Abstract
Aggressive systemic mastocytosis (ASM) and mast cell leukemia (MCL) are advanced hematopoietic neoplasms with poor prognosis. In these patients, neoplastic mast cells (MCs) are resistant against various drugs. We examined the effects of 2 demethylating agents, 5-azacytidine and decitabine on growth and survival of neoplastic MCs and the MC line HMC-1. Two HMC-1 subclones were used, HMC-1.1 lacking KIT D816V and HMC-1.2 exhibiting KIT D816V. Both agents induced apoptosis in HMC-1.1 and HMC-1.2 cells. Decitabine, but not 5-azacytidine, also produced a G2/M cell-cycle arrest in HMC-1 cells. Drug-induced apoptosis was accompanied by cleavage of caspase-8 and caspase-3 as well as FAS-demethylation and FAS–re-expression in neoplastic MCs. Furthermore, both demethylating agents were found to synergize with the FAS-ligand in inducing apoptosis in neoplastic MCs. Correspondingly, siRNA against FAS was found to block drug-induced expression of FAS and drug-induced apoptosis in HMC-1 cells. Neither 5-azacytidine nor decitabine induced substantial apoptosis or growth arrest in normal MCs or normal bone marrow cells. Together, 5-azacytidine and decitabine exert growth-inhibitory and proapoptotic effects in neoplastic MCs. These effects are mediated through “FAS–re-expression” and are augmented by the FAS-ligand. Whether epigenetic drugs produce antineoplastic effects in vivo in patients with ASM and MCL remains to be determined.
Introduction
Systemic mastocytosis (SM) is a myeloid neoplasm characterized by pathologic accumulation of mast cells (MCs) in one or more extracutaneous organs.1,2 Indolent and aggressive variants of SM have been described.1-5 In most patients, the transforming KIT mutation D816V is detectable independent of the category of SM.6-8 Patients with indolent SM (ISM) have an excellent prognosis with normal or near-normal life expectancy.1-5,9,10 In patients with aggressive SM (ASM) and mast cell leukemia (MCL), the prognosis is grave,1-5,9,10 and responses to conventional drugs and most targeted drugs are poor.1-4,9,11-15 During the past few years, several attempts have been made to define new molecular targets in neoplastic MCs and to establish new treatment concepts for these patients.16,17
Abnormal DNA methylation and histone-acetylation are frequently observed in various neoplasms and supposedly contribute to disease evolution and drug resistance.18-20 In hematologic malignancies, epigenetic abnormalities have been reported in acute and chronic leukemias as well as in myelodysplastic syndromes (MDS).21-23 Especially in patients with MDS, abnormal methylation of the genome has been described.21,22 Correspondingly, epigenetically active drugs such as 5-azacytidine and 5-aza-2′deoxycytidine (decitabine) reportedly act as antineoplastic agents in these patients.23-25 More recent data suggest that demethylating agents also exert beneficial effects in patients in whom neoplastic cells exhibit myeloproliferative and myelodysplastic features, such as chronic myelomonocytic leukemia (CMML), and sometimes even in patients with advanced myeloproliferative neoplasms (MPN).26,27 Advanced SM is a myeloid neoplasm that often presents with myeloproliferative features and sometimes with bone marrow (BM) dysplasia.1-5 In addition, advanced SM may coexist with MDS (SM-MDS), CMML, or a JAK2-mutated MPN.28
However, so far, no data on the effects of demethylating agents on neoplastic cells in advanced SM or SM-AHNMD are available. In addition, little is known about methylation patterns in neoplastic MCs. We explored the effects of 2 widely used demethylating agents, 5-azacytidine and decitabine, on growth and survival of neoplastic MCs, and examined the mechanism(s) of action of these drugs.
Methods
Drugs, monoclonal antibodies, and other reagents
PKC412 (midostaurin) was kindly provided by Dr J. Roesel and Dr P. W. Manley (Novartis). 5-azacytidine, decitabine, recombinant human FAS-ligand, and propidium iodide (PI) were purchased from Sigma-Aldrich, RPMI 1640 medium and FCS from PAA Laboratories, IMDM from Gibco Life Technologies, and 3H-thymidine from Perkin Elmer. The phycoerythrin (PE)–labeled monoclonal antibody (mAb) WM15 (CD13), DX2 (CD95), 104D2 (CD117), N6B6 (CD164), C92605 (anti-active caspase-3), pS473 (anti-pAkt), and the Alexa fluor647-labeled mAb pY694 (anti-pSTAT5), and N7548 (anti-p-S6) were from BD Biosciences, annexin V–FITC from Biosciences, and FITC-labeled CLBGran/12 mAb (CD63) from Beckman Coulter.
Isolation and culture of neoplastic cells
Primary neoplastic cells were obtained by BM aspiration (diagnostic samples) in 6 patients with SM (ISM, n = 4; ASM, n = 1; SM-AHNMD, n = 1) after informed consent was given. In patients with ASM and SM-AHNMD, a majority of peripheral blood and BM leukocytes were found to carry KIT D816V, confirming previous observations.4,6 The patients' characteristics are shown in supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Diagnoses were established according to published criteria.3-5 Ficoll-isolated BM mononuclear cells (MNCs) were cultured in RPMI 1640 medium containing 10% FCS and antibiotics (5% CO2, 37°C). Normal BM cells were obtained from 3 lymphoma patients without BM involvement (routine staging). Cultured normal MCs were generated from CD133+ cord blood progenitors using IL-3, IL-6, and stem cell factor (SCF) as described.29 All studies were approved by the Local Ethics Committee of the Medical University of Vienna. The MC line HMC-130 was kindly provided by Dr J. H. Butterfield (Mayo Clinic). Two subclones were used, HMC-1.1 harboring KIT V560G, and HMC-1.2 harboring KIT V560G as well as KIT D816V.31 HMC-1 cells were cultured in IMDM with 10% FCS and antibiotics. Additional cell lines used in this study were HL60, U937, KG1, KU812, and K562. These cell lines were cultured in RPMI 1640 medium with 10% FCS and antibiotics (5% CO2, 37°C).
Treatment with pharmacologic inhibitors and measurement of proliferation
Primary neoplastic MCs and HMC-1 cells were cultured in control medium or in various concentrations of 5-azacytidine (1-20μM), decitabine (1-20μM), or PKC412 (50-110nM) for 48, 72, or 96 hours. In select experiments, demethylating agents were applied in the absence or presence of FAS-ligand (1 ng/mL) or PKC412 (200nM). Proliferation was determined by 3H-thymidine as reported32,33 with the following modification: before addition of 3H-thymidine, cells were washed in phosphate-buffered saline (PBS) because in control experiments we found that 5-azacytidine competes with 3H-thymidine in the uptake process (supplemental Figure 1). All experiments were performed in triplicates. In a separate set of experiments, combinations of PKC412 and demethylating agents at suboptimal concentrations (with fixed ratio of drug-concentrations) were applied on HMC-1 cells.
