ZNF676, and RSPH10B2 (Table 1). In all patients, mutations were heterozygous and presumably clonally represented in the leukemic population. Four additional variants predicted by the SAVI algorithm were not confirmed by Sanger sequencing as somatic mutations; in 1 instance, the normal DNA harbored the variant, and in the other 3 instances, the variant was not present in the tumor DNA (Table 1).
List of candidate somatic mutations identified in the AML index patient
| Chromosome . | Position . | Gene . | R/V . | AA . | Tumor . | Normal . | P . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| (F+R)/T . | % . | (F+R)/T . | % . | T err . | N err . | Germ. . | |||||
| Somatic mutations confirmed by Sanger sequencing | |||||||||||
| 3 | 174317684 | SPATA16 | C/T | V178I | (38 + 38)/182 | 42 | (0 + 2)/138 | 1 | 2.8 × 10−78 | .77 | 1.6 × 10−61 |
| 6 | 7529633 | DSP | C/T | P2380L | (33 + 19)/100 | 52 | (1 + 0)/91 | 1 | 1.6 × 10−60 | .84 | 3.5 × 10−51 |
| 2 | 25312132 | DNMT3A | G/A | P660S | (26 + 20)/95 | 48 | (0 + 0)/88 | 0 | 8.2 × 10−52 | 1 | 8.1 × 10−52 |
| 1 | 70488756 | SFRS11 | G/C | E413D | (29 + 18)/114 | 41 | (0 + 1)/117 | 1 | 1 × 10−48 | .91 | 5.8 × 10−42 |
| 5 | 21788048 | CDH12 | A/C | M647R | (11 + 18)/67 | 43 | (1 + 1)/81 | 2 | 2.0 × 10−31 | .48 | 5.4 × 10−22 |
| 11 | 56856812 | SSRP1 | G/A | R211C | (12 + 11)/49 | 47 | (0 + 0)/43 | 0 | 3.0 × 10−26 | 1 | 2.9 × 10−26 |
| X | 39818006 | BCOR | C/T | G513R | (6 + 13)/48 | 40 | (0 + 1)/40 | 3 | 3.5 × 10−20 | .55 | 5.3 × 10−14 |
| X | 21784900 | YY2 | C/T | S126L | (2 + 14)/39 | 41 | (0 + 0)/29 | 0 | 1.6 × 10−17 | 1 | 1.6 × 10−17 |
| 18 | 42814666 | TCEB3B | C/T | R323Q | (2 + 12)/29 | 48 | (0 + 0)/32 | 0 | 9.6 × 10−17 | 1 | 9.4 × 10−17 |
| X | 39808770 | BCOR | −/AT | R1089HfsX25 | (8 + 6)/35 | 40 | (0 + 0)/37 | 0 | 2.6 × 10−15 | 1 | 2.5 × 10−15 |
| 2 | 25315524 | DNMT3A | C/T | G607D | (6 + 1)/11 | 64 | (0 + 0)/10 | 0 | 3.9 × 10−10 | 1 | 3.9 × 10−10 |
| 7 | 6787044 | RSPH10B2 | G/A | G537S | (3 + 5)/39 | 21 | (0 + 0)/37 | 0 | 9.0 × 10−07 | .49 | 8.4 × 10−7 |
| 19 | 22155572 | ZNF676 | T/A | S263C | (15 + 35)/142 | 35 | (0 + 0)/139 | 0 | 1.3 × 10−47 | 1 | 1.3 × 10−47 |
| Candidate somatic mutations not confirmed by Sanger sequencing in the leukemic cell DNA | |||||||||||
| 2 | 86112970 | POLR1A | G/C | A1403G | (1 + 10)/56 | 20 | (0 + 1)/33 | 3 | 1.3 × 10−8 | .49 | 6.2 × 10−5 |
| 3 | 195336860 | HES1 | G/A | splice | (4 + 1)/9 | 56 | (0 + 0)/7 | 0 | 3.8 × 10−7 | 1 | 3.7 × 10−7 |
| 22 | 36351755 | GGA1 | A/C | T316P | (1 + 5)/16 | 38 | (0 + 0)/20 | 0 | 4.3 × 10−7 | 1 | 4.2 × 10−7 |
| Germline variant (present both in leukemic and normal cell DNA) | |||||||||||
| 1 | 208924107 | KCNH1 | −/GA | (8 + 2)/19 | 53 | (1 + 0)/13 | 8 | 8.0 × 10−13 | .23 | 2.0 × 10−6 | |
| Chromosome . | Position . | Gene . | R/V . | AA . | Tumor . | Normal . | P . | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| (F+R)/T . | % . | (F+R)/T . | % . | T err . | N err . | Germ. . | |||||
| Somatic mutations confirmed by Sanger sequencing | |||||||||||
| 3 | 174317684 | SPATA16 | C/T | V178I | (38 + 38)/182 | 42 | (0 + 2)/138 | 1 | 2.8 × 10−78 | .77 | 1.6 × 10−61 |
| 6 | 7529633 | DSP | C/T | P2380L | (33 + 19)/100 | 52 | (1 + 0)/91 | 1 | 1.6 × 10−60 | .84 | 3.5 × 10−51 |
| 2 | 25312132 | DNMT3A | G/A | P660S | (26 + 20)/95 | 48 | (0 + 0)/88 | 0 | 8.2 × 10−52 | 1 | 8.1 × 10−52 |
| 1 | 70488756 | SFRS11 | G/C | E413D | (29 + 18)/114 | 41 | (0 + 1)/117 | 1 | 1 × 10−48 | .91 | 5.8 × 10−42 |
| 5 | 21788048 | CDH12 | A/C | M647R | (11 + 18)/67 | 43 | (1 + 1)/81 | 2 | 2.0 × 10−31 | .48 | 5.4 × 10−22 |
| 11 | 56856812 | SSRP1 | G/A | R211C | (12 + 11)/49 | 47 | (0 + 0)/43 | 0 | 3.0 × 10−26 | 1 | 2.9 × 10−26 |
| X | 39818006 | BCOR | C/T | G513R | (6 + 13)/48 | 40 | (0 + 1)/40 | 3 | 3.5 × 10−20 | .55 | 5.3 × 10−14 |
| X | 21784900 | YY2 | C/T | S126L | (2 + 14)/39 | 41 | (0 + 0)/29 | 0 | 1.6 × 10−17 | 1 | 1.6 × 10−17 |
| 18 | 42814666 | TCEB3B | C/T | R323Q | (2 + 12)/29 | 48 | (0 + 0)/32 | 0 | 9.6 × 10−17 | 1 | 9.4 × 10−17 |
| X | 39808770 | BCOR | −/AT | R1089HfsX25 | (8 + 6)/35 | 40 | (0 + 0)/37 | 0 | 2.6 × 10−15 | 1 | 2.5 × 10−15 |
| 2 | 25315524 | DNMT3A | C/T | G607D | (6 + 1)/11 | 64 | (0 + 0)/10 | 0 | 3.9 × 10−10 | 1 | 3.9 × 10−10 |
| 7 | 6787044 | RSPH10B2 | G/A | G537S | (3 + 5)/39 | 21 | (0 + 0)/37 | 0 | 9.0 × 10−07 | .49 | 8.4 × 10−7 |
| 19 | 22155572 | ZNF676 | T/A | S263C | (15 + 35)/142 | 35 | (0 + 0)/139 | 0 | 1.3 × 10−47 | 1 | 1.3 × 10−47 |
| Candidate somatic mutations not confirmed by Sanger sequencing in the leukemic cell DNA | |||||||||||
| 2 | 86112970 | POLR1A | G/C | A1403G | (1 + 10)/56 | 20 | (0 + 1)/33 | 3 | 1.3 × 10−8 | .49 | 6.2 × 10−5 |
| 3 | 195336860 | HES1 | G/A | splice | (4 + 1)/9 | 56 | (0 + 0)/7 | 0 | 3.8 × 10−7 | 1 | 3.7 × 10−7 |
| 22 | 36351755 | GGA1 | A/C | T316P | (1 + 5)/16 | 38 | (0 + 0)/20 | 0 | 4.3 × 10−7 | 1 | 4.2 × 10−7 |
| Germline variant (present both in leukemic and normal cell DNA) | |||||||||||
| 1 | 208924107 | KCNH1 | −/GA | (8 + 2)/19 | 53 | (1 + 0)/13 | 8 | 8.0 × 10−13 | .23 | 2.0 × 10−6 | |
F, R and T indicate the number of variant reads observed in the forward strand (F) or in the reverse strand (R) and the number of total reads (T) covering that position; R/V, reference/variant nucleotide; P, p-value associated to the probability that the variant is detected due to a sequencing error in the tumor (T err), a sequencing error in the normal (N err), and a germline variant (Germ.).
Identification of recurrent BCOR mutations in AML
The mutated genes identified in the index patient (Table 1) included DNMT3A, which has been previously reported to be mutated in AML,13 and 3 additional genes, BCOR, YY2, and SSRP1, which we selected for further mutational screening because of their biologic functions and/or putative implication in AML pathogenesis. The BCOR gene—initially identified as a BCL6 transcriptional corepressor27 —was recently found to translocate with RARA in acute promyelocytic leukemia.28 The YY2 gene29 shares many structural and functional features with YY1, the transcriptional repression activity of which is under control of the NPM1 gene,30 which is frequently mutated in AML.3 The SSRP1 gene, corresponding to FACT (facilitates chromatin transcription), was selected because it is involved in transcriptional mechanisms through chromatin remodeling.31
We then performed deep-sequencing analyses of all exons of BCOR, DNMT3A, YY2, and SSRP1 in an initial set of 30 AMLs that showed the same genetic characteristics of our AML index patient. None of the 30 samples was mutated for YY2, and only 1 of 16 AML patients carried a SSRP1 mutation. In contrast, DNMT3A was mutated in 4 of 30 patients (13.3%) and BCOR in 5 of 30 patients (16.6%). Because the characteristics of DNMT3A mutations have been described previously,13 we next focused on BCOR, a gene that has not been reported previously to be mutated in AML.
BCOR mutations are mainly distributed in CN-AML
Further studies were organized in 2 phases (Figure 1). The aim of phase 1 was to establish the frequency of BCOR mutations in AML patients that have genetic features similar to those of the AML index patient: a normal karyotype in the absence of NPM1, CEBPA, FLT3-ITD mutations, and MLL-PTD (supplemental Table 11). To address this issue, we extended the analysis of BCOR mutations performed in the first cohort of 30 patients (see “Identification of recurrent BCOR mutations in AML”) to an additional 51 CN-AML patients (n = 82 samples, including the index patient) that were selected for carrying the same genetic features as the AML index patient. In a subsequent step, the patients were retrospectively characterized and found to carry IDH1 and IDH2 mutations in 3 and 16 patients, respectively (supplemental Table 11). BCOR-disruptive mutations, ie nonsense mutations, out-of-frame small indels, and consensus splice-site mutations (see “Characteristics of BCOR mutations in AML”) were detected in 14/82 (17.1%) in this cohort of patients.
Flow chart outlining the different phases of the study. Two phases of the study were performed. In phase 1, the identification of candidate somatic mutations by WES was carried out in an index patient. BCOR mutations were subsequently searched for in 82 CN-AML patients with the same genotype as the AML index patient. In phase 2 of the study, additional patient cohorts and associations between BCOR and other molecular mutations were investigated.
Flow chart outlining the different phases of the study. Two phases of the study were performed. In phase 1, the identification of candidate somatic mutations by WES was carried out in an index patient. BCOR mutations were subsequently searched for in 82 CN-AML patients with the same genotype as the AML index patient. In phase 2 of the study, additional patient cohorts and associations between BCOR and other molecular mutations were investigated.
The aim of phase 2 (Figure 1) was to assess the real frequency of BCOR mutations in unselected patients of CN-AML. For this purpose, we analyzed 262 unselected CN-AML patients from an independent Italian cohort characterized for mutations in NPM1, FLT3-ITD and DNMT3A. BCOR-disruptive mutations were found in 10/262 (3.8%) patients (Figure 1). Of the 10 BCOR-mutated patients, 3 were reinvestigated for mutations in CEBPA, MLL-PTD, and IDH1. Interestingly, they were demonstrated to be wild-type for these genes as well as for NPM1 and FLT3-ITD, thus paralleling the molecular background of the AML index patient subjected to WES.
To assess the frequency of BCOR mutations in AML with abnormal karyotype, we studied 131 AML carrying cytogenetic abnormalities. In particular, 96 AML patients belonged to the category of “AML with recurrent cytogenetic abnormalities” of WHO 2008, including t(8;21)(q22;q22) (n = 30), inv(16)(p13q22) (n = 40), t(15;17)(q22;21) (n = 10), t(11q23)/MLL (n = 13), t(6;9)(p23;q24) (n = 2), and t(3;3)(q21;q26.2) (n = 1). Thirty-five AML patients showed other abnormal karyotypes, including+8 (n = 8), complex (n = 7), del(7q) (n = 5), t(8;16)(p11;p13) (n = 3), and 1 patient each of +5, +13, +21, del(Yq), del(20q), i(17q), t(2;12), t(2;17), t(16;19), t(9;22), t(1;3), and t(2;14). None of these patients carried BCOR disruptive mutations.
In summary, BCOR disruptive mutations were relatively rare in unselected CN-AML patients (3.8%), whereas they accounted for a significant fraction (17.1%) of the least genetically characterized subgroup of CN-AML, those carrying no NPM1, CEBPA, FLT3-ITD, or IDH1 mutations and no MLL-PTD (such as the AML index patient).
Characteristics of BCOR mutations in AML
The BCOR gene is located on chromosome Xp11.4 and is targeted by disruptive germline mutations that cause the X-linked oculo- facio-cardio-dental (OFCD) syndrome in heterozygous females and prenatal lethality in hemizygous males.32
We identified BCOR mutations in a total of 26 AML patients (14 female and 12 male), including 2 patients with NPM1 mutations (see “BCOR mutations are mutually exclusive with NPM1 mutations”). BCOR mutations were scattered across the whole length of the coding sequence and included 6 nonsense mutations, 15 small frame-shift insertions/deletions (one of which was observed in 2 patients), and 4 mutations of consensus splice-sites (2 of which targeted the same splice-site in 2 distinct patients; Figure 2 and Table 2). This disruptive mutation pattern is similar to that of BCOR germline mutations causing the OFCD syndrome.32 However, BCOR mutations in AML were of somatic origin, as confirmed by their absence in the germline DNA of all 7 AML patients studied at the time of complete remission (including 1 patient with a consensus splice-site mutation), and in line with the observation that the OFCD syndrome was not apparent in any of our 14 female AML patients. The mutations appeared to be present in a major leukemic clone based on the visual inspection of chromatograms (patients analyzed by Sanger sequencing), or based on the proportion of mutated reads (patients analyzed by 454 amplicon sequencing), relative to the percentage of leukemic cells in the analyzed samples and to the sex of the patient.
BCOR disruptive mutations in AML. (A) Schematic representation of the BCOR protein and its domains. (B) Type and distribution of BCOR disruptive mutations (out-of-frame small indels are depicted as rectangles, nonsense mutations as ovals, and consensus splice-site mutations as [splice site mut.]) along the BCOR coding exons (numbered boxes with a width proportional to the exon nucleotide length) and the exon/intron junctions (short horizontal black lines before and after each exon). Mutations are shown above or below the exons based on whether they were observed in the German or Italian cohorts, respectively. The number 2 appended to the Arg1547X oval denotes that this mutation was found in 2 distinct patients of the German cohort. (C) Chromatograms of a representative clonal, somatic out-of-frame indel (delC) observed by direct DNA Sanger sequencing in the BM sample of a female AML patient at disease onset (containing approximately 70% blast cells), but not in her matched normal sample taken at the time of complete remission.
BCOR disruptive mutations in AML. (A) Schematic representation of the BCOR protein and its domains. (B) Type and distribution of BCOR disruptive mutations (out-of-frame small indels are depicted as rectangles, nonsense mutations as ovals, and consensus splice-site mutations as [splice site mut.]) along the BCOR coding exons (numbered boxes with a width proportional to the exon nucleotide length) and the exon/intron junctions (short horizontal black lines before and after each exon). Mutations are shown above or below the exons based on whether they were observed in the German or Italian cohorts, respectively. The number 2 appended to the Arg1547X oval denotes that this mutation was found in 2 distinct patients of the German cohort. (C) Chromatograms of a representative clonal, somatic out-of-frame indel (delC) observed by direct DNA Sanger sequencing in the BM sample of a female AML patient at disease onset (containing approximately 70% blast cells), but not in her matched normal sample taken at the time of complete remission.
In AML male patients, BCOR mutations disrupted the single copy of the gene. In female AML patients, the mutations targeted the functional allele too, as indicated by their detection in all females studied by cDNA-based amplicon deep-sequencing. Moreover, in the majority of these female patients, the high proportion of mutated reads, relative to the percentage of leukemic cells in the analyzed samples, pointed to the exclusive expression of the mutated allele (Table 2) arguing against escape of this gene from X-chromosome inactivation; a phenomenon known to involve approximately 20% of human X-chromosome genes and leading to their biallelic expression.33
Details of BCOR-disruptive mutations observed in AML patients
| Patient ID . | Cohort . | Sex . | Exon . | Mutation* . | Predicted consequence . | Analysis type† . | Blasts, %§ . | Mutated reads, % . | Coverage, fold . |
|---|---|---|---|---|---|---|---|---|---|
| 1‡ | Munich | F | 7 | 39808770insAT | Arg1089HisfsX25 | gDNA/WES | 83 | 40 | 35 |
| 2 | Munich | M | 15 | 4981C > T | Arg1661X | cDNA/454 | 72 | 97 | 968 |
| 3 | Munich | M | 12 | 4639C > T | Arg1547X | cDNA/454 | 3 | 64 | 315 |
| 4 | Munich | M | 4 | 2920_2923dupGGTG | Asp975GlyfsX2 | cDNA/454 | 60 | 92 | 1126 |
| 5 | Munich | M | 4 | 1116delCinsGG | Ser369TrpfsX12 | cDNA/454 | 57 | 94 | 971 |
| 6 | Munich | F | 14 | 4925C > G | Ser1642X | gDNA/454 and cDNA/454 | 84 | 25 and 97 | 783 and 1479 |
| 7 | Munich | M | 4 | 1024C > T | Arg342X | gDNA/454 and cDNA/454 | 25 | 96 and 44 | 637 and 1859 |
| 8 | Munich | F | 9 | 4257_4258delTG | Cys1363GlnfsX78 | gDNA/454 and cDNA/454 | 60 | 40 and 40 | 536 and 2064 |
| 9 | Munich | F | 4 | 2814dupC | Tyr939LeufsX7 | gDNA/454 and cDNA/454 | 67 | 46 and 90 | 861 and 1492 |
| 10 | Munich | M | 9 | 4272dupG | Gly1376IlefsX65 | gDNA/454 and cDNA/454 | 90 | 30 and 26 | 196 and 2064 |
| 11 | Munich | F | 11 | 4440–1G > A | Consensus splice site mutation | gDNA/454 and cDNA/454 | 96 | 43 and 85 | 529 and 1025 |
| 12 | Munich | F | 4 | 865dupT | Trp289LeufsX12 | gDNA/454 and cDNA/454 | 59 | 35 and 93 | 553 and 915 |
| 13 | Munich | F | 14 | 4936dupC | Leu1646ProfsX6 | gDNA/454 and cDNA/454 | 93 | 27 and 32 | 151 and 1479 |
