Key Points
Fibrin αC389-402 binds a cleft in the β-sandwich domain of FXIII-A2* exposed only after cleavage of the activation peptide.
Binding of fibrin αC389-402 to FXIII-A2* regulates fibrin cross-linking and thus clot stabilization and fibrinolysis.
Abstract
Formation of a stable fibrin clot is dependent on interactions between factor XIII and fibrin. We have previously identified a key residue on the αC of fibrin(ogen) (Glu396) involved in binding activated factor XIII-A2 (FXIII-A2*); however, the functional role of this interaction and binding site(s) on FXIII-A2* remains unknown. Here we (1) characterized the functional implications of this interaction; (2) identified by liquid-chromatography–tandem mass spectrometry the interacting residues on FXIII-A2* following chemical cross-linking of fibrin(ogen) αC389-402 peptides to FXIII-A2*; and (3) carried out molecular modeling of the FXIII-A2*/peptide complex to identify contact site(s) involved. Results demonstrated that inhibition of the FXIII-A2*/αC interaction using αC389-402 peptide (Pep1) significantly decreased incorporation of biotinamido-pentylamine and α2-antiplasmin to fibrin, and fibrin cross-linking, in contrast to Pep1-E396A and scrambled peptide controls. Pep1 did not inhibit transglutaminase-2 activity, and incorporation of biotinyl-TVQQEL to fibrin was only weakly inhibited. Molecular modeling predicted that Pep1 binds the activation peptide cleft (AP-cleft) within the β-sandwich domain of FXIII-A2* localizing αC cross-linking Q366 to the FXIII-A2* active site. Our findings demonstrate that binding of fibrin αC389-402 to the AP-cleft is fundamental to clot stabilization and presents this region of FXIII-A2* as a potential site involved in glutamine-donor substrate recognition.
Introduction
Factor XIII-A2B2 (FXIII-A2B2) is a 325 000-Da protransglutaminase, which, when activated, is responsible for covalently cross-linking fibrin chains and inhibitors of fibrinolysis to fibrin, thereby stabilizing the fibrin clot and making it resistant to degradation.1 FXIII-A2B2 is a heterotetramer composed of 2 identical catalytic A subunits noncovalently bound to 2 carrier FXIII-B subunits.2-4 FXIII-A2B2 is converted to the enzymatically active form by thrombin-catalyzed hydrolysis of the Arg37-Gly38 peptide bond at the amino terminus of the FXIII-A2 subunit,5 resulting in the release of a 37–amino acid (aa) activation peptide.6 In the presence of calcium, thrombin-cleaved FXIII-A2B2 subunits dissociate to yield FXIII-B2 and activated FXIII-A2 (FXIII-A2*).7-9 This process is significantly enhanced in the presence of fibrinogen, an effect that has been ascribed to the AαC region 242-424.10,11 Fibrinogen is a 340 000-Da glycoprotein composed of 2 sets of disulfide-linked nonidentical polypeptide chains: Aα, Bβ, and γ.12,13 Thrombin catalyzes the polymerization of fibrinogen to fibrin by sequentially cleaving fibrinopeptide A and fibrinopeptide B initiating lateral aggregation of protofibrils and fiber formation.14-16 FXIII-A2* stabilizes the forming protofibril by introducing cross-links between adjacent γ chains.17 Cleavage of fibrinopeptide B causes the release of the αC from the central E region enabling FXIII-A2* to cross-link adjacent glutamine residues on the AαC at positions Q221, Q237, Q328, and Q366,18 thereby increasing both the mechanical strength of the developing fiber and resistance to fibrinolysis.19
In addition to cross-linking γ and Aα chains for clot stabilization, FXIII-A2* has a key role in cross-linking proteins involved in clot formation and fibrinolysis to the fibrin(ogen) αC regions. These include plasminogen activator inhibitor 2,20 thrombospondin,21 von Willebrand factor,22 and fibronectin.23 The principal inhibitor of plasmin, α2-antiplasmin,24,25 is also cross-linked to fibrin AαC residue Lys303 by FXIII-A2* making the fibrin clot resistant to degradation.20,26 Recently, Fraser et al27 demonstrated that resistance of a fibrin clot to lysis was primarily dependent on FXIII-A2* cross-linking of α2-antiplasmin to fibrin AαC Lys303, rather than fibrin-fibrin cross-linking. We have recently identified a key residue on the αC of fibrin (AαGlu396) involved in the binding of the FXIII-A2* subunit.28 Here, we demonstrate the importance of this interaction with respect to FXIII-A2 activity and its cross-linking functions. In addition, using synthetic peptides of fibrin αC region 389-402, we have localized the fibrin αC binding region on FXIII-A2* to an interaction site within the β-sandwich domain of FXIII-A2*, specifically the activation peptide cleft (AP-cleft), and provide molecular modeling on the predicted interaction. Our findings suggest that the interaction between the AP-cleft of FXIII-A2* and the fibrin αC region 389-402 has a fundamental role in regulating fibrin cross-linking, and thus clot stabilization and fibrinolysis, and furthermore presents a novel role for the FXIII-A2* AP-cleft in glutamine-donor substrate recognition.
Materials and methods
Expression and purification of rFXIII-A2, recombinant fibrinogen, and plasma-derived FXIII-A2B2
Recombinant FXIII-A2 (rFXIII-A2) was expressed and purified as previously detailed by Smith et al.28 In brief, FXIII-A2 was expressed in Escherichia coli as a glutathione S-transferase (GST)–fusion protein and purified using GST-affinity chromatography. The GST tag was proteolytically removed using an on-column cleavage approach to yield pure FXIII-A2. A plasma-derived concentrate of FXIII-A2B2, (Fibrogammin P; ZLBBehring, Sussex, United Kingdom) was separated from albumin using gel filtration chromatography. Recombinant fibrinogen (α,β,γ)2 was obtained from stably transfected Chinese hamster ovary (CHO) cells (a generous gift from Susan Lord) and purified by ammonium sulfate precipitation and IF-1 affinity chromatography as previously described.29-31 The purified fibrinogen was dialyzed against 100 mM NaCl, 50 mM tris(hydroxymethyl)aminomethane (Tris) at pH 7.4 and stored at −80°C. All proteins were analyzed for purity using 4% to 12% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE).
Activation of rFXIII-A2 and FXIII-A2B2
rFXIII-A2 and FXIII-A2B2 were activated using 5 U/mL human α-thrombin with 1.5 mM calcium for 2 hours at 37°C, unless stated otherwise. When removal of thrombin was necessary, biotinylated-thrombin (Merck Millipore, Darmstadt, Germany) was used at 5 U/mL for 2 hours at 37°C. Biotinylated-thrombin was removed using streptavidin agarose according to the manufacturer’s protocol (Merck). Following activation, the FXIII-A2 (FXIII-A2*) was centrifuged at 500g for 5 minutes to remove precipitate, and protein concentration determined by measurement at an absorbance of 280 nm applying the extinction coefficient 1.58 or 1.31 for a solution of 1 mg/mL of rFXIII-A2* or FXIII-A2B2, respectively.
Synthetic peptide development
Six synthetic peptides of fibrinogen αC region 389-402 were synthesized by Biomatik: Pep1; Pep1-E396A; Pep1-Cys; Cys-Pep1; and 2 blind-scrambled peptides, Pep1-X and Pep1-Y. See Table 1. Peptide purity was >95% as determined by high-performance liquid chromatography analysis. Pep1-Cys and Cys-Pep1 were designed with opposing terminal-end Cys residues to enable chemical cross-linking to rFXIII-A2* using a cross-linker for cysteine-lysine conjugation as detailed subsequently.
Synthetic peptides of fibrin αC residues 389-402 used in this investigation
| Fibrin αC peptide name . | Fibrin αC peptide 389-402 sequence and modification . |
|---|---|
| Pep1 | PDWGTFEEVSGNVS |
| Pep1-Cys* | PDWGTFEEVSGNVSC |
| Cys-Pep1* | CPDWGTFEEVSGNVS |
| Pep1-E396A† | PDWGTFEAVSGNVS |
| Pep1-X‡ | EPNVEGFGWSSVTD |
| Pep1-Y‡ | EGTWGPSNDSVEFV |
| Fibrin αC peptide name . | Fibrin αC peptide 389-402 sequence and modification . |
|---|---|
| Pep1 | PDWGTFEEVSGNVS |
| Pep1-Cys* | PDWGTFEEVSGNVSC |
| Cys-Pep1* | CPDWGTFEEVSGNVS |
| Pep1-E396A† | PDWGTFEAVSGNVS |
| Pep1-X‡ | EPNVEGFGWSSVTD |
| Pep1-Y‡ | EGTWGPSNDSVEFV |
Peptide employed in LC-MS/MS. Terminal cysteine residues highlighted in bold and underlined.
Control peptide. The point mutation AαE396A highlighted in bold and underlined.
Blind-scrambled peptide.
Peptide inhibition of the rFXIII-A2*/fibrinogen αC interaction by ELISA
rFXIII-A2 and recombinant fibrinogen αC fragment 1 (α233-425) were expressed and purified as detailed by Smith et al.28 Binding of the synthetic peptides to rFXIII-A2* and the ability of the peptides to inhibit the rFXIII-A2*/fibrinogen αC interaction was determined by enzyme-linked immunosorbent assay (ELISA). In brief, a 96-well plate was coated with 5 µg/mL recombinant fibrinogen αC fragment 1 in 50 mM sodium carbonate, pH 9.6, at 4°C overnight. The wells were blocked with 300 µL 1% (weight to volume ratio) bovine serum albumin for 90 minutes at 37°C. rFXIII-A2 was activated with 5 U/mL biotinylated-thrombin and 1.5 mM CaCl2 for 2 hours at 37°C, and the biotinylated-thrombin removed. rFXIII-A2* (500 nM) was incubated with increasing molar concentrations of the individual peptides (Pep1, Pep1-E396A, Pep1-Cys, Cys-Pep1: [0.05-500 µM]) for 30 minutes at room temperature. Control rFXIII-A2* incubations were carried out in the absence of any peptide. One hundred microliters of the rFXIII-A2*/peptide mix was added to the fibrinogen αC–coated wells and incubated for 1 hour at 37°C. Following washing, sheep anti-human FXIII-A horseradish peroxidase (HRP) conjugate (dilution 1/10 000 [Enzyme Research Laboratories, Swansea, United Kingdom]) was added to detect bound rFXIII-A2*. Detection of the HRP-conjugated antibody was performed using chromogenic substrate: 1,2-phenylenediamine dihydrochloride (DakoCytomation, Dako, Ely, UK) measured at an absorbance of 490 nm. The data generated for each peptide were normalized against the FXIII-A2* control sample and converted into percentage of FXIII-A2* bound to αF1 in the presence of each peptide.
FXIII-A2 biotinamido-pentylamine incorporation activity assay with peptide inhibition
The FXIII-A2–mediated incorporation of biotinamido-pentylamine to recombinant fibrinogen in the presence and absence of Pep1 was determined using a 5-(biotinamido) pentylamine incorporation assay.28 This assay was also used to (1) screen Pep1 for its ability to inhibit guinea pig liver tissue TG-2 (Sigma Aldrich, St. Louis, MO); (2) determine the effect of Pep1 on the FXIII-A2–mediated incorporation of biotinyl-TVQQEL glutamine-donor to lysine-acceptor sites on plates coated with fibrin and poly-L-lysine; and (3) confirm that Pep1 is not inhibiting thrombin. Details of each methodology used can be found in the supplemental data (see the Blood Web site).
α2-Antiplasmin incorporation assay with peptide inhibition
The effect of Pep1 on the FXIII-A2–mediated cross-linking of α2-antiplasmin to fibrin was determined using a method modified from that described by Dunn et al.32 In brief, a microtiter plate coated with recombinant fibrin (40 µg/mL) was incubated with nonactivated rFXIII-A2 (1.1 µg/mL: final concentrations given are per well) in the presence and absence of increasing molar concentrations of Pep1 (6.95-55.2 µM). The reaction mix contained 0.1 mM dithiothreitol (DTT), 10 µg/mL α2-antiplasmin (Calbiochem, Nottingham, UK), 1 U/mL human α-thrombin (Calbiochem), and 5 mM calcium in Tris-buffered saline, pH 7.4. At 20-minute intervals (up to 80 minutes), the reaction was stopped with 0.2 M EDTA. After washing, a goat anti-human α2-antiplasmin HRP-conjugated IgG antibody (Enzyme Research Laboratories) was added to each well, and cross-linked α2-antiplasmin detected using a chromogenic substrate 1,2-phenylenediamine dihydrochloride (OPD) at an absorbance of 490 nm. Controls included (1) Tris-buffered saline, pH 7.4 (no rFXIII-A2*); (2) Pep1-E396A at 55.2 µM; and (3) rFXIII-A2* minus the peptide (n = 3). Further details of this method can be found in the supplemental data.
SDS-PAGE analysis displaying the effect of Pep1 on FXIII-A2–mediated cross-linking of fibrin chains
Preactivated FXIII-A2B2 (4.4 µg/mL) was incubated with 735 µM of Pep1 for 30 minutes at room temperature. The FXIII-A2B2/Pep1 mix was added to 1.47 µM (0.5 mg/mL) recombinant fibrinogen, followed by an activation mix containing human α-thrombin (0.05 U/mL) and CaCl2 (1.5 mM). The sample was incubated at 37°C and stopped with NuPAGE 10× SDS-PAGE reducing buffer with NuPAGE loading dye (Invitrogen, Carlsbad, CA, USA) and boiled for 15 minutes at given time points 0 to 180 minutes. Samples were run on a 4% to 12% 2-[Bisamino]-2–1,3-propanediol NuPAGE gel with NuPAGE 3-(N-morpholino)propanesulfonic acid running buffer (150 V, 85 minutes). Gels were stained with GelCode blue (Pierce, Rockport, IL) and imaged using a Kodak Image Station 2000R (Labtech International, Uckfield, UK) (n = 3). For each time course, the gel bands corresponding to the γ chains, αC, γ-γ dimers, and γ-α hybrids were measured by densitometry in the presence and absence of Pep1 to highlight the effect of the peptide on clot stabilization by rFXIII-A2*. Net intensities for each band were blank subtracted against the background signal and plotted against time. Results are expressed as means: standard error of the mean (SEM). Control samples included 0.5 mg/mL recombinant fibrinogen containing 0.05 U/mL thrombin, 1.5 mM calcium, and 4.4 µg/mL preactivated FXIII-A2B2 incubated with 735 µM of (1) Pep1; (2) Pep1-E396A; (3) Pep1-X; and (4) Pep1-Y. Each sample was incubated at 37°C for 30 minutes to determine the effect on fibrin clot formation.
Chemical cross-linking of modified synthetic peptides to rFXIII-A2*
Pep1-Cys and Cys-Pep1 were chemically cross-linked to rFXIII-A2* using a short-chain (6.8 Å) succinimidyl 3-(2-pyridyldithio)-propionate (SPDP; Thermo Scientific, Waltham, MA) cross-linker for cysteine-to-lysine conjugation forming a cleavable disulfide bond with cysteine sulfhydryls. Twenty-five microliters of 20 mM SPDP cross-linker was added to 2.5 mg of Pep1-Cys and Cys-Pep1 in phosphate-buffered saline, pH 7.4, and incubated for 30 minutes at room temperature. The SPDP-modified peptide samples were injected onto a 5-mL polyacrylamide desalting gravity flow column (Thermo Scientific) and washed with 5 column volumes of phosphate-buffered saline, pH 7.4, to elute the SPDP peptide, removing any nonreacted cross-linker. rFXIII-A2* (1 mg) was added to the eluted SPDP-peptide samples and incubated for 16 hours at 4°C on a roller mixer to allow for conjugation as detailed in the manufacturer’s instructions for SPDP chemical cross-linkers. The cross-linked rFXIII-A2*/peptide samples were analyzed by liquid chromatography–tandem mass spectrometry (LC-MS/MS) (Bioscience Technology Facility, University of York, York, United Kingdom) to identify the SPDP-modified lysine residues on rFXIII-A2* localizing the αC peptide binding region.
LC-MS/MS
In-gel tryptic digestion was performed on the rFXIII-A2*/peptide complex. Samples were acidified in 0.1% aqueous trifluoroacetic acid and loaded onto a nanoAcquity UPLC system (Waters, Milford, MA) equipped with a nanoAcquity Symmetry C18, 5-μm trap (180 μm × 20 mm, Waters) and a nanoAcquity BEH130 1.7-μm C18 capillary column (75 m × 250 mm, Waters). The nanoLC system was interfaced with a maXis LC-MS/MS System (Bruker Daltonics, Billerica, MA) with a nanoelectrospray source fitted with a steel emitter needle (180 μm optical density × 30 μm internal diameter, Proxeon). Positive electrospray ionization mass spectrometry and tandem mass spectrometry spectra were acquired using the AutoMSMS mode. Instrument control, data acquisition, and processing were performed using Compass 1.3 SP1 software (microTOF control, Hystar, and DataAnalysis; Bruker Daltonics). Tandem mass spectral data were submitted to database searching using a locally running copy of the Mascot program (version 2.3; Matrix Science Ltd., London, UK) through the Bruker ProteinScape interface (version 2.1). A further variable modification named SPDP-carbamidomethyl (CAM) was added to detect the presence of the alkylated cross-linker moiety. This modification allowed for the mass addition of 145.0 Da on any rFXIII-A2* lysine residue, as would be predicted by the addition of the SPDP cross-linker, subsequently alkylated with iodoacetamide. Peptides with an expect value of ≤.05 were considered significant. A detailed methodology is included in the supplemental data (supplemental Figure 5).
Size exclusion chromatography–multiangle laser light scattering (SEC-MALLS)
SEC-MALLS was carried out on the rFXIII-A2 subunit to confirm its dimeric structure. In brief, 130.6 µg rFXIII-A was injected onto a Superdex 200 10/300 gel filtration column fitted to a Wyatt HELEOS-II multiangle light-scattering detector and a Wyatt rEX refractive index detector linked to a Shimadzu high-performance liquid chromatography system, with UV detection at 280 nm (Wyatt Technology Corporation, Santa Barbara, CA). The molecular weight (mwt) of the rFXIII-A was estimated using the Zimm fit method with degree 1. Details of this method can be found in the supplemental data.
Molecular modeling and structural analysis of the rFXIII-A2*/Pep1-Cys complex
Structural analysis of nonactivated FXIII-A2 was based on the crystal structure of the human cellular FXIII-A2 homodimer (Protein Data Bank [PDB] code 1F13).33 Molecular modeling of rFXIII-A2 in its active conformation was based on the crystal structure of TG-2 in its extended active conformation (PDB code 2Q3Z),34 in the absence of any crystal structure for FXIII-A2*. Pep1-Cys and Cys-Pep1 peptides were then modeled with the predicted rFXIII-A2* conformation. Molecular modeling was performed using the Maestro Modeling suite from Schrodinger Inc. using an analogous approach to that previously described by Komaromi et al.35 In brief, minimization was performed with the OPLS_2005 force field within a simulated aqueous environment. The peptides were constructed within the Maestro interface and then linked in silico to the relevant chemically modified lysine sites (K446, K257, and K113) using the SPDP-CAM cross-linker. The most favorable site in terms of overall energy of the complex was identified.
Molecular modeling of the extended fibrin αC fragment residues Q328-S402
An extended fragment of the fibrin αC including residues Gln328-Ser402 in complex with FXIII-A2* was modeled. The αC fragment (residues 389-402) was docked in the FXIII-A2* AP-cleft, and the remaining αC fragment was manually threaded through the FXIII-A2* structure. The crystal structure of the fibrin αC region remains undetermined; however, investigations into the folding status of the αC connector region demonstrate characteristics consistent with that of a poly(L-proline) type 2 conformation, which typically forms an extended left-handed helical conformation.36 Therefore, in this investigation, a largely α-helical conformation was adopted for modeling the extended 75-aa αC fragment 328-402. Molecular modeling was performed using the Maestro Modeling suite from Schrodinger Inc.
Results
Inhibition of the rFXIII-A2*/fibrinogen αC interaction using synthetic peptides
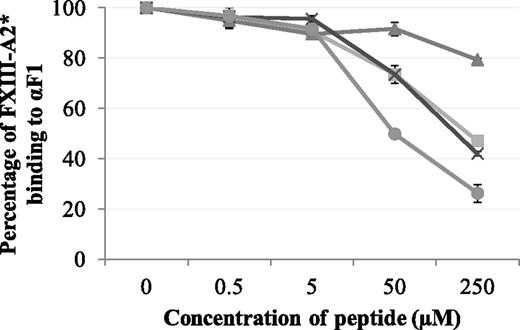
Synthetic peptides Pep1, Pep1-E396A, Pep1-Cys, and Cys-Pep1 were screened for their ability to inhibit the binding of rFXIII-A2* to the recombinant fibrin αC fragment 1 (residues 233-425). Of the 4 peptides, Pep1, Pep1-Cys, and Cys-Pep1 inhibited the interaction to a similar degree. Therefore, Pep1-Cys and Cys-Pep1 were validated candidates for chemical cross-linking to FXIII-A2*. As predicted, Pep1-E396A, lacking the key αC-binding residue Glu396, did not inhibit the interaction to the same degree (Table 1; Figure 1).
Competitive inhibition assay displaying the effect of synthetic peptides Pep1 (▪), Pep1-E396A (▲), Pep1-Cys (X), and Cys-Pep1 (●) on the binding of rFXIII-A2* to recombinant αC fragment 1 (Aα233-425). rFXIII-A2* (0.5 µM) was preincubated with increasing concentrations of each peptide (0.5-250 µM) and added to wells coated with recombinant fibrin αC fragment 233-425 (αF1). Bound rFXIII-A2* was detected using an HRP-conjugated anti–FXIII-A antibody and OPD substrate at an absorbance of 490 nm. The optical density was converted to percentage of FXIII-A2 binding to αF1. Samples were performed in triplicate (n = 3). Error bars show ± 1 standard deviation (SD).
Competitive inhibition assay displaying the effect of synthetic peptides Pep1 (▪), Pep1-E396A (▲), Pep1-Cys (X), and Cys-Pep1 (●) on the binding of rFXIII-A2* to recombinant αC fragment 1 (Aα233-425). rFXIII-A2* (0.5 µM) was preincubated with increasing concentrations of each peptide (0.5-250 µM) and added to wells coated with recombinant fibrin αC fragment 233-425 (αF1). Bound rFXIII-A2* was detected using an HRP-conjugated anti–FXIII-A antibody and OPD substrate at an absorbance of 490 nm. The optical density was converted to percentage of FXIII-A2 binding to αF1. Samples were performed in triplicate (n = 3). Error bars show ± 1 standard deviation (SD).
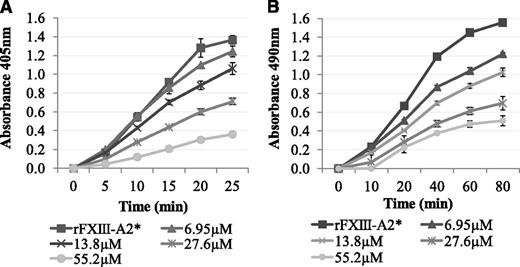
The effect of Pep1 on the FXIII-A2–mediated incorporation of biotinamido-pentylamine and α2-antiplasmin to fibrin
It was determined whether Pep1 was able to inhibit the rFXIII-A2*–mediated cross-linking of biotinamido-pentylamine to fibrin. The results demonstrated a dose-dependent decrease in rFXIII-A2* cross-linking of biotinamido-pentylamine to fibrin in the presence of Pep1 (Figure 2A). In contrast, Pep1-E396A did not inhibit rFXIII-A2* cross-linking, confirming that GluAα396 of the peptide, and of the fibrin αC, is a key site in the binding of rFXIII-A2* (supplemental Figure 1A).
The effect of Pep1 on the rFXIII-A2*–mediated incorporation of biotinamido-pentylamine and α2-antiplasmin to fibrin. (A) Biotinamido-pentylamine incorporation assays: rFXIII-A2*/Pep1 mixtures (final concentrations per well, 0.5 µg/mL and 6.95-55.2 µM, respectively) were added to fibrinogen-coated wells. The reactions were activated by adding DTT (0.1 mM), biotinamido-pentylamine (0.3 mM), CaCl2 (10 mM), and human α-thrombin (1 U/mL). Reactions were stopped at 5-minute intervals by addition of 200 mM EDTA. Incorporated biotinamido-pentylamine was detected at an absorbance of 405 nm (n = 3). Error bars show ± 1 SD. (B) α2-Antiplasmin incorporation assays: An rFXIII-A2*/Pep1 mix (final concentrations per well, 1.1 µg/mL and 6.95-55.2 µM, respectively) was added to fibrin-coated wells. An activation mix containing 0.1 mM DTT, 5 mM CaCl2, 1 U/mL human α-thrombin, and 10 µg/mL α2-antiplasmin was added, and the reaction stopped at 20-minute intervals using 200 mM EDTA. Cross-linked α2-antiplasmin was detected using an α2-antiplasmin HRP-conjugated antibody and OPD substrate at an absorbance of 490 nm. Blank wells were absent of rFXIII-A2*. Samples were performed in triplicate (n = 3). Error bars show ± 1 SD.
The effect of Pep1 on the rFXIII-A2*–mediated incorporation of biotinamido-pentylamine and α2-antiplasmin to fibrin. (A) Biotinamido-pentylamine incorporation assays: rFXIII-A2*/Pep1 mixtures (final concentrations per well, 0.5 µg/mL and 6.95-55.2 µM, respectively) were added to fibrinogen-coated wells. The reactions were activated by adding DTT (0.1 mM), biotinamido-pentylamine (0.3 mM), CaCl2 (10 mM), and human α-thrombin (1 U/mL). Reactions were stopped at 5-minute intervals by addition of 200 mM EDTA. Incorporated biotinamido-pentylamine was detected at an absorbance of 405 nm (n = 3). Error bars show ± 1 SD. (B) α2-Antiplasmin incorporation assays: An rFXIII-A2*/Pep1 mix (final concentrations per well, 1.1 µg/mL and 6.95-55.2 µM, respectively) was added to fibrin-coated wells. An activation mix containing 0.1 mM DTT, 5 mM CaCl2, 1 U/mL human α-thrombin, and 10 µg/mL α2-antiplasmin was added, and the reaction stopped at 20-minute intervals using 200 mM EDTA. Cross-linked α2-antiplasmin was detected using an α2-antiplasmin HRP-conjugated antibody and OPD substrate at an absorbance of 490 nm. Blank wells were absent of rFXIII-A2*. Samples were performed in triplicate (n = 3). Error bars show ± 1 SD.
Pep1 was also used to determine if the rFXIII-A2*/fibrin αC interaction plays a role in the incorporation of α2-antiplasmin to fibrin αC residue Lys303. The results demonstrated a dose-dependent decrease in α2-antiplasmin cross-linking to αC residue Lys303 on full-length fibrin in the presence of Pep1 (Figure 2B). Control experiments demonstrated that Pep1-E396A and 2 additional scrambled peptides of Pep1 (Pep1-X and Pep1-Y) did not inhibit the incorporation of α2-antiplasmin (supplemental Figure 1B-C), highlighting the specificity of this interaction with Pep1.
To confirm that Pep1 was specifically inhibiting rFXIII-A2* and not thrombin, the FXIII-A2 biotinamido-pentylamine incorporation activity assay was carried out using FXIII-A2 preactivated with biotinylated-thrombin. Following removal of the biotinylated-thrombin, the assay was performed as described. The results demonstrated that Pep1 inhibited the rFXIII-A2*–mediated cross-linking of biotinamido-pentylamine to fibrin and was therefore acting directly on rFXIII-A2* rather than nonspecifically inhibiting thrombin (supplemental Figure 2).
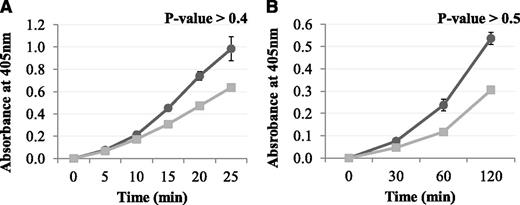
The effect of Pep1 on the FXIII-A2–mediated incorporation of a biotinyl-TVQQEL glutamine-donor peptide to lysine-acceptor sites on fibrin and poly-L-lysine
The results above demonstrated that Pep1 inhibited the FXIII-A2–mediated cross-linking of a low-mwt lysine analog to fibrin. The reaction was then carried out in the reverse orientation to determine the effect of Pep1 on the cross-linking of a low-mwt glutamine-donor peptide biotinyl-TVQQEL to lysine-acceptor sites on fibrin and poly-L-lysine. Under these conditions, Pep1 only weakly inhibited the incorporation of biotinyl-TVQQEL to fibrin and poly-L-lysine (Figure 3A-B), in contrast to biotinamido-pentylamine, which was strongly inhibited (Figure 2A). These results suggest that Pep1 is binding to a region on FXIII-A2* involved in glutamine substrate recognition.
The effect of Pep1 on the rFXIII-A2*–mediated incorporation of glutamine-donor peptide biotinyl-TVQQEL to lysine-acceptor sites on plates coated with (A) fibrin and (B) poly-L-lysine in the presence of Pep1. A master mix containing 0.5 mM biotinyl-TVQQEL peptide, 0.1 mM DTT, 10 mM calcium, 1 U/mL human α-thrombin in 100 mM NaCl, 50 mM Tris at pH 7.4 was added to wells coated with fibrin (A) or poly-L-lysine (B) and containing 5.5 µg/mL FXIII-A2 in the presence (▪) or absence (●) of Pep1 (final concentration, 55.2 µM). Samples were performed in triplicate (n = 3), and the results are expressed as means (SEM).
The effect of Pep1 on the rFXIII-A2*–mediated incorporation of glutamine-donor peptide biotinyl-TVQQEL to lysine-acceptor sites on plates coated with (A) fibrin and (B) poly-L-lysine in the presence of Pep1. A master mix containing 0.5 mM biotinyl-TVQQEL peptide, 0.1 mM DTT, 10 mM calcium, 1 U/mL human α-thrombin in 100 mM NaCl, 50 mM Tris at pH 7.4 was added to wells coated with fibrin (A) or poly-L-lysine (B) and containing 5.5 µg/mL FXIII-A2 in the presence (▪) or absence (●) of Pep1 (final concentration, 55.2 µM). Samples were performed in triplicate (n = 3), and the results are expressed as means (SEM).
The effect of Pep1 on the FXIII-A2–mediated cross-linking of fibrin γ and α chains
Pep1 was used as a competitive inhibitor to examine the effect of the rFXIII-A2*/fibrin αC interaction on fibrin γ- and α-chain cross-linking and subsequent clot stabilization. Direct comparisons of FXIII-A2B2 preactivated in the presence and absence of Pep1 demonstrated that blocking the rFXIII-A2*/fibrin αC interaction caused a statistically significant delay in the formation of γ dimers (γ dimer formation over time in the presence and absence of Pep1 = P < .01, and the decrease in the γ and αC over time in the presence and absence of Pep1 = P < .03 and P < .01, respectively). In addition, the cross-linking of fibrin γ-α chains was significantly delayed, and α-α–chain cross-linking was almost absent when compared with samples without Pep1 (γ-α cross-linking over time in the presence and absence of Pep1 = P < .004; net intensities for α-α cross-linking were too low to undertake formal analysis) (Figure 4). Control experiments demonstrated that Pep1-E396A and scrambled peptides Pep1-X and Pep1-Y did not inhibit the FXIII-A2–mediated cross-linking of fibrin γ and α chains, highlighting the specificity of this interaction with Pep1 αC sequence 389-402 (supplemental Figure 3).
SDS-PAGE analysis displaying the effect of Pep1 on rFXIII-A2*–mediated cross-linking of fibrin γ-γ, α-α, and α-γ. (A) An FXIII-A2B2/Pep1 mix (4.4 µg/mL and 735 µM Pep1, respectively) was added to 1.47 µM recombinant fibrinogen, followed by an activation mix containing human α-thrombin (0.05 U/mL) and CaCl2 (1.5 mM). The samples were incubated at 37°C for set time points (0-180 minutes), and the reaction stopped using NuPAGE 10× SDS-PAGE reducing buffer with NuPAGE loading dye and boiled for 15 minutes. The SDS-PAGE image shown is representative of 3 independent experiments (n = 3). (B) SDS-PAGE gel densitometry displaying reduction in the fibrin γC and αC over time in the presence (+) and absence (-) of Pep1. (*γ-chain reduction +/− Pep1, P < .03; **αC reduction +/− Pep1, P < .01). (C) SDS-PAGE gel densitometry displaying the formation of γ dimers (left axis) and γ-α hybrids (right axis) over time in the presence (+) and absence (-) of Pep1 (P = .01). Net intensities were converted to fold increase over time point 0. γ-Dimer formation in the presence and absence of Pep1 gave a P value of .01. γ-α–Hybrid formation in the presence and absence of Pep1 gave a P value of .004. (All experiments were a total of n = 3; the results are expressed as means [SEM].)
SDS-PAGE analysis displaying the effect of Pep1 on rFXIII-A2*–mediated cross-linking of fibrin γ-γ, α-α, and α-γ. (A) An FXIII-A2B2/Pep1 mix (4.4 µg/mL and 735 µM Pep1, respectively) was added to 1.47 µM recombinant fibrinogen, followed by an activation mix containing human α-thrombin (0.05 U/mL) and CaCl2 (1.5 mM). The samples were incubated at 37°C for set time points (0-180 minutes), and the reaction stopped using NuPAGE 10× SDS-PAGE reducing buffer with NuPAGE loading dye and boiled for 15 minutes. The SDS-PAGE image shown is representative of 3 independent experiments (n = 3). (B) SDS-PAGE gel densitometry displaying reduction in the fibrin γC and αC over time in the presence (+) and absence (-) of Pep1. (*γ-chain reduction +/− Pep1, P < .03; **αC reduction +/− Pep1, P < .01). (C) SDS-PAGE gel densitometry displaying the formation of γ dimers (left axis) and γ-α hybrids (right axis) over time in the presence (+) and absence (-) of Pep1 (P = .01). Net intensities were converted to fold increase over time point 0. γ-Dimer formation in the presence and absence of Pep1 gave a P value of .01. γ-α–Hybrid formation in the presence and absence of Pep1 gave a P value of .004. (All experiments were a total of n = 3; the results are expressed as means [SEM].)
Identification of the binding domain on FXIII-A2* for fibrin αC 389-402 by LC-MS/MS
Chemical cross-linking of the Pep1-Cys peptide to FXIII-A2* was detected by the addition of the 145.0-Da SPDP-CAM cross-linker (subsequently alkylated with iodoacetamide) on any lysine residue on the FXIII-A2* fragments. The results of the LC-MS/MS demonstrated that αC Pep1-Cys was chemically cross-linked specifically to FXIII-A2* Lys446 of an FXIII-A2* fragment 446-462. In order to confirm the Pep1-Cys chemical cross-linking site on FXIII-A2*, and to determine whether the C-terminal cysteine on Pep1-Cys was interfering with binding, an additional peptide, Cys-Pep1 with an N-terminal cysteine, was employed. The N-terminal Cys-peptide was chemically cross-linked to FXIII-A2* using the same methodology as applied for Pep1-Cys. The LC-MS/MS results for Cys-Pep1 identified 3 FXIII-A2* chemically modified lysine residues: (1) Lys446 of FXIII-A2* fragment 446-462, the same site identified previously with Pep1-Cys; (2) Lys257 of FXIII-A2* fragment 253-260; and (3) Lys113 of FXIII-A2* fragment 108-129. Because of varying sequence coverage by LC-MS/MS (Pep1-Cys, 48%; Cys-Pep1, 39%), sites 2 and 3 were only identified using Cys-Pep1 (Table 2).
| Fibrin αC peptide used . | rFXIII-A2 SPDP-modified fragment identified . | rFXIII-A2 fragment region . | rFXIII-A2 SPDP-modified lysine . | Mass error (Da) . | Ion score . | Expect . |
|---|---|---|---|---|---|---|
| Pep1-Cys* | KDGTHVVENVDATHIGK | 446-462 | 446 | 0.0105 | 20 | 0.011 |
| Cys-Pep1† | KDGTHVVENVDATHIGK | 446-462 | 446 | 0.00158 | 15 | 0.031 |
| GNPIKVSR | 253-260 | 257 | –0.0005 | 41 | 8.40E-05 | |
| YPQENKGTYIPVPIVSELQSGK | 108-129 | 113 | –0.00089 | 18 | 0.015 |
| Fibrin αC peptide used . | rFXIII-A2 SPDP-modified fragment identified . | rFXIII-A2 fragment region . | rFXIII-A2 SPDP-modified lysine . | Mass error (Da) . | Ion score . | Expect . |
|---|---|---|---|---|---|---|
| Pep1-Cys* | KDGTHVVENVDATHIGK | 446-462 | 446 | 0.0105 | 20 | 0.011 |
| Cys-Pep1† | KDGTHVVENVDATHIGK | 446-462 | 446 | 0.00158 | 15 | 0.031 |
| GNPIKVSR | 253-260 | 257 | –0.0005 | 41 | 8.40E-05 | |
| YPQENKGTYIPVPIVSELQSGK | 108-129 | 113 | –0.00089 | 18 | 0.015 |
SPDP-modified lysine residues are shown in bold and underlined in the amino acid fragment sequence. Sequence coverage by LC-MS/MS was 48% for Pep1-Cys and 39% for Cys-Pep1.
Pep1-Cys PDWGTFEEVSGNVSC.
Cys-Pep1 CPDWGTFEEVSGNVS.
Verifying that bacterially expressed FXIII-A2 exists as a dimer
The structure of the rFXIII-A2 was confirmed using SEC-MALLS to determine whether the rFXIII-A2 expressed in E. coli for this investigation exists as a dimer or a monomer. The results showed a peak at 27.8 minutes for light scattering, refractive index, and UV absorption at 280 nm consistent with a dimer of rFXIII-A2 at ∼175 kDa (supplemental Figure 4). Therefore, molecular modeling was based on the homodimer crystal structure of FXIII-A2 (PDB code 1F13).
Molecular modeling of the FXIII-A2*/Pep1-Cys and Cys-Pep1 complex
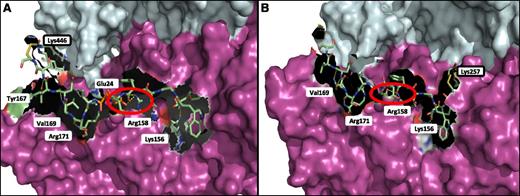
The FXIII-A2* dimer was modeled in complex with Pep1-Cys and Cys-Pep1 to identify and characterize the potential binding region on FXIII-A2* in relation to the chemically modified lysine residues previously identified by LC-MS/MS. The results indicated that Pep1-Cys and Cys-Pep1 were both binding to a cleft in the β-sandwich domain, exposed only after cleavage of the FXIII-A2* activation peptide, termed the AP-cleft. A number of FXIII-A2* contact sites were identified for the αC peptides; specifically, 4 sites were recognized for both peptides, including Lys156, Arg158, Val169, and Arg171 (Figure 5A). Cys-Pep1 LC-MS/MS analysis revealed 2 additional modified lysine sites, Lys257 and Lys113 (Table 2). Molecular modeling of the additional chemically cross-linked lysine sites identified the same AP-cleft binding region on FXIII-A2*. Therefore, only the model of Cys-Pep1 chemically cross-linked to FXIII-A2* Lys257 is given (Figure 5B). In addition, αC Glu396 residue (residue E8 in Pep1 corresponds to E396 in the fibrin αC), previously identified as a key residue in the interaction with FXIII-A2*,28 was predicted to form a salt bridge interaction specifically with FXIII-A2* Arg158 when either αC Pep1-Cys or Cys-Pep1 was employed in the LC-MS/MS (Figure 5A-B).
Molecular mapping of rFXIII-A2* chemically cross-linked to Pep1-Cys and Cys-Pep1 synthetic peptides. (A) rFXIII-A2* chemically cross-linked via Lys446 to Pep1-Cys (PDWGTFEEVSGNVSC). The chemically cross-linked lysine at rFXIII-A2* residue 446 is shown (black rectangle) in addition to potential attachment sites involving key FXIII-A2 residues Tyr167, Val169, Arg171, Glu24, Arg158, and Lys156. (B) rFXIII-A2* chemically cross-linked via Lys257 to Cys-Pep1 (CPDWGTFEEVSGNVS). Cys-Pep1 was also found to be chemically cross-linked to Lys446 in addition to Lys113 and Lys257. Chemically cross-linked Lys257 is shown (black rectangle). Potential attachment sites involved in the binding of Cys-Pep1 include Val169, Arg171, Arg158, and Lys156. The salt bridge interaction between Glu396 of the αC and Arg158 of rFXIII-A2* is shown by the red circles. Both Pep1-Cys and Cys-Pep1 were modeled in the same orientation. Molecular modeling was performed using the Maestro Modeling suite from Schrodinger Inc.
Molecular mapping of rFXIII-A2* chemically cross-linked to Pep1-Cys and Cys-Pep1 synthetic peptides. (A) rFXIII-A2* chemically cross-linked via Lys446 to Pep1-Cys (PDWGTFEEVSGNVSC). The chemically cross-linked lysine at rFXIII-A2* residue 446 is shown (black rectangle) in addition to potential attachment sites involving key FXIII-A2 residues Tyr167, Val169, Arg171, Glu24, Arg158, and Lys156. (B) rFXIII-A2* chemically cross-linked via Lys257 to Cys-Pep1 (CPDWGTFEEVSGNVS). Cys-Pep1 was also found to be chemically cross-linked to Lys446 in addition to Lys113 and Lys257. Chemically cross-linked Lys257 is shown (black rectangle). Potential attachment sites involved in the binding of Cys-Pep1 include Val169, Arg171, Arg158, and Lys156. The salt bridge interaction between Glu396 of the αC and Arg158 of rFXIII-A2* is shown by the red circles. Both Pep1-Cys and Cys-Pep1 were modeled in the same orientation. Molecular modeling was performed using the Maestro Modeling suite from Schrodinger Inc.
Molecular modeling of the extended fibrin αC fragment residues Q328-S402
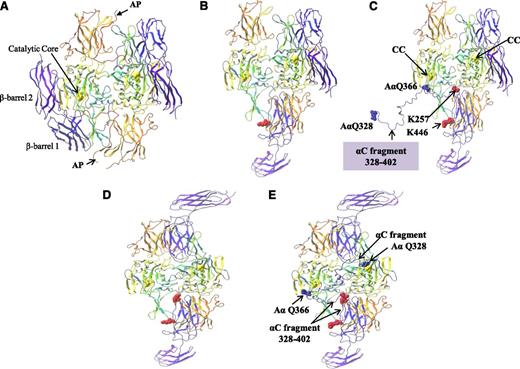
The binding of the αC region 389-402 to the FXIII-A2* AP-cleft was examined further to determine if this interaction served as a potential “docking” site to localize the αC cross-linking glutamines (specifically, Gln366 and Gln328) to the FXIII-A2* active site. In order to address this, an extended fragment of the fibrin αC including residues Gln328-Ser402 was docked at residues 389-402, and the remaining αC fragment was manually threaded through the FXIII-A2* structure. Structural analysis revealed that when the αC region 389-402 was bound to the FXIII-A2* AP-cleft, the αC glutamine AαQ366 was within close proximity to the active site Cys314 of 1 FXIII-A* monomer (Figure 6A-C). This model assumes that only 1 of 2 active sites present in the dimer is exposed at any one time. In addition, the modeling was repeated using the “fully open” form in which both active sites are exposed. This model demonstrated that if both FXIII-A2* active sites were exposed, the αC cross-linking site Q366 was in close proximity to 1 active site, whereas further modeling of the αC sequence revealed that Q328 was able to reach the active site of the second monomer (Figure 6D-E). Although the modeling presented here is a prediction of the expected route that the αC fragment might take through the FXIII-A2* in order to localize the αC Q366 and Q328 to the active sites, the modeling demonstrates that docking of the αC fragment at the FXIII-A2* AP-cleft localizes the FXIII-A2* active sites to the αC cross-linking glutamines in either a half open or fully open conformation.
Molecular modeling of rFXIII-A2* bound to the extended fibrin αC fragment residues 328-402. (A) FXIII-A2 in its nonactive form complete with activation peptides (AP). (B) Activated FXIII-A2 (half open) following cleavage of the activation peptide. (C) Activated FXIII-A2 (half open) shown bound to the fibrin αC fragment 328-402. rFXIII-A2* cross-linking site AαQ366 is shown adjacent to 1 rFXIII-A2* active site C314 residue (CC, yellow ball). The rest of the αC fragment up to AαQ328 is unbound. (D) Activated FXIII-A2 (fully open) following cleavage of the activation peptide. (E) Activated FXIII-A2 (fully open) following cleavage of the activation peptide, shown bound to the fibrin αC fragment 328-402. Both AαQ366 and AαQ328 rFXIII-A2* cross-linking sites are adjacent to both rFXIII-A2* active site C314 residues (yellow ball). The crystal structures (PDB codes 1F13 and 2Q3Z) are shown as ribbons. Molecular modeling was performed using the Maestro Modeling suite from Schrodinger Inc. using an analogous approach to that previously described by Komaromi et al.35
Molecular modeling of rFXIII-A2* bound to the extended fibrin αC fragment residues 328-402. (A) FXIII-A2 in its nonactive form complete with activation peptides (AP). (B) Activated FXIII-A2 (half open) following cleavage of the activation peptide. (C) Activated FXIII-A2 (half open) shown bound to the fibrin αC fragment 328-402. rFXIII-A2* cross-linking site AαQ366 is shown adjacent to 1 rFXIII-A2* active site C314 residue (CC, yellow ball). The rest of the αC fragment up to AαQ328 is unbound. (D) Activated FXIII-A2 (fully open) following cleavage of the activation peptide. (E) Activated FXIII-A2 (fully open) following cleavage of the activation peptide, shown bound to the fibrin αC fragment 328-402. Both AαQ366 and AαQ328 rFXIII-A2* cross-linking sites are adjacent to both rFXIII-A2* active site C314 residues (yellow ball). The crystal structures (PDB codes 1F13 and 2Q3Z) are shown as ribbons. Molecular modeling was performed using the Maestro Modeling suite from Schrodinger Inc. using an analogous approach to that previously described by Komaromi et al.35
The effect of Pep1 on the TG-2–mediated incorporation of biotinamido-pentylamine to fibrin
TG-2 is an enzyme that has near-identical active site topology to FXIII-A2 but which lacks the AP-cleft.35 Therefore, to further confirm that the FXIII-A2* AP-cleft is involved in the binding of Pep1, we employed a TG-2 activity assay. The results demonstrated that Pep1 did not inhibit TG-2–mediated incorporation of biotinamido-pentylamine to fibrin (Figure 7), supporting our molecular modeling, which predicts that Pep1 is binding specifically to the AP-cleft on FXIII-A2*.
The effect of Pep1 on the TG-2–mediated incorporation of biotinamido-pentylamine to fibrin TG-2 (11 µg/mL) was added to fibrin-coated wells followed by increasing concentrations of Pep1 (10-100 µM). A reaction mix containing DTT (0.1 mM), biotinamido-pentylamine (0.3 mM), CaCl2 (10 mM), and human α-thrombin (1 U/mL) in 100 mM 2-[Bisamino]-2–1,3-propanediol, pH 7.4, 0.0% triton X-100 was added to each well, and TG-2 activity was taken after 7 minutes. Samples were performed in triplicate (n = 3), and the results are expressed as means (SEM).
The effect of Pep1 on the TG-2–mediated incorporation of biotinamido-pentylamine to fibrin TG-2 (11 µg/mL) was added to fibrin-coated wells followed by increasing concentrations of Pep1 (10-100 µM). A reaction mix containing DTT (0.1 mM), biotinamido-pentylamine (0.3 mM), CaCl2 (10 mM), and human α-thrombin (1 U/mL) in 100 mM 2-[Bisamino]-2–1,3-propanediol, pH 7.4, 0.0% triton X-100 was added to each well, and TG-2 activity was taken after 7 minutes. Samples were performed in triplicate (n = 3), and the results are expressed as means (SEM).
Discussion
We have previously shown that rFXIII-A2* binds to the fibrin αC region 389-403 (dissociation constant 2.35 µM), an interaction that may be involved in enhancing FXIII activity.28 However, the corresponding binding sites on FXIII-A2* and the functional significance of the interaction have not been determined. Therefore, the aim of the present study was to (1) identify the functional effects of the rFXIII-A2*/αC interaction on the cross-linking of the fibrin chains and incorporation of α2-antiplasmin; (2) identify the binding region on FXIII-A2* for the αC region 389-402; and (3) further characterize the interaction at a molecular level.
Our studies demonstrated that a 14-aa peptide (Pep1) spanning the αC region of fibrin residues (389-402) specifically competed with binding of the αC region to FXIII-A2*. Furthermore, we have shown that Pep1 inhibits the ability of FXIII-A2* (1) to cross-link biotinamido-pentylamine to glutamine residues on fibrin; (2) to form cross-links between fibrin chains; and (3) to incorporate α2-antiplasmin, which renders fibrin clots resistant to fibrinolysis. However, Pep1 did not inhibit the cross-linking of a biotinyl-glutamine–donor peptide to acceptor lysine residues on fibrin, which suggests that Pep1 is not exerting its effects by occupying the FXIII-A2* active site, but rather by occupying a second site within the enzyme. Further support for this view comes from the observation that Pep1 does not inhibit the cross-linking of biotinamido-pentylamine to fibrin by TG-2, an enzyme that has near-identical active site topology to FXIII-A2. Scrambled peptides of Pep1 did not inhibit γ-γ–chain cross-linking or α2-antiplasmin cross-linking to fibrin, demonstrating that the action of Pep1 was specific and suggesting that inhibition of enzyme activity resulted from the binding of Pep1 to the same site to which the intact αC region binds.
The catalytic mechanism of transglutaminases involves attack of the catalytic cysteine thiolate on the amide group of a target glutamine to form a thioester linkage with resultant release of ammonia. Subsequently, the reactive thioester moiety transfers this glutamyl unit to an amine to form an amide linkage or to water to hydrolyze glutamine to glutamate.37-39 The peptide context and stereochemistry of the glutamine residue are critical for recognition by FXIII-A2, whereas the requirement for the amine is very relaxed,40 and in the case of protein-protein cross-linking, selection may depend primarily on which lysine residue happens to be in proximity. Implicit within this mechanism is the prediction that there will be a preferred localization within FXIII-A2 to accommodate the protein presenting the donor glutamine. We therefore suggest that Pep1 binds to a site within FXIII-A2* that is identical, or close to, the site that binds the fibrin chains when the enzyme-fibrin acyl intermediate forms. This is supported by our finding that the short 6-aa biotinylated glutamine-donor peptide can be accommodated within the enzyme while Pep1 is bound, allowing the acyl intermediate to form and cross-linking to occur.
To identify the binding site on rFXIII-A2* for the αC region 389-402, chemical cross-linking of rFXIII-A2* was undertaken with derivatives of Pep1 extended to include a cysteine residue. Analysis by LC-MS/MS identified 3 separate lysine residues on FXIII-A2* to which Pep1-Cys or Cys-Pep1 could be chemically cross-linked (Lys446, Lys257, and Lys113), all of which localized within the β-sandwich domain, specifically the cleft vacated following thrombin cleavage and dissociation of the activation peptide. Identification of the binding site within the AP-cleft explains our previous observation that the fibrin αC binds specifically to thrombin and calcium-activated but not zymogen FXIII-A2.
To rationalize the binding interaction, molecular modeling of Pep1 bound within the AP-cleft was undertaken. Although major conformational changes must occur in the FXIII-A2 molecule upon activation, the conformation of the active site cleft itself has been presumed to be relatively unchanged, except for removal of the activation peptide. The energy-minimized conformation calculated for Pep1 within the AP-cleft predicted various contacts, including a salt bridge between αC glutamic acid residue 396 and FXIII-A2* Arg158. The predicted salt bridge may account for our previous finding that Glu396 is a key residue involved in the binding of the αC region to FXIII-A2*.28
Molecular modeling of an extended segment of the fibrin αC residues Q328-S402 was used to address whether localization of the fibrin αC region residues 389-402 to the AP-cleft is consistent with cross-linking of distal αC glutamine residues AαQ366 and AαQ328. The αC region is predicted to follow closely with the surface of FXIII-A2* aligning fibrin AαQ366 to the active site of 1 FXIII-A* monomer and could localize AαQ328 to the active site of the second FXIII-A monomer if simultaneous activation of both monomers in the FXIII-A2 dimer were possible. This is, however, a more speculative undertaking because a crystal structure of the activated form of FXIII-A2 is not available. However, in common with previous work by Komaromi et al,35 we have used the radiograph crystal structure of TG-2 in its open/active conformation as the starting geometry from which to build the model for FXIII-A2* in its active conformation. Despite limited homology in their primary sequences, the crystal structures of the major domains of nonactivated TG-2 and of each monomer within the FXIII-A2 zymogen justify the assumption that the activated conformations will also be similar. The model we obtained for the activated conformation of FXIII-A2* closely resembles that derived by Komaromi et al. As also recognized by these workers, there are, however, limitations to using TG-2 as the structural basis for our modeling. The monomeric TG-2 model does not entirely align to form the dimeric structure of active FXIII-A2*; therefore, cuts in the structure at the junction between the β sandwich and core domains are necessary to allow the β barrels to be fitted. In addition, reconstruction of the unresolved regions of TG-2 is necessary for the modeling. Despite this, a credible working structure was obtained, which we have shown to be consistent with the deduction from the enzyme assays that, by occupying the vacated AP-cleft, Pep1 is able to block fibrin αC cross-linking.
It is interesting to note that inhibition of the FXIII-A2*/fibrin αC389-402 interaction using Pep1 caused a dose-dependent decrease in the incorporation of α2-antiplasmin to fibrin and inhibition of fibrin γ-chain cross-linking. These findings provide preliminary indications that the FXIII-A2* AP-cleft may well serve as a glutamine-donor substrate recognition domain for not only the fibrin αC but also different substrates, including α2-antiplasmin and the γ chains of fibrin. Confirmation of these interactions using mutagenesis of full-length fibrinogen variants lacking the αC-binding site, coupled with enzyme assays tailored to examine specific cross-linking at αC sites, will help to elucidate critical mechanisms involved in the molecular regulation of clot stabilization.
The results of this study have identified and characterized a binding interaction between the fibrinogen αC and FXIII-A2*. Thrombin-mediated cleavage of the FXIII-A2 subunit activation peptide is an absolute prerequisite for this interaction to take place, which acts to coordinate the thrombin-mediated events of fibrinogen cleavage and FXIII-A2 activation in the developing thrombus. This elegant interaction both contributes to limiting thrombus formation to the site of vascular damage and has potentially important local effects on the activation of FXIII-A2 and on cross-linking of α2-antiplasmin to fibrin. The intricate processes involved in clot formation have important implications for our understanding of the pathophysiology of thrombosis and the broader role of coagulation in tissue repair. Our findings provide further details on some of the processes involving FXIII/fibrin(ogen) interactions and raise the possibility of small-molecule approaches to interrupt these interactions in the management of thrombotic disorders.
The online version of this article contains a data supplement.
There is an Inside Blood commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Prof Susan Lord for the generous provision of CHO cells expressing fibrinogen; Boothby for technical assistance; and Dr Adam Dowle for expertise in mass spectrometry (Bioscience Technology Facility, University of York, UK).
This work was supported by the British Heart Foundation (program grant RG/08/004/25 292).
Authorship
Contribution: K.A.S. wrote the manuscript, assisted in study design, carried out laboratory work, and analyzed data; R.J.P. assisted in writing the manuscript and in study design and analyzed data; C.A.A. carried out the molecular modeling and analyzed data; J.M.B. carried out the laboratory work; P.J.A. made the recombinant FXIII-A2 expression constructs; E.J.C. expressed recombinant fibrinogen from CHO cells for use in functional studies; S.N.-P. carried out the laboratory work; P.A.C. assisted in writing the manuscript; R.A.S.A. assisted in study design; C.W.G.F. carried out the molecular modeling, assisted in study design, and analyzed data; H.P. assisted in study design; P.J.G. provided overall supervision, assisted in study design, and assisted in writing the manuscript; and all authors critically reviewed the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Peter J. Grant, Division of Cardiovascular and Diabetes Research, Leeds Institute for Genetics Health and Therapeutics, University of Leeds, Clarendon Way, Leeds LS2 9JT, United Kingdom; e-mail: p.j.grant@leeds.ac.uk; and for molecular modeling contact Colin W.G. Fishwick, School of Chemistry, Division of Cardiovascular and Diabetes Research, Clarendon Way, University of Leeds, Leeds LS2 9JT, United Kingdom; e-mail c.w.g.fishwick@leeds.ac.uk.

![Figure 4. SDS-PAGE analysis displaying the effect of Pep1 on rFXIII-A2*–mediated cross-linking of fibrin γ-γ, α-α, and α-γ. (A) An FXIII-A2B2/Pep1 mix (4.4 µg/mL and 735 µM Pep1, respectively) was added to 1.47 µM recombinant fibrinogen, followed by an activation mix containing human α-thrombin (0.05 U/mL) and CaCl2 (1.5 mM). The samples were incubated at 37°C for set time points (0-180 minutes), and the reaction stopped using NuPAGE 10× SDS-PAGE reducing buffer with NuPAGE loading dye and boiled for 15 minutes. The SDS-PAGE image shown is representative of 3 independent experiments (n = 3). (B) SDS-PAGE gel densitometry displaying reduction in the fibrin γC and αC over time in the presence (+) and absence (-) of Pep1. (*γ-chain reduction +/− Pep1, P < .03; **αC reduction +/− Pep1, P < .01). (C) SDS-PAGE gel densitometry displaying the formation of γ dimers (left axis) and γ-α hybrids (right axis) over time in the presence (+) and absence (-) of Pep1 (P = .01). Net intensities were converted to fold increase over time point 0. γ-Dimer formation in the presence and absence of Pep1 gave a P value of .01. γ-α–Hybrid formation in the presence and absence of Pep1 gave a P value of .004. (All experiments were a total of n = 3; the results are expressed as means [SEM].)](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/121/11/10.1182_blood-2012-07-446393/3/m_2117f4.jpeg?Expires=1769256848&Signature=rXn0qhxXYUdrBlHgc~t0qyWl~4NYXs27ZTIVqVMUNX7FScqFzaoQ~a9CEjIrSBOd0U9RiXVTeV0MBvD0QUqMWGlMozeNxZMzREfXV2VkVApr9NF4VueGzaPR4r8aBXLmK6iiNIn3PC2~iWTimyIep3cpgDkEmfVD~TT7YDnjpqtTpzHDA8CTSGWkWjdCSRqreXQxsYGo24XUX5MCaIH9cr4M-YQ3PDTddqmCmVt8qXKe4~eAJgupsxLavAX~dyI0L3dgmeoJZ17Rgs3q9HSrnqIL-rNzteRUMlW-ZkPaADiVaKP5EQTFe69QNylQYJhQpCckQupYf9pXDilGRixW-g__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 7. The effect of Pep1 on the TG-2–mediated incorporation of biotinamido-pentylamine to fibrin TG-2 (11 µg/mL) was added to fibrin-coated wells followed by increasing concentrations of Pep1 (10-100 µM). A reaction mix containing DTT (0.1 mM), biotinamido-pentylamine (0.3 mM), CaCl2 (10 mM), and human α-thrombin (1 U/mL) in 100 mM 2-[Bisamino]-2–1,3-propanediol, pH 7.4, 0.0% triton X-100 was added to each well, and TG-2 activity was taken after 7 minutes. Samples were performed in triplicate (n = 3), and the results are expressed as means (SEM).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/121/11/10.1182_blood-2012-07-446393/3/m_2117f7.jpeg?Expires=1769256848&Signature=eiZTdaVhVfX1Q~SUPOY-w7~U973qhM5U4MIQiI9L~ka78gJZfBzgXuP4Ti1MLTcSlSQcvsSg6JNLx2Nh-eDf80HjqDUf5LYRkX3Ymew3Qdxnnsd7XPqX7a3boHeAkvSjudvb2oQTqi3dP-HpuULl0VrD0g9TVLcCTSUEJqK6Xsb6tmdbD2AtSDUHgVWBCHARF3fKjBaaez7vPfM7nvWiLNtR3ufirzONPYjkwoekJuaz6t~eFQbMik56WVt0FYY7l5TQ~sT4EmCWqycEM4cQrjAVjTOXBa2k55pzFJ~aWcUvBqC~K74PEdGEpSYD0GdkLk1eC0m-w-wEZGALH29NUQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal