Abstract
Proteasome inhibition with bortezomib is a validated approach to the treatment of multiple myeloma, but drug resistance often emerges and limits its utility in the retreatment setting. To begin to identify some of the mechanisms involved, we developed bortezomib-resistant myeloma cell lines that, unlike previously reported models, showed no β5 subunit mutations. Instead, up-regulation of the insulin-like growth factor (IGF)–1 axis was identified, with increased autocrine and paracrine secretion of IGF-1, leading to increased activation of the IGF-1 receptor (IGF-1R). Exogenous IGF-1 reduced cellular sensitivity to bortezomib, whereas pharmacologic or small hairpin RNA–mediated IGF-1R suppression enhanced bortezomib sensitivity in cell lines and patient samples. In vitro studies with OSI-906, a clinically relevant dual IGF-1R and insulin receptor inhibitor, showed it acted synergistically with bortezomib, and potently resensitized bortezomib-resistant cell lines and patient samples to bor-tezomib. Importantly, OSI-906 in combination with bortezomib also overcame bor-tezomib resistance in an in vivo model of myeloma. Taken together, these data support the hypothesis that signaling through the IGF-1/IGF-1R axis contributes to acquired bortezomib resistance, and provide a rationale for combining bortezomib with IGF-1R inhibitors like OSI-906 to overcome or possibly prevent the emergence of bortezomib-refractory disease in the clinic.
Introduction
Multiple myeloma is a malignancy of immunoglobulin-secreting clonal plasma cells that is most often found in the bone marrow.1,2 Modulation of the activity of the ubiquitin-proteasome pathway with the small molecule proteasome inhibitor bortezomib (VELCADE) has been validated as a rational therapeutic strategy for this disease3,4 both in the front-line and relapsed/refractory settings. Despite these and other advances, myeloma remains an incurable disease characterized by decreasing response durations with each subsequent salvage therapy.5 This is mediated in part through both intrinsic and acquired drug resistance, the latter of which emerges during and after bortezomib therapy.6 Response rates in patients with previously bortezomib-sensitive disease are typically decreased on drug rechallenge7–9 and may be as low as 23% among patients who had achieved at least a partial remission previously.7 These findings indicate a need for an understanding of the molecular basis for bortezomib resistance.
Proteasome inhibition acutely activates multiple inducible chemoresistance pathways that reduce the efficacy of bortezomib. One example is the antiapoptotic Akt pathway that can be activated by proteasome inhibitors,10 and suppression of this pathway can induce chemosensitization to bortezomib.11–13 Another possible mechanism aiding in acquired resistance to bortezomib may be the development of mutations in the bortezomib-binding pocket of the β5 proteasome subunit, or increased expression of β5 itself.14–16 However, β5 proteasome subunit mutations have not to date been identified in myeloma patients who are clinically resistant to bortezomib,17 and proteasome activity differences have not been found in gene resequencing studies of bortezomib-treated myeloma patients.18 These findings together suggest that other mechanisms may contribute to clinical bortezomib resistance.
To further elucidate mechanisms of bortezomib resistance, we developed human-derived multiple myeloma cell lines with a 4-fold or greater resistance to bortezomib. Our bortezomib-resistant (BR) models consistently displayed up-regulation of insulin-like growth factor (IGF)–1 and/or IGF-1 receptor (IGF-1R; CD221) transcripts and protein levels. Pharmacologic inhibition of the IGF-1 signaling axis, as well as small hairpin (sh) RNA–mediated IGF-1R suppression, preferentially induced apoptosis in BR cells over drug-naive parental cells, and restored bortezomib sensitivity in both cell lines and patient samples. Combinations of the IGF-1R inhibitor OSI-906 and bortezomib were able to suppress myeloma xenograft tumor growth, whereas OSI-906 or bortezomib alone had negligible activity in this setting. These data indicate that combination therapies targeting IGF-1R signaling in conjunction with bortezomib may be attractive and viable approaches for patients with clinical resistance to bortezomib, and possibly other proteasome inhibitors.
Methods
Development of BR cells
RPMI 8226, OPM-2, ANBL-6, and KAS-6/1 drug-naive myeloma cell lines, and their BR counterparts, were cultured as described previously.19,20 BR cells were developed by exposing parental cells to serially increased drug concentrations. Cell line authentication was performed by our Cell Line Characterization Core using short tandem repeat profiling. Patient samples were collected under an MD Anderson Cancer Center Institutional Review Board–approved protocol after consent was obtained in accordance with the Declaration of Helsinki Protocol. Mononuclear cells from bone marrow aspirates or peripheral blood samples were isolated by density gradient centrifugation over Ficoll-Paque Plus (Amersham Biosciences). Malignant cells were isolated by immunomagnetic bead–positive selection in a Midi MACS LS column (Miltenyi Biotec).
Cell culture, measurement of proteasome activity, immunoblotting, cell viability, apoptosis assays, and enzyme-linked immunosorbent assays
These assays are detailed in supplemental Methods (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Gene expression profiling
The Illumina TotalPrep RNA Amplification kit (Ambion) was used to generate amplified, biotinylated cRNA from 300 ng of total RNA from wild-type and BR cells by the Eberwine procedure. cRNA (750 ng) from BR cell lines on a 10- to 15-day drug holiday were hybridized overnight to Illumina HT-12 BeadArrays, stained with streptavidin-Cy3 (Amersham-Pharmacia Biotech), and scanned on a BeadArray Reader (Illumina) at the Biomarker Core Laboratory at the University of Texas Health Science Center at Houston. Bead-level data were extracted from GenomeStudio (Illumina) files and processed using open-source and custom software, following the approach of Dunning et al.21 Values were corrected by the model-based background correction method that uses values for negative control probes to estimate and remove the nonspecific signal component for each transcript probe,22 and quantile-normalized.23 We excluded the ∼ 20% of probes reported not to be perfect or good matches to actual transcripts,24 and log2-transformed the probe-level data, which are the median values for each probe after discarding outliers, as in GenomeStudio.
Measurement of bortezomib and its metabolites in cell lysates, IGF-1R gene expression and silencing, PSMB5 exon 2 resequencing, and animal studies
Please consult supplemental Methods for additional details about these studies.
Results
Induction of bortezomib resistance in myeloma cell lines
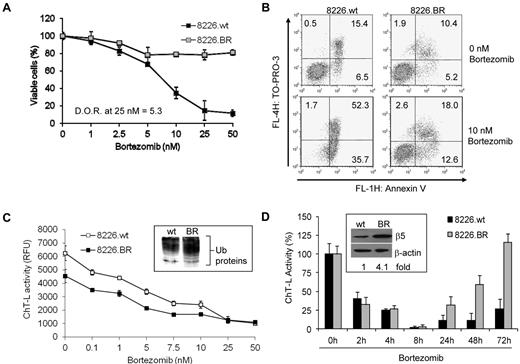
Plasma cells from both patients, and in established myeloma cell lines, are typically extremely sensitive to bortezomib-induced apoptosis, with IC50 values in the 0.5 to 5nM range. By exposing cell lines initially to low bortezomib doses, and then slowly increasing its concentration, we isolated BR myeloma cells that displayed significant decreases in their sensitivity to the antiproliferative effects of bortezomib. For example, drug-naive RPMI 8226 (8226.wt) cells exposed continuously to 25nM bortezomib for 24 hours experienced an 85% decrease in viability, whereas only a 22% loss of viability was seen in 8226.BR cells, indicating a 5.3-fold increase in resistance (Figure 1A). Similar levels of resistance were observed in ANBL-6.BR cells (supplemental Figure 1). Likewise, when 8226.wt and 8226.BR cells were exposed to 10nM drug, bortezomib induced the appearance of annexin-V and TO-PRO-3–positive cells, indicative of cell death, only in 8226.wt cells (Figure 1B).
BR myeloma cells remain sensitive to bortezomib treatment and recover proteasome activity. (A) The degree of resistance (D.O.R.) of cells tolerant of bortezomib, compared with their drug-naive counterparts, was evaluated by examining live cell populations using the WST-1 proliferation assay after exposure to bortezomib for 24 hours. Data shown are representative from triplicate experiments. (B) Apoptosis was evaluated in 8226.wt and 8226.BR cells by staining with annexin-V and TO-PRO-3. Data shown are representative from duplicate experiments. (C) Dose-dependent effects of bortezomib on the proteasome ChT-L activity were measured using cellular extracts. Inset, ANBL-6.wt and ANBL-6.BR cells propagated in the absence of bortezomib were probed for their content of ubiquitin-protein conjugates. (D) Time-dependent effects of bortezomib (10nM) on proteasome ChT-L activity were examined in 8226.wt and 8226.BR cells. Inset, expression of the basal level of the β5 proteasome subunit was examined by Western blotting and quantified by densitometry after normalization for loading using β-actin as a control. Data are representative from experiments performed in triplicate, and error bars indicate SD.
BR myeloma cells remain sensitive to bortezomib treatment and recover proteasome activity. (A) The degree of resistance (D.O.R.) of cells tolerant of bortezomib, compared with their drug-naive counterparts, was evaluated by examining live cell populations using the WST-1 proliferation assay after exposure to bortezomib for 24 hours. Data shown are representative from triplicate experiments. (B) Apoptosis was evaluated in 8226.wt and 8226.BR cells by staining with annexin-V and TO-PRO-3. Data shown are representative from duplicate experiments. (C) Dose-dependent effects of bortezomib on the proteasome ChT-L activity were measured using cellular extracts. Inset, ANBL-6.wt and ANBL-6.BR cells propagated in the absence of bortezomib were probed for their content of ubiquitin-protein conjugates. (D) Time-dependent effects of bortezomib (10nM) on proteasome ChT-L activity were examined in 8226.wt and 8226.BR cells. Inset, expression of the basal level of the β5 proteasome subunit was examined by Western blotting and quantified by densitometry after normalization for loading using β-actin as a control. Data are representative from experiments performed in triplicate, and error bars indicate SD.
BR cells retain sensitivity to proteasome inhibition
Two recent studies of BR models found no decrease in basal proteasome activity in leukemia-derived cell lines,14,16 and identified mutations in the β5 proteasome subunit that precluded bortezomib binding as contributing factors to drug resistance. We therefore evaluated the chymotrypsin-like (ChT-L) proteasome activity catalyzed by the β5 subunit.25 Relative fluorescence values using the fluorogenic substrate Suc-LLVY-AMC showed that both 8226.wt and 8226.BR cells retained ChT-L activity but that 8226.BR cells exhibited a 27% decrease in basal activity compared with 8226.wt cells (Figure 1C). This decreased proteasome activity was associated with a higher basal level of intracellular ubiquitin-protein conjugates (Figure 1C inset). A similar decrease in basal activity was found in ANBL-6.BR cells over their drug-naive counterparts (supplemental Figure 2A). Notably, unlike in previously reported resistance models,14,16 bortezomib decreased ChT-L activity in both drug-naive and BR cells (Figure 1C and supplemental Figure 2A), indicating the absence of proteasome mutations that precluded bortezomib binding. Finally, resequencing of the catalytic region of β5 (PSMB5, exon 2) in 8666.BR and ANBL-6.BR cells confirmed the absence of active site mutations (data not shown).
Given the lower levels of basal proteasome activity in BR cells, it was of interest to examine the recovery of proteasome function after bortezomib treatment. Although 8226.BR cells had a similar percentage of proteasome inhibition compared with 8226.wt cells at early time points, the 8226.BR cells recovered ChT-L activity more rapidly later in the time course, with full activity returning 72 hours after bortezomib exposure (Figure 1D). It should be noted that the suppression of the ChT-L activity was not affected by IGF-1 supplementation (data not shown). This more rapid kinetics of proteasome activity recovery also was found in ANBL-6.BR cells (supplemental Figure 2B). These findings suggested the possibility that enhanced drug efflux was contributing to proteasome recovery; therefore, we determined intracellular bortezomib concentrations. Interestingly, just the opposite was seen, in that bortezomib accumulated to substantially higher levels in the 8226.BR cells and was retained for a longer period (Table 1). For example, at 24 hours, bortezomib concentrations were up to 18-fold higher in 8226.BR cells compared with their 8226.wt counterparts. To further evaluate the possibility that multidrug resistance mediators were involved in bortezomib resistance, we examined whether inhibition of P-glycoprotein with verapamil could sensitize 8226.BR cells to bortezomib. Similar levels of cell death were observed in 8226.wt cells treated with bortezomib in the presence or absence of verapamil (supplemental Figure 2C), whereas 8226.BR sensitivity was not impacted at lower bortezomib levels and only modestly increased at the highest bortezomib concentrations tested (supplemental Figure 2D). We then examined the expression of β5, and we did find up to a 4-fold increase in β5 protein expression in 8226.BR (Figure 1D inset) and ANBL-6.BR cells (supplemental Figure 2E). These increased levels of β5 may account for the higher intracellular accumulation and longer retention of bortezomib in the drug-resistant cells.
Intracellular bortezomib concentrations in RPMI 8226.wt and 8226.BR
| Cell line . | Time after treatment, h . | Bortezomib, ng/mL . | Bortezomib, ng/1 × 106 cells . |
|---|---|---|---|
| 8226.wt | 0 | n.d. | n.d. |
| 1 | 0.13 | 0.16 | |
| 3 | 0.32 | 0.40 | |
| 6 | 0.38 | 0.48 | |
| 24 | 0.19 | 0.24 | |
| 48 | n.d. | n.d. | |
| 8226.BR | 0 | n.d. | n.d. |
| 1 | 0.52 | 0.65 | |
| 3 | 2.46 | 3.08 | |
| 6 | 3.45 | 4.31 | |
| 24 | 3.43 | 4.29 | |
| 48 | 2.01 | 2.51 |
| Cell line . | Time after treatment, h . | Bortezomib, ng/mL . | Bortezomib, ng/1 × 106 cells . |
|---|---|---|---|
| 8226.wt | 0 | n.d. | n.d. |
| 1 | 0.13 | 0.16 | |
| 3 | 0.32 | 0.40 | |
| 6 | 0.38 | 0.48 | |
| 24 | 0.19 | 0.24 | |
| 48 | n.d. | n.d. | |
| 8226.BR | 0 | n.d. | n.d. |
| 1 | 0.52 | 0.65 | |
| 3 | 2.46 | 3.08 | |
| 6 | 3.45 | 4.31 | |
| 24 | 3.43 | 4.29 | |
| 48 | 2.01 | 2.51 |
wt indicates wild-type; BR, bortezomib-resistant cells; and n.d., not detected (less than limit of detection).