| 14 | Munich | M | 12 | 4639C > T | Arg1547X | gDNA/454 | 18 | 20 | 715 |
| 93 | Munich | M | 10 | 4428 + 1G > A | Consensus splice site mutation | gDNA/454 and cDNA/454 | 78 | 98 and 28 | 556 and 745 |
| 136 | Munich | F | 15 | 5011A > T | Lys1671X | gDNA/454 and cDNA/454 | 91 | 42 and 91 | 659 and 859 |
| 197 | Perugia | F | 4 | 104639insA | Gly886ArgfsX30 | gDNA/Sanger | 90 | n.a. | n.a. |
| 169 | Perugia | M | 4 | 104361C > T | Gln793X | gDNA/Sanger | 30 | n.a. | n.a. |
| 406II | Perugia | F | 11 | 120035delC | Asn1485LysfsX5 | gDNA/Sanger | 70 | n.a. | n.a. |
| 326 | Perugia | M | 4 | 104181_104182delAC | Thr733AlafsX5 | gDNA/Sanger | 90 | n.a. | n.a. |
| 258 | Perugia | F | 4 | 102717_102720delCTCT | Leu245ThrfsX19 | gDNA/Sanger | 100 | n.a. | n.a. |
| 644 | Perugia | F | 4 | 104004delC | His674MetfsX41 | gDNA/Sanger | 80 | n.a. | n.a. |
| 447 | Perugia | M | 7 | 112835_112844delCCTCCCGCAG | Pro1115ThrfsX41 | gDNA/Sanger | 67 | n.a. | n.a. |
| 139 | Perugia | F | 4 | 103183delG | Gly400AlafsX41 | gDNA/Sanger | 75 | n.a. | n.a. |
| 110 | Perugia | F | 12 | 121963G → A | Consensus splice site mutation | gDNA/Sanger | 85 | n.a. | n.a. |
| 119 | Perugia | M | 11 | 120008G → C | Consensus splice site mutation | gDNA/Sanger | 32 | n.a. | n.a. |
| Patient ID . | Cohort . | Sex . | Exon . | Mutation* . | Predicted consequence . | Analysis type† . | Blasts, %§ . | Mutated reads, % . | Coverage, fold . |
|---|---|---|---|---|---|---|---|---|---|
| 1‡ | Munich | F | 7 | 39808770insAT | Arg1089HisfsX25 | gDNA/WES | 83 | 40 | 35 |
| 2 | Munich | M | 15 | 4981C > T | Arg1661X | cDNA/454 | 72 | 97 | 968 |
| 3 | Munich | M | 12 | 4639C > T | Arg1547X | cDNA/454 | 3 | 64 | 315 |
| 4 | Munich | M | 4 | 2920_2923dupGGTG | Asp975GlyfsX2 | cDNA/454 | 60 | 92 | 1126 |
| 5 | Munich | M | 4 | 1116delCinsGG | Ser369TrpfsX12 | cDNA/454 | 57 | 94 | 971 |
| 6 | Munich | F | 14 | 4925C > G | Ser1642X | gDNA/454 and cDNA/454 | 84 | 25 and 97 | 783 and 1479 |
| 7 | Munich | M | 4 | 1024C > T | Arg342X | gDNA/454 and cDNA/454 | 25 | 96 and 44 | 637 and 1859 |
| 8 | Munich | F | 9 | 4257_4258delTG | Cys1363GlnfsX78 | gDNA/454 and cDNA/454 | 60 | 40 and 40 | 536 and 2064 |
| 9 | Munich | F | 4 | 2814dupC | Tyr939LeufsX7 | gDNA/454 and cDNA/454 | 67 | 46 and 90 | 861 and 1492 |
| 10 | Munich | M | 9 | 4272dupG | Gly1376IlefsX65 | gDNA/454 and cDNA/454 | 90 | 30 and 26 | 196 and 2064 |
| 11 | Munich | F | 11 | 4440–1G > A | Consensus splice site mutation | gDNA/454 and cDNA/454 | 96 | 43 and 85 | 529 and 1025 |
| 12 | Munich | F | 4 | 865dupT | Trp289LeufsX12 | gDNA/454 and cDNA/454 | 59 | 35 and 93 | 553 and 915 |
| 13 | Munich | F | 14 | 4936dupC | Leu1646ProfsX6 | gDNA/454 and cDNA/454 | 93 | 27 and 32 | 151 and 1479 |
| 14 | Munich | M | 12 | 4639C > T | Arg1547X | gDNA/454 | 18 | 20 | 715 |
| 93 | Munich | M | 10 | 4428 + 1G > A | Consensus splice site mutation | gDNA/454 and cDNA/454 | 78 | 98 and 28 | 556 and 745 |
| 136 | Munich | F | 15 | 5011A > T | Lys1671X | gDNA/454 and cDNA/454 | 91 | 42 and 91 | 659 and 859 |
| 197 | Perugia | F | 4 | 104639insA | Gly886ArgfsX30 | gDNA/Sanger | 90 | n.a. | n.a. |
| 169 | Perugia | M | 4 | 104361C > T | Gln793X | gDNA/Sanger | 30 | n.a. | n.a. |
| 406II | Perugia | F | 11 | 120035delC | Asn1485LysfsX5 | gDNA/Sanger | 70 | n.a. | n.a. |
| 326 | Perugia | M | 4 | 104181_104182delAC | Thr733AlafsX5 | gDNA/Sanger | 90 | n.a. | n.a. |
| 258 | Perugia | F | 4 | 102717_102720delCTCT | Leu245ThrfsX19 | gDNA/Sanger | 100 | n.a. | n.a. |
| 644 | Perugia | F | 4 | 104004delC | His674MetfsX41 | gDNA/Sanger | 80 | n.a. | n.a. |
| 447 | Perugia | M | 7 | 112835_112844delCCTCCCGCAG | Pro1115ThrfsX41 | gDNA/Sanger | 67 | n.a. | n.a. |
| 139 | Perugia | F | 4 | 103183delG | Gly400AlafsX41 | gDNA/Sanger | 75 | n.a. | n.a. |
| 110 | Perugia | F | 12 | 121963G → A | Consensus splice site mutation | gDNA/Sanger | 85 | n.a. | n.a. |
| 119 | Perugia | M | 11 | 120008G → C | Consensus splice site mutation | gDNA/Sanger | 32 | n.a. | n.a. |
n.a. indicates not applicable to direct Sanger sequencing. In the chromatogram of these samples, the size of the mutated peak relative to the proportion of leukemic cells was consistent with a clonal event.
Numbers are according to transcript-ID ENST00000378444 (for samples analyzed in Munich) and according to NG_008880.1 (for samples analyzed in Perugia).
In 10 cases, the disruptive mutation was validated with either genomic DNA or cDNA.
Index patient.
Blast percentage is given according to the diagnostic report; the percentage of leukemic cells actually present can be greater in the samples used for sequencing because the latter were mostly Ficoll-enriched with mononuclear leukemic cells before nucleic acid extraction. In the chromatogram of these samples, the size of the mutated peak relative to the proportion of leukemic cells was consistent with a clonal event.
In conclusion, BCOR mutations appear to be clonal, somatic, disruptive events that target the only functional allele not only in male, but also in female AML patients.
Expression of BCOR mRNA and protein in BCOR-mutated AML
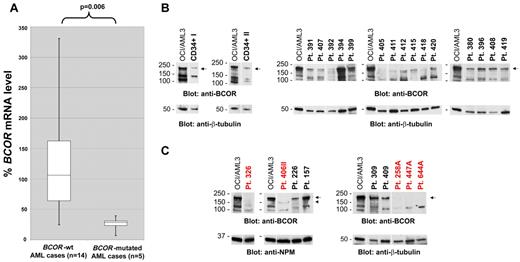
Because most BCOR mutations are predicted to trigger nonsense-mediated mRNA decay, we used quantitative RT-PCR to quantify BCOR mRNA levels in 5 patients harboring frame-shift mutations that introduced premature stop codons (before the second last exon) in comparison with 14 patients devoid of BCOR mutations. Interestingly, in all 5 BCOR-mutated AML patients, BCOR mRNA levels were substantially reduced to a mean of 22% of the levels detected in the 14 control AML patients (range, 6.9%-39.3%; Figure 3A; P = .006 by Wilcoxon 2-sample test of ΔCt values). This observation points to nonsense-mediated mRNA decay as one likely mechanism for BCOR gene inactivation by these mutations.
BCOR mRNA and protein expression in AML patients carrying BCOR disruptive mutations. (A) Box plots of BCOR mRNA levels quantified by real-time RT-PCR in 5 primary AML patients with BCOR disruptive mutations introducing premature stop codons (before the second last exon) compared with 14 BCOR-unmutated AML patients. All of the 5 BCOR-mutated AML patients display substantially decreased BCOR mRNA levels (P = .006 by Wilcoxon 2-sample test of ΔCt values). (B) Western blot analysis of BCOR protein expression in normal donor CD34+ hematopoietic cells (CD34+ I and CD34+ II, left panels) and in primary AML cells isolated from 15 patients with wild-type BCOR gene (right panels) showing a specific protein band of variable intensity corresponding to full-length BCOR (192 kDa predicted MW; arrow). (C) Western blot analysis of primary AML cells isolated from 5 patients harboring BCOR disruptive mutations (labeled in red: patient 326 with Thr733AlafsX5; patient 406II with Asn1485LysfsX5; patient 258A with Leu245ThrfsX19; patient 447A with Pro1115ThrfsX41; and patient 644A with His674MetfsX41). As a comparison, full-length BCOR expression in lysates from further 4 AML patients (patients 226, 157, 309, and 409) all devoid of BCOR mutations is shown. In the top panels, a specific protein band corresponding to full-length BCOR (192 kDa predicted MW) is observed (arrow) in wild-type BCOR AML, but not in BCOR-mutated AML. In patient 406II, a new faint band is detected (arrowhead) that likely corresponds to a truncated BCOR protein (162 kDa predicted MW). (B-C) The OCI/AML3 cell line not carrying BCOR mutations was used as a positive control for full-length BCOR protein expression. Protein lysate loading was evaluated by blotting the membranes with an anti-NPM1 or anti–β-tubulin Ab. Vertical lines have been inserted to indicate repositioned gel lanes.
BCOR mRNA and protein expression in AML patients carrying BCOR disruptive mutations. (A) Box plots of BCOR mRNA levels quantified by real-time RT-PCR in 5 primary AML patients with BCOR disruptive mutations introducing premature stop codons (before the second last exon) compared with 14 BCOR-unmutated AML patients. All of the 5 BCOR-mutated AML patients display substantially decreased BCOR mRNA levels (P = .006 by Wilcoxon 2-sample test of ΔCt values). (B) Western blot analysis of BCOR protein expression in normal donor CD34+ hematopoietic cells (CD34+ I and CD34+ II, left panels) and in primary AML cells isolated from 15 patients with wild-type BCOR gene (right panels) showing a specific protein band of variable intensity corresponding to full-length BCOR (192 kDa predicted MW; arrow). (C) Western blot analysis of primary AML cells isolated from 5 patients harboring BCOR disruptive mutations (labeled in red: patient 326 with Thr733AlafsX5; patient 406II with Asn1485LysfsX5; patient 258A with Leu245ThrfsX19; patient 447A with Pro1115ThrfsX41; and patient 644A with His674MetfsX41). As a comparison, full-length BCOR expression in lysates from further 4 AML patients (patients 226, 157, 309, and 409) all devoid of BCOR mutations is shown. In the top panels, a specific protein band corresponding to full-length BCOR (192 kDa predicted MW) is observed (arrow) in wild-type BCOR AML, but not in BCOR-mutated AML. In patient 406II, a new faint band is detected (arrowhead) that likely corresponds to a truncated BCOR protein (162 kDa predicted MW). (B-C) The OCI/AML3 cell line not carrying BCOR mutations was used as a positive control for full-length BCOR protein expression. Protein lysate loading was evaluated by blotting the membranes with an anti-NPM1 or anti–β-tubulin Ab. Vertical lines have been inserted to indicate repositioned gel lanes.
To assess the consequences of BCOR mutations at the protein level, 5 BCOR-mutated AML patients with available material for protein analysis were studied by Western blotting, and compared with CD34+ hematopoietic cells from 2 normal donors and 19 AML patients devoid of BCOR mutations. Both normal CD34+ hematopoietic cells and AML cells lacking BCOR mutations showed a clear band, of variable intensity, corresponding to the full-length BCOR protein (192 kDa predicted molecular weight [MW]; Figure 3B). In contrast, this band was absent in all the BCOR-mutated AML samples (Figure 3C). In one BCOR-mutated patient (patient 406II; Figure 3C), a new band corresponding to a lower-MW protein, which likely represents the new truncated protein product (predicted MW, 162 kDa), was detected at much lower intensity than the full-length BCOR protein observed in BCOR-unmutated AML patients. BCOR protein expression was therefore consistent with down-regulation of BCOR mRNA expression (Figure 3A). Therefore, at least in the 5 patients we investigated, BCOR mutations were associated with the absence of full-length BCOR and lack or low expression of a truncated BCOR protein.
BCOR mutations are mutually exclusive with NPM1 mutations
Because BCOR mutations appear to associate with CN-AML and NPM1 and CEBPA mutations define WHO 20082 provisional entities, we next investigated their relationship. None of the 10 CEBPA-mutated AML patients studied were found to carry BCOR mutations. We also analyzed 10 AML patients with MLL-PTD, another genetic lesion recurrently observed in CN-AML, and 1 patient was shown to harbor a BCOR mutation.
Associations between BCOR and NPM1 mutations were investigated in a total of 197 NPM1-mutated CN-AML patients (139 of the unselected Italian series and 58 German patients selected for this purpose). BCOR mutations were found only in 2 of 197 (1%) NPM1-mutated patients, as opposed to 24 of 213 (11.3%) NPM1-wild-type CN-AML patients (Fisher exact test P < .001; Table 3 block A). The same significant result was obtained when the Italian and German patients were analyzed separately (Table 3 blocks A1 and A2, respectively). These findings suggest that the simultaneous occurrence of NPM1 and BCOR mutations is uncommon.
Relationship of BCOR disruptive mutations with NPM1, FLT3-ITD, DNMT3A, and RUNX1 mutations
| Block . | BCOR status . | Patients, n . | P* . | ||
|---|---|---|---|---|---|
| Total (I+G) | NPM1 wt | NPM1 mut | |||
| A | Mutated | 26 | 24 | 2 | < .001 |
| Wild-type | 384 | 189 | 195 | ||
| Total (I) | NPM1 wt | NPM1 mut | |||
| A1 | Mutated | 10 | 9 | 1 | .006 |
| Wild-type | 240 | 102 | 138 | ||
| Total (G) | NPM1 wt | NPM1 mut | |||
| A2 | Mutated | 16 | 15 | 1 | .01 |
| Wild-type | 144 | 87 | 57 | ||
| Total (I) | NPM1 wt/FLT3 wt | NPM1 wt/FLT3-ITD | |||
| B | Mutated | 10 | 10 | 0 | .2 |
| Wild-type | 96 | 77 | 19 | ||
| Total (I + G) | NPM1 wt/DNMT3A wt | NPM1 wt/DNM3A mut | |||
| C | Mutated | 23 | 13 | 10 | .001 |
| Wild-type | 172 | 148 | 24 | ||
| Total (I) | NPM1 wt/DNMT3A wt | NPM1 wt/DNMT3A mut | |||
| C1 | Mutated | 9 | 5 | 4 | .063 |
| Wild-type | 102 | 85 | 17 | ||
| Total (G) | NPM1 wt/DNMT3A wt | NPM1 wt/DNMT3A mut | |||
| C2 | Mutated | 14 | 8 | 6 | .006 |
| Wild-type | 70 | 63 | 7 | ||
| Total (G) | NPM1 wt/RUNX1 wt | NPM1 wt/RUNX1 mut | |||
| D | Mutated | 18† | 10 | 8 | .07 |
| Wild-type | 86 | 67 | 19 | ||
| Block . | BCOR status . | Patients, n . | P* . | ||
|---|---|---|---|---|---|
| Total (I+G) | NPM1 wt | NPM1 mut | |||
| A | Mutated | 26 | 24 | 2 | < .001 |
| Wild-type | 384 | 189 | 195 | ||
| Total (I) | NPM1 wt | NPM1 mut | |||
| A1 | Mutated | 10 | 9 | 1 | .006 |
| Wild-type | 240 | 102 | 138 | ||
| Total (G) | NPM1 wt | NPM1 mut | |||
| A2 | Mutated | 16 | 15 | 1 | .01 |
| Wild-type | 144 | 87 | 57 | ||
| Total (I) | NPM1 wt/FLT3 wt | NPM1 wt/FLT3-ITD | |||
| B | Mutated | 10 | 10 | 0 | .2 |
| Wild-type | 96 | 77 | 19 | ||
| Total (I + G) | NPM1 wt/DNMT3A wt | NPM1 wt/DNM3A mut | |||
| C | Mutated | 23 | 13 | 10 | .001 |
| Wild-type | 172 | 148 | 24 | ||
| Total (I) | NPM1 wt/DNMT3A wt | NPM1 wt/DNMT3A mut | |||
| C1 | Mutated | 9 | 5 | 4 | .063 |
| Wild-type | 102 | 85 | 17 | ||
| Total (G) | NPM1 wt/DNMT3A wt | NPM1 wt/DNMT3A mut | |||
| C2 | Mutated | 14 | 8 | 6 | .006 |
| Wild-type | 70 | 63 | 7 | ||
| Total (G) | NPM1 wt/RUNX1 wt | NPM1 wt/RUNX1 mut | |||
| D | Mutated | 18† | 10 | 8 | .07 |
| Wild-type | 86 | 67 | 19 | ||
I indicates the Italian unselected CN-AML series; G, German selected CN-AML series comprising: 82 patients with the genotype NPM1wt/CEBPAwt/IDH1wt/FLT3-ITD– negative/MLL-PTD–negative, 58 cases with NPM1 mutations, 10 cases with MLL-PTD, and 10 selected cases with biallelic CEBPA mutations; wt, wild-type; and mut, mutated.
By Fisher exact test.
Including 3 NPM1wt/BCOR-mut cases of the Italian series analyzed for RUNX1 (2 mutated and 1 wild-type).
We then searched for a possible association of BCOR-mutated AML with FLT3-ITD in the unselected Italian cohort of 111 NPM1-unmutated CN-AML patients evaluated for BCOR, in which the FLT3-ITD status was available for 106 patients. Interestingly, FLT3-ITD was present in none of the 10 BCOR-mutated patients, as opposed to 19 of 96 (19.8%) BCOR-unmutated patients (Table 3 block B). By Fisher exact test, P was not significant (.2), likely because of the low total number of BCOR-mutated patients.
BCOR mutations associate with DNMT3A mutations
The finding that, like in the index patient, BCOR disruptive mutations clustered in a subgroup of CN-AML patients without NPM1 mutations, and that BCOR and DNMT3A mutations coexisted in the index patient, prompted us to investigate the recurrence of DNMT3A mutations among BCOR-mutated compared with BCOR-unmutated CN-AML patients (all devoid of NPM1 mutations). When considering the Italian and German series together, DNMT3A mutations were detected in 10 of 23 (43.5%) BCOR-mutated patients as opposed to 24 of 172 (13.9%) BCOR-unmutated patients, a statistically significant difference (P = .001 by Fisher exact test; Table 3 block C). A similar result was obtained when the Italian and German series were analyzed separately (Table 3 blocks C1 and C2, respectively).
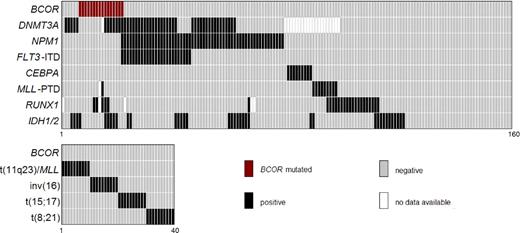
Because RUNX1 mutations play a role in CN-AML,26 we further investigated their relationship with BCOR mutations among CN-AML patients without NPM1 mutations (n = 104). This included 101 patients from the German series (15 with and 86 without BCOR mutations) and 3 of 10 Italian BCOR-mutated patients with material available for analysis. RUNX1 mutations were detected in 8 of 18 (44.4%) BCOR-mutated patients. In contrast, among the 86 BCOR-unmutated patients, only 22.1% (19/86) had RUNX1 mutations (P = .07 by Fisher exact test; Table 3 block D). The relationships between BCOR and other genetic mutations in the German selected series of 160 CN-AML patients for whom an extensive molecular genetic characterization was available are shown in Figure 4.
Correlation pattern between molecular mutations and karyotype. Data are given for associations between BCOR mutations and DNMT3A, NPM1, FLT3-ITD, CEBPA, MLL-PTD, RUNX1, and IDH1 and IDH2 mutations in 160 patients (top panel). The association between BCOR mutations and entity-defining cytogenetic abnormalities (40 patients) is shown in the bottom panel.
Correlation pattern between molecular mutations and karyotype. Data are given for associations between BCOR mutations and DNMT3A, NPM1, FLT3-ITD, CEBPA, MLL-PTD, RUNX1, and IDH1 and IDH2 mutations in 160 patients (top panel). The association between BCOR mutations and entity-defining cytogenetic abnormalities (40 patients) is shown in the bottom panel.
Associations between BCOR mutations and clinical parameters and survival
No significant associations were observed between BCOR disruptive mutations and sex, age, WBC count, hemoglobin, or platelets (supplemental Table 13). Analysis of the impact of BCOR disruptive mutations on the outcome of CN-AML (n = 160; German cohort) showed that BCOR-mutated patients had an inferior survival compared with the BCOR wild-type patients (n = 16 vs n = 144; alive at 2 years, 28.0% vs 66.3%; P = .024), and a trend for a worse event-free survival (n = 16 vs n = 144; event-free survival at 2 years, 12% vs 47.6%; P = .083). Further limiting this cohort to 82 patients with wild-type status for NPM1, FLT3-ITD, CEBPA, and MLL-PTD, a trend remained toward an inferior survival in BCOR-mutated patients compared with BCOR wild-type patients (n = 14 vs n = 68; alive at 2 years, 16.5% vs 57.1%; P = .111), and a worse event-free survival (n = 14 vs n = 68; event-free survival at 2 years, 0% vs 43.8%; P = .068).
In the Italian cohort (n = 262 patients; supplemental Table 12), no significant association was detected between BCOR disruptive mutations and overall survival. However, a significantly shorter event-free survival was observed in BCOR-mutated patients (n = 10 vs n = 252; event-free survival at 2 years, 0% vs 38.3%; P = .014; supplemental Table 13).
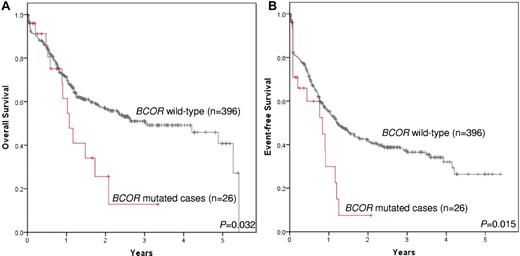
When combining the German and Italian cohorts, a significant association between BCOR-disruptive mutations and a shorter overall survival (n = 26 vs n = 396; alive at 2 years, 25.6% vs 56.7%; P = .032; Figure 5A) and event-free survival (n = 26 vs n = 396; event-free survival at 2 years, 7.5% vs 41.4%; P = .015; Figure 5B) was detected.
Impact of BCOR mutations on survival. Data are shown for overall survival (n = 26 vs n = 396; alive at 2 years, 25.6% vs 56.7%; P = .032; A) and event-free survival (n = 26 vs n = 396; event-free survival at 2 years, 7.5% vs 41.4%; P = .015; B).
Impact of BCOR mutations on survival. Data are shown for overall survival (n = 26 vs n = 396; alive at 2 years, 25.6% vs 56.7%; P = .032; A) and event-free survival (n = 26 vs n = 396; event-free survival at 2 years, 7.5% vs 41.4%; P = .015; B).
Other parameters significantly associated with survival in the combined cohort included age, hemoglobin, and NPM1 and FLT3 mutation status. In the multivariable analyses, BCOR disruptive mutations did reveal an independent association with event-free survival in the Italian cohort (P = .04), but not with overall survival in the different patient groups (supplemental Table 14). Further analyses on individual and combined cohorts are available online (supplemental Table 14).
Discussion
Next-generation sequencing technologies represent a promising method for discovering novel genetic lesions in human neoplasms. Recently, this approach identified recurrent mutations of IDH1 and DNMT3A in a significant proportion of CN-AML patients.11-13,34,35 To maximally increase the chance of identifying novel genetic lesions, we selected a patient with CN-AML for WES who was devoid of NPM1, CEBPA, and FLT3-ITD mutations and of MLL-PTD, and was subsequently found to be unmutated for IDH1. Using this strategy, we identified recurrent mutations affecting the DNMT3A and BCOR genes.
Whereas alterations of DNMT3A have been described previously in AML,13,34,35 we report here for the first time recurrent clonal and somatic BCOR mutations in AML. The BCOR gene is located on p11.4 of chromosome X and encodes an ubiquitously expressed nuclear protein,27,36 which was initially identified as a corepressor that interacts with the transcriptional repressor BCL627 through the BCOR-BCL6–binding domain (BCORBBD).37 Subsequently, BCOR has also been found to suppress the activity of transcription factors other than BCL6 by participating in the formation of large multiprotein complexes. BCOR is a key transcriptional regulator of early embryonic development,38 mesenchymal stem cell function,39 and hemopoiesis.38 Germline BCOR mutations are responsible for the OFCD syndrome,32 which is inherited in an X-linked pattern with presumed male lethality and is characterized by microphthalmia, dysmorphic appearance, dental abnormalities (radiculomegaly), hammer-toe deformity, and cardiac defects.32
Our novel finding of BCOR mutations in AML is in agreement with other studies pointing to a role of BCOR in leukemogenesis. BCOR was identified as a fusion partner of RAR-α in a single acute promyelocytic leukemia patient with 45,-Y,t(X;17)(p11;q12) and unique morphological and clinical features (ie, rectangular cytoplasmic bodies and multiple relapses after chemotherapy plus all-trans retinoic acid).28 Moreover, BCOR isoforms can interact with the mixed-lineage leukemia fusion partner AF9 (MLLT3) and modulate its transcriptional activity.40
The implication of BCOR mutations in AML is supported by their disruptive character: nonsense or conserved splice-site mutations and out-of-frame insertions/deletions introducing premature stop codons, which are scattered throughout the whole coding sequence. In both male and female AML patients, BCOR mutations consistently targeted the only functional allele. Moreover, BCOR mutations were associated with a decrease in BCOR mRNA levels, an absence of full-length BCOR protein, and a lack or low expression of a truncated BCOR protein. These features conform to those characteristics of a loss-of-function mutation in a tumor-suppressor gene. Finally, similar mutations are considered to inactivate BCOR gene function when they occur in the germline of patients with OFCD syndrome.32
The distribution of BCOR disruptive mutations among the cytogenetic/molecular categories of AML is also interesting. No such mutations were observed in patients with abnormal cytogenetics, including recurrent genetic lesions according to WHO 2008 (n = 96), as well as intermediate-risk and adverse karyotypes (n = 35). However, because of the low frequency of BCOR disruptive mutations and the relatively low number of patients analyzed for each cytogenetic category, a definitive picture of the frequency of such mutations among the entire spectrum of AML genotypes will require the study of additional patients.
Because the AML index patient used for WES carried a normal karyotype, we mainly focused our search for BCOR mutations in patients with a similar genomic profile. Interestingly, BCOR disruptive mutations were mostly enriched in the least characterized subgroup of CN-AML, those with germline NPM1, FLT3-ITD, IDH1, and MLL genes, mimicking the genotype of the AML index patient used for WES. The minimal overlap of BCOR and NPM1 mutations suggests a contribution of mutated BCOR to AML development through a pathway different from that mediated by mutated NPM1. Similarly, FLT3-ITD, a frequent genetic lesion in CN-AML, does not appear to play a major cooperating role in BCOR-mutated AML.
Conversely, our finding that approximately 50% of BCOR- mutated patients harbored mutations of the DNMT3A gene suggests that these 2 mutations may cooperate to induce AML, possibly acting through interference with epigenetic mechanisms. Interestingly, DNMT3A encodes for a methyltransferase enzyme catalyzing the addition of methyl groups to CpG dinucleotides.13 Moreover, BCOR augments transcriptional repression by interacting with class I and II HDACs, the polycomb group protein PCGF1/NSPC1, and the histone demethylase FBXL10 through its C-terminal region,41,42 which implies that BCOR may suppress gene transcription by epigenetic mechanisms.39,43 Association of BCOR disruptive mutations with mutations of the transcription factor RUNX1, a key regulator of hematopoiesis (44.4% of patients), is also interesting and warrants further investigations.
Finally, our findings may have clinical implications. Whereas prognostic assessment is possible for a significant proportion of CN-AML patients, based on analysis of NPM1, CEBPA, and FLT3 genes, a fraction of CN-AML patients still lack prognostic molecular biomarkers. In the present study, we demonstrate that BCOR mutations occur in CN-AML with limited overlap to other genetic mutations and that they may confer an inferior prognosis. Therefore, the analysis for BCOR mutations may improve the capabilities for prognostication in CN-AML, and its clinical value should be further investigated in larger studies.
The online version of this article contains a data supplement.
This work was presented in part by B.F. at the joint Japanese Society of Hematology (JSH)/European Hematology Association (EHA) Symposium during the EHA Congress in London, June 11, 2011.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors are grateful to Dr Geraldine A. Boyd for editing the paper; Claudia Tibidò for secretarial assistance; Dr Magne Osteras and Dr Nicolas Gonzalez for technical help in library preparation, exome capture, and massively parallel sequencing; and Dr Loich Baerlocher and Dr Julien Prados for excellent bioinformatic support. They also thank the following colleagues for providing samples and information about some AML patients included in the study: Dr Andreas Grüneisen, Vivantes Klinikum Neukölln, Berlin, Germany (n = 7 patients); Dr Francesco Mannelli, Institute of Hematology, University of Firenze, Italy (n = 7 patients); Prof Dr Yon-Dschun Ko, Johanniter Krankenhaus, Bonn, Germany (n = 5 patients); Prof Dr Winfried Gassmann, St Marien-Krankenhaus Siegen GmbH, Siegen, Germany (n = 5 patients); Dr Giuseppe Avvisati, Institute of Hematology, Campus Biomedico University, Rome, Italy (n = 6 patients); Dr Giovanna Meloni, Institute of Hematology, Università “La Sapienza” University, Rome, Italy (n = 6 patients); and Dr Alfonso Maria D'Arco, Institute of Hematology, Nocera Inferiore, Italy (n = 3 patients).
This work was supported by grants from the Associazione Italiana Ricerca Cancro (AIRC).
Authorship
Contribution: V.G. performed next-generation amplicon sequencing screening and gene expression profiling, and contributed to writing the manuscript; E.T. designed the WES strategy, selected BCOR as a gene to pursue, supervised the BCOR and DNMT3A mutation analysis in all Italian patients and the BCOR mRNA expression analysis, and wrote the manuscript; A.B.H., V.T., J.C., and R. Rabadan designed the SAVI algorithm, carried out the biostatistical analyses on the AML index patient, and contributed to writing the manuscript; A.K. contributed to next-generation amplicon sequencing screening, performed microarray analysis, and contributed to writing the manuscript; W.K., S. Schnittger, and C.H. characterized patient samples, including the index patient, according to molecular analyses, cytogenetics, FISH, and immunophenotyping, provided clinical information, and contributed to writing the manuscript; A.S.-R. analyzed all Sanger sequences from Italian AML patients; H.-U.K. and M.D. supported the microarray analysis and performed the GEO database submission; S. Schindela performed primer design and validation for next-generation deep sequencing; R.B., O.S., and A.R. from Northern Italy Leukemia Group provided leukemic samples for molecular analysis of BCOR and DNMT3A, information about the mutational status of other genes, and clinical data; V.A.W. performed Sanger sequencing validation of bioinformatic analyses; M.P.M. collected genotypic and clinical data of the Italian AML cohort, performed with R. Rossi all studies on protein expression of BCOR, and contributed to writing the manuscript; S.B. performed studies on mRNA expression of BCOR; L.D.C., G.S., F.D.R., F.F., M.S., and A.L. from various Italian hematologic centers provided patient samples and immunophenotypic, cytogenetic, molecular, and clinical information; K.G., H.S., R.P., K.-A.K., and D.O. collected data and clinical information; L.F. supervised the technical aspects of WES; L.P. supervised bioinformatic analysis validation, helped to organize the sequencing of all Italian patients, and contributed to writing the manuscript; T.H. performed cytomorphology, supervised the MLL cohort analyses, and contributed to writing the manuscript; and B.F. had the original idea for the study, led the project, and wrote the manuscript.
Conflict-of-interest disclosure: C.H., S. Schnittger, W.K., and T.H. have equity ownership of MLL Munich Leukemia Laboratory GmbH. A.K., V.G., and S. Schindela are employed by MLL Munich Leukemia Laboratory GmbH. The remaining authors declare no competing financial interests.
Correspondence: Prof Brunangelo Falini, Institute of Hematology, University of Perugia, Ospedale S Maria della Misericordia, S Andrea delle Fratte, 06132 Perugia, Italy; e-mail: faliniem@unipg.it.
References
Author notes
V.G. and E.T. contributed equally to this work.


![Figure 2. BCOR disruptive mutations in AML. (A) Schematic representation of the BCOR protein and its domains. (B) Type and distribution of BCOR disruptive mutations (out-of-frame small indels are depicted as rectangles, nonsense mutations as ovals, and consensus splice-site mutations as [splice site mut.]) along the BCOR coding exons (numbered boxes with a width proportional to the exon nucleotide length) and the exon/intron junctions (short horizontal black lines before and after each exon). Mutations are shown above or below the exons based on whether they were observed in the German or Italian cohorts, respectively. The number 2 appended to the Arg1547X oval denotes that this mutation was found in 2 distinct patients of the German cohort. (C) Chromatograms of a representative clonal, somatic out-of-frame indel (delC) observed by direct DNA Sanger sequencing in the BM sample of a female AML patient at disease onset (containing approximately 70% blast cells), but not in her matched normal sample taken at the time of complete remission.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/118/23/10.1182_blood-2011-07-365320/4/m_zh89991182870002.jpeg?Expires=1765897075&Signature=qk7ZcrfWW8r8Qyb1sJneKOzlsbveLbpET9fdq176TULOtFmTECP1NygoymBcsbERJf5nus0J5IjvhNRk9Nv98IUFNZaRsY4WR0gQSssfelcgUuA8HAWY1BLQm0ALZIT~PEUXj8IsbpLmqAYmsUzbuJA9zM37UvD8OL9aYzqVs-dV7Rq-trbWiisGJorYptchnRphDzwHLfP0-IN6VnD26hd88zPUubVaH2xCv-Xx69RILUnVxfApIkEGKpctZdv3Ep74UnI0vdFVKjRaZidVeMRhWrunIZJ0Wf4xVyAE3LpCnFM4I4vZ1897Q6yi03qCDzSiU9vBt5f8DJJKKTewHA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)