Western blot experiments
HMC-1 cells were incubated with control medium, 5-azacytidine (5μM) or decitabine (5μM) for 48 hours. Then, Western blotting (WB) was performed as described32,33 using a polyclonal antibody against cleaved caspase-3 or mAb 18C8 against cleaved caspase-8 (Cell Signaling Technology). To confirm equal loading, a polyclonal antibody against β-actin (Sigma-Aldrich) was used. Antibody-reactivity was made visible by donkey antirabbit immunoglobulin (Ig)G and lumingen PS-3 detection reagent (GE-Healthcare).
Evaluation of apoptosis by morphology and TUNEL assay
HMC-1 cells were incubated with various concentrations of 5-azacytidine (1-20μM), decitabine (1-20μM), or control medium at 37°C for 48 or 96 hours. In select experiments, drugs were applied in the absence or presence of FAS-ligand (1 ng/mL). The percentage of apoptotic cells was quantified on Wright-Giemsa–stained cytospin slides. In TUNEL (terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling) assay experiments, cells were cultured in medium without or with demethylating agents (5-azacytidine, 5μM or decitabine, 5μM) for 96 hours. The TUNEL assay was performed as reported32,33 using the In situ cell death detection kit-fluorescein (Roche Diagnostics). Cells were analyzed with a Nikon Eclipse E 800 fluorescence microscope or a Carl Zeiss Imager A1 microscope (Carl Zeiss).
Flow cytometry experiments
To determine the effects of 5-azacytidine and decitabine on expression of surface antigens (CD13/aminopeptidase-N, CD63/LAMP-3, CD95/FAS, CD117/KIT, and CD164/endolyn) or cytoplasmic molecules (pAkt, pS6, pSTAT3, pSTAT5, activated caspase-3) in HMC-1 cells, flow cytometry was performed on a FACScalibur (Becton Dickinson). Cells were incubated in control medium, 5-azacytidine (1-20μM), or decitabine (1-20μM) at 37°C for 4, 24, or 48 hours (signaling molecules), or for 48 or 96 hours (surface molecules and caspase-3). Before staining with mAbs against cytoplasmic molecules, HMC-1 cells were fixed in 2% formaldehyde and permeabilized using ice-cold methanol (−20°C, 10 minutes). Apoptosis was measured by combined annexin V/propidium iodide staining32-34 after exposure of MCs to 5-azacytidine or decitabine (each 1-20μM) for 48 or 96 hours (37°C). In select experiments, HMC-1 cells were cultured in the presence or absence of FAS-ligand (1 ng/mL) or PKC412 (70-200nM) before being analyzed for cell viability. Cell-cycle distribution was analyzed as reported.34 In these experiments, HMC-1 cells were incubated with 5-azacytidine or decitabine (each 1-20μM) for 96 hours (37°C).
Quantitative RT-PCR
HMC-1 cells were incubated with or without 5-azacytidine (5μM) or decitabine (5μM) at 37°C for 48 or 96 hours. Then, RNA was isolated using Omniscript Reverse Transcriptase kit or RNeasy MinElute Cleanup Kit (QIAGEN) according to the manufacturer's instructions. Real-time polymerase chain reaction (PCR) for FAS, p16, p21, Bid, Puma, Noxa, Bad, Bim, and ABL was performed using specific primers (supplemental Table 2). mRNA levels were quantified on a 7900HT Fast real-time PCR System (Applied Biosystems) using iTAq SYBR Green Supermix with ROX (Bio-Rad). FAS, p16, p21, Bid-, Puma-, Noxa-, Bad-, and Bim mRNA expression levels were expressed as percentage of ABL mRNA. In select experiments, expression of transcripts specific for FAS, p16, and p21 were quantified by quantitative (q) PCR using TaqMan Gene Expression assay using the ABI PRISM-7000 system (Applied Biosystems) as reported.34 GAPDH served as housekeeping gene. Results were expressed as fold-induction compared with medium control.
Isolation of genomic DNA and methylation-specific PCR
Genomic DNA was isolated from untreated and drug-exposed HMC-1 cells and untreated normal BM cells by digestion with proteinase K, followed by phenol-chloroform extraction and ethanol precipitation.35 Genomic DNA (1 μg) was modified by treatment with sodium bisulfite using QIAGEN EpiTect Bisulfite kit according to the manufacturer′s instructions. The 5′ methylation status of p15 (CDKN2B), p16 (CDKN2A), p21 (CDKN1A), and FAS was determined by methylation-specific PCR (MSP). Primer sequences and PCR conditions for p15 and p16 were the same as previously described.36 MSP primer sequences for p21 were designed using Methyl Primer Express Version 1.0 (Applied Biosystems) and are shown in supplemental Table 3. Primer sequences and MSP conditions for evaluation of FAS were prepared according to methPrimerDB (http://medgen.ugent.be/methprimerdb/index.php, ID:136). EpiTect methylated control DNA (QIAGEN) was used as positive control for methylated alleles. Water blanks were used as negative control. PCR products were resolved on a 2% agarose gel and stained with GelRed (Biotium).
Bisulfite genomic sequencing of the FAS promoter
Part of the FAS CpG island (CGI) was amplified using methylation-insensitive PCR primers. Primer sequences were designed using Methyl Primer Express Version 1.0 software and are shown in supplemental Table 3. PCR was performed on sodium bisulfite converted genomic DNA. PCR products were gel-purified and cloned using TOPO TA Cloning Kit for Sequencing (Invitrogen). PCR amplification for sequencing was performed using M13 primers. Three clones from each cell sample were sequenced. In total, 18 CpG sites located between nucleotide (nt) 90750233 and nt 90750773 of chromosome 10 were analyzed for methylation. The percentage of methylation was calculated as number of methylated CpG sites relative to the total number of CpG sites. Differences in the extent of methylation between HMC-1 cells and control BM cells were assessed statistically by applying the Fisher exact test and the χ2 test.
EpiTect methyl qPCR array analysis and methylation-sensitive high resolution melting analysis
Genomic DNA (1 μg) from HMC-1.2 cells and normal BM cells was digested using EpitTect methyl DNA restriction Kit (QIAGEN) according to the manufacturer's instructions. Apoptosis EpiTect methyl qPCR arrays and tumor suppressor gene EpiTect methyl qPCR Arrays (QIAGEN) were applied. Real-time PCR was performed using RT2 SYBR Green ROX qPCR Mastermix (QIAGEN) according to the manufacturer′s recommendations in a 96-well ABI StepOne system (Applied Biosystems). Cycle threshold (Ct) values were used to calculate the percentages of hypermethylated (HM), unmethylated (UM), and intermediately methylated (IM) DNA according to the recommendation of the manufacturer (QIAGEN). For methylation-sensitive high resolution melting (MS-HRM) analyses, sodium bisulfite-treated genomic DNA was used. Primer sequences (supplemental Table 3) were designed using Methyl Primer Express Version 1.0 software. MS-HRM analyses were performed using EpiTect HRM PCR kit in a RotorGeneQ cycler (QIAGEN) as described.37
Application of siRNA against FAS/CD95
siRNAs directed against FAS or against luciferase (supplemental Table 2) were synthesized in 2′-deprotected, duplexed, and desalted form by Dharmacon and transfected into HMC-1.1 and HMC-1.2 cells using lipofectin (Invitrogen) as described.33 In brief, cells were exposed to lipofectin (75nM) and siRNA (200nM) in serum-free IMDM at 37°C for 4 hours. The siRNA-induced knockdown of FAS (CD95) in HMC-1 cells was confirmed by flow cytometry. siRNA-treated cells were cultured in control medium in the presence or absence of 5-azacytidine (5μM) or decitabine (5μM) for 30 hours, and were then examined for signs of apoptosis and expression of active caspase-3 by flow cytometry.
Statistical analyses
To determine the significant differences in growth and survival of cells after exposure to control medium or inhibitors, the student t test for dependent samples was applied. The Fisher exact test and the χ2 test were applied to define difference in methylation of the FAS promoter in normal and neoplastic cells. Results were considered statistically significant when P is less than .05.
Results
5-azacytidine and decitabine induce apoptosis in neoplastic MCs
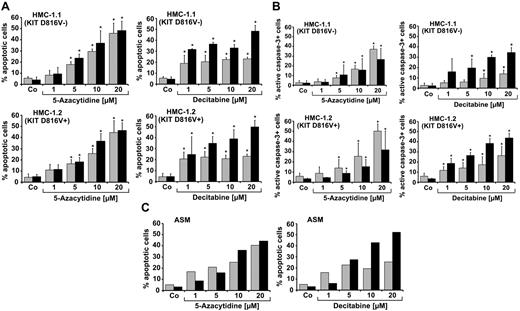
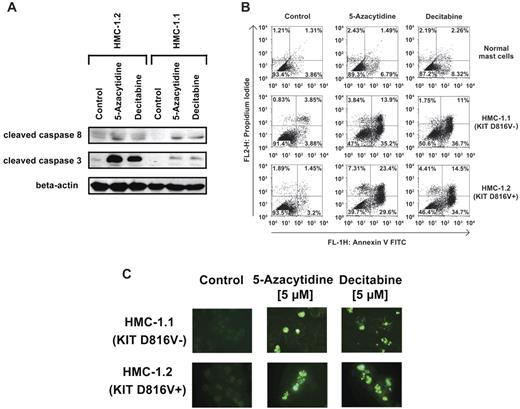
As assessed by light microscopy, 5-azacytidine and decitabine were found to induce apoptosis in HMC-1.1 cells and HMC-1.2 cells (Figure 1A). The effects of both drugs were dose-dependent. In case of decitabine, drug effects were more pronounced after 96 hours than after 48 hours (Figure 1A). The dose-dependent effects of 5-azacytidine and decitabine on survival of HMC-1 cells were confirmed by measuring active caspase-3 levels by flow cytometry (Figure 1B). We were also able to show that 5-azacytidine and decitabine induce apoptosis in primary neoplastic MCs in a patient with ASM (Figure 1C). In our WB experiments, 5-azacytidine and decitabine induced cleavage of caspase-3 and caspase-8 in both HMC-1 subclones (Figure 2A). Moreover, we were able to confirm drug-induced apoptosis in HMC-1 cells by annexin V staining (Figure 2B) and by a TUNEL assay (Figure 2C). In normal cultured MCs, the demethylating agents did not induce apoptosis (Figure 2B).
5-azacytidine and decitabine induce apoptosis in neoplastic MCs. (A-B) HMC-1.1 cells and HMC-1.2 cells were incubated in control medium (Co) or in medium containing various concentrations (1-20μM) of 5-azacytidine or decitabine at 37°C for 48 hours (gray bars) or 96 hours (black bars). After incubation, apoptotic cells were quantified by either light microscopy (A) or by flow cytometry using an antibody against active caspase-3 (B). Results show the percentage of apoptotic cells and active caspase-3-positive cells, and represent the mean ± SD of 5 independent experiments (*P < .05 compared with control). (C) BM-derived MCs from a patient with ASM were cultured in the presence or absence (Co) of various concentrations of 5-azacytidine or decitabine at 37°C for 48 hours (gray bars) or 96 hours (black bars). Results show the percentage of apoptotic cells determined by light microscopy.
5-azacytidine and decitabine induce apoptosis in neoplastic MCs. (A-B) HMC-1.1 cells and HMC-1.2 cells were incubated in control medium (Co) or in medium containing various concentrations (1-20μM) of 5-azacytidine or decitabine at 37°C for 48 hours (gray bars) or 96 hours (black bars). After incubation, apoptotic cells were quantified by either light microscopy (A) or by flow cytometry using an antibody against active caspase-3 (B). Results show the percentage of apoptotic cells and active caspase-3-positive cells, and represent the mean ± SD of 5 independent experiments (*P < .05 compared with control). (C) BM-derived MCs from a patient with ASM were cultured in the presence or absence (Co) of various concentrations of 5-azacytidine or decitabine at 37°C for 48 hours (gray bars) or 96 hours (black bars). Results show the percentage of apoptotic cells determined by light microscopy.
5-azacytidine and decitabine induce expression of active caspase-8 and active caspase-3 in neoplastic MCs. (A) HMC-1.1 and HMC-1.2 cells were incubated in control medium or in medium containing 5-azacytidine (5μM) or decitabine (5μM) at 37°C for 96 hours. Then, Western blotting was performed using antibodies against cleaved caspase-8 and cleaved caspase-3. To confirm equal loading, a polyclonal antibody against β-actin was applied. (B) Cord blood progenitor-derived normal MCs and HMC-1 cells were cultured in the presence of control medium (left panels), 5-azacytidine (5μM; middle panels), or decitabine (5μM; right panels) at 37°C for 96 hours. Then, cells were analyzed by annexin V/PI staining. Results show the percentage of annexin V/PI-positive cells determined by flow cytometry. (C) HMC-1.1 cells and HMC-1.2 cells were incubated in control medium (Co) or in medium containing 5-azacytidine (5μM) or decitabine (5μM) at 37°C for 96 hours. After incubation, the presence of apoptotic cells was determined by TUNEL assay.
5-azacytidine and decitabine induce expression of active caspase-8 and active caspase-3 in neoplastic MCs. (A) HMC-1.1 and HMC-1.2 cells were incubated in control medium or in medium containing 5-azacytidine (5μM) or decitabine (5μM) at 37°C for 96 hours. Then, Western blotting was performed using antibodies against cleaved caspase-8 and cleaved caspase-3. To confirm equal loading, a polyclonal antibody against β-actin was applied. (B) Cord blood progenitor-derived normal MCs and HMC-1 cells were cultured in the presence of control medium (left panels), 5-azacytidine (5μM; middle panels), or decitabine (5μM; right panels) at 37°C for 96 hours. Then, cells were analyzed by annexin V/PI staining. Results show the percentage of annexin V/PI-positive cells determined by flow cytometry. (C) HMC-1.1 cells and HMC-1.2 cells were incubated in control medium (Co) or in medium containing 5-azacytidine (5μM) or decitabine (5μM) at 37°C for 96 hours. After incubation, the presence of apoptotic cells was determined by TUNEL assay.
5-azacytidine and decitabine promote expression of FAS in neoplastic MCs
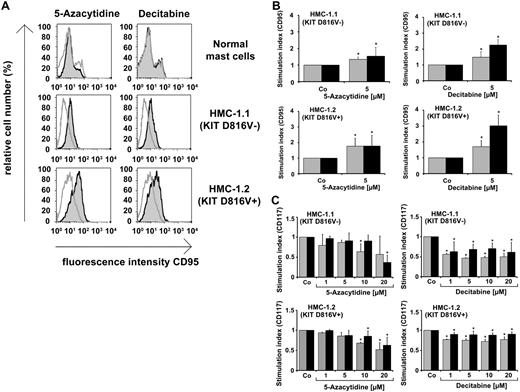
As determined by flow cytometry, 5-azacytidine and decitabine were found to promote expression of FAS/CD95 in HMC-1.1 and HMC-1.2 cells (Figure 3A), whereas PKC412 showed no effects on FAS expression (not shown). FAS-promoting effects of both demethylating drugs were seen after 48 and 96 hours (Figure 3B), and were confirmed by qPCR (supplemental Table 4). The other surface molecules examined (CD13, CD63, CD117, CD164) were not up-regulated by the 2 demethylating agents applied. Rather, 5-azacytidine and decitabine were found to slightly down-regulate the expression of KIT/CD117 in HMC-1.1 and HMC-1.2 cells (Figure 3C). 5-azacytidine and decitabine failed to promote FAS expression in normal MCs (Figure 3A). We next asked whether 5-azacytidine or decitabine would cause deactivation of KIT-downstream signaling molecules in neoplastic MCs. However, no effects of 5-azacytidine or decitabine on expression of pAkt, pS6, pSTAT3, or pSTAT5 in HMC-1 cells were seen (supplemental Figure 2).
Effects of 5-azacytidine and decitabine on expression of FAS (CD95) in neoplastic MCs. (A) Cord blood progenitor-derived MCs and HMC-1 cells were cultured in control medium (open histograms) or in the presence of 5-azacytidine (5μM) or decitabine (5μM; gray histograms) at 37°C for 96 hours. Thereafter, CD95 expression was analyzed by flow cytometry. As visible, the demethylating agents promoted FAS expression in neoplastic MCs but not in normal MCs. (B) HMC-1 cells were cultured in the absence (Co) or presence of 5-azacytidine or decitabine (each 5μM) for 48 hours (gray bars) or 96 hours (black bars). Then, expression of CD95 was analyzed by flow cytometry. Drug-induced up-regulation of CD95 was calculated from mean fluorescence intensities (MFIs) obtained with treated (MFIstim) and untreated (MFIco) cells, and is expressed as stimulation index = SI (MFIstim:MFIco). Results show SI values and represent the mean ± SD of 3 independent experiments (*P < .05 compared with control). (C) HMC-1 cells were exposed to various concentrations of 5-azacytidine or decitabine (1-20μM) at 37°C for 48 hours (gray bars) or 96 hours (black bars). Then, cells were washed and stained with antibodies against CD117/KIT and analyzed by flow cytometry. Drug-induced down-regulation of KIT was expressed as SI (MFIstim:MFIco). Results represent the mean ± SD of 3 independent experiments (*P < .05 compared with control).
Effects of 5-azacytidine and decitabine on expression of FAS (CD95) in neoplastic MCs. (A) Cord blood progenitor-derived MCs and HMC-1 cells were cultured in control medium (open histograms) or in the presence of 5-azacytidine (5μM) or decitabine (5μM; gray histograms) at 37°C for 96 hours. Thereafter, CD95 expression was analyzed by flow cytometry. As visible, the demethylating agents promoted FAS expression in neoplastic MCs but not in normal MCs. (B) HMC-1 cells were cultured in the absence (Co) or presence of 5-azacytidine or decitabine (each 5μM) for 48 hours (gray bars) or 96 hours (black bars). Then, expression of CD95 was analyzed by flow cytometry. Drug-induced up-regulation of CD95 was calculated from mean fluorescence intensities (MFIs) obtained with treated (MFIstim) and untreated (MFIco) cells, and is expressed as stimulation index = SI (MFIstim:MFIco). Results show SI values and represent the mean ± SD of 3 independent experiments (*P < .05 compared with control). (C) HMC-1 cells were exposed to various concentrations of 5-azacytidine or decitabine (1-20μM) at 37°C for 48 hours (gray bars) or 96 hours (black bars). Then, cells were washed and stained with antibodies against CD117/KIT and analyzed by flow cytometry. Drug-induced down-regulation of KIT was expressed as SI (MFIstim:MFIco). Results represent the mean ± SD of 3 independent experiments (*P < .05 compared with control).
5-azacytidine and decitabine revert DNA hypermethylation in HMC-1 cells and lead to demethylation of the FAS promoter and thus to FAS expression
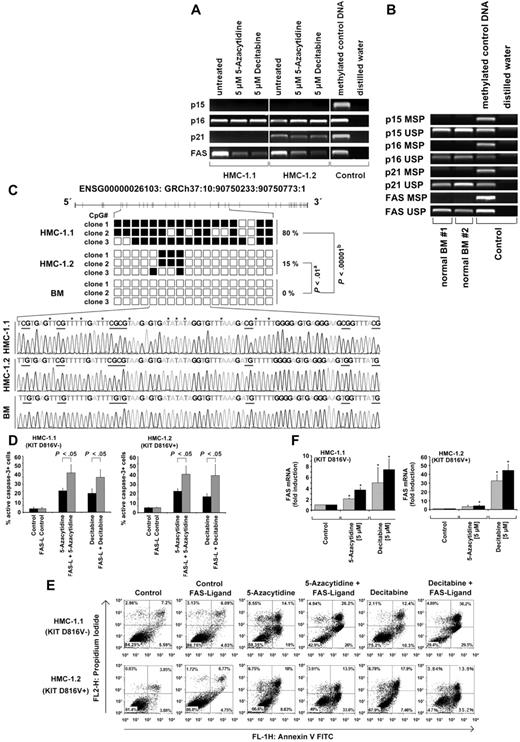
As determined by MSP, p16 and FAS were found to be hypermethylated in HMC-1.1 cells and HMC-1.2 cells (Figure 4A) but not in normal BM cells (Figure 4B). Interestingly, p21 was found to be hypermethylated in HMC-1.2 cells but not in HMC-1.1 cells, and p15 was neither hypermethylated in HMC-1.1 nor in HMC-1.2 cells (Figure 4A). Bisulfite genomic sequencing (BGS) confirmed that the FAS promoter is hypermethylated in HMC-1 cells but not in normal BM cells (Figure 4C). Both 5-azacytidine and decitabine were found to induce demethylation of FAS in HMC-1.1 cells and HMC-1.2 cells (Figure 4A). By contrast, no effects of these drugs on methylation of p16 or p21 were seen (Figure 4A). In both qPCR protocols applied, we were able to show that 5-azacytidine and decitabine promote FAS mRNA expression in HMC-1 cells (Figure 4D, supplemental Table 4). To define the specificity of these drug effects, we examined the effects of 5-azacytidine and decitabine on expression of other proapoptotic molecules known to play a role in survival of neoplastic MCs. We found that Bad and Bid are not up-regulated by exposure to 5-azacytidine or decitabine in these experiments, and that decitabine slightly promotes the expression of p21 mRNA, Noxa mRNA, and Bim mRNA, in both HMC-1 subclones (supplemental Tables 4-5).
Effects of 5-azacytidine and decitabine on the 5′methylation status of FAS and FAS mRNA expression levels in neoplastic MCs. (A) HMC-1 cells were exposed to control medium (untreated), 5-azacytidine (5μM) or decitabine (5μM) for 96 hours. Then, the 5′methylation status of p15, p16, p21, and FAS was determined by MSP. EpiTect-methylated control DNA was used as positive-control. Vertical lines have been inserted to indicate a repositioned gel lane. (B) 5′methylation status of p15, p16, p21, and FAS in normal BM cells in 2 donors (#1 and #2) determined by MSP. To verify efficient sodium bisulfite conversion, we also performed unmethylated specific PCR (USP) for p15, p16, p21, and FAS. EpiTect-methylated and unmethylated control DNA were used as positive-control. (C) BGS of a part of the FAS CGI was performed as previously described using HMC-1.1 cells, HMC-1.2 cells, and normal BM cells. Black squares indicate methylated cytosines at CpG sites, and white squares represent unmethylated cytosines at CpG sites. Whereas in HMC-1.1 cells, 80% of all cytosines at CpG sites analyzed were methylated, 15% of all cytosines at CpG sites analyzed were methylated in HMC-1.2 cells. No methylation was detected in normal BM cells. The percentage of methylation and the significance by Fisher exact test (A) and χ2 test (B) are shown. The bottom panel shows representative chromatograms from BGS in HMC-1.1 cells, HMC-1.2 cells, and normal BM cells. Sites for methylation are underlined. Asterisks indicate cytosines that were converted to thymine. (D) HMC-1.1 and HMC-1.2 cells were incubated in control medium, 5-azacytidine (5μM) or decitabine (5μM) at 37°C for 48 hours (gray bars) or 96 hours (black bars). Thereafter, FAS mRNA expression was analyzed by qPCR. GAPDH served as a reference gene. Results show the fold-increase of mRNA expression and represent the mean ± SD of 3 independent experiments (*P < .05 compared with control). (E) HMC-1 cells were incubated with control medium, 5-azacytidine, or decitabine (each 5μM) in the absence or presence of FAS-ligand (1 ng/mL) for 96 hours. Then, annexin V/PI staining was performed. Results show the percentage of annexin V/PI+ cells. (F) HMC-1 cells were incubated in control medium, 5-azacytidine, or decitabine (each 5μM) in the absence (black bars) or presence (gray bars) of FAS-ligand (1 ng/mL) for 96 hours. Then, the numbers (percentage) of active caspase-3–positive cells were assessed by flow cytometry. Results show the mean ± SD of 3 independent experiments.
Effects of 5-azacytidine and decitabine on the 5′methylation status of FAS and FAS mRNA expression levels in neoplastic MCs. (A) HMC-1 cells were exposed to control medium (untreated), 5-azacytidine (5μM) or decitabine (5μM) for 96 hours. Then, the 5′methylation status of p15, p16, p21, and FAS was determined by MSP. EpiTect-methylated control DNA was used as positive-control. Vertical lines have been inserted to indicate a repositioned gel lane. (B) 5′methylation status of p15, p16, p21, and FAS in normal BM cells in 2 donors (#1 and #2) determined by MSP. To verify efficient sodium bisulfite conversion, we also performed unmethylated specific PCR (USP) for p15, p16, p21, and FAS. EpiTect-methylated and unmethylated control DNA were used as positive-control. (C) BGS of a part of the FAS CGI was performed as previously described using HMC-1.1 cells, HMC-1.2 cells, and normal BM cells. Black squares indicate methylated cytosines at CpG sites, and white squares represent unmethylated cytosines at CpG sites. Whereas in HMC-1.1 cells, 80% of all cytosines at CpG sites analyzed were methylated, 15% of all cytosines at CpG sites analyzed were methylated in HMC-1.2 cells. No methylation was detected in normal BM cells. The percentage of methylation and the significance by Fisher exact test (A) and χ2 test (B) are shown. The bottom panel shows representative chromatograms from BGS in HMC-1.1 cells, HMC-1.2 cells, and normal BM cells. Sites for methylation are underlined. Asterisks indicate cytosines that were converted to thymine. (D) HMC-1.1 and HMC-1.2 cells were incubated in control medium, 5-azacytidine (5μM) or decitabine (5μM) at 37°C for 48 hours (gray bars) or 96 hours (black bars). Thereafter, FAS mRNA expression was analyzed by qPCR. GAPDH served as a reference gene. Results show the fold-increase of mRNA expression and represent the mean ± SD of 3 independent experiments (*P < .05 compared with control). (E) HMC-1 cells were incubated with control medium, 5-azacytidine, or decitabine (each 5μM) in the absence or presence of FAS-ligand (1 ng/mL) for 96 hours. Then, annexin V/PI staining was performed. Results show the percentage of annexin V/PI+ cells. (F) HMC-1 cells were incubated in control medium, 5-azacytidine, or decitabine (each 5μM) in the absence (black bars) or presence (gray bars) of FAS-ligand (1 ng/mL) for 96 hours. Then, the numbers (percentage) of active caspase-3–positive cells were assessed by flow cytometry. Results show the mean ± SD of 3 independent experiments.
Methylation analyses of other apoptosis-related and tumor suppressor antigens
In a next step, we determined the methylation status of 24 classic apoptosis-associated genes and 24 classic tumor suppressor genes (TSGs) in HMC-1.2 cells and in control (normal BM) cells using a commercially available methylation qPCR array. The 48 genes analyzed are shown in supplemental Figure 3. Seven apoptosis-associated genes (CIDEB, GADD45A, HRK, TNFRSF25, BIK, BID, and TNFRSF21) and 6 TSG (NEUROG1, CDH1, GSTP1, CDH13, TP73, and WIF1) were found to be methylated in HMC-1.2 cells but not in normal BM cells, suggesting that these genes are aberrantly hypermethylated in neoplastic MCs. Next, 2 genes (CIDEB and NEUROG1) hypermethylated in HMC-1.2 cells but not in normal BM cells and 1 gene (PDLIM4) hypermethylated in both cell types, were subjected to MS-HRM analyses to confirm qPCR array results (supplemental Figure 3). Methylation qPCR data and MS-HRM data were found to be highly comparable (Pearson correlation coefficient = 0.984; P < .001). In a next step, we examined the effects of 5-azacytidine, and decitabine on the methylation status of these genes. Using MS-HRM analysis, we observed a (slight) demethylating effect of 5-azacytidine on CIDEB, NEUROG1, and PDLIM4 (reduction of methylation by approximately 10%), and a slight demethylating effect (approximately 6%) of decitabine on CIDEB.
5-azacytidine and decitabine cooperate with FAS-ligand in inducing apoptosis in neoplastic MCs
5-azacytidine and decitabine were found to cooperate with FAS-ligand in producing apoptosis in HMC-1 cells (Figure 4E-F, supplemental Figure 4). However, interestingly only a slight cooperative effect was seen when proliferation was analyzed (supplemental Figure 5). In the absence of demethylating agents, FAS-ligand did not induce apoptosis (Figure 4E-F). We also examined whether FAS-ligand and the demethylating agents tested would exert cooperative antineoplastic effects in other myeloid neoplasms. However, in all other cell line models examined, including 3 acute myeloid leukemia (AML) cell lines (HL60, U937, KG1) and 2 chronic myeloid leukemia (CML) cell lines (K562, KU812), no cooperative effects of the drug combinations tested were seen (supplemental Figures 6-7).
siRNA-induced knockdown of FAS in neoplastic MCs leads to resistance against 5-azacytidine and decitabine
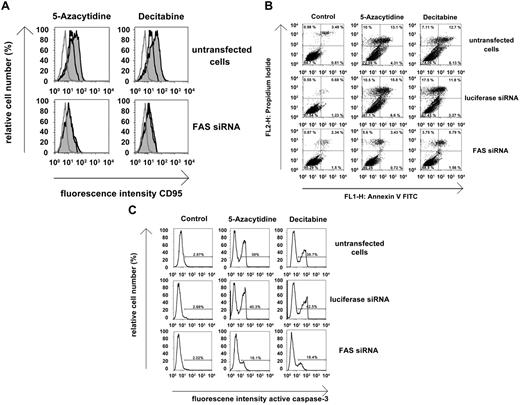
To further confirm that FAS is a relevant death regulator involved in drug-induced growth inhibition and apoptosis in neoplastic MCs, we performed experiments using FAS-specific siRNA. The siRNA-induced knockdown of FAS was confirmed by flow cytometry (supplemental Figure 8). In both HMC-1 subclones, transfection with siRNA was found to lead to resistance against 5-azacytidine and decitabine. In fact, both drugs were unable to induce FAS expression (Figure 5A) or apoptosis (Figure 5B-C) in cells transfected with FAS-specific siRNA (Figure 5A-C). Moreover, FAS siRNA-transfected cells were found to be unresponsive against the growth-inhibitory effects of 5-azacytidine and decitabine (not shown). By contrast, FAS siRNA-transfected cells were still responsive to PKC412 in the same way as nontransfected cells (not shown).
Effects of FAS siRNA on drug-induced FAS expression in HMC-1 cells and responsiveness against demethylating agents. (A) HMC-1.2 cells were kept in control medium (untransfected cells) or were transfected with siRNA against FAS (200nM) using lipofectin. Cells were then incubated in control medium (black-lined open histograms), 5-azacytidine, or decitabine (each 5μM, gray histograms) at 37°C for 30 hours. Thereafter, CD95 expression was analyzed by flow cytometry. Expression of FAS was compared with staining reactions produced by isotype-matched control antibodies (gray-lined open histograms). (B-C) HMC-1.2 cells were kept in control medium (untransfected cells) or were transfected with siRNA against luciferase (200nM) or against FAS (200nM) using lipofectin. After 1 hour, cells were incubated in control medium, 5-azacytidine, or decitabine (each 5μM) at 37°C for 30 hours. After incubation, annexin V/PI staining (B) or active caspase-3 staining (C) was performed by flow cytometry. Results show the percentage of annexin V/PI-positive cells determined by flow cytometry (B), and the percentage of active caspase-3–positive cells (C).
Effects of FAS siRNA on drug-induced FAS expression in HMC-1 cells and responsiveness against demethylating agents. (A) HMC-1.2 cells were kept in control medium (untransfected cells) or were transfected with siRNA against FAS (200nM) using lipofectin. Cells were then incubated in control medium (black-lined open histograms), 5-azacytidine, or decitabine (each 5μM, gray histograms) at 37°C for 30 hours. Thereafter, CD95 expression was analyzed by flow cytometry. Expression of FAS was compared with staining reactions produced by isotype-matched control antibodies (gray-lined open histograms). (B-C) HMC-1.2 cells were kept in control medium (untransfected cells) or were transfected with siRNA against luciferase (200nM) or against FAS (200nM) using lipofectin. After 1 hour, cells were incubated in control medium, 5-azacytidine, or decitabine (each 5μM) at 37°C for 30 hours. After incubation, annexin V/PI staining (B) or active caspase-3 staining (C) was performed by flow cytometry. Results show the percentage of annexin V/PI-positive cells determined by flow cytometry (B), and the percentage of active caspase-3–positive cells (C).
5-azacytidine and decitabine exert differential effects on cell cycle progression and proliferation in HMC-1 cells
We next examined drug effects on cell-cycle progression and proliferation in HMC-1 cells. As visible in Figure 6A, decitabine was found to induce a G2/M cell cycle arrest in HMC-1 cells, whereas no effects were seen with 5-azacytidine. Corresponding results were obtained in a proliferation assay. In particular, as determined in proliferation experiments, decitabine was found to inhibit 3H-thymidine uptake, and thus the proliferation of HMC-1 cells in a dose-dependent manner with comparable IC50 values obtained in the 2 subclones (1-5μM; Figure 6B), whereas 5-azacytidine showed less pronounced growth-inhibitory effects in HMC-1 cells (IC50: 10-20μM; Figure 6B). We also confirmed the growth-inhibitory effects of 5-azacytidine and decitabine in primary neoplastic MCs. As visible in Figure 6C, 5-azacytidine and decitabine were found to inhibit 3H-thymidine uptake in primary neoplastic cells in all patients examined, including one with ISM, one with ASM, and one with SM-CMML. In the patients with ASM and SM-AHNMD, most cells in the test sample were found to express KIT D816V. Similar to HMC-1 cells, the effects of decitabine on proliferation of primary MCs were more pronounced compared with effects seen with 5-azacytidine (Figure 6C). In normal BM cells, no growth-inhibitory effects of 5-azacytidine or decitabine were found (Figure 6D).
Effects of 5-azacytidine and decitabine on proliferation of neoplastic MCs and normal BM cells. (A) HMC-1.1 and HMC-1.2 cells were incubated in control medium (Co) or various concentrations of 5-azacytidine or decitabine (37°C, 96 hours). Thereafter, cell-cycle distribution was measured by flow cytometry using PI. The percentage of cells in G0/G1 phase (black bars), G2/M phase (gray bars), and S phase (open bars) are shown. Results represent the mean ± SD of 3 independent experiments (*P < .05 compared with control). (B) HMC-1 cells were incubated in control medium (Co) or various concentrations of 5-azacytidine or decitabine at 37°C for 48 hours. Thereafter, 3H-thymidine uptake was determined. Results show the percentage of 3H-thymidine uptake compared with control (Co on x axis = 100%) and represent the mean ± SD of 5 independent experiments (*P < .05 compared with control). (C-D) Primary neoplastic cells from patients with systemic mastocytosis, SM (ISM, SM-CMML, and ASM [C]), or normal BM cells (D) were incubated in control medium (Co) or various concentrations of 5-azacytidine or decitabine (37°C, 48 hours). Then, 3H-thymidine uptake was measured. Results show percent 3H-thymidine uptake compared with control. Data obtained in HMC-1 cells (B) represent the mean ± SD from 3 independent experiments, and data obtained with primary cells (C-D) represent the mean ± SD from triplicates (*P < .05 compared with control).
Effects of 5-azacytidine and decitabine on proliferation of neoplastic MCs and normal BM cells. (A) HMC-1.1 and HMC-1.2 cells were incubated in control medium (Co) or various concentrations of 5-azacytidine or decitabine (37°C, 96 hours). Thereafter, cell-cycle distribution was measured by flow cytometry using PI. The percentage of cells in G0/G1 phase (black bars), G2/M phase (gray bars), and S phase (open bars) are shown. Results represent the mean ± SD of 3 independent experiments (*P < .05 compared with control). (B) HMC-1 cells were incubated in control medium (Co) or various concentrations of 5-azacytidine or decitabine at 37°C for 48 hours. Thereafter, 3H-thymidine uptake was determined. Results show the percentage of 3H-thymidine uptake compared with control (Co on x axis = 100%) and represent the mean ± SD of 5 independent experiments (*P < .05 compared with control). (C-D) Primary neoplastic cells from patients with systemic mastocytosis, SM (ISM, SM-CMML, and ASM [C]), or normal BM cells (D) were incubated in control medium (Co) or various concentrations of 5-azacytidine or decitabine (37°C, 48 hours). Then, 3H-thymidine uptake was measured. Results show percent 3H-thymidine uptake compared with control. Data obtained in HMC-1 cells (B) represent the mean ± SD from 3 independent experiments, and data obtained with primary cells (C-D) represent the mean ± SD from triplicates (*P < .05 compared with control).
Cooperative effects of demethylating agents and PKC412 on growth of neoplastic MCs
Because demethylating agents did not alter KIT expression or KIT downstream signaling in HMC-1 cells, we screened for cooperative or even synergistic antineoplastic effects of the KIT D816V-blocker PKC412 and the demethylating agents tested. We found that PKC412, when applied at a constant dose (200nM), promotes the growth-inhibitory effects of both 5-azazytidine and decitabine in HMC-1 cells (supplemental Figure 9). These effects were mostly additive but not synergistic. Moreover, no synergistic effects of 5-azacytidine or decitabine and PKC412 on survival of HMC-1 cells were seen (supplemental Figure 9).
Discussion
Demethylating agents are increasingly used to treat patients with high risk MDS or other advanced myeloid neoplasms.21-27 Although advanced SM is considered a myeloproliferative neoplasm and is sometimes accompanied by an overt MDS or CMML,3-5,28 the disease has so far not been studied in the context of epigenetic mechanisms or epigenetically active drugs. Here, we describe that 5-azacytidine and decitabine induce apoptosis in neoplastic MCs, and that both drugs induce FAS demethylation and surface expression of FAS in MCs. Moreover, both drugs were found to inhibit proliferation and to cooperate with the FAS-ligand in producing apoptosis in neoplastic MCs. The proapoptotic effects of 5-azacytidine and decitabine were seen in HMC-1 cells as well as in primary neoplastic MCs, but not in normal human MCs. These data would be in favor of new treatment concepts using demethylating agents in advanced SM, that is, ASM and MCL.
Apoptosis-inducing effects of 5-azacytidine and decitabine were demonstrable by microscopy, annexin V staining, and by a TUNEL assay. Moreover, we were able to show that both drugs induce caspase-3 and caspase-8 cleavage in HMC-1 cells. The drug concentrations required to produce apoptosis were found to be in a pharmacologically meaningful range, and were similar to concentrations required for induction of apoptosis in other myeloid cells.38-40 An interesting observation was that 5-azacytidine and decitabine induce apoptosis in both HMC-1 subclones, namely HMC-1.1 cells lacking KIT D816V and HMC-1.2 cells expressing KIT D816V, with comparable efficacy. This is of importance because KIT D816V is expressed in neoplastic MCs in a majority of patients with advanced SM6-8 and introduces resistance against KIT tyrosine kinase blockers.31,32 In all, the application of demethylating agents in ASM and MLC may be an interesting approach. In this regard it is also important to state that neither 5-azacytidine nor decitabine were found to induce apoptosis in normal MCs or normal BM cells.
So far, little is known about the methylation profile of neoplastic MCs in systemic mastocytosis or MCL. In this study, we found that a number of classic apoptosis-related genes, including FAS, and several TSG, including p16 and p21, are hypermethylated in HMC-1 cells. Interestingly, p16 and FAS were found to be hypermethylated in both HMC-1 subclones, whereas p21 was found to be hypermethylated only in HMC-1.2 cells, but not in HMC-1.1 cells.
Demethylating agents exert growth-inhibitory and proapoptotic effects on cancer cells through multiple mechanisms, including demethylation and subsequent reexpression of critical TSGs or modulation of other critical target genes.18-20,38-40 In this study, we asked whether the methylation-status of the identified, hypermethylated TSG or apoptosis-related genes in HMC-1 cells is modulated by 5-azacytidine and/or decitabine. As assessed by MS-HRM analysis, 5-azacytidine was found to exert slight demethylating effects on CIDEB, NEUROG1, and PDLIM4, and decitabine produced a mild demethylating effect on CIDEB. However, the most impressive observation was, that both drugs exert profound demethylating effects on FAS in both HMC-1 subclones. We also found that 5-azacytidine and decitabine induce reexpression of the FAS protein in HMC-1 cells. Two other proapoptotic death regulators, Bim and Noxa, were also found to increase on exposure to decitabine in HMC-1 cells, but these drug effects did not reach statistical significance. All in all, these results suggest that hypermethylated FAS may be a major target of 5-azacytidine and decitabine in neoplastic MCs.
To confirm this hypothesis, we asked whether FAS reexpression in neoplastic MCs is of functional significance and responsible for drug-induced apoptosis. To address this question, we applied siRNA against FAS. The siRNA-induced knockdown of FAS in HMC-1 cells resulted in resistance against 5-azacytidine and decitabine. In particular, siRNA-transfected cells were no longer able to up-regulate FAS and to undergo apoptosis on exposure to 5-azacytidine or decitabine. These data strongly suggest that 5-azacytidine and decitabine exert proapoptotic effects on MCs via FAS reexpression, and that FAS is a critical death regulator in neoplastic MCs. Interestingly, only a few data are available about FAS hypermethylation and FAS reexpression induced by demethylating agents in other cancer types. In one study, FAS has been described to be hypermethylated in colon cancer cells, and decitabine was found to lead to demethylation of FAS in neoplastic cells.41 To the best of our knowledge, our report is the first to demonstrate epigenetic regulation of FAS in neoplastic MCs, and that demethylating agents induce FAS expression and thereby apoptosis in these cells.
A number of previous and more recent data suggest that decitabine induces a cell-cycle arrest in various neoplastic cells, whereas 5-azacytidine is usually less effective.38,42-47 In line with these observations, we found that decitabine, but not 5-azacytidine, is capable of inducing cell cycle arrest in HMC-1.1 and HMC-1.2 cells. Moreover, we found that decitabine, and less effectively 5-azacytidine, inhibit the proliferation of HMC-1 cells. The effects of decitabine on growth of neoplastic MCs were dose-dependent and were also seen in primary neoplastic MCs obtained from patients with indolent SM, ASM, and SM with associated CMML. Again IC50 values were found to be within a pharmacologically meaningful range. An important point in our experiments was to adapt the 3H-thymidine uptake assay to the drug-type analyzed. In fact, in pilot experiments we found that 5-azacytidine directly interferes with 3H-thymidine uptake. Therefore, cells had to be washed thoroughly before adding 3H-thymidine. However, after thorough washing, results were comparable in all assays and all control conditions analyzed.
Recent data suggest that interactions between FAS and the FAS-ligand support apoptosis in neoplastic cells.41,48,49 We asked whether the FAS-ligand promotes drug-induced apoptosis in neoplastic MCs. In these experiments we found that the FAS-ligand per se does not induce growth inhibition or apoptosis in neoplastic MCs. However, the FAS-ligand was found to promote 5-azacytidine–induced and decitabine-induced apoptosis in neoplastic MCs. These data suggest that FAS is an inducible death regulator in neoplastic MCs, which may act as a more potent proapoptotic cofactor in drug-exposed cells when interacting with the FAS-ligand, an observation that may have clinical implications. We therefore asked whether the cooperative inhibitory effects of the FAS-ligand and demethylating agents on cell growth is a general phenomenon common to all types of myeloid neoplasms. However, we found that the cooperative effects of the FAS-ligand with 5-azacytidine and decitabine are specific for neoplastic MCs, as no such cooperative effects were seen in the other myeloid leukemia models analyzed, that is, AML and CML.
As ASM and MCL are highly drug-resistant neoplasms, several attempts have been made to develop new effective therapeutic concepts.11-17 One approach is to apply kinase blockers capable of suppressing the kinase activity of KIT D186V, such as PKC412 (midostaurin). We asked whether 5-azacytidine and decitabine would down-regulate expression of KIT D816V. However, no effects of the demethylating agents on KIT expression were found. Next, we asked whether 5-azacytidine or decitabine would cooperate with PKC412 in producing growth inhibition or apoptosis in neoplastic MCs. Indeed, when applied at a fixed concentration of 200nM, PKC412 was found to promote the growth-inhibitory effects of 5-azacytidine and decitabine on HMC-1 cells in an additive fashion. However, no synergistic drug interactions, neither on growth nor on survival of HMC-1 cells, were seen in this study.
In advanced SM including ASM and MCL, researchers are seeking novel drugs that may be capable of counteracting the devastating growth of MCs in ASM and MCL. In our study, pharmacologically meaningful concentrations of 5-azacytidine and decitabine were found to induce apoptosis in HMC-1 cells, including the HMC-1.2 subclone that exhibits the drug-resistant KIT mutant D816V. To the best of our knowledge this is the first study reporting antineoplastic effects of demethylating agents on human neoplastic MCs. In addition, our report is the first to describe that the death regulator FAS is hypermethylated in neoplastic MCs and that 5-azacytidine and decitabine can induce demethylation and thus reexpression of FAS. Whether these effects can be confirmed in clinical trials in patients with ASM and MCL, remains to be determined.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This study was supported by Austrian Science Fund (FWF) grants P21173-B13 and SFB 04611; Vienna Science and Technology Fund (WWTF) grant LS07-019; and Jubiläumsfonds der Österreichischen Nationalbank grant 13068.
Authorship
Contribution: V.G. performed key staining, cell growth, and siRNA experiments and wrote parts of the paper; H.H. contributed cell growth and flow cytometry staining experiments; G.H. and S.Z-M. performed MSP, bisulfite genomic sequencing, EpiTect methyl qPCR array analyses, methylation-sensitive melting curve analyses, and qPCR experiments; B.P. contributed Western blot and qPCR experiments; E.H. contributed TUNEL staining experiments; K.B. contributed cell isolation and culture as well as flow cytometry experiments; K.S. and W.P. performed 3H-thymidine uptake experiments; S.C.-R. performed qPCR experiments; I.M. cultured and provided primary human cord blood–derived MCs; H.K. performed qPCR and siRNA experiments; and P.V. contributed logistic and budget support and wrote and approved the paper.
Conflicts-of-interest disclosure: P.V. is consultant of a Novartis PKC412 trial and received a Research Grant from Novartis. The remaining authors declare no competing financial interests.
Correspondence: Peter Valent, Dept of Internal Medicine I, Division of Hematology & Hemostaseology, Medical University of Vienna, Waehringer Guertel 18-20, A-1090 Vienna, Austria; e-mail: peter.valent@meduniwien.ac.at.

![Figure 6. Effects of 5-azacytidine and decitabine on proliferation of neoplastic MCs and normal BM cells. (A) HMC-1.1 and HMC-1.2 cells were incubated in control medium (Co) or various concentrations of 5-azacytidine or decitabine (37°C, 96 hours). Thereafter, cell-cycle distribution was measured by flow cytometry using PI. The percentage of cells in G0/G1 phase (black bars), G2/M phase (gray bars), and S phase (open bars) are shown. Results represent the mean ± SD of 3 independent experiments (*P < .05 compared with control). (B) HMC-1 cells were incubated in control medium (Co) or various concentrations of 5-azacytidine or decitabine at 37°C for 48 hours. Thereafter, 3H-thymidine uptake was determined. Results show the percentage of 3H-thymidine uptake compared with control (Co on x axis = 100%) and represent the mean ± SD of 5 independent experiments (*P < .05 compared with control). (C-D) Primary neoplastic cells from patients with systemic mastocytosis, SM (ISM, SM-CMML, and ASM [C]), or normal BM cells (D) were incubated in control medium (Co) or various concentrations of 5-azacytidine or decitabine (37°C, 48 hours). Then, 3H-thymidine uptake was measured. Results show percent 3H-thymidine uptake compared with control. Data obtained in HMC-1 cells (B) represent the mean ± SD from 3 independent experiments, and data obtained with primary cells (C-D) represent the mean ± SD from triplicates (*P < .05 compared with control).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/119/18/10.1182_blood-2011-09-382770/4/m_zh8999129046006a.jpeg?Expires=1767708348&Signature=XU3fUk~pQPG4y4ZDA0bt4wyWbCr6i7nX0ly4eReatHxkHOT4QKo43EDhHS~T3asCsQxM2Jw2OgSrIt74krS1o6RpLLWX8xhewxLYc4VXHkilJdB85eyPRMAJb97DQ3t~GCxxCEyG4~AAHBLdOZL3WyJDQ9zGc28-xo4JZ6jKnCqwJKY-5ySbBT1Q6orI-o-VSUT56IrkbgXqpGGg504b12Rbp8ZxM~QyckhLHDtxP-oF-msmPXzP9IrqUtWiNTJIxUPFBTDaIDvVa2mp8OWVk5I0Jd4IutBbvhMt1ESgixZuUzho97F-NghBKVcdNpvj0UwiZ64WqxqDDar4M8XOcw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal