Abstract
Constitutive activation of FLT3 by internal tandem duplication (ITD) is one of the most common molecular alterations in acute myeloid leukemia (AML). FLT3/ITD mutations have also been observed in myelodysplastic syndrome patients both before and during progression to AML. Previous work has shown that insertion of an FLT3/ITD mutation into the murine Flt3 gene induces a myeloproliferative neoplasm, but not progression to acute leukemia, suggesting that additional cooperating events are required. We therefore combined the FLT3/ITD mutation with a model of myelodysplastic syndrome involving transgenic expression of the Nup98-HoxD13 (NHD13) fusion gene. Mice expressing both the FLT3/ITD and NHD13 transgene developed AML with 100% penetrance and short latency. These leukemias were driven by mutant FLT3 expression and were susceptible to treatment with FLT3 tyrosine kinase inhibitors. We also observed a spontaneous loss of the wild-type Flt3 allele in these AMLs, further modeling the loss of the heterozygosity phenomenon that is seen in human AML with FLT3-activating mutations. Because resistance to FLT3 inhibitors remains an important clinical issue, this model may help identify new molecular targets in collaborative signaling pathways.
Introduction
The FMS-like tyrosine kinase-3 (FLT3) receptor plays a critical role in early hematopoiesis. In humans, FLT3 is expressed in CD34+ hematopoietic stem/progenitor cells (HSPCs), with decreased expression in more differentiated cell types.1 Constitutive activation of FLT3 by internal tandem duplication (ITD) mutations is one of the most common molecular alterations in acute myeloid leukemia (AML), occurring in approximately 25% of adult AML cases and 15% of pediatric AML cases.2 The presence of FLT3/ITD mutations is associated with decreased overall survival in both adult and pediatric AML patients.3-5 To understand the roles of FLT3/ITD mutations in acute leukemia, we have previously generated a knock-in mouse model in which an 18-bp ITD mutation, isolated from a patient with AML, was inserted into the homologous juxtamembrane domain of the murine Flt3 gene.6 Heterozygous Flt3WT/ITD mice develop a myeloproliferative neoplasm (MPN) and die within 6 to 20 months. However, no signs of acute leukemia are observed over the lifetime of these mice, indicating that additional cooperating events are required for leukemic progression.
In keeping with FLT3's crucial role in hematopoiesis, FLT3 mutations are associated not only with acute leukemia, but also with myelodysplastic syndrome (MDS). MDS is a group of clonal disorders characterized by ineffective hematopoiesis, refractory cytopenias, and frequent progression to leukemia.7 Genetic data provide evidence that MDS and the later development of leukemia in these patients are characterized by a step-wise genetic progression.8,9 In a study of 82 MDS patients who went on to develop myeloid leukemias, 6% of patients had FLT3/ITD mutations at the time of presentation with MDS and an additional 10% of patients acquired FLT3 mutations during the progression to AML. Patients with FLT3/ITD mutations progressed more rapidly to AML than those without the mutation and had a significantly shorter survival. These studies indicate that the acquisition of FLT3 mutations is an important driver in the transformation of MDS to AML.
Another group of mutations that are involved in the development of hematopoietic neoplasms are translocations involving the gene NUP98. NUP98 has been observed in fusions with at least 20 different partner genes, the most common of which are the Adb-type HOX genes.10,11 These translocations appear to result in the dysregulation of several genes regulating development and are associated with a variety of hematopoietic disorders, including MDS, AML, T-cell acute lymphoblastic leukemia, and chronic myeloid leukemia.12 Oncogenic fusions involving Nup98 and HoxD13 have been detected in patients developing acute myelomonocytic and erythroid leukemias.13,14 We have previously reported a transgenic model in which the Nup98-HoxD13 (NHD13) fusion gene was expressed under control of hematopoietic-specific vav regulatory elements.15 These mice developed a highly penetrant disease with the phenotype of MDS, including ineffective hematopoiesis characterized by cytopenias and dysplasia, and eventual progression to leukemia. However, as was seen in the murine model of FLT3/ITD, these NHD13 mice had a long latency before the development of overt disease, suggesting the need for additional cooperative events.
There is increasing clinical and experimental evidence that FLT3 activating mutations are associated with NUP98 translocations and other mutations regulating HOX gene expression.16,17 Patients with leukemias expressing high levels of HOX mRNA correlate with higher levels of FLT3 mRNA and an increased incidence of FLT3 activating mutations.18,19 The association between FLT3 mutations and translocations involving NUP98 was supported by a recent study in which a high rate of FLT3/ITD mutations (56%) was observed in patients with NUP98-related leukemias, regardless of the specific fusion partners.20 In addition, the overexpression of WT FLT3 has been found to induce AMLs in mice expressing Nup98-Hox fusions. In contrast to the overexpression of WT FLT3, the use of a knock-in model of a FLT3 activating mutation may provide a more accurate model of human disease because studies of cell lines and primary cells with FLT3 activating mutations have suggested that the constitutively activated receptor may have different signaling properties compared with WT FLT3.21-23 This model also has potential advantages over retroviral expression models in that the FLT3/ITD allele is expressed under control of the endogenous promoter, thus ensuring expression at the appropriate levels and in the proper cell populations.
Despite the clear association between FLT3-activating mutations and NUP98 translocations, the molecular mechanism of the interaction between these 2 proteins has not been explored. To further characterize this interaction, we generated a mouse that expresses both FLT3/ITD and the NHD13 fusion protein.
Methods
Mice
FLT3/ITD knock-in mice were generated as previously reported.6 Mice were bred with CMV-cre transgenic mice (The Jackson Laboratory) to induce excision of the PGK-Neo cassette. Nup98-HoxD13 mice were generated as previously reported.15 All mice were housed in microisolator cages in a pathogen-free animal facility. Primers used to confirm the presence of the FLT3/ITD and NHD13 mutations are given in supplemental Methods (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). All animal experiments were performed according to protocols approved by the Animal Care and Use Committee of Johns Hopkins University in accordance with guidelines set forth by the National Institutes of Health.
Complete peripheral blood cell count and cytology
A total of 50 μL of peripheral blood was collected from murine retro-ocular vessels and subjected to complete blood cell counting, and a white blood cell (WBC) differential was performed manually or using the Hemavet950 system (Drew Scientific). Peripheral blood smears and bone marrow cytospins were visualized using a modified Wright-Giemsa stain (Sigma-Aldrich). Representative histopathology images were acquired using a Zeiss Axioskop upright microscope system (Carl Zeiss).
Flow cytometric analysis and cell sorting
Flow cytometric analysis and cell sorting were performed as described previously.6 For hematopoietic stem/progenitor cell staining, bone marrow was isolated from 2-month-old wild-type (WT), NHD13, FLT3/ITD, and FLT3/ITD-NHD13 littermates and stained with a modified lineage cocktail of 6 μL each CD3, Ter119, CD19, and Gr-1 antibodies. Phenotypic definitions used to define the compartments are: KSL: Lin−Sca-1hic-KIThi; LT-HSC: Lin−Sca-1hic-KIThiCD34−CD135−; ST-HSC: Lin−Sca-1hic-KIThiCD34+CD135−; MPP: Lin−Sca-1hic-KIThiCD34+CD135+; CMP: Lin−Sca-1−c-KIThiCD34+CD16mid; GMP: Lin−Sca-1−c-KIThiCD34+CD16hi; GMP-L: Lin−Sca-1−c-KIThiCD34−CD16hi; MEP: Lin−Sca-1−c-KIThiCD34−CD16−. All data were analyzed by FACSDIVA (BD Biosciences) or FlowJo Version 9.3.3 analysis software (TreeStar). A detailed record of antibodies used is available in supplemental Methods.
Quantitative RT-PCR analysis
Quantitative RT-PCR was performed using an iCycler iQ multicolor real-time PCR system (Bio-Rad). The levels of transcripts were normalized based on that of S16. The primer sequences used for Hox gene and Meis1 quantitative PCR are listed in supplemental Table 1.
Limiting dilution transplantation experiments
Transplantation of leukemic FLT3/ITD-NHD13 bone marrow was performed as described previously.6 Limiting dilution experiments consisted of cohorts of mice transplanted with 1 × 106, 1 × 105, 1 × 104, 1 × 103, 100, or 10 bulk bone marrow cells with 5 × 105 CD45.1 total bone marrow helper cells injected for each mouse. The frequency of leukemic initiating cells (LICs) in each population was calculated using L-Calc Version 1.1 software (StemCell Technologies).
Colony formation assays and liquid culture
Sorted B220+CD19−Mac-1−/Gr-1− precursors were plated at a concentration of 1 × 104 cells per 35-mm dish in M3434 methylcellulose (StemCell Technologies). Colonies were scored after 7 days incubation at 37°C and 5% CO2. This population was grown in liquid cultures in RPMI 20% FBS supplemented with 10 ng/mL IL-3, 10 ng/mL IL-6, and 100 ng/mL SCF (PeproTech) for 2 weeks. Functional macrophage activity was evaluated by imaging the phagocytosis of FITC-labeled Saccharomyces cerevisiae at room temperature using a Nikon Eclipse E600 microscope system.
Myeloperoxidase expression
Myeloperoxidase expression assay was performed according to protocols described previously.24 Cells were resuspended in FACS buffer and analyzed on the FACSAria II.
Sorafenib treatment and Western blotting
Cells derived from the spleen of leukemic mice were treated for 30 minutes with either 10nM or 20nM sorafenib. Cells were lysed with CelLytic M lysis reagent (Sigma-Aldrich) containing 1:100 P8340 protease inhibitory cocktail and 1mM sodium orthovanadate (Sigma-Aldrich). A detailed record of antibodies used is available in supplemental Methods. In vivo sorafenib treatment of transplant recipients was initiated 4 days after injection of primary leukemic cells. Mice were given 10 mg/kg sorafenib or vehicle once a day for 2 months.
FLIVO assay
In vivo apoptosis was evaluated using the FAM-FLIVO Apoptosis Kit (ImmunoCytochemistry Technologies). Mice were injected with 100 μL of 1× FLIVO reagent by tail vein injection. After 1 hour, mice were killed and bone marrow was subjected to flow cytometric analysis.
Statistical analysis
Data are expressed as the mean ± SEM or SD, where applicable. Differences between groups were analyzed by Student t test. Values of P < .05 were considered to be significant.
Results
FLT3/ITD and Nup98-HoxD13 mutations cooperate to induce acute leukemia with high penetrance
Heterozygous double-transgenic mice expressing the knock-in FLT3/ITD mutation and NHD13 transgene were generated on both C57Bl/6N and FVB/N backgrounds (Figure 1A-B). Offspring were genotyped using PCR amplification of the NHD13 transgene and FLT3/ITD mutations from tail DNA (Figure 1C). The FLT3/ITD-NHD13 mice on the FVB/N background developed acute leukemia with 100% penetrance and a mean survival of 95 ± 32 days (Figure 1D) compared with 281 ± 94 days and 372 ± 84 days for the NHD13 alone and FLT3/ITD alone mice, respectively (both P < .0001, n = 20, log-rank test). FLT3/ITD-NHD13 mice generated on the C57Bl/6N background developed leukemia with a longer latency of 143 ± 37 days (n = 20) but still had a significantly shorter survival compared with the single mutants alone; 260 ± 75 days and 332 ± 68 days for the NHD13 alone and FLT3/ITD alone mice, respectively (P < .0001, n = 20, Figure 1E).
FLT3/ITD and NHD13 mutations cooperate to induce acute leukemia with high penetrance. (A) Schematic of FLT3/ITD knock-in construct and NHD13 transgene. An 18-bp sequence derived from a patient with AML was inserted into exon 14 of murine Flt3. (B) The Nup98-HoxD13 transgene contains the N-terminal GLFG repeats and the DNA binding homeodomain of HoxD13. (C) Examples of PCR genotyping results to detect the FLT3/ITD mutation and presence of the NHD13 transgenes from tail snip DNA. (D-E) Kaplan-Meier survival curves of WT, NHD13, FLT3/ITD, and FLT3/ITD-NHD13 mice (n = 20 in each group) for both the FVB/N and C57Bl/6N strains.
FLT3/ITD and NHD13 mutations cooperate to induce acute leukemia with high penetrance. (A) Schematic of FLT3/ITD knock-in construct and NHD13 transgene. An 18-bp sequence derived from a patient with AML was inserted into exon 14 of murine Flt3. (B) The Nup98-HoxD13 transgene contains the N-terminal GLFG repeats and the DNA binding homeodomain of HoxD13. (C) Examples of PCR genotyping results to detect the FLT3/ITD mutation and presence of the NHD13 transgenes from tail snip DNA. (D-E) Kaplan-Meier survival curves of WT, NHD13, FLT3/ITD, and FLT3/ITD-NHD13 mice (n = 20 in each group) for both the FVB/N and C57Bl/6N strains.
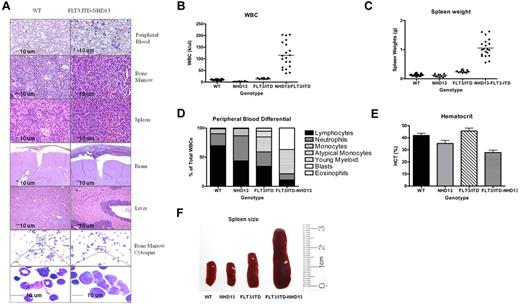
Mice were killed when they presented with ruffled coats and lethargy. Diagnosis of the hematologic disease of sick mice was determined by flow cytometry, modified Wright-Giemsa staining of the peripheral blood and bone marrow, and H&E staining of organs (Figure 2A-E). Analysis of blood, bone marrow, and organs revealed numerous blasts circulating in the peripheral blood, sheets of blasts replacing the marrow, and substantial leukemic infiltrates in the spleen, liver, and brain (Figure 2A). Peripheral blood counts obtained from 3-month old littermates demonstrated elevated WBC counts for the FLT3/ITD-NHD13 mice (115.2 ± 55.8 ×109/L) compared with WT (9.7 ± 2.0 ×109/L), NHD13 (3.2 ± 1.7 ×109/L), and FLT3/ITD (13.4 ± 2.5 ×109/L) littermate groups (Figure 2B). The spleen weights were significantly higher in the FLT3/ITD-NHD13 mice (1.05 ± 0.3 g, P < .0001, n = 20) compared with WT (0.130 ± 0.04 g, n = 10), NHD13 (0.117 ± 0.04 g, n = 10), or FLT3/ITD (0.239 ± 0.04 g, n = 10) mice at 3 months (Figure 2C). WBC differential analysis demonstrated that the FLT3/ITD-NHD13 mice had significantly elevated fractions of atypical monocytes and blasts, with a decreased percentage of lymphocytes and neutrophils (Figure 2D) and a reduced hematocrit compared with WT littermates (Figure 2E). An example of the typical spleen sizes of each genotype is shown (Figure 2F).
Organ histology and peripheral blood counts at 3-month time point. (A) The peripheral blood in FLT3/ITD-NHD13 mice contains numerous circulating blasts, and the normal bone marrow is replaced by sheets of blasts. Leukemic infiltrates are present in the spleen, brain, and liver. Bone marrow cytospins in the FLT3/ITD-NHD13 mice show abundant agranular blasts that have a scant to moderate amount of basophilic cytoplasm. Images for the modified Wright-Giemsa stain were acquired at room temperature using a Zeiss Axioskop upright microcope system (Carl Zeiss) with Achroplan 5×/0.16 NA, 10×/0.3 NA, and 40×/0.6 NA objectives and were photographed with an AxioCam camera (Carl Zeiss) and Axiovision Version 4.0 software (Carl Zeiss). (B) Comparison of WBC counts, (C) spleen weights, (D) differential count, (E) and hematocrit. (F) Example of spleen size in 3-month-old littermates representing each genotype.
Organ histology and peripheral blood counts at 3-month time point. (A) The peripheral blood in FLT3/ITD-NHD13 mice contains numerous circulating blasts, and the normal bone marrow is replaced by sheets of blasts. Leukemic infiltrates are present in the spleen, brain, and liver. Bone marrow cytospins in the FLT3/ITD-NHD13 mice show abundant agranular blasts that have a scant to moderate amount of basophilic cytoplasm. Images for the modified Wright-Giemsa stain were acquired at room temperature using a Zeiss Axioskop upright microcope system (Carl Zeiss) with Achroplan 5×/0.16 NA, 10×/0.3 NA, and 40×/0.6 NA objectives and were photographed with an AxioCam camera (Carl Zeiss) and Axiovision Version 4.0 software (Carl Zeiss). (B) Comparison of WBC counts, (C) spleen weights, (D) differential count, (E) and hematocrit. (F) Example of spleen size in 3-month-old littermates representing each genotype.
Peripheral blood samples taken from FLT3/ITD-NHD13 mice over time indicate that these mice develop abnormal WBC counts (> 20 ×109/L) at 8 to 10 weeks of age. To determine whether these mice progress through an MDS-like stage before the development of leukemia, cohorts of 4 mice per genotype were examined for dysplastic changes in the peripheral blood and bone marrow at one month of age. Peripheral blood smears and bone marrow cytospins showed marked polychromasia, hypersegmented neutrophils, hypolobated megakaryocytes, and multinucleated erythroid precursors in both the NHD13 and FLT3/ITD-NHD13 mice (supplemental Figure 1Ai-iv). Peripheral blood counts indicated a decrease in total WBC, monocyte, and lymphocyte counts in the NHD13 alone mice, with the FLT3/ITD-NHD13 mice showing an intermediate count between the FLT3/ITD and NHD13 groups (supplemental Figure 1B-D). An assay for in vivo apoptosis indicated that the NHD13 group had the highest percentage of apoptotic cells, with FLT3/ITD having significantly fewer apoptotic cells and FLT3/ITD-NHD13 having levels comparable to WT (supplemental Figure 1E). These results indicate that the FLT3/ITD-NHD13 mice do progress through a phase with many of the features of MDS but appear to have a less severe phenotype than what is observed in the NHD13 alone mice.
Mice with FLT3/ITD-NHD13 mutations develop AML
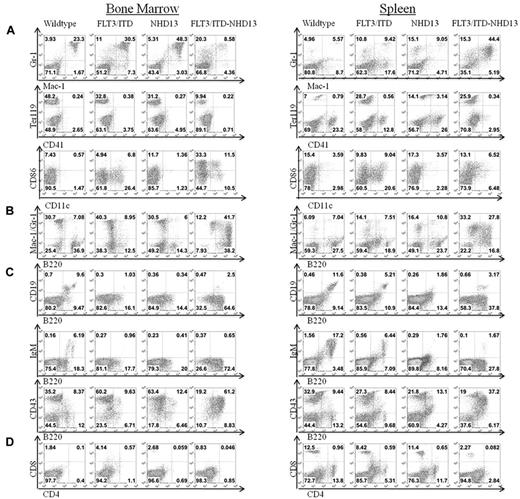
Flow cytometric analysis was performed on bone marrow and splenocytes from mice of each genotype to characterize the immunophenotype of the disease (Figure 3). Cells were analyzed for the surface expression of mature myeloid markers, including granulocyte/monocyte (Gr-1, Mac-1), erythrocyte (Ter119, CD41a), and dendritic cell markers (CD86, CD11c; Figure 3A). FLT3/ITD-NHD13 mice had decreased numbers of Mac-1+/Gr-1+ cells compared with WT or single mutants but showed an increase in the Gr-1 single positive population, indicating an increase in granulocyte and/or monocyte precursors. FLT3/ITD-NHD13 mice had a decreased Ter119+CD41− population in the bone marrow, which includes mature erythrocytes and erythroid precursor cells, but an increase in this population in the spleen of both FLT3/ITD and FLT3/ITD-NHD13 mice, suggesting extramedullary hematopoiesis. The most striking finding, however, was the increase in expression of the receptor CD45R/B220 in all FLT3/ITD-NHD13 mice. B220+ cells accounted for 75% to 90% of the bulk marrow population, with 40% of them additionally expressing the myeloid markers Mac-1 or Gr-1 (Figure 3B). Because B220 is commonly used as a marker of B-cell development, bone marrow and spleen were stained for markers of B-cell maturation (CD19, IgM; Figure 3C). Despite the significant increase in B220 expression, there were decreased B220+CD19+ or B220+IgM+ cells in both the bone marrow and spleen of FLT3/ITD-NHD13 mice compared with WT mice. Lymphocyte development was also examined by staining for markers of T-cell development (CD4/CD8; Figure 3D). FLT3/ITD-NHD13 mice had decreased numbers of mature T cells in both the bone marrow and spleen. Taken together, the flow cytometric analysis indicates an AML with high expression of B220 and defective lymphoid development. Cell sorting and transplantation were used to further define the identity of the leukemia-initiating population.
Mice with FLT3/ITD-NHD13 mutations develop AML. Comparison of cell surface staining of FLT3/ITD-NHD13 mice to WT, NHD13, and FLT3/ITD littermates at a 3-month time point. Bone marrow and spleen were evaluated for the expression of (A) mature myeloid markers, (B) coexpression of Mac-1/Gr-1 and B220, (C) B-cell markers, and (D) T-cell markers.
Mice with FLT3/ITD-NHD13 mutations develop AML. Comparison of cell surface staining of FLT3/ITD-NHD13 mice to WT, NHD13, and FLT3/ITD littermates at a 3-month time point. Bone marrow and spleen were evaluated for the expression of (A) mature myeloid markers, (B) coexpression of Mac-1/Gr-1 and B220, (C) B-cell markers, and (D) T-cell markers.
Limiting dilution transplantation of leukemic cells reveals the frequency and cell surface expression of the leukemia-initiating population
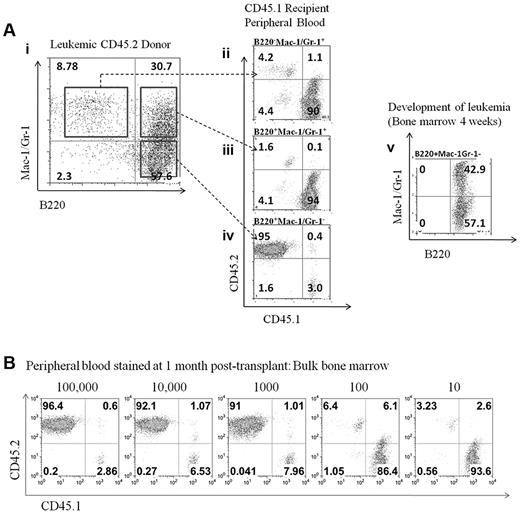
To determine which population within the bone marrow could transplant the disease, bulk bone marrow was sorted into 3 populations (B220+Mac-1+/Gr-1+, B220+Mac-1−/Gr-1−, and B220−Mac-1+/Gr-1+; Figure 4Ai). The ability of these cells to competitively repopulate the bone marrow was evaluated. Only the B220+Mac-1−/Gr-1− population was capable of significant engraftment as evaluated by the percentage of CD45.2+ cells in the peripheral blood 1 month after transplantation (Figure 4Aiv). The development of leukemia in mice receiving the B220+Mac-1−/Gr-1− was evaluated by flow cytometry and recapitulated the features of the transplant donor; more than 90% of cells expressed B220, and both the B220+Mac-1+/Gr-1+and B220+Mac-1−/Gr-1− populations were present (Figure 4Av). A summary of 3 independent transplant experiments is given in Table 1. Limiting dilution transplantation to determine the frequency of the LIC in the bulk bone marrow was performed (Figure 4B). The frequency of LICs was calculated from 3 independent transplantation experiments according to Poisson statistics using the L-Calc program and determined to be 1:200 bulk bone marrow cells. Because the B220+Mac-1−/Gr-1− population includes the multipotent progenitor (MPP), common myeloid progenitor (CMP), granulocyte/macrophage progenitor (GMP), granulocyte/macrophage progenitor-like (GMP-L), and megakaryocyte/erythroid progenitor (MEP) populations (supplemental Figure 2), each fraction was isolated by cell sorting and was transplanted to recipient mice to determine the potential to engraft (Table 2). The highest engraftment potential was found in the MPP population (1:20 cells), with the CMP and GMP populations having a reduced ability to engraft and generate leukemia (1:4000 and 1:7000 cells, respectively). The MEP, GMP-L, and B220+CD19+ populations did not engraft in any recipient mice.
Limiting dilution transplantation of leukemic cells reveals the frequency and cell surface expression of the leukemic initiating population. (Ai) Sorting of populations from bulk bone marrow in a leukemic donor mouse. Bulk bone marrow was sorted into 3 populations (B220+Mac-1+/Gr-1+, B220+Mac-1−/Gr-1−, and B220−Mac-1+/Gr-1+) and transplanted to lethally irradiated recipients. (Aii-iv) Peripheral blood from transplant recipients evaluated for the CD45.2/CD45.1 ratio at 1 month after transplantation. (Av) Development of leukemia in the bone marrow of mice receiving the B220+Mac-1−/Gr-1− population. (B) Limiting dilution transplantation was used to determine the frequency of the LIC in bulk bone marrow.
Limiting dilution transplantation of leukemic cells reveals the frequency and cell surface expression of the leukemic initiating population. (Ai) Sorting of populations from bulk bone marrow in a leukemic donor mouse. Bulk bone marrow was sorted into 3 populations (B220+Mac-1+/Gr-1+, B220+Mac-1−/Gr-1−, and B220−Mac-1+/Gr-1+) and transplanted to lethally irradiated recipients. (Aii-iv) Peripheral blood from transplant recipients evaluated for the CD45.2/CD45.1 ratio at 1 month after transplantation. (Av) Development of leukemia in the bone marrow of mice receiving the B220+Mac-1−/Gr-1− population. (B) Limiting dilution transplantation was used to determine the frequency of the LIC in bulk bone marrow.
Summary of transplanted populations and fraction of mice developing leukemia
| Population transplanted . | Fraction of recipients developing leukemia (latency in days) . |
|---|---|
| Group 1: B220−Mac-1+Gr-1+ | 0/20 |
| Group 2:B220+Mac-1+Gr-1+ | 0/20 |
| Group 3: B220+Mac-1−/Gr-1− | 20/20 (30 ± 2) |
| Population transplanted . | Fraction of recipients developing leukemia (latency in days) . |
|---|---|
| Group 1: B220−Mac-1+Gr-1+ | 0/20 |
| Group 2:B220+Mac-1+Gr-1+ | 0/20 |
| Group 3: B220+Mac-1−/Gr-1− | 20/20 (30 ± 2) |
Data represent 3 independent experiments.
Summary of transplanted population, development of leukemia, and frequency of leukemic-initiating population
| Experimental design . | No. of mice developing leukemia (latency in days) . | |||||||
|---|---|---|---|---|---|---|---|---|
| No. of cells transplanted . | No. of mice per group . | Bulk . | MPP . | CMP . | GMP . | GMP-L . | MEP . | B220+CD19+ . |
| 1 000 000 | 2 | 2 (27 ± 2) | 2 (21 ± 2) | 2 (33 ± 1) | 2(33 ± 0) | 0 | 0 | 0 |
| 100 000 | 4 | 4 (33 ± 1) | 4 (35 ± 1) | 4 (34 ± 0) | 4 (42 ± 2) | 0 | 0 | 0 |
| 10 000 | 6 | 6 (39 ± 2) | 6 (33 ± 1) | 6 (76 ± 13) | 5 (61 ± 8) | 0 | 0 | 0 |
| 1000 | 6 | 6 (40 ± 1) | 6 (42 ± 7) | 0 | 0 | 0 | 0 | 0 |
| 100 | 6 | 2 (152 + 20) | 6 (71 ± 3) | 0 | 0 | 0 | 0 | 0 |
| 10 | 6 | 0 | 2 (85 ± 5) | 0 | 0 | 0 | 0 | 0 |
| Frequency of LIC in sorted population | 1:234 | 1:22 | 1:4,341 | 1:7,212 | NA | NA | NA | |
| Experimental design . | No. of mice developing leukemia (latency in days) . | |||||||
|---|---|---|---|---|---|---|---|---|
| No. of cells transplanted . | No. of mice per group . | Bulk . | MPP . | CMP . | GMP . | GMP-L . | MEP . | B220+CD19+ . |
| 1 000 000 | 2 | 2 (27 ± 2) | 2 (21 ± 2) | 2 (33 ± 1) | 2(33 ± 0) | 0 | 0 | 0 |
| 100 000 | 4 | 4 (33 ± 1) | 4 (35 ± 1) | 4 (34 ± 0) | 4 (42 ± 2) | 0 | 0 | 0 |
| 10 000 | 6 | 6 (39 ± 2) | 6 (33 ± 1) | 6 (76 ± 13) | 5 (61 ± 8) | 0 | 0 | 0 |
| 1000 | 6 | 6 (40 ± 1) | 6 (42 ± 7) | 0 | 0 | 0 | 0 | 0 |
| 100 | 6 | 2 (152 + 20) | 6 (71 ± 3) | 0 | 0 | 0 | 0 | 0 |
| 10 | 6 | 0 | 2 (85 ± 5) | 0 | 0 | 0 | 0 | 0 |
| Frequency of LIC in sorted population | 1:234 | 1:22 | 1:4,341 | 1:7,212 | NA | NA | NA | |
NA indicates not applicable.
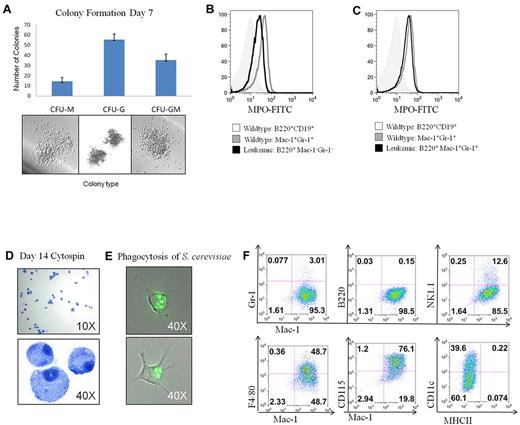
Because leukemia stem cells have been described that have both myeloid and lymphoid differentiation potential, the transplantable B220+Mac-1−/Gr-1− population was further analyzed to determine whether it had features of immature myeloid and/or B-lymphoid cells. Although this population expressed TdT and had early D to J immunoglobulin rearrangements, the cells did not display any B-cell specific transcription factor expression, cell surface markers consistent with early immature B cells, or show any signs of V-DJ rearrangements, which are definitive for B-lymphoid cells (supplemental Figure 3). Sorted B220+Mac1−/Gr-1− cells did generate myeloid colonies (CFU-M, CFU-G, and CFU-GM) when cultured in myeloid methylcellulose conditions (Figure 5A) but failed to generate B-cell colonies when cultured under lymphoid culture conditions despite multiple attempts (data not shown). This population also failed to generate CFU-GEMM colonies despite giving rise to other early myeloid colonies. The B220+Mac-1−/Gr-1− LICs expressed the myeloid specific enzyme myeloperoxidase (Figure 5B), with even higher expression in the more differentiated B220+Mac-1+/Gr-1+ population (Figure 5C). In addition, after 2 weeks in liquid culture, these cells generated mature macrophages expressing Mac-1, F4/80, and CD115 and were functional as assessed by their ability to phagocytose Saccharomyces cerevisiae (Figure 5D-F). Taken together, these results indicate that, although these leukemias initiate from a cell with the phenotypic properties of a multipotent progenitor, the LICs in this model only have the capacity for myeloid differentiation.
Analysis of myeloid lineage properties and in vitro differentiation potential. (A) Sorted B220+Mac1−/Gr-1− cells were evaluated for the ability to generate myeloid colonies in M3434 methylcellulose. (B) MPO expression in the B220+Mac-1−/Gr-1−, compared with WT controls. (C) MPO expression in the B220+Mac-1+/Gr-1+ population compared with WT controls. (D) B220+Mac-1−/Gr-1− cells from FLT3/ITD-NHD13 mice were grown in liquid culture with myeloid cytokines and evaluated for differentiation ability and (E) functional activity. Images were acquired at room temperature using a Nikon TE 2000-E microscope system (Nikon) with a Nikon Plan APO VC 100×/1.40 oil objective and Nikon EZ-C1 Version 3.5 software. (F) Cells differentiated in liquid culture were stained for markers of macrophage differentiation.
Analysis of myeloid lineage properties and in vitro differentiation potential. (A) Sorted B220+Mac1−/Gr-1− cells were evaluated for the ability to generate myeloid colonies in M3434 methylcellulose. (B) MPO expression in the B220+Mac-1−/Gr-1−, compared with WT controls. (C) MPO expression in the B220+Mac-1+/Gr-1+ population compared with WT controls. (D) B220+Mac-1−/Gr-1− cells from FLT3/ITD-NHD13 mice were grown in liquid culture with myeloid cytokines and evaluated for differentiation ability and (E) functional activity. Images were acquired at room temperature using a Nikon TE 2000-E microscope system (Nikon) with a Nikon Plan APO VC 100×/1.40 oil objective and Nikon EZ-C1 Version 3.5 software. (F) Cells differentiated in liquid culture were stained for markers of macrophage differentiation.
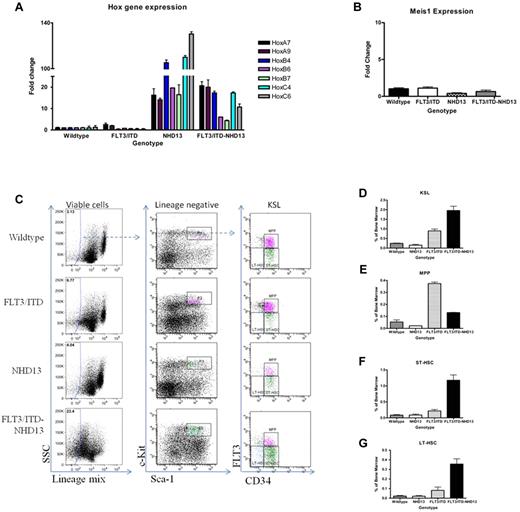
Mice with FLT3/ITD-NHD13 mutations demonstrate altered Hox gene expression and stem cell expansion
Translocations involving Nup98 and a homeodomain-containing fusion partner have been shown to cause dysregulation of homeobox genes, either through direct binding or indirect mechanisms. To study the Hox gene expression levels that might contribute in this leukemia model, RNA was extracted from the total bone marrow of mice before development of disease. A 2-month-old time point was selected based on the fact that at this age FLT3/ITD-NHD13 mice have an average WBC count of less than 20 ×109/L, normal numbers of blasts in the bone marrow, and no evidence of leukemic infiltration of organs. A list of candidate Hox genes was generated based on microarray results (data not shown) comparing bulk bone marrow from NHD13 and WT mice, and analyzed by quantitative RT-PCR (Figure 6A). The expression level of the Hox gene regulator Meis1 was evaluated because this gene is frequently up-regulated in patients with AML (Figure 6B). Fold changes relative to the WT control mice were determined, and a Student t test was used to identify statistically significant changes in gene expression. HoxA7, HoxA9, HoxB4, HoxB6, HoxC4, and HoxC6 were all up-regulated in the FLT3/ITD-NHD13 mice compared with the age-matched WT or FLT3/ITD mice. The overexpression of these genes appeared to be primarily driven by the NHD13 transgene, although there was also an increase in HoxA7 and HoxA9 expression in the FLT3/ITD mice. There was no significant change in Meis1 expression levels observed in the FLT3/ITD-NHD13 mice compared with the WT, NHD13, or FLT3/ITD alone mice.
Mice with FLT3/ITD-NHD13 mutations demonstrate altered Hox gene expression and stem cell expansion. (A) Quantitative RT-PCR analysis was performed on RNA extracted from the total bone marrow of 2-month-old WT/NHD13, FLT3/ITD, and FLT3/ITD-NHD13 C57Bl/6N littermates; Hox gene expression represents an average of 4 mice per genotype. (B) Meis1 expression of 2-month-old WT/NHD13, FLT3/ITD, and FLT3/ITD-NHD13 C57Bl/6N littermates. (C) Examples of sorted HSPC populations isolated from bone marrow are given for mice of each genotype. Bone marrow was stained for the cell surface markers characterizing the (D) KSL, (E) MPP, (F) ST-HSC, and (G) LT-HSC populations. Values in the graphs represent average percentages of these populations in the bone marrow from 3 independent experiments.
Mice with FLT3/ITD-NHD13 mutations demonstrate altered Hox gene expression and stem cell expansion. (A) Quantitative RT-PCR analysis was performed on RNA extracted from the total bone marrow of 2-month-old WT/NHD13, FLT3/ITD, and FLT3/ITD-NHD13 C57Bl/6N littermates; Hox gene expression represents an average of 4 mice per genotype. (B) Meis1 expression of 2-month-old WT/NHD13, FLT3/ITD, and FLT3/ITD-NHD13 C57Bl/6N littermates. (C) Examples of sorted HSPC populations isolated from bone marrow are given for mice of each genotype. Bone marrow was stained for the cell surface markers characterizing the (D) KSL, (E) MPP, (F) ST-HSC, and (G) LT-HSC populations. Values in the graphs represent average percentages of these populations in the bone marrow from 3 independent experiments.
Because Hox genes are known to play an important role in stem cell self-renewal, we examined whether the Hox gene overexpression initiated by expression of the NHD13 transgene combined with the strong proliferative signal from constitutive FLT3/ITD signaling resulted in an increase in the frequency of cells with the immunophenotype of HSPCs. Bone marrow was isolated from 2-month-old littermates and stained for the cell surface markers characterizing the KSL, MPP, ST-HSC, and LT-HSC populations (Figure 6C-G). FLT3/ITD-NHD13 and FLT3/ITD mice both had a significant increase in the percentage of KSLs (WT, 0.25% ± 0.03%; NHD13, 0.15% ± 0.07%; FLT3/ITD, 0.9% ± 0.17%; and FLT3/ITD-NHD13, 1.96% ± 0.38%; P < .0001; Figure 6D). FLT3/ITD mice showed increased numbers of MPPs with a smaller but significant increase in the FLT3/ITD-NHD13 cohort (WT, 0.05% ± 0.03%; NHD13, 0.02% ± 0.00%; FLT3/ITD, 0.37% ± 0.02%; FLT3/ITD-NHD13, 0.13% ± 0.01%; P < .0001; Figure 6E). FLT3/ITD-NHD13 mice had an increase in the short-term HSCs (WT, 0.09% ± 0.03%; NHD13, 0.09% ± 0.05%; FLT3/ITD, 0.22% ± 0.08%; FLT3/ITD-NHD13, 1.18% ± 0.29%; P < .0001; Figure 6F), and long-term HSCs (WT 0.02% ± 0.01%; NHD13, 0.02% ± 0.01%; FLT3/ITD, 0.08% ± 0.06%; FLT3/ITD-NHD13, 0.36% ± 0.09%; P < .001; Figure 6G). The FLT3/ITD-NHD13 mice also had increased frequencies of CMP and GMP-like populations at this time point (supplemental Figure 5). These data are consistent with an expansion of HSPCs in the FLT3/ITD-NHD13 mice compared with mice with either FLT3/ITD or NHD13 mutations alone.
The leukemias derived from FLT3/ITD-NHD13 mice are driven by FLT3/ITD signaling with frequent loss of the WT Flt3 allele
A significant number of AML patients with FLT3/ITD-positive leukemias present with loss the WT allele of FLT3.25,26 To examine whether this also occurred in the FLT3/ITD-NHD13 mice, DNA from the bone marrow of leukemic mice was evaluated for a change in the allelic ratio of Flt3. Somewhat surprisingly, loss of the WT allele of Flt3 occurred in the bone marrow in 100% of leukemic mice (Figure 7A). This loss was also evident in the leukemic cells infiltrating the liver and spleen and was clearly somatic as it did not occur in DNA from the tail of the same mice. FLT3/ITD knock-in mice without the NHD13 transgene were also evaluated for loss of heterozygosity (LOH). Loss of the WT allele did occur in these mice but was a much less frequent event, occurring in only 2 of 30 mice. Both FLT3/ITD mice with LOH were older than 14 months and had developed a lethal MPN.
The leukemias derived from FLT3/ITD-NHD13 mice are driven by FLT3/ITD signaling with frequent loss of the WT Flt3 allele. (A) PCR for a region of Flt3 encompassing the ITD mutation. Leukemic bone marrow, liver, and spleen compared with control tail DNA to assay for loss of the WT Flt3 allele. (B) Loss of heterozygosity in sorted HSPC populations. (Ci) Transplant recipients treated with either sorafenib or vehicle for 4 weeks. Percent engraftment as ratio of CD45.2/CD45.1 in peripheral blood at 2 weeks and 4 weeks after initiation of sorafenib treatment. (Cii-iii) Example of peripheral blood stained to evaluate for the presence of blasts in mice treated with vehicle or sorafenib for 1 month. (D) Primary leukemic cells treated in vitro with either 10nM or 20nM sorafenib and probed for phosphorylation of FLT3, STAT5, AKT, and MAPK along with total protein of each. (E) Gene expression profiling for Flt3 and Meis1 in 10 NHD13 alone mice developing AML.
The leukemias derived from FLT3/ITD-NHD13 mice are driven by FLT3/ITD signaling with frequent loss of the WT Flt3 allele. (A) PCR for a region of Flt3 encompassing the ITD mutation. Leukemic bone marrow, liver, and spleen compared with control tail DNA to assay for loss of the WT Flt3 allele. (B) Loss of heterozygosity in sorted HSPC populations. (Ci) Transplant recipients treated with either sorafenib or vehicle for 4 weeks. Percent engraftment as ratio of CD45.2/CD45.1 in peripheral blood at 2 weeks and 4 weeks after initiation of sorafenib treatment. (Cii-iii) Example of peripheral blood stained to evaluate for the presence of blasts in mice treated with vehicle or sorafenib for 1 month. (D) Primary leukemic cells treated in vitro with either 10nM or 20nM sorafenib and probed for phosphorylation of FLT3, STAT5, AKT, and MAPK along with total protein of each. (E) Gene expression profiling for Flt3 and Meis1 in 10 NHD13 alone mice developing AML.
The ratio of mutant to WT FLT3 has prognostic significance and can be used identify the stage at which leukemic transformation occurs. Previous work with AML patient samples has shown that CD34+/CD38− enriched fractions have the same mutant to WT ratio as the bulk unsorted bone marrow, supporting the hypothesis that FLT3 mutations are present in leukemic stem cells.27 To determine whether the LOH event observed in our model also occurred early in hematopoiesis, DNA was extracted from cells with the immunophenotype of MPP, CMP, GMP, GMP-L, MEP, and B220+CD19+ cells sorted from a leukemic mouse to look for changes in the mutant to WT Flt3 ratio. In all mice examined, a change in allelic ratio was detected in the MPP, CMP, and GMP sorted populations (Figure 7B; Table 3). The MEP and B220+CD19+ populations had no change in allelic ratio compared with the tail DNA control and are thus most likely not derived from the leukemia-initiating population (supplemental Figure 4). These data suggest that, similar to patients, loss of the WT allele in FLT3/ITD-NHD13 mice occurs in HSPCs and provides a selective advantage to these cells during leukemogenesis.
Sorted HSPC populations evaluated for change in the allelic ratio of Flt3
| Sorted population . | Change in allelic ratio . |
|---|---|
| MPP | Yes |
| CMP | Yes |
| GMP | Yes |
| GMP-L | No |
| MEP | No |
| B220+CD19+ | No |
| Sorted population . | Change in allelic ratio . |
|---|---|
| MPP | Yes |
| CMP | Yes |
| GMP | Yes |
| GMP-L | No |
| MEP | No |
| B220+CD19+ | No |
To study the dependence of the leukemias on FLT3/ITD signaling, primary transplant recipients of leukemic FLT3/ITD-NHD13 bone marrow were treated with sorafenib, a tyrosine kinase inhibitor with activity against FLT3/ITD. Sorafenib has been found to reduce the percentage of leukemic blasts in the peripheral blood and bone marrow of patients with FLT3/ITD-positive AML in a phase 1 clinical trial.28 Mice were treated with 10 mg/kg sorafenib or vehicle control once a day by oral gavage for 4 weeks and evaluated for the engraftment of CD45.2+ leukemic cells by FACS analysis (Figure 7Ci) and by morphology for the appearance of blasts in the peripheral blood (Figure 7Cii-iii). Two weeks after initiation of sorafenib treatment, the percentage of CD45.2+ leukemic cells was reduced in the peripheral blood mononuclear cells of treatment recipients (15.6% ± 3.5%) compared with the group receiving vehicle control treatment (30.0% ± 6.9%, P < .085). CD45.2+ leukemic cells were almost completely lacking from the peripheral blood mononuclear cells of the recipients after 4 weeks (0.01% ± 0.001%) of treatment, whereas they constituted 71.7% ± 7.1% of the cells of the vehicle control treated group. Peripheral blood from the sorafenib-treated cohort was routinely evaluated until 6 months after treatment with no evidence of disease. These data suggest that the expansion of leukemic cells in the FLT3/ITD-NHD13 mice is very much dependent on constitutively activated FLT3/ITD signaling. To evaluate the leukemic cells' sensitivity to FLT3 inhibition, leukemic cells harvested from mice transplanted with bulk leukemic cells were treated in vitro for one hour with either 10nM or 20nM sorafenib and probed for phosphorylation of FLT3, STAT5, AKT, and MAPK (Figure 7D). FLT3 autophosphorylation was completely inhibited after treatment with 10nM sorafenib. Meanwhile, the phosphorylation status of STAT5, AKT, and MAPK, downstream targets of FLT3 signaling, was also greatly reduced, indicating their dependence on FLT3/ITD signaling in the leukemic cells.
As previously noted, 50% of mice that carry only the NHD13 mutation also develop acute leukemia, although with a much longer latency than the FLT3/ITD-NHD13 mice. To determine whether Flt3 overexpression was important in the leukemias arising in NHD13 mice alone, RNA was extracted from the bone marrow of NHD13 mice developing AML and Flt3 mRNA expression was evaluated by quantitative RT-PCR (Figure 7E). Three of the 10 mice had elevated Flt3 expression, and this correlated with increased expression of Meis1, suggesting that the activation of Flt3 expression through MEIS1 may be an alternate method of leukemic transformation in the absence of FLT3-activating mutations.
Discussion
We report here a novel model combining a FLT3/ITD mutation with an NHD13 fusion that produces a highly penetrant acute leukemia with only a modest lag. This differs from previous models of FLT3 induced disease that relied on retroviral transduction of murine bone marrow or transgenic expression using exogenous promoters.29,30 Although each of these other models leads to a rapidly fatal myeloproliferative neoplasm, they often express the mutation at high levels or induce expression in cell fractions that would not normally express FLT3. In contrast, the FLT3/ITD-NHD13 model, which is the subject of this report, expresses FLT3/ITD under control of the endogenous promoter, providing a more biologically relevant model because the mutation is expressed at normal levels and is activated in the proper cell lineages during murine hematopoiesis.
Initially, the leukemia seen in this model appeared to express markers characteristic of both primitive myeloid and lymphoid development. Several murine models of hematopoietic malignancy have reported leukemias that coexpress cell surface markers of both the myeloid and lymphoid lineages, which were interpreted as biphenotypic leukemias, usually using B220 as evidence of B-cell differentiation.31,32 However, B220 appears early in hematopoietic differentiation and can also be expressed on many different activated cell types. Moreover, there is previous evidence of a connection between FLT3 signaling and the expansion of myeloid progenitors expressing B220.33,34 Although leukemias in these mice expressed the enzyme terminal deoxynucleotidyl transferase (TdT) and had D to J rearrangements of the immunoglobulin heavy chain locus, neither of these characteristics is specific to lymphocyte development and sometimes observed in myeloid leukemias.35 This phenomenon may be the result of the expression of recombination activating genes, which has been documented in the MPP and CLP populations before loss of myeloid differentiation potential.36 V to DJ rearrangements, which are specific to B-lymphocytes, did not occur in this model. In addition, the B220+ LICs had no lymphoid developmental potential in vitro and showed no cell surface expression of additional B-lineage markers. Sorted B220+CD19+ cells, which include B cells as early as the immature pro-B stage, showed no ability to engraft in lethally irradiated recipients and no loss of the WT allele of Flt3 (supplemental Figure 5). The absence of LOH in this population suggests that HSPCs that have lost the WT allele of Flt3 are restricted to myeloid development and do not give rise to B-lymphoid progenitors.
In this model, we observed overexpression of several Hox genes, including HoxA7, HoxA9, HoxB4, HoxB6, HoxB7, HoxC4, and HoxC6. The overexpression of clustered Hox genes appears to be primarily driven by the NHD13 transgene, although there was a significant up-regulation of HoxA7 and HoxA9 expression in the FLT3/ITD single mutant mice. Many of these genes have been shown to play an important role in HSC self-renewal and are up-regulated in cases of acute leukemia. For example, HoxA cluster genes are up-regulated in MLL and CALM-AF10 related acute lymphoblastic leukemias.37 Within the HoxB cluster, studies have shown that murine bone marrow transduced with HoxB4 undergoes expansion of primitive HSCs while retaining the repopulating potential.38,39 Similarly, overexpression of HoxB6 in murine bone marrow causes immortalization of a myelomonocytic precursor in vitro and the eventual development of AML in mice.40,41 A gene expression signature characterized by high expression of both Hox A and B clustered genes has been identified in patients with NUP98 translocations, providing further evidence that the FLT3/ITD-NHD13 mice accurately model human disease.42 In this study, we also noted the overexpression of Meis1 and Flt3 in a subset of NHD13 alone mice that spontaneously developed AML. Meis1 is frequently up-regulated in patients with AML and has been shown to cooperate with Nup98 fusions to induce the rapid development of AML in bone marrow transplantation models.43 In contrast to the NHD13 alone mice, no leukemic FLT3/ITD-NHD13 mice showed elevated Meis1 expression levels, which may indicate less selective pressure to activate FLT3-dependent signaling through alternative mechanisms in the case where FLT3 activating mutations are already present.
Although FLT3 mutations are typically categorized as class I mutations, recent data have suggested that the FLT3/ITD mutation also contributes to a block in differentiation.44,45 A subset of NHD13 alone mice is known to develop differentiated leukemias of both the myeloid and lymphoid lineages.15 However, when combined with the FLT3/ITD, mice developed strictly myeloid leukemias with minimal differentiation. Because the FLT3/ITD knock-in is expressed under the endogenous murine Flt3 promoter, this lineage restriction may be driven by expression of the Flt3 promoter in HSPCs. These data provide further evidence that FLT3/ITD mutations play a role in the regulation of differentiation and lineage restriction of the leukemic stem cell.
An intriguing finding in this model is the high frequency of LOH resulting in the loss of the WT Flt3 allele. A significant number of AML patients with FLT3/ITD mutations present with loss of the WT allele of FLT3 and additional patients acquire this loss at the time of relapse.26 Hemizygosity at the FLT3 locus in FLT3/ITD mutant AML patients is associated with an adverse prognosis compared with patients with an intact WT allele.25 This suggests that there is strong selective pressure to lose the WT allele driven by FLT3/ITD expression. Experimental murine models using the FLT3/ITD knock-in also show a more aggressive MPN in mice that are hemizygous for the ITD mutation compared with heterozygous mice.46,47 In the experiments described by Li et al,47 replacing the lost WT Flt3 allele reduced the aggressiveness of the MPN, suggesting that, in the context of FLT3/ITD malignancy, the WT allele acts as a tumor suppressor. The frequent LOH seen in the FLT3/ITD-NHD13 mouse provides additional evidence that this transgenic mouse serves as an accurate model to further investigate the molecular pathways that underlie the development of leukemias with activating FLT3 mutations. It is also the first model to our knowledge that generates an LOH event spontaneously and with such high frequency.
In conclusion, these data demonstrate that FLT3/ITD functions as a driving mutation, in cooperation with the NUP98-HOXD13 fusion oncoprotein, to cause AML with minimal differentiation. AML arising from MDS is a notoriously difficult disease to treat; therefore, a better understanding of its underlying biology may lead to improved therapeutic options. This transgenic mouse is one of the first models of MDS-related AML and will provide a means to explore the molecular pathways that underlie these hematopoietic neoplasms.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the National Cancer Institute (CA90668, CA70970), Leukemia & Lymphoma Society, the Giant Food Pediatric Cancer Research Fund, and the Intramural Research Program of the National Institutes of Health, National Cancer Institute. D.S. is also supported by the Kyle Haydock Professorship.
National Institutes of Health
Authorship
Contribution: S.G. designed and performed experiments, analyzed data, and wrote the first draft; L.L., B.N., and A.D. designed and performed experiments; D.H. and R.N. performed experiments and analyzed data; M.J.B. and S.D. analyzed data; and D.S., C.S., and P.A. designed experiments, analyzed data, and revised the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Donald Small, Sidney Kimmel Comprehensive Cancer Center at Johns Hopkins, CRB Rm 251, 1650 Orleans St, Baltimore, MD 21231; e-mail: donsmall@jhmi.edu.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal