Abstract
MiR-125b-1 maps at 11q24, a chromosomal region close to the epicenter of 11q23 deletions in chronic lymphocytic leukemias (CLLs). Our results establish that both aggressive and indolent CLL patients show reduced expression of miR-125b. Overexpression of miR-125b in CLL-derived cell lines resulted in the repression of many transcripts encoding enzymes implicated in cell metabolism. Metabolomics analyses showed that miR-125b overexpression modulated glucose, glutathione, lipid, and glycerolipid metabolism. Changes on the same metabolic pathways also were observed in CLLs. We furthermore analyzed the expression of some of miR-125b–target transcripts that are potentially involved in the aforementioned metabolic pathways and defined a miR-125b–dependent CLL metabolism-related transcript signature. Thus, miR-125b acts as a master regulator for the adaptation of cell metabolism to a transformed state. MiR-125b and miR-125b–dependent metabolites therefore warrant further investigation as possible novel therapeutic approaches for patients with CLL.
Introduction
One way tumor cells gain advantage over nontransformed cells is by reorienting their metabolic activity toward the synthesis of nucleotides, lipids, and proteins needed for cell growth and proliferation. This is mainly done by increasing the rate of glycolysis even in aerobic conditions (ie, the Warburg effect)1,2 and decreasing the activity of the tricarboxylic acid (TCA) cycle, which provides tumor cells with both the energy (ie, ATPs) and carbon and nitrogen donors subsequently used for the synthesis of needed macromolecules.3-7 This assumption has led to the development of metabolomics, which is defined as the quantitative measurement of all low-molecular-weight metabolites in an organism at a specified time under specific environmental conditions. Indeed, different analytical techniques used to explore the chemical fingerprint of the samples recently have been shown to be effective tools for disease diagnosis, biomarker screening, and characterization of biologic pathways in cancer.8
miRNAs are short, noncoding RNAs that regulate cell homeostasis, including proliferation, differentiation, response to signaling molecules, and metabolism.9,10 Increased or decreased expression of oncogenic or tumor-suppressor miRNAs has been associated with cancer.11,12 Being the homologue of Caenorhabditis elegans lin-4, miR-125b is one of the evolutionary most conserved miRNAs.13 Depending on the context, miR-125b behaves as either a tumor suppressor or an oncogenic miRNA.14-17 In humans, miR-125b-1 maps on chromosome 11q24, a region known for its recurrent chromosomal abnormalities observed in different types of lymphoid or myeloid malignancies, including chronic lymphocytic leukemia (CLL).18,19 The expression of miR-125b decreases shortly after LPS stimulation of RAW264.7 macrophages, suggesting that it might have anti-inflammatory effects.20 In contrast, LPS increases the expression of miR-155, a proinflammatory miRNA that is highly expressed in several types of cancers, including CLL.20-22 Given that ± 25% of tumors have been linked to the presence of a proinflammatory environment,23,24 these results suggest that some tumors might arise from the concurrent up-regulation of miR-155 and down-regulation of miR-125b, as, for example, in breast cancer.25
To determine whether miR-125b might be implicated in CLL, we analyzed miR-125b and miR-155 expression in a panel of patients with either an indolent or aggressive form of CLL. Our report establishes that miR-125b down-regulation in both forms of CLL is associated with the up-regulation of several transcripts encoding enzymes or regulators of cell metabolism, a significant proportion of whose represent putative miR-125b targets, and that the down-regulation of miR-125b in CLL is associated with metabolic adaptation to cancer transformation.
Methods
Patients
In accordance with the Declaration of Helsinki, in the current study, 80 patients with CLL (40 indolent [IND] and 40 aggressive [AGG] in addition to 20 healthy donors [HD]) were enrolled in the CLL Research Consortium after they provided written informed consent (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). The institutional review board of The Ohio State University (OSU) provided approval for this research. PBMCs were isolated by density gradient centrifugation with the use of Ficoll-Paque Plus (Amersham Biosciences). The PBMCs obtained from these patients were > 98% leukemic CD5+/CD19+ B cells. Purified B cells from different healthy donors were purchased from Astarte Biologics and from Sanguine BioSciences.
Cell culture and transfection
Cell lines used on this study were purchased from ATCC. MEC1 and MEC2 CLL cell lines were developed in Dr Cappio's laboratory.26 Cells were maintained in culture following standard procedures. Cells in suspension were electroporated with the AMAXA kit. HEK-293 and MEG-01 cells were transfected with Lipofectamine 2000 (Invitrogen). MicroRNAs used for transfection were purchased from Ambion. For metabolite profiling, cells were frozen 48 hours after the transfection.
Affymetrix microarrays analyses
Affymetrix analyses were performed at the Micro-Array facility at OSU. Data were submitted to MIAME ArrayExpress database, with accession no. E-MTAB-1131.
Preparation of clones and luciferase reporter assays
The 3′-untranslated region (UTR) of the IKZF2, IKZF3, and IKZF4 genes was cloned downstream of the Luciferase gene in the XbaI site of the pGL3-Control vector (Promega). Luciferase assays were performed 48 hours after transfection, as previously described.27 The 3′-UTRs of GSS, LIPA, and PCTP cloned downstream of Renilla luciferase gene were purchased from Switchgear Genomics and used according to the manufacturer's instructions. Luciferase assays for IKZF genes were performed in MEG-01 cells, whereas those for other genes were performed in HEK-293 cells.
Metabolite analyses
Metabolite profiling analysis of all samples was performed by Metabolon Inc, as previously described.28 Two types of statistical analysis were performed: (1) significance tests and (2) classification analysis. (1) For pair-wise comparisons, a Welch 2-sample t test was used to identify biochemicals that differed significantly between experimental groups. Only the biochemicals that achieved statistical significance (P ≤ .05), as well as those approaching significance (.05 < P < .1) were considered. (2) For classification, Random Forests (RFs) analyses were performed. RFs provide an estimate of how well individuals can be classified in a new dataset into each group, in contrast to a t test, which tests whether the unknown means for 2 populations are different. RFs create a set of classification trees on the basis of continual sampling of the experimental units and compounds. Then, each observation is classified by the majority votes from all the classification trees. Statistical analyses were performed with the program R (http://cran.r-project.org/). The metabolite profiles within each group of samples were compared to assess (1) the capacity to distinguish between disease groups by global metabolite profiles and (2) biochemicals important to the classification. RF analyses predicted the statistical differences among the 3 groups with 71% accuracy, an accuracy that increased to 93% when IND and AGG patients were pooled before comparison with HD patients. For the cell lines, global biochemical profiles were determined in each of the indicated cell lines after miR-125b overexpression in relation to miR-Control.
Pair-wise differences in expression in the cell line data were tested by the use of 2-tailed t tests with Satterthwaite variance estimation. An estimate of the false-discovery rate (q-value) was used for calculation that takes into account the multiple comparisons normally occurring in metabolomic-based studies. For example, when analyzing 200 compounds, one would expect to see approximately 10 compounds meeting the P ≤ .05 cut-off by random chance. The q-value describes the false-discovery rate; a low q-value (q < 0.10) is an indication of high confidence in a result. Although a greater q-value indicates diminished confidence, it does not necessarily rule out the significance of a result. Other lines of evidence may be taken into consideration when determining whether a result merits further scrutiny. Such evidence may include (1) significance in another dimension of the study, (2) inclusion in a common pathway with a highly significant compound, or (3) residing in a similar functional biochemical family with other significant compounds.
In addition to producing a metric of predictive accuracy, RF analysis produces an associated list of biochemicals ranked in order of their importance to the classification scheme. The top 30 biochemicals for these RF comparisons were selected for disease groups. Instrument variability was determined by calculating the median relative SD for the internal standards that were added to each sample before injection into the mass spectrometers. Overall process variability was determined by calculating the median relative SD for all endogenous metabolites (ie, noninstrument standards) present in 100% of the Client Matrix samples, which are technical replicates of pooled client samples.
Quantitative real-time PCR
MiRNA and gene expression quantitative real-time PCR (qRT-PCR) assays were performed with the use of TaqMan MicroRNA Assays or Gene Expression Assays (both from Applied Biosystems), respectively. Values were normalized with RNU44 for miRNAs and OAZ1 for genes. Real-time PCRs were run in triplicate for each cDNAs. Because miR-125b-1 and miR-125b-2 are 100% identical, they are collectively referred to as miR-125b. Assay IDs were as follows: (1) for miRNAs: 1094: RNU44, small nucleolar RNA, used as a normalizer; 449: hsa-miR-125b; 2623: hsa-miR-155; (2) for genes: Hs00427923_m1*: OAZ1, used as a normalizer; Hs00287264_m1: ACSS1 (CDS); Hs00609286_m1: GSS (CDS); Hs00606086_m1: HK2 (CDS); Hs01548815_m1: LIPA (CDS); Hs00221886_m1: PCTP (CDS); Hs01561850_m1: PDK1 (CDS); Hs01682761_m1: SCD1 (CDS); Hs01086099_m1: AKT2 (CDS); Hs01085579_m1*: EIF2C2 (AGO-2); Hs00222230_m1: FADS3 (CDS); Hs00222230_m1.
Western blots
ACSS1, GNPDA1, GSS, HK2, IKZF4, LIPA, PCTP, PDK1, and TP53 antibodies were purchased from Santa Cruz Biotechnology. Peroxisome proliferator-activated receptor alpha (PPARα) antibody was from Cayman Chemical. AKT2 and α-tubulin antibodies were from Cell Signaling Technology. GLS1, ALOX5, FADS3, and vinculin antibodies were from Abcam. SCD1 antibody was a gift from Dr J. B. Demoulin (Université catholique de Louvain Brussels, Belgium).29
Results and discussion
MiR-125b is down-regulated in CLL
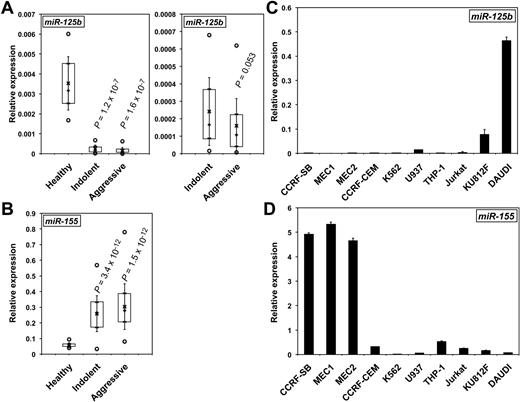
MiR-125b levels were measured by qRT-PCR in the lymphocytes of HD patients and purified B cells of HD, IND, and AGG CLL patients (supplemental Table 1), including some with del(17p). MiR-125b expression was reduced in both CLL forms compared with total lymphocytes from HD patients (Figure 1A) or purified B cells of HD, irrespective of del(17p) (supplemental Figure 1A and supplemental Figure 2A). Of note, miR-125b levels were slightly lower in AGG than in IND CLLs (P = .053; Figure 1A). In contrast, miR-155 levels in the same patients were greater than in HD cells (Figure 1B and supplemental Figure 1B), in agreement with previous reports.21 Interestingly the paralogue miR-125a may possibly be down-regulated only in the aggressive form of CLL, suggesting that miR-125a might be a marker of aggressiveness (supplemental Figure 1C). We then measured the expression of miR-125b and miR-155 in CLL-derived cell lines (MEC1 and MEC2) and other leukemic B, T, or monocytic cell lines. As expected, miR-125b levels were low in MEC cells (Figure 1C). In contrast, miR-155 showed an inverse expression, with the greatest levels found in MEC1 and MEC2 cells (Figure 1D) and lowest in DAUDI, as previously reported for Burkitt lymphoma.30 Altogether, these results show that CLL is characterized by low miR-125b and high miR-155 expression compared with HD and that miR-125b expression further decreases during the transformation to aggressive CLL.
MiR-125b expression decreases in CLL patients and in CLL-derived cell lines. The relative levels of miR-125b (A) and miR-155 (B) in patients with CLL were determined by qRT-PCR. P values (Student t test) are given for indolent (n = 35) and aggressive (n = 38) versus healthy (n = 15; A, left) and for indolent versus aggressive (A, right). Boxes include values from the first to the third quartiles; o indicates extreme data points; +, median; x, mean ± SD. The relative levels of miR-125b (C) and miR-155 (D) in CLL cell lines (MEC1 and MEC2), a B-lymphoblastoid leukemia cell line (CCRF-SB), Burkitt lymphoma DAUDI B lymphoblasts, T cells (CCRF-CEM, Jurkat, and K812F), and monocytes (THP-1 and U937) were determined by qRT-PCR. qRT-PCRs were run in triplicate with the use of 3 different cDNAs (C-D).
MiR-125b expression decreases in CLL patients and in CLL-derived cell lines. The relative levels of miR-125b (A) and miR-155 (B) in patients with CLL were determined by qRT-PCR. P values (Student t test) are given for indolent (n = 35) and aggressive (n = 38) versus healthy (n = 15; A, left) and for indolent versus aggressive (A, right). Boxes include values from the first to the third quartiles; o indicates extreme data points; +, median; x, mean ± SD. The relative levels of miR-125b (C) and miR-155 (D) in CLL cell lines (MEC1 and MEC2), a B-lymphoblastoid leukemia cell line (CCRF-SB), Burkitt lymphoma DAUDI B lymphoblasts, T cells (CCRF-CEM, Jurkat, and K812F), and monocytes (THP-1 and U937) were determined by qRT-PCR. qRT-PCRs were run in triplicate with the use of 3 different cDNAs (C-D).
MiR-125b overexpression modifies the expression of transcripts encoding enzymes and other factors involved in cell metabolism
MiRNAs often down-regulate the levels of their target transcripts. Therefore, we analyzed the consequences of miR-125b overexpression on MEC2 cell transcriptome by using Affymetrix microarrays. Overall, the expression of 765 coding transcripts was modified with high level of significance (P < .0005). As many as 420 of these 765 transcripts (ie, ± 55%) were down-regulated, including TP53INP1, VDR, TP53, PRDM1, and IRF4, all validated targets of miR-125b (supplemental Table 2). Accordingly, TargetScan analyses (http://www.targetscan.org/) showed that the 3′-UTR of 186 down-regulated transcripts contains at least one miR-125b target site, suggesting that they may represent actual miR-125b targets (supplemental Table 2). Interestingly, 60 of the 186 transcripts down-regulated after miR-125b overexpression (ie, ± 32%) encode enzymes, a majority implicated either in lipid, phospholipid, carbohydrate, amino acid, or nucleotide metabolism (supplemental Table 2). In addition, many transcripts modulated by miR-125b expression with .0005 ≤ P < .005 also encode enzymes and contain miR-125b target site(s). Furthermore, ± 23% of transcripts predicted by TargetScan software to represent miR-125b targets also encode enzymes compared with ± 15% for miR-155. Therefore, we hypothesized that by potentially altering the levels of many metabolic enzymes, miR-125b down-regulation in CLL might translate into cancer-related metabolic changes.
MiR-125b targets transcripts encoding enzymes and factors implicated in metabolism
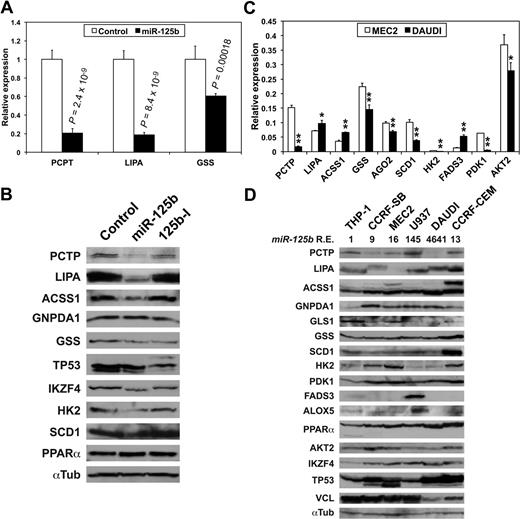
We then checked the effects of miR-125b on luciferase constructs containing the 3′-UTR of some of the transcripts of supplemental Table 2. MiR-125b overexpression in HEK-293 cells decreased the expression of luciferase constructs containing the 3′-UTR of PCTP, LIPA, and GSS (Figure 2A, supplemental Table 2). In contrast, in DAUDI cells, where miR-125b was highly expressed (Figure 1C), a miR-125b inhibitory RNA (miR-125-I) slightly but significantly increased luciferase activity produced from the 3 same constructs (supplemental Figure 3). Furthermore, luciferase assays showed that the 3′-UTRs of IKZF2, IKZF3, and IKZF4 also are targeted by miR-125b (supplemental Figure 4), confirming Affymetrix analyses (supplemental Table 2) and suggesting that miR-125b might impact hematopoiesis by targeting Ikaros transcripts.31 The members of the Ikaros family of transcription factors are hematopoietic-specific factors involved in the regulation of lymphocyte development, and their deregulated expression is observed in different hematopoietic malignancies, including CLL.32 Overexpressing miR-125b in MEC2 cells decreased the levels of PCTP, LIPA, ACCS1, GSS, HK2 and SCD1, IKZF4, and TP53, in agreement with Affymetrix analysis, while not affecting GNPDA1 and slightly increasing PPARα levels (Figure 2B). Given the weak miR-125b expression in MEC2 cells, miR-125-I had no effect (Figure 2B). In conclusion, miR-125b potentially targets many of the transcripts encoding enzymes and metabolic regulators of supplemental Table 2 and might thus have a global impact on CLL patient metabolism.
miR-125b targets transcripts encoding enzymes and regulators of cell metabolism. (A) Luciferase assays showing miR-125b effects on the expression of luciferase constructs containing the indicated 3′-UTR in HEK-293 cells. Values represent mean ± SD (n = 4). P values (Student t test) are given for miR-125b versus control. (B) The effects of miR-125b and of an antisense miR-125b inhibitory RNA (125b-I) on the expression of the indicated enzymes and factors in MEC2 cells were analyzed on Western blots. A representative normalizer (αTub, ie, α-tubulin) is shown. (C) The relative levels of the indicated transcripts in MEC2 and DAUDI cell lines were determined by qRT-PCRs run in triplicate from 3 different cDNAs. Values represent mean ± SD (n = 3). * and **, DAUDI significantly different from MEC2. *P < .05, **P < .027. (D) The relative levels of the indicated enzymes and factors in cell lines of different origins were assessed on Western blots. miR-125b relative expression levels (miR-125b R.E.) are shown underneath the name of the cell lines, with THP-1 cells used as a reference.
miR-125b targets transcripts encoding enzymes and regulators of cell metabolism. (A) Luciferase assays showing miR-125b effects on the expression of luciferase constructs containing the indicated 3′-UTR in HEK-293 cells. Values represent mean ± SD (n = 4). P values (Student t test) are given for miR-125b versus control. (B) The effects of miR-125b and of an antisense miR-125b inhibitory RNA (125b-I) on the expression of the indicated enzymes and factors in MEC2 cells were analyzed on Western blots. A representative normalizer (αTub, ie, α-tubulin) is shown. (C) The relative levels of the indicated transcripts in MEC2 and DAUDI cell lines were determined by qRT-PCRs run in triplicate from 3 different cDNAs. Values represent mean ± SD (n = 3). * and **, DAUDI significantly different from MEC2. *P < .05, **P < .027. (D) The relative levels of the indicated enzymes and factors in cell lines of different origins were assessed on Western blots. miR-125b relative expression levels (miR-125b R.E.) are shown underneath the name of the cell lines, with THP-1 cells used as a reference.
Patients with CLL show evidence of a cancer-specific metabolic signature
We then conducted a Metabolon metabolomics analysis in IND and AGG patients to determine whether their lower levels of endogenous miR-125b compared with HDs translated into metabolic changes. Most of the significant alterations (P < .05) in biochemical levels corresponded to increases: 108 of 141 in IND versus HD and 96 of 131 in AGG versus HD. In contrast, 25 of 28 changes in AGG versus IND were decreases, indicating that metabolic activity tends to be greater in IND than in AGG samples (Figure 3, supplemental Table 3).
Heat map of biochemical changes in CLL patients. Values give the ratios of metabolite levels, determined with the Metabolon platform, between the indicated phenotypes (healthy donors [n = 20], indolent [n = 40], and aggressive [n = 40] CLL patients). This table is an extract of the data for all metabolites levels shown in supplemental Table 3. Statistical comparisons were done with the use of the Welch 2-sample t test. All data were normalized with the Bradford protein concentration.
Heat map of biochemical changes in CLL patients. Values give the ratios of metabolite levels, determined with the Metabolon platform, between the indicated phenotypes (healthy donors [n = 20], indolent [n = 40], and aggressive [n = 40] CLL patients). This table is an extract of the data for all metabolites levels shown in supplemental Table 3. Statistical comparisons were done with the use of the Welch 2-sample t test. All data were normalized with the Bradford protein concentration.
IND and AGG patients showed lower levels of glucose and glucose 6-phosphate than HD patients (Figure 3, supplemental Table 3). In addition, there were also greater levels of glycolytic intermediates 3-phosphoglycerate, 2-phosphoglycerate, and lactate. These changes are consistent with a hallmark of cancer, namely the Warburg effect.1,2 Accordingly, citrate levels were lower in both the IND and AGG, findings consistent with limited entry of glycolytic end products into the TCA cycle, although the differences reached statistical significance only in the comparison of IND versus HD. Additional TCA intermediates (succinate, fumarate, and malate) were significantly greater in both IND and AGG, suggesting an anaplerotic incorporation of amino acids into the TCA cycle for energy production. Expectedly, there were also greater levels of essential, medium- and long-chain fatty acids (FAs) in IND and AGG in contrast to control patients, as well as greater levels of monoacylglycerols and glycerol, which further suggests increased lipolysis to support FA synthesis and membrane remodeling. The levels of polyamines such as spermine and spermidine, typically synthesized in small quantities by healthy living cells, were high in patients with CLL. There were also indications of altered arginine metabolism in IND and AGG, possibly to support the observed increase of polyamine synthesis.
Elevated levels of both oxidized and reduced glutathione in IND and AGG samples and lower levels of cysteine, a key precursor for glutathione synthesis, were also characteristics of CLL patients (Figure 3, supplemental Table 3). Of note, elevated glutathione levels in tumor cells have been associated with resistance to chemotherapy. Furthermore, it was recently reported that BM stromal cells promote GSH metabolism in CLL cells and enhance the leukemia cell survival and drug resistance.33 Metabolomics analyses suggested that S-lactoylglutathione; the polyamines spermine and spermidine; citrulline; and 12-HETE can be used as potential CLL-biomarkers (Figure 3, supplemental Table 3, supplemental Figure 5), whereas creatine and creatinine can discriminate between IND and AGG CLLs (Figure 3, supplemental Figure 6). The levels of mutagenic betaine aldehyde were greater in patients with CLL, especially those with del(17p) (Figure 3, supplemental Figure 7), suggesting that patients with del(17p) are at greater risk of mutation. Because 74% of aldehyde dehydrogenases (ALDH) map in CLL-fragile sites (supplemental Table 4), with ALDH3A1 and ALDH3A2 on 17p11.2, they might be lacking in del(17p) CLLs.
Overall, CLL cells showed strong evidence of metabolic adaptation to proliferation. Of note, although miR-125b levels were lower in AGG patients (Figure 1A), metabolite levels were generally greater in patients with IND CLL (Figure 3, supplemental Table 3), suggesting that miR-125b effects on metabolism, and consequently during cancer transformation, might be dose- or context-dependent. Glutathione increase in CLL cells suggested that they are predisposed to respond to therapies that generate oxidative stress to induce apoptosis. In addition, elevated FA synthesis and enhanced lypolysis in CLLs suggest that FA synthesis inhibitors might be relevant to CLL treatment.
Metabolic consequences of miR-125b overexpression
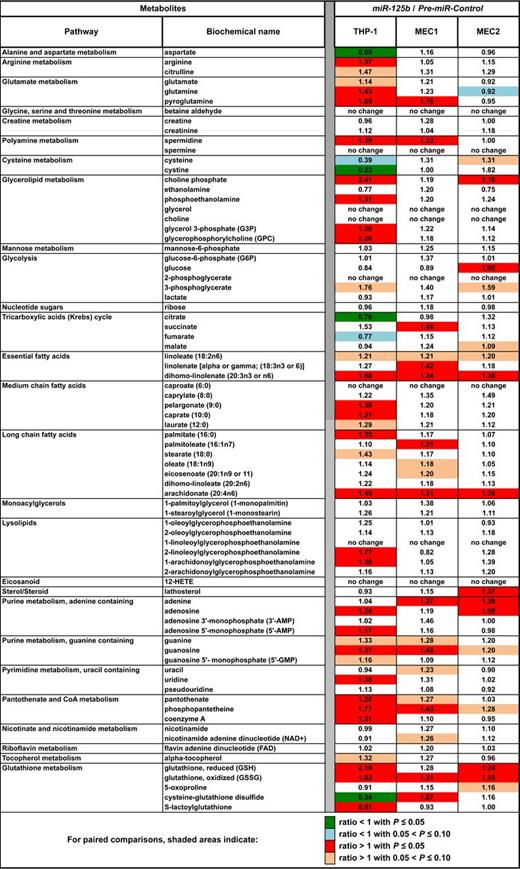
The aforementioned results prompted us to do metabolomics profiling after miR-125b overexpression in MEC1, MEC2, and THP-1 cell lines. High miR-125b levels mostly increased (P < .05) metabolite levels (Figure 4, supplemental Table 5). Fold changes generally were greater in THP-1 and lower in MEC2 cells, ie, those with lowest and highest miR-125b endogenous levels, respectively. MiR-125b overexpression was generally associated with changes in lipid and glycerolipid metabolism, coenzyme A, and glutathione synthesis. In particular, miR-125b overexpression in MEC1 and MEC2 cells led to changes opposite to those found in CLLs for aspartate, citrulline, glutamine, cysteine, glucose, lathosterol, guanine, guanosine, uracil, or cysteine-glutathione disulfide, which suggests that metabolic changes in patients with CLL were linked to miR-125b down-regulation and that greater miR-125b levels might oppose Warburg effect by decreasing glucose consumption (Figure 4, supplemental Table 5). Nevertheless, other metabolites did not follow this rule, possibly because of miR-125b–dependent dose effects. The fact that most of statistically significant changes occurred in THP-1 cells, with the lowest miR-125b endogenous levels, further suggests that miR-125b effects might be dose-dependent. Regardless, metabolomics analyses showed that miR-125b affects the same metabolic pathways in CLLs and cell lines, supporting its role as a metabolic regulator.
Heat map of biochemical changes in the indicated cell lines after miR-125b overexpression. Values give the ratios of metabolite levels, determined by use of the Metabolon platform, between miR-125b versus miR-Control in THP-1, MEC1, and MEC2 cell lines (n = 4 in each case). This table is an extract of the data for all metabolites levels shown in supplemental Table 5. Pair-wise differences in expression within each cell line were tested with a 2-tailed t test with Satterthwaite variance estimation.
Heat map of biochemical changes in the indicated cell lines after miR-125b overexpression. Values give the ratios of metabolite levels, determined by use of the Metabolon platform, between miR-125b versus miR-Control in THP-1, MEC1, and MEC2 cell lines (n = 4 in each case). This table is an extract of the data for all metabolites levels shown in supplemental Table 5. Pair-wise differences in expression within each cell line were tested with a 2-tailed t test with Satterthwaite variance estimation.
Expression of key metabolic factors varies according to miR-125b levels
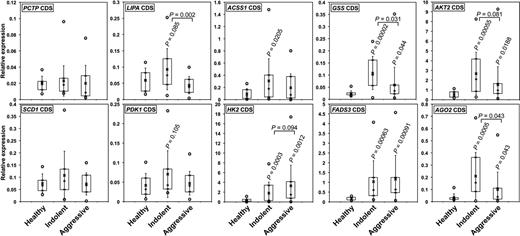
Beside the transcripts presented in supplemental Table 2, the expression of transcripts encoding metabolic enzymes and regulators such as HK2, PDK1, SCD1, FADS3, GLS1, ALOX5, IGF1R, AKT2, and PPARα, all containing miR-125b target sites (supplemental Table 6), also was significantly affected by miR-125b overexpression in MEC2 cells (P < .005). We therefore compared their levels in MEC2 and DAUDI cells. Although PCTP, GSS, SCD1, HK2, PDK1, and AKT2 levels were expectedly lower in DAUDI than in MEC2 cells, those of LIPA, ACSS1, and FADS3 were actually greater (Figure 2C). The levels of the corresponding proteins in MEC2 and DAUDI cells were in good agreement with qRT-PCR results (Figure 2D). However, LIPA, ACSS1, TP53, and PPARα levels were greater in DAUDI cells. It must be noted that LIPA, ACSS1, and TP53 had different isoforms expressed in the studied cells (Figure 2B, D), whereas PPARα was previously up-regulated after miR-125b overexpression in MEC2 cells (Figure 2B). The aforementioned results strongly suggest that these key metabolic enzymes and regulators represent actual miR-125b targets.
The expression of miR-125b putative target transcripts encoding key effectors of metabolism is modified in CLLs
We subsequently analyzed the expression of the aforementioned transcripts in CLLs. In most cases, transcript levels increased in IND but decreased again in AGG CLLs (Figure 5), in agreement with the fact that metabolite accumulation was previously found higher in IND than in AGG CLLs (Figure 3). These results again suggest that effects of miR-125b may be dose-dependent, a phenomenon reported for MYC as well.34 On the basis of these facts, we can hypothesize that the loss of miR-125b expression might contribute to the initial steps of CLL development, especially by allowing Warburg effect. At later stages, although miR-125b levels decrease further, the activity of other regulators, possibly aimed at limiting miR-125b deleterious effects, may oppose a further increase of cell metabolism. Interestingly, transcripts encoding AGO2, a component of the RISC complex, presented a similar pattern of expression, suggesting that miR-125b might possibly affect the expression of several transcripts indirectly. Of note, the levels of AGO2 transcripts, also predicted targets of miR-125b, decreased after overexpression of miR-125b (Affymetrix microarrays) and were lower in DAUDI than in MEC2 cells (Figure 2C).
The relative levels of miR-125b target transcripts encoding key enzymes or regulators of metabolism increase in patients with CLL. The relative levels of the coding sequences (CDS) of the indicated transcripts were determines by qRT-PCR in IND and AGG CLL patients as well as in HD. P values (Student t test) are given for Indolent (n = 20) and AGG (n = 20) versus HD (n = 10), or for AGG versus IND (above the bars). Boxes include values from the first to the third quartiles; o indicates extreme data points; +, median; and x, mean ± SD.
The relative levels of miR-125b target transcripts encoding key enzymes or regulators of metabolism increase in patients with CLL. The relative levels of the coding sequences (CDS) of the indicated transcripts were determines by qRT-PCR in IND and AGG CLL patients as well as in HD. P values (Student t test) are given for Indolent (n = 20) and AGG (n = 20) versus HD (n = 10), or for AGG versus IND (above the bars). Boxes include values from the first to the third quartiles; o indicates extreme data points; +, median; and x, mean ± SD.
Overall, our data suggest that different key metabolic enzymes, including PCTP, LIPA, GSS, ACSS1, HK2, SCD1, AKT2, and PDK1, are potential targets of miR-125b. We presume that by coordinately fine-tuning the expression of a large number of enzymes, miR-125b provides better accuracy than any other protein-coding genes in acting as a rheostat to regulate the metabolic intensity in a given cell. Furthermore, this finding suggests that the levels of miR-125b are fundamental for maintaining the correct metabolic balance in a given cell. In addition to targeting different metabolic enzymes, miR-125b potentially also targets 3 of IKZF transcripts (IKZF2, IKZF3, IKZF4). Thus, the physiologic consequences of miR-125b regulatory mechanisms certainly reach beyond glucose or lipid homeostasis in B cells and may consist of IKZFs-mediated control of cell survival, differentiation, and proliferation.
Finally, it is not surprising that one of the best-conserved miRNAs from C elegans to human acts as a rheostat of cell metabolism, considering that a balanced metabolism is fundamental for survival of any given organism. Therefore, any malfunction of this rheostat might favor the initiation of cell transformation.
There is an Inside Blood commentary on this article in this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Karoly and Dr Early at Metabolon for their help in metabolomic analyses, and Melissa Brown at OSU for her help in the gene expression analyses.
This work was supported by a R01CA151319 grant to C.M.C.
National Institutes of Health
Authorship
Contribution: E.T. and C.M.C. designed the strategy of the study and experimental flow; E.T. and J.-J.M. performed experiments and analyzed data; Z.L. performed miR-125a qRT-PCR analyses; S.V. performed Affymetrix analyses; L.Z.R. and T.J.K. performed patient analyses; and E.T., C.M.C., and J.-J.M. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Esmerina Tili or Carlo Maria Croce, Department of Molecular Virology, Immunology and Medical Genetics, OSU, Biomedical Research Tower, 460 W 12th Ave, Columbus, OH 43210; e-mail: esmerina.tili@gmail.com or carlo.croce@osumc.edu.
References
Author notes
E.T. and J.-J.M. contributed equally to this work.

![Figure 3. Heat map of biochemical changes in CLL patients. Values give the ratios of metabolite levels, determined with the Metabolon platform, between the indicated phenotypes (healthy donors [n = 20], indolent [n = 40], and aggressive [n = 40] CLL patients). This table is an extract of the data for all metabolites levels shown in supplemental Table 3. Statistical comparisons were done with the use of the Welch 2-sample t test. All data were normalized with the Bradford protein concentration.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/120/13/10.1182_blood-2012-03-415737/4/m_zh89991294770003.jpeg?Expires=1769152806&Signature=v3sQLDK0jTykwn6j8TRJMyk5wbNvb-Imkuz7L7DLKiaAXyvY9K3Sol9NuRvaU7-slgcaVQvUs0TryJZnzykD-UbIt5jFK5B4z7L4B-~T-QdLAmb77UNJwQLu-RhfdJ~5rTICITGy4LGnI5tgbCgF5tRkm~bCeG5MUIZ49W3-PPVDALWtz~e9F0WDZvUvfKxfyo6sGcltoZRdXoVrPOdaA1Vk5LknH6LLWD2YO4PKRyC62A1S~RTA3gfBaVWhO2ni1Q-EoqGXELyc9muNhY-E62T51HqKtwBzXRh6cK8WSNrtQbuRbpQE-HSfv3jed3k7qWAB4gHlttgnoJWEL706og__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)