One potential use of BR myeloma models is in the testing of novel strategies to overcome such a phenotype, and their utility would be further enhanced if they retained drug resistance in vivo. Testing of ANBL-6.BR and 8226.BR cells in murine xenograft models was therefore pursued, and we confirmed that these cells retained resistance to bortezomib. Both drug-naive 8226.wt (supplemental Figure 3A) and ANBL-6.wt (supplemental Figure 3B) xenografts showed sensitivity to treatment with bortezomib. However, 8226.BR (supplemental Figure 3C) and ANBL-6.BR (supplemental Figure 3D) cells showed a significantly reduced sensitivity to bortezomib. 8226.BR cells were especially resistant in this regard, as demonstrated by the lack of any reduction in tumor progression compared with vehicle-treated controls.
Genomic changes associated with stable bortezomib resistance
To further characterize these BR models, gene expression profiling was performed on ANBL-6.BR, OPM-2.BR, and 8226.BR cells. We identified the IGF-1/IGF-1R signaling axis as a dysregulated pathway in 3 BR cell lines by applying gene set enrichment analysis26 for each line, using sets of previously reported IGF-1–regulated genes. As shown in Table 2, each myeloma cell line showed significant enrichment in genes that were up- or down-regulated in MCF-7 breast carcinoma cells by acute stimulation with IGF-1.27 These gene sets were previously validated by various correlations, most notably with gene-expression-defined subtypes and outcome in breast carcinoma.27 Heat maps were made for the combined core genes from each gene set enrichment analysis comparison, and they showed that the cell lines were quite similar in their up- or down-regulation of genes regulated by IGF-1 in MCF-7 cells (supplemental Figure 4). In addition, using Ingenuity Pathway Analysis Version 14197757 software (Ingenuity Systems), we also identified the IGF-1 signaling pathway as up-regulated in BR cells (data not shown).
Significance data for gene set enrichment analysis comparisons between wild-type and BR cells for the myeloma lines studied using sets of IGF-1–regulated genes in MCF-7 breast carcinoma27
| Gene set . | Total no. of genes . | ES . | NES . | Nominal P value . | FDR q value . |
|---|---|---|---|---|---|
| Up in MCF-7 with IGF-1 | 377 | ||||
| OPM2* | 0.45 | 1.94 | .000 | .071 | |
| 8226 | 0.36 | 1.57 | .000 | .079 | |
| ANBL-6 | 0.38 | 1.59 | .000 | .214 | |
| Down in MCF-7 with IGF-1 | 374 | ||||
| OPM2* | −0.45 | −1.89 | .000 | .010 | |
| 8226 | −0.45 | −1.85 | .000 | .017 | |
| ANBL-6 | −0.37 | −1.51 | .000 | .206 |
| Gene set . | Total no. of genes . | ES . | NES . | Nominal P value . | FDR q value . |
|---|---|---|---|---|---|
| Up in MCF-7 with IGF-1 | 377 | ||||
| OPM2* | 0.45 | 1.94 | .000 | .071 | |
| 8226 | 0.36 | 1.57 | .000 | .079 | |
| ANBL-6 | 0.38 | 1.59 | .000 | .214 | |
| Down in MCF-7 with IGF-1 | 374 | ||||
| OPM2* | −0.45 | −1.89 | .000 | .010 | |
| 8226 | −0.45 | −1.85 | .000 | .017 | |
| ANBL-6 | −0.37 | −1.51 | .000 | .206 |
ES indicates enrichment score; NES, normalized enrichment score; and FDR, false discovery rate.
OPM2 was studied twice.
BR cell lines have increased IGF-1/IGF-1R expression
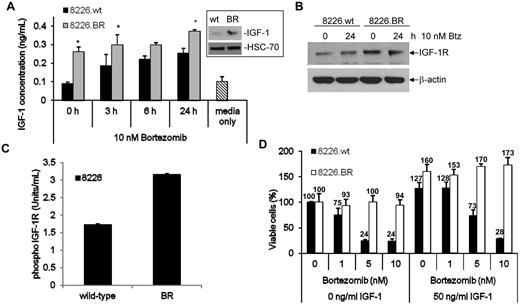
Given the role of IGF-1 as a growth and survival factor in myeloma,28–30 we next sought to determine whether the increased IGF-1/IGF-1R gene expression correlated with an increase in IGF-1 and IGF-1R at the protein level. First, levels of soluble IGF-1 were examined, and were found to be increased at baseline in 8226.BR cell culture supernatants compared with 8226.wt cells, at 0.26 ng/mL versus 0.09 ng/mL IGF-1, respectively (Figure 2A). Western blot analysis also confirmed increased levels of intracellular and membrane bound IGF-1 (Figure 2A inset). Exposure to 10nM bortezomib led to increases in soluble IGF-1 levels in both drug-naive and BR cells in a time-dependent manner, but 8226.BR cells consistently maintained higher levels of IGF-1 secretion (Figure 2A). Real-time PCR (qPCR) showed that 8226.BR, ANBL-6.BR, and OPM-2.BR all had greater levels of IGF-1 and IGF-1R mRNA (supplemental Figure 5). These data were confirmed in ANBL-6.wt and ANBL-6.BR cells (supplemental Figure 6A inset).
BR cells have increased expression and activation of the IGF-1/IGF-1R signaling pathway. (A) The levels of soluble IGF-1 present in cellular supernatants of cells grown in culture overnight were determined using an ELISA. The student's paired t test was used to determine statistical significance (*P < .05 compared with wild-type time points). Data shown are representative from triplicate experiments, and error bars denote ± SD. Basal levels of IGF-1 protein expression were confirmed by Western blotting (inset). (B) IGF-1R expression was determined by Western blotting in cells exposed to vehicle or 10nM bortezomib for 24 hours. (C) Activation of IGF-1R was determined in RPMI 8226 and OPM-2 drug-naive and BR cells using an ELISA. Data shown are representative of 2 experiments, and error bars denote ± SD. Concentrations were determined using a standard curve with activated IGF-1R supplied by the manufacturer. (D) 8226.wt and 8226.BR cells were propagated for 24 hours in media supplemented with 50 ng/mL of human recombinant IGF-1 (right) or IGF-1–free media (left), followed by bortezomib addition for an additional 24 hours, and viable populations were determined using the WST-1 reagent. Data shown are representative from triplicate experiments, and error bars are ± SD.
BR cells have increased expression and activation of the IGF-1/IGF-1R signaling pathway. (A) The levels of soluble IGF-1 present in cellular supernatants of cells grown in culture overnight were determined using an ELISA. The student's paired t test was used to determine statistical significance (*P < .05 compared with wild-type time points). Data shown are representative from triplicate experiments, and error bars denote ± SD. Basal levels of IGF-1 protein expression were confirmed by Western blotting (inset). (B) IGF-1R expression was determined by Western blotting in cells exposed to vehicle or 10nM bortezomib for 24 hours. (C) Activation of IGF-1R was determined in RPMI 8226 and OPM-2 drug-naive and BR cells using an ELISA. Data shown are representative of 2 experiments, and error bars denote ± SD. Concentrations were determined using a standard curve with activated IGF-1R supplied by the manufacturer. (D) 8226.wt and 8226.BR cells were propagated for 24 hours in media supplemented with 50 ng/mL of human recombinant IGF-1 (right) or IGF-1–free media (left), followed by bortezomib addition for an additional 24 hours, and viable populations were determined using the WST-1 reagent. Data shown are representative from triplicate experiments, and error bars are ± SD.
To determine whether these increased IGF-1 levels impacted on the phosphorylation and therefore activation status of IGF-1R, we first examined the expression of IGF-1R in BR cells and their drug-naive counterparts. Increased IGF-1R mRNA levels were found in 8226.BR cells by qPCR (data not shown). In addition, increased IGF-1R protein levels were found in 8226.BR cells, both in the absence and presence of bortezomib (Figure 2B), and similar results were obtained from Western blot analysis of ANBL-6.BR and ANBL-6.wt cells (data not shown). The activation status of IGF-1R was then studied using an ELISA to detect IGF-1R phosphorylated at tyrosine residues 1135 and 1136. Consistent with the hypothesis that secreted IGF-1 induced increased activity of IGF-1R, 8226.BR cells were found to have a 3.6-fold increased content of phospho-IGF-1R compared with 8226.wt cells (Figure 2C). Similar data were obtained in OPM-2.BR, where an ∼ 2-fold increase in phospho-IGF-1R content was seen over the OPM-2.wt parental cells (data not shown). Interestingly, exposure of both 8226.wt and 8226.BR cells to bortezomib resulted in further activation of IGF-1R (supplemental Figure 6B).
IGF-1 confers resistance to bortezomib
To determine whether increased IGF-1 secretion and IGF-1R activation contributed to bortezomib resistance, we challenged drug-resistant and parental cells with bortezomib with or without IGF-1. The addition of recombinant human IGF-1 to 8226.wt cells without bortezomib enhanced their viability, and IGF-1 allowed more myeloma cells to remain viable in the presence of low bortezomib concentrations (Figure 2D). However, at higher drug concentrations, such as 10nM, bortezomib overcame this protective effect in wild-type cells, and similar trends were seen in the OPM-2.wt model (supplemental Figure 6C). When the 8226.BR cells were similarly studied, these proved to be less responsive to bortezomib when supported by IGF-1–free media than the 8226.wt cells, as expected. Notably, addition of exogenous IGF-1 enhanced 8226.BR cell viability to an even greater extent than was the case for 8226.wt cells. Importantly, in the presence of IGF-1, 8226.BR cells were much more resistant to bortezomib than parental 8226.wt cells, and more resistant than 8226.BR cells without exogenous IGF-1 (Figure 2D). To confirm these findings, OPM-2.wt and OPM-2.BR cells were similarly stimulated with exogenous IGF-1, and qualitatively analogous results were obtained. Although IGF-1 protected OPM-2.wt cells at low bortezomib concentrations, this effect could be overcome by 10nM of this proteasome inhibitor (supplemental Figure 6C), and the extent of protection was much greater in the OPM-2.BR cells. IGF-1 supplementation alone had no effect on ChT-L activity in 8226.wt or 8226.BR cells, and only when combined with bortezomib was an effect observed (supplemental Figure 6D).
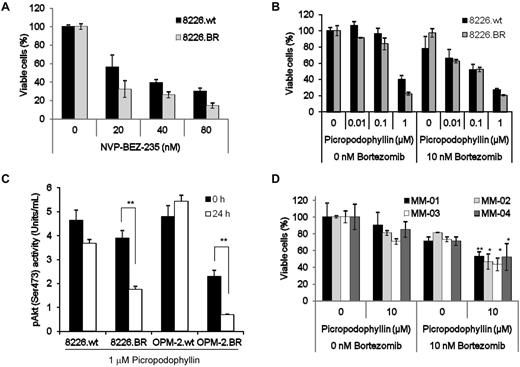
Phosphoinositide 3-kinase (PI3K) and the mammalian target of rapamycion (mTOR) are both important growth and survival signaling kinases located downstream of IGF-1/IGF-1R; therefore, it was of interest to determine the impact of their inhibition on these models using NVP-BEZ-235, a dual inhibitor of PI3K and mTOR.31 NVP-BEZ-235 showed activity against 8226.wt (Figure 3A), KAS-6/1.wt (supplemental Figure 7A), and OPM-2.wt (supplemental Figure 7B) myeloma models, as measured by its ability to reduce cell viability. The same was true for their drug-resistant counterparts, and indeed, NVP-BEZ-235 seemed to be more active against all the BR cell lines, suggesting a greater dependence for survival on the IGF-1/IGF-1R pathway in these BR cells. Notably, qPCR showed increased Akt mRNA expression in 3 BR cell lines (supplemental Figure 5). Measurement of the effect of NVP-235 on the ChT-L activity of the proteasome revealed no effect and indicated that NVP-BEZ-235 did not interact with the proteasome (data not shown).
Inhibition of Akt, mTOR, and IGF-1R induces cell death in BR cells. (A) 8226.wt and 8226.BR cells were exposed to the indicated concentrations of NVP-BEZ-235 for 24 hours, and viability was assessed using the WST-1 reagent. Data shown are representative of 3 independent experiments, and errors bars denote ± SD. (B) RPMI 8226.wt and 8226.BR cells were treated with increasing concentrations of PPP with (right) or without (left) simultaneous addition of 10nM bortezomib for 24 hours, and assessed by WST-1. Data points are representative from triplicate experiments, and error bars indicate SD. (C) The effect of PPP on Akt kinase activity was determined using RPMI 8226 and OPM-2 drug-naive and BR cell lines treated with 1μM PPP for 24 hours. After each experiment, cell lysates were prepared, and 25 mg of protein from each was probed for the levels of phospho-Akt in duplicate using an ELISA. Data shown are the mean of 2 experiments performed in duplicate ± SD. (D) CD138+ plasmacytes from patients were treated with increasing concentrations of PPP without (left) or with (right) simultaneous addition of 10nM bortezomib. Viable cell populations were evaluated using the WST-1 assay (*P < .05, **P < .01). All data points are the mean of experiments performed in triplicate ± SD.
Inhibition of Akt, mTOR, and IGF-1R induces cell death in BR cells. (A) 8226.wt and 8226.BR cells were exposed to the indicated concentrations of NVP-BEZ-235 for 24 hours, and viability was assessed using the WST-1 reagent. Data shown are representative of 3 independent experiments, and errors bars denote ± SD. (B) RPMI 8226.wt and 8226.BR cells were treated with increasing concentrations of PPP with (right) or without (left) simultaneous addition of 10nM bortezomib for 24 hours, and assessed by WST-1. Data points are representative from triplicate experiments, and error bars indicate SD. (C) The effect of PPP on Akt kinase activity was determined using RPMI 8226 and OPM-2 drug-naive and BR cell lines treated with 1μM PPP for 24 hours. After each experiment, cell lysates were prepared, and 25 mg of protein from each was probed for the levels of phospho-Akt in duplicate using an ELISA. Data shown are the mean of 2 experiments performed in duplicate ± SD. (D) CD138+ plasmacytes from patients were treated with increasing concentrations of PPP without (left) or with (right) simultaneous addition of 10nM bortezomib. Viable cell populations were evaluated using the WST-1 assay (*P < .05, **P < .01). All data points are the mean of experiments performed in triplicate ± SD.
Suppression of IGF-1R resensitizes resistant cells to bortezomib
Targeting the IGF-1 receptor itself is another potentially attractive approach to overcome bortezomib resistance. This was evaluated first using picropodophyllin (PPP), a cyclolignan that suppresses IGF-1R activity and downstream pathways, in part through a murine double minute 2- and β-arrestin-1–mediated mechanism.32 When 8826.wt and 8226.BR cells were treated with PPP, a reduction in viability was seen in both models (Figure 3B), and this was greater in the BR clones. A qualitatively similar finding was obtained in the OPM-2.wt and OPM-2.BR cells (supplemental Figure 7C), possibly because of the greater dependence of the resistant cells on IGF-1R signaling for survival. Bortezomib was then added to PPP, and this regimen was used in parallel against the 8226 (Figure 3B) and OPM-2 (supplemental Figure 7C) models. Evaluation of PPP's effect on the proteasome was measured in 8226.wt and 8226.BR cells and showed no effect, indicating that PPP is not a proteasome inhibitor (data not shown). At PPP concentrations of 0.01 and 0.1μM, the combination of PPP and 10nM bortezomib was more effective than comparable concentrations of PPP alone. To evaluate the downstream consequences of PPP treatment, its effects on Akt activation status were evaluated using an ELISA to detect phospho-Akt levels. Treatment of both 8226.wt and 8226.BR cells with PPP resulted in decreased levels of phospho-Akt (Figure 3C), but this reduction was greater in 8226.BR cells, at 2.6-fold, than was the case in 8226.wt cells, where only a 1.2-fold decrease was seen. Qualitatively similar findings were obtained in studies of OPM-2.wt and OPM-2.BR cells (Figure 3C). CD138+ plasma cells isolated from 4 patients who progressed after receiving regimens containing bortezomib were then tested for sensitivity to PPP alone, or in the presence of bortezomib. Cells isolated from all 4 patients had a minimal response to PPP or BTZ alone, but when the 2 were combined, viable cell populations dropped significantly, with an additional 27% to 37% reduction over PPP and 18% to 34% reduction over bortezomib (Figure 3D). Studies into the mechanism of cell death showed that drug-naive cell lines exposed to PPP underwent apoptosis, as judged by the disappearance of the full-length p116 PARP, and the appearance of the p85 PARP caspase cleavage product (supplemental Figure 7D). Similar effects were seen in the BR cells, indicating that PPP worked by activating type I, caspase-mediated cell death.
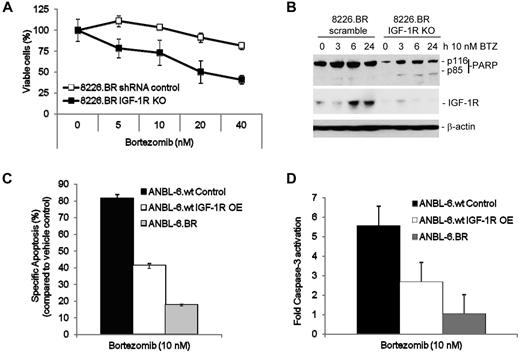
In that PPP may exert proapoptotic effects through pathways not dependent on IGF-1R,32 it was of interest to confirm more directly that blockade of this axis resensitized BR cells to bortezomib. 8226.wt and 8226.BR cells were therefore infected with Lentiviral constructs directing expression of a control shRNA, or shRNAs directed at IGF-1R, and then treated with bortezomib. 8226.BR cells with IGF-1R knockdown (IGF-1R KO) readily underwent apoptosis, whereas the 8226.BR cells infected with the scrambled control were still bortezomib resistant (Figure 4A). It is notable that the 8226.BR IGF-1R KO cells had similar levels of cell death as 8226.wt cells treated with a scrambled control shRNA Lentivirus (data not shown). In addition, increasing multiplicity of infection of the IGF-1R shRNA in 8226.BR cells led to increasing sensitivity to bortezomib (data not shown). Western blotting was then performed to evaluate the success of IGF-1R suppression, and although 8226.BR cells infected with control Lentiviral particles contained detectable levels of the IGF-1 receptor, these were substantially reduced by specific IGF-1R shRNAs (Figure 4B). The loss of IGF-1R expression in 8226.BR IGF-1R KO cells led to a moderate decrease in phospho-Akt compared with the shRNA scrambled control cells. In addition, loss of IGF-1R expression led to decreased sensitivity to bortezomib's ability to inhibit Akt activity (supplemental Figure 8A-B). However, knockout of IGF-1R had no effect on bortezomib's proteasome inhibitory properties (supplemental Figure 8C-D). To further probe the potential role of IGF-1R as an important contributor to bortezomib resistance, ANBL-6.wt cells were infected with a Lentivirus directing IGF-1R expression. Overexpression of IGF-1R was confirmed by a significant increase in IGF-1R surface membrane expression (data not shown) and resulted in a 40% decrease in apoptotic cells (Figure 4C), as well as a 2-fold decrease in caspase-3 activation (Figure 4D) after bortezomib exposure, compared with ANBL-6.wt vector control cells.
IGF-1R signaling mediates resistance to bortezomib. Expression of IGF-1Rα/β was suppressed in RPMI 8226.wt and 8226.BR cells by infecting them with Lentiviral vectors expressing receptor-specific shRNAs. These cells (1 × 104/well) were then plated in 96-well plates, bortezomib was added at the indicated concentrations for 24 hours, and either the WST-1 assay (A) or Western blotting (B) was performed. Data presented are representative from experiments performed in triplicate. ANBL-6.wt cells overexpressing IGF-1R protein (IGF-1R OE) were treated with bortezomib for 24 hours, and levels of apoptosis (C) and activated caspase-3 (D) were measured using fluorescence-activated cell sorting analysis. IGF-1R–overexpressing cell results were then compared with vector control ANBL-6.wt cells. Data shown are the mean of duplicate experiments ± SD.
IGF-1R signaling mediates resistance to bortezomib. Expression of IGF-1Rα/β was suppressed in RPMI 8226.wt and 8226.BR cells by infecting them with Lentiviral vectors expressing receptor-specific shRNAs. These cells (1 × 104/well) were then plated in 96-well plates, bortezomib was added at the indicated concentrations for 24 hours, and either the WST-1 assay (A) or Western blotting (B) was performed. Data presented are representative from experiments performed in triplicate. ANBL-6.wt cells overexpressing IGF-1R protein (IGF-1R OE) were treated with bortezomib for 24 hours, and levels of apoptosis (C) and activated caspase-3 (D) were measured using fluorescence-activated cell sorting analysis. IGF-1R–overexpressing cell results were then compared with vector control ANBL-6.wt cells. Data shown are the mean of duplicate experiments ± SD.
IGF-1R inhibitor OSI-906 resensitizes BR cells to bortezomib
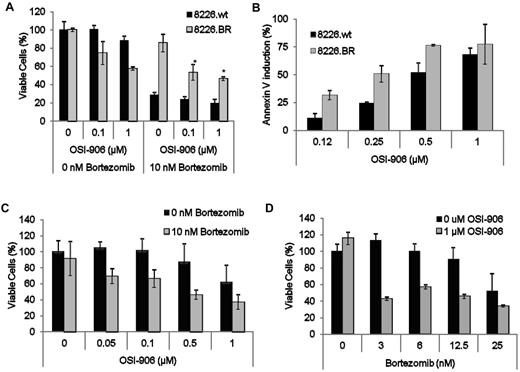
Because modulation of IGF-1R activity seemed to overcome bortezomib resistance, it was of interest to develop an approach that could be translated to the clinic. We therefore next evaluated whether OSI-906, a clinically relevant small molecule inhibitor of both IGF-1R and the insulin receptor tyrosine kinase, could sensitize BR cells. OSI-906 alone preferentially induced cell death in 8226.BR cells, whereas drug-naive cell populations were relatively spared (Figure 5A). Simultaneous addition of 10nM bortezomib and increasing concentrations of OSI-906 enhanced the amount of cell death in 8226.BR cells. For example, 0.1μM OSI-906 alone induced 25% death in 8226.BR cells, whereas addition of bortezomib increased this to 47% (Figure 5A). Similar levels of enhanced cell death were observed in ANBL-6.BR cells (supplemental Figure 9A). In addition, OSI-906 preferentially induced apoptosis in 8226.BR cells, as measured by annexin-V staining (Figure 5B), and in ANBL-6.BR cells (supplemental Figure 8B). Next, we determined whether OSI-906 alone would have an effect on MDA-MM-002, a cell line developed from the pleural effusion of a patient with advanced, clinically bortezomib-refractory myeloma. MDA-MM-002 cells showed only slight decreases in the viable cell population when treated with 1μM OSI-906 alone (Figure 5C). However, when combined with bortezomib, there was an enhanced, dose-dependent decrease in the viable cell population (Figure 5C). Similarly, MDA-MM-002 cells treated with 10nM bortezomib showed a minimal loss of viability, whereas significant increases in cell death were found with increasing concentrations of OSI-906 (Figure 5D).
Suppression of IGF-1R with OSI-906 sensitizes 8226.BR cells to bortezomib. (A) RPMI 8226.wt and 8226.BR cells were simultaneously treated with increasing concentrations of OSI-906 alone (left), or with OSI-906 and 10nM bortezomib combined (right) for 24 hours, followed by measurement of live cells with WST-1. Error bars represent the SD of triplicate experiments. (B) Induction of apoptosis was assessed in single samples of 8226.wt and 8226.BR cell lines treated with OSI-906 for 24 hours and stained with annexin-V–FITC and TOPRO-3. Multiple myeloma cells (2 × 104) isolated from a pleural effusion of a patient displaying bortezomib resistance were cotreated with increasing concentrations of bortezomib and 1μM OSI-906 (C), or increasing concentrations of OSI-906 and 10nM bortezomib alone (D) for 24 hours, and examined using the WST-1 assay. Data presented are representative of triplicate experiments, and error bars are ± SD.
Suppression of IGF-1R with OSI-906 sensitizes 8226.BR cells to bortezomib. (A) RPMI 8226.wt and 8226.BR cells were simultaneously treated with increasing concentrations of OSI-906 alone (left), or with OSI-906 and 10nM bortezomib combined (right) for 24 hours, followed by measurement of live cells with WST-1. Error bars represent the SD of triplicate experiments. (B) Induction of apoptosis was assessed in single samples of 8226.wt and 8226.BR cell lines treated with OSI-906 for 24 hours and stained with annexin-V–FITC and TOPRO-3. Multiple myeloma cells (2 × 104) isolated from a pleural effusion of a patient displaying bortezomib resistance were cotreated with increasing concentrations of bortezomib and 1μM OSI-906 (C), or increasing concentrations of OSI-906 and 10nM bortezomib alone (D) for 24 hours, and examined using the WST-1 assay. Data presented are representative of triplicate experiments, and error bars are ± SD.
To further evaluate this apparently synergistic effect, cells were treated with both drugs and subjected to isobologram analysis (Table 3). Compared with OSI-906 or bortezomib alone, the combination demonstrated a greater antiproliferative effect. Moreover, statistical analysis indicated that a high degree of synergy was present between the 2 drugs at most concentrations tested, with the few outlying concentrations exhibiting an additive effect.
Combination index analysis of a regimen of bortezomib and OSI-906 in drug-naive and BR myeloma models
| . | OSI-906, μM . | Bortezomib, nM . | ||
|---|---|---|---|---|
| 3 . | 6 . | 12 . | ||
| ANBL-6.wt | ||||
| 0.05 | 0.45 | 0.52 | 1.0 | |
| 0.1 | 0.57 | 0.55 | 0.97 | |
| 0.5 | 0.46 | 0.56 | 1.1 | |
| 1 | 0.41 | 0.51 | 1.1 | |
| ANBL-6.BR | ||||
| 0.05 | 0.26 | 0.43 | 0.61 | |
| 0.1 | 0.24 | 0.42 | 0.6 | |
| 0.5 | 0.23 | 0.4 | 0.59 | |
| 1 | 0.21 | 0.39 | 0.56 | |
| 8226.wt | ||||
| 0.05 | 0.38 | 0.44 | 0.62 | |
| 0.1 | 0.55 | 0.5 | 0.68 | |
| 0.5 | 0.35 | 0.34 | 0.58 | |
| 1 | 0.46 | 0.42 | 0.66 | |
| 8226.BR | ||||
| 0.05 | 1.0 | 0.49 | 0.63 | |
| 0.1 | 1.3 | 0.63 | 0.63 | |
| 0.5 | 1.2 | 0.84 | 0.5 | |
| 1 | 0.36 | 0.69 | 0.48 | |
| . | OSI-906, μM . | Bortezomib, nM . | ||
|---|---|---|---|---|
| 3 . | 6 . | 12 . | ||
| ANBL-6.wt | ||||
| 0.05 | 0.45 | 0.52 | 1.0 | |
| 0.1 | 0.57 | 0.55 | 0.97 | |
| 0.5 | 0.46 | 0.56 | 1.1 | |
| 1 | 0.41 | 0.51 | 1.1 | |
| ANBL-6.BR | ||||
| 0.05 | 0.26 | 0.43 | 0.61 | |
| 0.1 | 0.24 | 0.42 | 0.6 | |
| 0.5 | 0.23 | 0.4 | 0.59 | |
| 1 | 0.21 | 0.39 | 0.56 | |
| 8226.wt | ||||
| 0.05 | 0.38 | 0.44 | 0.62 | |
| 0.1 | 0.55 | 0.5 | 0.68 | |
| 0.5 | 0.35 | 0.34 | 0.58 | |
| 1 | 0.46 | 0.42 | 0.66 | |
| 8226.BR | ||||
| 0.05 | 1.0 | 0.49 | 0.63 | |
| 0.1 | 1.3 | 0.63 | 0.63 | |
| 0.5 | 1.2 | 0.84 | 0.5 | |
| 1 | 0.36 | 0.69 | 0.48 | |
BR indicates bortezomib resistant; and wt, wild-type.
The combination index (CI) is a quantitative measure of the degree of drug interaction, with a CI < 1 indicating synergy, a CI = 1 indicating additive effects, and a CI > 1 indicating antagonism.
Combination OSI-906 and bortezomib suppresses 8226.BR tumor growth in vivo
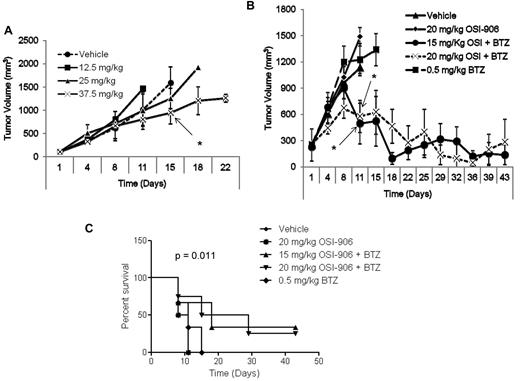
Because OSI-906 showed such potent activity in cell line models of bortezomib resistance, its activity was next tested in xenograft models of BR multiple myeloma. First, a pilot study was used to identify effective doses of OSI-906 that were administered intraperitoneally, and twice weekly on days 1 and 4 until tumor volume necessitated euthanasia. Xenografts consisting of 8226.BR tumors grew rapidly in all dose and vehicle cohorts (n = 5) except at the highest dose used (37.5 mg/kg), where a modest decrease in the rate of tumor growth was observed (Figure 6A). For combination studies, doses of 15 and 20 mg/kg OSI-906 were selected, doses that by themselves were ineffective (Figure 6A), to reduce any potential for overlapping toxicities with bortezomib, dosed at 0.5 mg/kg. Control cohorts were treated with either vehicle alone, 20 mg/kg OSI-906 alone, or 0.5 mg/kg bortezomib alone using the same dose, route, and schedule. No dose-limiting side effects were noticed with any of the treatments, all of which the mice tolerated well. Combination treatment with either 15 or 20 mg/kg OSI-906 and bortezomib proved very efficacious at first slowing tumor growth and then reducing tumor volume (Figure 6B). Indeed, 2 mice from each combination cohort seemed to experience complete tumor regression based on the absence of palpable tumor starting on day 11 and extending until termination of the study, on day 42. No such benefit was found in the cohorts treated with vehicle, 20 mg/kg OSI-906 alone, or 0.5 mg/kg bortezomib alone. These data implicate IGF-1R suppression using OSI-906 as an attractive approach to resensitize 8226.BR cells to bortezomib in vivo. The survival curves indicated that the OSI-906 and bortezomib combination significantly prolonged the life of the animals receiving this treatment over single-agent bortezomib or OSI-906 alone (Figure 6C).
OSI-906 overcomes bortezomib resistance in vivo. Animal studies were performed in C.B-17 severe combined immunodeficiency male mice with flank xenografts of 8226.BR cells. Drugs were administered via intraperitoneal (i.p.) injections on days 1 and 4 of each week. Tumors were measured on each day of treatment by a researcher who was blinded to the treatment assignments, and volumes were calculated using the equation 1/2 (length × width2). (A) Pilot dose escalation studies were performed with OSI-906, in which mice received drug weekly on days 1 and 4 (n = 5). (B) Combination regimens of OSI-906 and bortezomib were tested in cohorts of tumor-bearing animals randomized to either vehicle, OSI-906 alone, 15 mg/kg OSI-906 + 0.5 mg/kg bortezomib, 20 mg/kg OSI-906 + 0.5 mg/kg bortezomib, or 0.5 mg/kg bortezomib (n = 6).
OSI-906 overcomes bortezomib resistance in vivo. Animal studies were performed in C.B-17 severe combined immunodeficiency male mice with flank xenografts of 8226.BR cells. Drugs were administered via intraperitoneal (i.p.) injections on days 1 and 4 of each week. Tumors were measured on each day of treatment by a researcher who was blinded to the treatment assignments, and volumes were calculated using the equation 1/2 (length × width2). (A) Pilot dose escalation studies were performed with OSI-906, in which mice received drug weekly on days 1 and 4 (n = 5). (B) Combination regimens of OSI-906 and bortezomib were tested in cohorts of tumor-bearing animals randomized to either vehicle, OSI-906 alone, 15 mg/kg OSI-906 + 0.5 mg/kg bortezomib, 20 mg/kg OSI-906 + 0.5 mg/kg bortezomib, or 0.5 mg/kg bortezomib (n = 6).
Xenograft tissues were excised and probed by Western blotting for ubiquitin conjugates that showed that bortezomib alone induced accumulation of ubiquitinated proteins. However, when bor-tezomib was combined with OSI-906, there was a decrease in the amount of ubiquitinated substrates in the 8226.BR tumors (supplemental Figure 10A). To further explore this phenomenon, we treated both 8226.wt and 8226.BR cells in vitro with 2.5 or 5μM OSI-906 with and without bortezomib. In the 8226.wt cells, there was an accumulation of ubiquitinated substrates in all samples treated with bortezomib. However, in the 8226.BR cells, a similar pattern to the in vivo data were observed, in that a decreased amount of ubiquitinated proteins were found (supplemental Figure 10B). Further investigation into this phenomenon is warranted, but we hypothesize that a compensatory mechanism may be activated to overcome OSI-906– and bortezomib-induced apoptosis through the activation of deubiquitinating enzymes to prevent the formation of aggresomes.
Discussion
IGF-1 produced by plasma cells, as well as by the marrow microenvironment, is a critical mediator of several downstream effects that contribute to multiple myeloma pathobiology. The IGF-1 receptor itself has been found to be overexpressed in myeloma,33,34 and this aberrant expression, as well as higher IGF-1 levels themselves, have been related to disease progression, severity, and prognosis.30,34–37 Our current studies revealed evidence that increased IGF-1 signaling through enhanced IGF-1 secretion and IGF-1R activation was associated with the phenotype of resistance to bortezomib (Figure 2). In addition, gene expression profiling confirmed that genes acutely activated by IGF-1 stimulation were chronically expressed in our BR cell lines (Table 2). Furthermore, blockade of downstream targets such as PI3K and mTOR could, to some extent, overcome this resistance (Figure 3). Pharmacologic (Figure 3) or genetic (Figure 4) suppression of IGF-1R also sensitized cell lines and patient samples to proteasome inhibitor therapy. Finally, the IGF-1R tyrosine kinase inhibitor OSI-906 synergized with bortezomib to enhance myeloma cell death (Table 3) and overcame bortezomib resistance in vivo (Figure 6). OSI-906 in combination also extended overall survival, albeit at a more modest level, and could be explained by variability in drug absorption and steady-state levels of concentration in the tissues. Thus, these data strongly implicate dysregulation of the IGF-1/IGF-1R axis in acquired bortezomib resistance.
Recent reports predominantly focusing on nonmyeloma model systems have indicated a possible contribution from several mechanisms to bortezomib resistance, including massive overexpression of β5,16 as well as induction of other proteasome subunits.38,39 Consistent with this finding, we also detected an increase in β5 expression in 8226.BR (Figure 1) and ANBL-6.BR cells (supplemental Figure 3), but β5 mRNA levels were not consistently increased. Notably, there are at least 2 levels of control in proteasome biogenesis, including subunit protein expression and subunit recruitment for proteasome assembly. Proteasome biogenesis occurs in a stepwise manner, with the formation of an α subunit ring that attracts the proteasome maturation protein POMP.40 The β subunits are translated with a prosequence41,42 essential for assembly and are then targeted to POMP for the final formation of a 16S hemiproteasome, composed of an α-ring and a β-ring, 2 of which ultimately are combined to form an active 20S proteasome. Therefore, it is possible that the alterations in proteasome biogenesis in the BR cells, rather than increased transcription and translation, are responsible for the higher expression of β5 subunits.
Massive overexpression of β5 has been proposed to contribute to resistance through the ability of this presumably freely occurring subunit to compete for bortezomib binding,16 possibly thereby sparing the β5 subunits within the proteasome from the effects of this inhibitor. It is possible that the β5 overexpression we encountered may have contributed to the BR phenotype. However, the level of induction we observed was much more modest than that reported previously, suggesting that, at a minimum, this was likely to be only a small contributor. More importantly, our studies of interventions targeting the IGF-1/IGF-1R signaling axis validated this pathway as a contributor to resistance in the absence of any effects on total β5 subunit expression, suggesting that these 2 mechanisms are independent of each other.
An alternative mechanism of resistance may be because of mutations in the β5 subunit, as recently reported by 2 independent groups.14–16 Lu et al and Oerlemans et al developed BR leukemic and myelomonocytic cell lines, respectively, that display point mutations in the bortezomib-binding site of the β5 subunit. However, examination of our BR myeloma cell lines showed that they remained sensitive to bortezomib-mediated proteasome inhibition in a dose- and time-dependent manner (Figure 1). These findings suggest that these models have developed without mutation of the bortezomib binding site, although it may be possible that other point mutations lie in the β5 subunit. For instance, a point mutation in the β5 subunit prosequence would prevent new proteasome assembly, and this has already been identified as a high-frequency mutation in myeloma patients.18
Another mechanism of resistance that impacts on the efficacy of several drugs is overexpression or increased activity of ATP-cassette binding proteins such as P-glycoprotein. Some preclinical studies have suggested that bortezomib sensitivity may be influenced by modulation of the function of these gene products.43,44 Others, however, have not shown any impact on bortezomib of these multidrug resistance protein pumps,45,46 and bortezomib has not been felt to be a substrate for the efflux pump action of these transporters.47 We also examined whether P-glycoprotein might be an active contributor to our BR models, but we found only a slight sensitization effect, and this only at very high bortezomib concentrations (supplemental Figure 2D), suggesting a modest, if any role for this mechanism.
Lastly, our examination into the intracellular levels of bor-tezomib showed a prodigious increase of bortezomib concentrations in the BR cells (Table 1). One possible reason for this increase may be because of decreased proteasome biogenesis, because if the β5 subunits are not being incorporated into proteasomes but remain functionally capable of binding inhibitors, they may act as a sink for bortezomib, as hypothesized by Oerlemans et al.16 Consistent with the possibility that there is decreased proteasome biogenesis, data showed that the BR cell lines have reduced basal levels of ChT-L activity (Figure 1).
Although IGF-1 signaling induces the activation of Akt, the possibility that this is the only downstream kinase activated by IGF-1 is unlikely. In fact, it seems likely that other downstream signaling pathways are also activated, such as the Janus kinase and signal transducers and activators of transcription. These pathways propagate survival and growth signaling, are known to play key roles in cancer neoplasia and tumor progression,48 and can be activated by growth factors such as IGF-1.49 Further study of these other signaling pathways that lie downstream of IGF-1 are needed, as these also may be possible targets whose inhibition may achieve similar resensitization to bortezomib.
Multiple myeloma is characterized clinically by a good initial response to therapy, but even patients who achieve a complete remission after standard-dose or high-dose approaches incorporating bortezomib inexorably relapse with disease that becomes progressively more chemoresistant. IGF-1 signaling is probably not the only mechanism by which plasma cells evade bortezomib-induced cell death. However, the near abrogation of bortezomib resistance through suppression of IGF-1R, and decreased survival outcomes of patients with increased IGF-1R expression, support a key role for this pathway. In addition to the important role of IGF-1 in myeloma biology, the findings herein provide an excellent rationale for studies targeting IGF-1 signaling in combination with bortezomib as an approach to overcome, or possibly even prevent outgrowth of resistance to bortezomib in myeloma patients.
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the Flow Cytometry and Cellular Imaging Core Facility, the DNA Analysis Core, Cell Line Characterization Core, and the Pharmacology and Analytical Facility funded by the MD Anderson Cancer Center Support grant (NCI CA16672). D.J.K. acknowledges support from the National Cancer Institute (1K99 CA149140) and the Diane and John Grace Family Foundation Fund. R.Z.O. acknowledges support from the Leukemia & Lymphoma Society (6096-07), the Multiple Myeloma Research Foundation, and the National Cancer Institute in the form of The MD Anderson Cancer Center SPORE in Multiple Myeloma (P50 CA142509). The authors also acknowledge support from the Brock Family Myeloma Research Fund.
National Institutes of Health
Authorship
Contribution: D.J.K. performed the majority of the experiments, analyzed the data, prepared the figures, and wrote the manuscript; Z.B. provided valuable assistance with OSI-906 pilot and combination animal studies, as well as editing of the manuscript; R.J.J. assisted with data analysis and manuscript preparation; R.W. performed studies assessing bortezomib resistance in vivo; C.C.B. provided valuable assistance in data analysis; W.M. and R.E.D. performed gene expression profiling on cell lines and provided bioinformatical studies; P.L. performed histopathology studies; H.W. created the IGF-1R expression vector; T.L.M. determined intracellular concentration levels of bortezomib; C.W. and V.B. provided biostatistical analyses; M.W., S.K.T., J.J.S., and D.M.W. provided primary patient samples; and R.Z.O. provided research guidance and supervision, and assisted in writing and proofing of the manuscript.
Conflict-of-interest disclosure: R.Z.O. serves on an advisory board for Millennium, the Takeda Oncology Company, which markets bortezomib. The remaining authors declare no competing financial interests.
Correspondence: Robert Z. Orlowski, Department of Lymphoma and Myeloma, The University of Texas MD Anderson Cancer Center, 1515 Holcombe Blvd, Unit 429, Houston, TX 77030-4009, e-mail: rorlowsk@mdanderson.org.