Key Points
The complication of stroke is common in patients with SCA, and there is a genetic component.
We have performed a large-association study to identify 2 genetic variants that protect patients with SCA from stroke.
Abstract
Stroke is a devastating complication of sickle cell anemia (SCA), occurring in 11% of patients before age 20 years. Previous studies of sibling pairs have demonstrated a genetic component to the development of cerebrovascular disease in SCA, but few candidate genetic modifiers have been validated as having a substantial effect on stroke risk. We performed an unbiased whole-genome search for genetic modifiers of stroke risk in SCA. Genome-wide association studies were performed using genotype data from single-nucleotide polymorphism arrays, whereas a pooled DNA approach was used to perform whole-exome sequencing. In combination, 22 nonsynonymous variants were identified and represent key candidates for further in-depth study. To validate the association of these mutations with the risk for stroke, the 22 candidate variants were genotyped in an independent cohort of control patients (n = 231) and patients with stroke (n = 57) with SCA. One mutation in GOLGB1 (Y1212C) and another mutation in ENPP1 (K173Q) were confirmed as having significant associations with a decreased risk for stroke. These mutations were discovered and validated by an unbiased whole-genome approach, and future studies will focus on how these functional mutations may lead to protection from stroke in the context of SCA.
Introduction
Cerebrovascular disease is perhaps the most devastating complication for children with sickle cell anemia (SCA), comprising a spectrum of clinical manifestations including overt stroke, transient ischemic attacks, silent infarcts, and frequent neurocognitive decline. Longitudinal cohort data from the National Heart, Lung, and Blood Institute (NHLBI–sponsored Cooperative Study of Sickle Cell Disease (CSSCD) have shown that between 5% and 10% of patients with SCA will experience a clinically overt stroke during childhood, whereas another 20% to 30% will have silent infarctions that are detectable by magnetic resonance imaging but without clinical correlation.1 The peak incidence of stroke in SCA occurs between ages 2 and 7 years, with most strokes being ischemic and having a high rate of recurrence in patients after the first episode. The severity of the anemia and a history of recent or recurrent acute chest syndrome are both independent risk factors for the development of stroke.1,2
The most clinically useful prognostic tool currently available for predicting cerebrovascular disease and primary stroke prevention is periodic screening of children using transcranial Doppler (TCD) ultrasonography.3 TCD measures the time-averaged mean velocity (TAMV) in the distal internal carotid arteries and middle cerebral arteries. Children with abnormal TCD results (defined as TAMV ≥200 cm/s) have a much higher risk for the development of primary stroke than those with normal TCD velocities (TAMV <170 cm/s). The maximal risk threshold of 200 cm/s is not absolute because children with conditional TCD velocities (TAMV 170-199 cm/s) also have an elevated risk for the development of stroke.4 The limitations of TCD to accurately identify all patients with SCA who will experience cerebrovascular complications, as well as the reluctance of some investigators and families to commit to a long-term transfusion program, expose the need for a more sensitive and specific panel of biomarkers for stroke prediction.
Previous evidence from analyses of sibling-pairs reported a genetic contribution to stroke risk in SCA, where a child with SCA had an increased risk for stroke if they had siblings who had experienced an overt stroke.5,6 Several retrospective studies have identified specific single nucleotide polymorphisms (SNPs) associated with stroke in pediatric patients with SCA, using candidate gene approaches.7-14 However, the majority of these potential modifiers did not have a replicated association with stroke using independent validation cohorts.15 Only a few genetic modifiers have confirmed association with stroke, such as α-thalassemia trait being protective against stroke, but these do not explain the entire genetic contribution to stroke risk. Partially, this is because of previous association studies being restricted by the use of candidate gene approaches, as well as minimal subsequent validation attempts. It is highly probable that additional untested genes, or rare variants, are private to individuals and families, which modify individual stroke risk in SCA.
With methodologic advances in whole-genome sequencing and genotyping techniques, it is now possible and appropriate to screen the entire genome for variants that are associated with any phenotype of interest. In an attempt to better define the principal genetic modifiers affecting stroke risk in patients with SCA, we performed an unbiased genome-wide association study (GWAS) using a combination of polymorphism arrays and whole-exome sequencing (WES). Strengths of this study include a large discovery cohort, age-matched control patients, and an independent validation cohort; additionally, we focused on genetic variants with a functional effect on coding regions rather than testing for common nonfunctional polymorphisms. We found several nonsynonymous mutations associated with the risk for stroke that provide new insights into the pathophysiologic pathways for stroke development. Investigation of these genetic modifiers could provide potential targets for drug therapy and possibly generate a panel of biomarkers that could be used for prognostic assessment of any patient’s individual stroke risk.
Methods
Patients
Pediatric patients with SCA and documented evaluation of cerebrovascular disease were recruited from multiple US institutions for several different clinical trials. All patients and parents gave informed consent in accordance with the Declaration of Helsinki for sample collection and storage as appropriate for each particular study; all current genomic analyses were approved by the Baylor College of Medicine Institutional Review Board. A total of 677 children with SCA had genomic DNA samples analyzed in this GWAS: 109 were enrolled in the Hydroxyurea Study of Long-Term Effects (HUSTLE, NCT00305175), 132 were enrolled in the NHLBI-sponsored Stroke With Transfusions Changing to Hydroxyurea (SWiTCH, NCT00122980), 215 were recruited from the Cooperative Study of Sickle Cell Disease (CSSCD, NCT00005300), 104 were enrolled in the Comprehensive Sickle Cell Centers Collaborative Data Project (C-DATA, NCT00529061), and 117 patients were enrolled in the Stroke Prevention in Sickle Cell Anemia (STOP1, NCT00000592). Among these 677 patients with SCA, 177 patients had sustained an independently documented primary clinical stroke event and comprised the stroke cohort used for genotyping analyses. As the control nonstroke group, 335 children with SCA were identified who had no history of any cerebrovascular disease by clinical or radiologic criteria and minimal therapeutic intervention before enrollment in their respective study. An additional 48 patients from the CSSCD study had documented silent cerebral infarction, as detected on brain magnetic resonance imaging but without clinically manifested complications. They were not included in the discovery or validation analyses but comprised a subsequent small cohort for comparison. Finally, the remaining 117 patients were enrolled in the STOP1 study and had confirmed evidence of cerebrovascular disease, specifically highly elevated TCD velocities (TAMV ≥200 cm/s), but without having a primary clinical stroke before enrollment. As in the silent infarction cohort, this STOP cohort was not included in the discovery or validation analyses but was used solely for comparison.
Affymetrix SNP6.0 array genotyping
Germline DNA was extracted from peripheral blood using standard methods. Genotyping with Affymetrix Genome-Wide Human SNP Array 6.0, which includes more than 900 000 single-nucleotide polymorphism (SNP) markers, was performed using the manufacturer’s recommendations. Briefly, genomic DNA concentration was determined by fluorometry (Qubit dsDNA BR Assay Kit; Invitrogen), and 500 ng of each DNA sample was applied to the SNP Array 6.0 set (Affymetrix, Santa Clara, CA). Chips were scanned, and genotype calls were made using the Bayesian Robust Linear Multichip with Mahalanobis Distance algorithm. Quality-control metrics for subsequent analysis removed SNP markers with overall genotype calls of less than 95%, minor allele frequencies of less than 5%, or if SNP allele frequencies deviated from Hardy-Weinberg equilibrium. A total of 772 065 SNPs passed quality control. Principal component analysis (PCA), statistical analyses of association, and population stratification correction were conducted using the SNP & Variation Suite (SVS) 7.5 from Golden Helix (http://goldenhelix.com). Genome identity by descent was calculated by the SVS7.5 software. Pairs of DNA samples with identity by descent measures >0.2 were considered to be related, and only one of any related set of samples was included in subsequent analyses. The EIGENSTRAT method was used to correct for population stratification by PCA correction.16 The Cochran-Armitage trend test was used to analyze the association of each SNP with the dichotomous variable of stroke or nonstroke. Analyses were run under the assumption of an additive model, and a P value below P < 5 × 10−8 was required to pass a genome-wide significance threshold.
Exome capture and next-generation sequencing
Two stroke pools (n = 60 DNA samples per pool) and 2 control pools (n = 52 DNA samples per pool) were prepared with equimolar quantities of each individual sample (100 ng/µL). DNA concentrations were accurately determined using a picogreen fluorescent detection method (Quant-iT; Invitrogen). Equal amounts of DNA from each sample constituting a pool were manually combined before WES. Exome enrichment of the 4 genomic DNA pools (3 µg per DNA pool) was performed using the Agilent SureSelect Human All Exon kit capture library (G3370), according to the manufacturer’s protocol (SureSelect Human All Exon Illumina Paired-End protocol version 1.0.1). Captured DNAs were sequenced on an Illumina GAIIx sequencer (Illumina, San Diego, CA) with 100-bp pair-end reads. Three lanes of sequencing data were collected for each pooled sample; detailed coverage information is shown in Table 1. Image analyses and base calling were performed using the Illumina Genome Analyzer Pipeline software (GAPipeline version 1.5 or greater) with default parameters. Reads were aligned to a human reference sequence (UCSC assembly hg19, NCBI build 37), and genotypes were called at all positions where there were high-quality sequence bases (Phred-like Q25 or greater) at a minimal coverage of 20, using CLC Genomics Workbench v4.5.1 (CLC Bio, Cambridge, MA). For each DNA pool, ∼60% of a minimal 200 million reads were uniquely mapped to the targeted human exon regions to give average depth coverage of 172 times. Under such coverage, more than 60% of targeted regions were covered by 100 or more reads.
Capture efficiency and sequencing statistics for WES results
| . | DNA pool #1 . | DNA pool #2 . | DNA pool #3 . | DNA pool #4 . |
|---|---|---|---|---|
| Total read (k) | 205 303 | 205 335 | 200 960 | 209 335 |
| After trimmed (k) | 203 977 | 204 017 | 198 493 | 206 924 |
| Genome mapped (k) | 180 133 | 179 889 | 175 683 | 181 008 |
| On target (k) | 129 112 | 133 493 | 111 209 | 128 144 |
| Mean coverage (×) | 179.6 | 188.0 | 157.5 | 163.7 |
| % on target at 100× coverage | 62.73 | 66.30 | 59.86 | 61.21 |
| Pearson correlation with SNP 6.0 | 0.985 | 0.983 | 0.983 | 0.984 |
| . | DNA pool #1 . | DNA pool #2 . | DNA pool #3 . | DNA pool #4 . |
|---|---|---|---|---|
| Total read (k) | 205 303 | 205 335 | 200 960 | 209 335 |
| After trimmed (k) | 203 977 | 204 017 | 198 493 | 206 924 |
| Genome mapped (k) | 180 133 | 179 889 | 175 683 | 181 008 |
| On target (k) | 129 112 | 133 493 | 111 209 | 128 144 |
| Mean coverage (×) | 179.6 | 188.0 | 157.5 | 163.7 |
| % on target at 100× coverage | 62.73 | 66.30 | 59.86 | 61.21 |
| Pearson correlation with SNP 6.0 | 0.985 | 0.983 | 0.983 | 0.984 |
The DNA pools contained 52 control patients in DNA pool #1; 52 control patients in DNA pool #2; 60 patients with stroke in DNA pool #3; and 60 patients with stroke in DNA pool #4. At >100× coverage, ∼60% of all sequences were on target for each of the DNA pools. Compared with the individual genotypes obtained from the SNP 6.0 arrays, a strong correlation with genotype data was generated by WES for each of the DNA pools.
Accuracy of exome variant detection and comparison of variant frequencies
To assess the accuracy of variation detection and allele frequency estimation, we compared the pooled WES data to the SNP data obtained from individual DNA genotyping using the Affymetrix Genome-Wide Human SNP Array 6.0 (described above). Using a minimal coverage cutoff value of 20 times, we observed a high degree of correlation with individual genotyping across the 4 DNA pools (Table 1; supplemental Figure 1). For association testing, a total of 27 446 autosomal variants were identified, including 26 521 single-nucleotide variants (SNVs) and 925 insertion-deletion variants. All variants had a minor allele frequency (MAF) of >5%, had a sequence coverage of more than 50 times, and introduced a nonsynonymous change to the encoded protein. A 2-tailed Fisher exact test was used to compare the combined allele frequency estimates of the 2 stroke DNA pools vs the 2 control DNA pools for each variant, and a false discovery rate correction was performed to account for the number of contingency tables tested. For subsequent validation of any associations in an independent cohort, a 1-tailed Fisher exact test was used when the direction of the association was maintained (increased or decreased stroke risk) for the polymorphism. Any variant with a P value < .05 was considered significant. Poststudy analyses of the genetic power of the discovery and validation steps showed that this study was best suited in finding variants with >5% MAF and large effects on stroke development (supplemental Figure 2).
SNP genotyping and sequencing
Genomic DNA TaqMan polymerase chain reaction (PCR) was performed on a StepOne Plus instrument (Applied Biosystems, Foster City, CA). After 40 amplification cycles, threshold cycle values were automatically calculated, and the individual SNP genotypes were called by the StepOne software version 2.0 (Applied Biosystems). For genotyping by sequencing, PCR products were purified after PCR amplification using MinElute 96 UF Plates (QIAGEN, Germantown, MD), and Sanger sequencing was performed by Big Dye Terminator (version 3.1) chemistry using capillary electrophoresis on an Applied Biosystem 3730XL DNA Analyzer.
Results
Clinical characteristics
Patients were enrolled into their respective studies as previously described.17-21 Briefly, all patients with a history of clinically overt stroke (n = 177) were between 5 and 18 years old at enrollment and were receiving long-term transfusion therapy for prevention of secondary stroke. These patients had experienced a verified primary (index) clinical stroke event at a median age of 6 years and all before they were 15 years old. In contrast, the children selected as nonstroke control patients (n = 335) were at least 5 years old at enrollment, were without previous clinical stroke, and did not have any imaging evidence of silent infarcts before beginning any clinical treatment. The average age of the 2 cohorts at enrollment into their respective studies was not significantly different (12.1 ± 4.1 years for patients with stroke vs 12.9 ± 3.7 years for nonstroke control patients), and the sex distribution was also similar (54.2% boys for the stroke cohort vs 54.6% boys for the nonstroke control cohort).
SNP GWAS
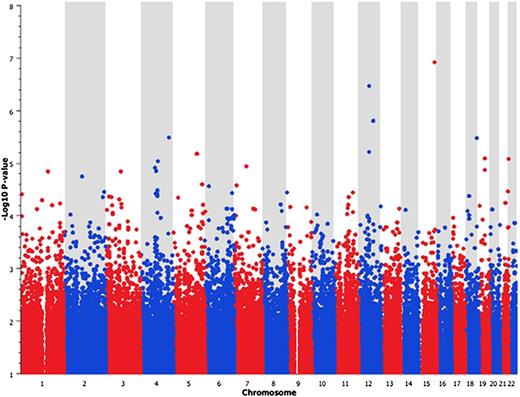
After quality-control procedures, we interrogated more than 770 000 SNP markers for any association with stroke, comparing the genotype results for all patients with stroke (n = 177) with the nonstroke cohort (n = 335). We corrected for population stratification using the EIGENSTRAT method to perform principal component correction for the first 5 principal components, and the quantile-quantile plot of the association data showed no early departure of the observed P values from the null (Figure 1). This trend suggests that our findings are unlikely to be influenced by poor genotyping, sample relatedness, or population stratification after PCA correction. A total of 139 SNPs were identified as being associated with stroke (P < .0001), but none of these SNP results passed a genome-wide significance threshold (P < 5 × 10−8). The Manhattan plot of the GWAS results shows peaks of significant association on chromosomes 12 and 15 (Figure 2). The SNP that had the strongest association with stroke resided in the GABA(A) receptors associated protein-like 3 pseudogene on chromosome 15. The top 10 SNP markers identified by GWAS are listed in Table 2. Although none of the P values achieved a genome-wide significance level (P < 5 × 10−8), these results offered suggestive signals of potential associations.
Quantile-quantile plot of the GWAS data. The quantile-quantile plot of the GWAS stroke association results is shown using ∼770 000 SNPs. The GWAS compared the 177 patients with stroke with the 335 control patients without stroke. The quantile-quantile plot showed good correlation between observed and expected P values, and no evidence of genomic inflation was found (mean λ = 1.07; median λ = 1.06).
Quantile-quantile plot of the GWAS data. The quantile-quantile plot of the GWAS stroke association results is shown using ∼770 000 SNPs. The GWAS compared the 177 patients with stroke with the 335 control patients without stroke. The quantile-quantile plot showed good correlation between observed and expected P values, and no evidence of genomic inflation was found (mean λ = 1.07; median λ = 1.06).
Manhattan plot of GWAS SNP6.0 array results. For each SNP analyzed, the significance of the association is given by the –Log10 (P value) as determined by the Cochran-Armitage test.
Manhattan plot of GWAS SNP6.0 array results. For each SNP analyzed, the significance of the association is given by the –Log10 (P value) as determined by the Cochran-Armitage test.
SNP markers identified by GWAS as being associated with risk for stroke
| SNP ID . | Associated gene . | Chromosome . | Position bp . | Alleles . | MAF in control patients (%) . | MAF in stroke patients (%) . | Odds ratio (95% CI) . | Significance P value . |
|---|---|---|---|---|---|---|---|---|
| rs3803539 | GABARAPL1 | 15 | 88692986 | [A/G] | 5.90 | 23.30 | 4.7 (3.1-7.1) | 2.71 × 10−7 |
| rs10123021 | LPPR1 | 9 | 102491889 | [A/C] | 29.50 | 33.60 | 1.3 (1.1-1.6) | 3.13 × 10−7 |
| rs17329620 | MKRN9P | 12 | 86674908 | [A/C] | 9.80 | 25.80 | 3.2 (2.2-4.5) | 5.52 × 10−7 |
| rs309247 | CCDC102B | 18 | 64534044 | [C/G] | 18.30 | 30.70 | 2.0 (1.5-2.7) | 9.77 × 10−7 |
| rs1481779 | ARHGAP24 | 4 | 86656079 | [C/T] | 19.50 | 33.60 | 2.1 (1.6-2.8) | 1.78 × 10−6 |
| rs16965445 | TSHZ3 | 19 | 36492060 | [A/G] | 9.90 | 19.60 | 2.2 (1.5-3.2) | 3.30 × 10−6 |
| rs4888761 | WWOX | 16 | 76780490 | [C/T] | 16.40 | 27.20 | 1.9 (1.4-2.6) | 4.17 × 10−6 |
| rs7219249 | EIF1 | 17 | 37086202 | [A/G] | 31.80 | 39.70 | 1.4 (1.1-1.8) | 5.80 × 10−6 |
| rs10516451 | DAPP1 | 4 | 100845572 | [C/T] | 13.20 | 21.10 | 1.8 (1.3-2.5) | 6.08 × 10−6 |
| rs572400 | AGPAT9 | 4 | 84669771 | [C/T] | 9.50 | 1.70 | 0.16 (0.1-0.4) | 9.81 × 10−6 |
| SNP ID . | Associated gene . | Chromosome . | Position bp . | Alleles . | MAF in control patients (%) . | MAF in stroke patients (%) . | Odds ratio (95% CI) . | Significance P value . |
|---|---|---|---|---|---|---|---|---|
| rs3803539 | GABARAPL1 | 15 | 88692986 | [A/G] | 5.90 | 23.30 | 4.7 (3.1-7.1) | 2.71 × 10−7 |
| rs10123021 | LPPR1 | 9 | 102491889 | [A/C] | 29.50 | 33.60 | 1.3 (1.1-1.6) | 3.13 × 10−7 |
| rs17329620 | MKRN9P | 12 | 86674908 | [A/C] | 9.80 | 25.80 | 3.2 (2.2-4.5) | 5.52 × 10−7 |
| rs309247 | CCDC102B | 18 | 64534044 | [C/G] | 18.30 | 30.70 | 2.0 (1.5-2.7) | 9.77 × 10−7 |
| rs1481779 | ARHGAP24 | 4 | 86656079 | [C/T] | 19.50 | 33.60 | 2.1 (1.6-2.8) | 1.78 × 10−6 |
| rs16965445 | TSHZ3 | 19 | 36492060 | [A/G] | 9.90 | 19.60 | 2.2 (1.5-3.2) | 3.30 × 10−6 |
| rs4888761 | WWOX | 16 | 76780490 | [C/T] | 16.40 | 27.20 | 1.9 (1.4-2.6) | 4.17 × 10−6 |
| rs7219249 | EIF1 | 17 | 37086202 | [A/G] | 31.80 | 39.70 | 1.4 (1.1-1.8) | 5.80 × 10−6 |
| rs10516451 | DAPP1 | 4 | 100845572 | [C/T] | 13.20 | 21.10 | 1.8 (1.3-2.5) | 6.08 × 10−6 |
| rs572400 | AGPAT9 | 4 | 84669771 | [C/T] | 9.50 | 1.70 | 0.16 (0.1-0.4) | 9.81 × 10−6 |
The top 10 SNP markers associated with stroke were identified by Cochran-Armitage testing on all stroke (n = 177) vs all control (n = 335) samples, assuming an additive model. None of the SNP markers have any predicted effects on gene function, but the associated gene is given for each SNP. The MAF is given for each of the phenotypes. The significance is the Cochran-Armitage P value, and the odds ratios with CIs are given for each associated variant.
Exome sequencing and validation
Because GWAS generally identifies linked genetic variants rather than actual disease-causing variants, we next performed WES to identify all variants affecting protein coding in every exon in the human genome. To minimize labor and cost, we took advantage of the dichotomous phenotype of interest to generate random DNA pools of stroke or control samples. Analysis of these pooled samples provided a rapid method to screen the entire exome for mutations linked with stroke. Similar pooling strategies have successfully identified variant enrichment in previous case-control studies, with subsequent direct genotyping of individual DNA samples required to verify any associations.22-24 We performed a 2-step genetic association analysis by randomly dividing our stroke cohort (n = 177) and our nonstroke control cohort (n = 335) into 2 discovery and validation sets. Our strategy was to identify variants associated with stroke by WES in the discovery group and then test specific candidate variants in the validation step. The discovery step compared 120 patients with stroke with 104 nonstroke control patients, whereas the validation step compared 57 stroke samples with 231 nonstroke controls. Within the discovery cohort, we performed exome sequencing of 2 pooled stroke groups (n = 60 samples per pool, 120 samples total) and 2 pooled control groups (n = 52 samples per pool, 104 samples total). We obtained ∼821 million reads, which generated, on average, 17.4 billion base pairs of sequence data per pool. At a threshold of 100 times coverage per base, ∼60% of the sequence data were on target for all 4 DNA pools (Table 1). There were 13 302 SNP markers on the Affymetrix SNP 6.0 arrays that were also present in the 50Mb-targeted WES regions. A total of 10 376 (78.0%) of these SNP markers genotyped individually by the SNP 6.0 arrays were also captured by the pooled WES at a coverage of more than 20 times. When the minor allele frequencies of these variants obtained by the genotyping methods were compared, there was a high degree of correlation for each DNA pool (Table 1, supplemental Figure 1).
We identified a total of 27 446 variants that introduced a nonsynonymous amino acid substitution. Using multiple contingency tables with adjustment for multiple testing, we found 300 variants that were significantly associated with stroke (P < .001). These variants included 294 SNVs and 6 insertion-deletion variants. Bioinformatic analyses using PolyPhen2 and SIFT algorithms revealed that 88 of these 300 mutations were predicted to have a deleterious effect on protein function.25,26 In combination with the results from the GWAS, 11 variants identified by WES were located within 250kb of at least 1 SNP identified by GWAS (Table 3). These variants represent excellent regions of the genome for future study, as they are all functional mutations. One of the variants identified by WES, with the closest proximity to an SNP identified by GWAS (1kb), was a SNV located in the PON1 gene. This variant has previously been associated with altered folate metabolism and ischemic stroke in several populations.27,28
Variants identified by WES that are located within 250kb of an SNP marker identified by GWAS
| Gene symbol . | Chromosome . | Position (bp) . | Variant identifier . | Amino acid change . | WES P value . | Distance to GWAS SNP . | GWAS P value . |
|---|---|---|---|---|---|---|---|
| PON1 | 7 | 94937446 | rs662 | Q192R | 9.0 × 10−4 | 1kb | 4.3 × 10−5 |
| ENPP1 | 6 | 132172368 | rs1044498 | K173Q | 7.7 × 10−5 | 3kb | 7.1 × 10−5 |
| CYP4F2 | 19 | 16008388 | rs3093105 | W12G | 4.1 × 10−4 | 44kb | 6.0 × 10−5 |
| INPPL1 | chr11 | 71948536 | rs11548491 | A1083G | 8.6 × 10−7 | 76kb | 8.8 × 10−5 |
| GOLGB1 | 3 | 121415720 | rs3732410 | Y1212C | 5.6 × 10−4 | 127kb | 9.3 × 10−5 |
| RSAD1 | 17 | 48557326 | rs2290862 | A119T | 6.5 × 10−5 | 129kb | 3.3 × 10−5 |
| PKD1L3 | 16 | 71983772 | rs1035543 | S1176R | 8.7 × 10−7 | 193kb | 6.1 × 10−5 |
| LOC100289165 | 8 | 124658210 | rs16898671 | R128C | 7.3 × 10−4 | 194kb | 7.6 × 10−5 |
| ARGFX | 3 | 121305086 | rs61750878 | T196I | 1.4 × 10−4 | 238kb | 9.3 × 10−5 |
| KRT5 | 12 | 52908917 | rs11549950 | S528G | 2.1 × 10−4 | 243kb | 1.9 × 10−5 |
| HSD3B1 | 1 | 120057246 | rs45609334 | T367N | 1.8 × 10−7 | 248kb | 2.7 × 10−5 |
| Gene symbol . | Chromosome . | Position (bp) . | Variant identifier . | Amino acid change . | WES P value . | Distance to GWAS SNP . | GWAS P value . |
|---|---|---|---|---|---|---|---|
| PON1 | 7 | 94937446 | rs662 | Q192R | 9.0 × 10−4 | 1kb | 4.3 × 10−5 |
| ENPP1 | 6 | 132172368 | rs1044498 | K173Q | 7.7 × 10−5 | 3kb | 7.1 × 10−5 |
| CYP4F2 | 19 | 16008388 | rs3093105 | W12G | 4.1 × 10−4 | 44kb | 6.0 × 10−5 |
| INPPL1 | chr11 | 71948536 | rs11548491 | A1083G | 8.6 × 10−7 | 76kb | 8.8 × 10−5 |
| GOLGB1 | 3 | 121415720 | rs3732410 | Y1212C | 5.6 × 10−4 | 127kb | 9.3 × 10−5 |
| RSAD1 | 17 | 48557326 | rs2290862 | A119T | 6.5 × 10−5 | 129kb | 3.3 × 10−5 |
| PKD1L3 | 16 | 71983772 | rs1035543 | S1176R | 8.7 × 10−7 | 193kb | 6.1 × 10−5 |
| LOC100289165 | 8 | 124658210 | rs16898671 | R128C | 7.3 × 10−4 | 194kb | 7.6 × 10−5 |
| ARGFX | 3 | 121305086 | rs61750878 | T196I | 1.4 × 10−4 | 238kb | 9.3 × 10−5 |
| KRT5 | 12 | 52908917 | rs11549950 | S528G | 2.1 × 10−4 | 243kb | 1.9 × 10−5 |
| HSD3B1 | 1 | 120057246 | rs45609334 | T367N | 1.8 × 10−7 | 248kb | 2.7 × 10−5 |
For all variants with significant association with stroke, the chromosomal position of each variant identified by WES (n = 300; P < .001) was compared with the location of all SNP markers (n = 139; P < .0001). We identified 11 variants by WES where there was at least 1 SNP marker within 250kb. The variants highlighted in dark gray are variants predicted by PolyPhen2 or SIFT to be deleterious.
To replicate the associations of the nonsynonymous variants with stroke, we selected 20 candidate SNVs and 2 insertion-deletion variants with the best statistical association with stroke risk (P < .001), and which had a predicted deleterious effect on protein function (Table 4). First, these 22 candidate variants were individually genotyped in the DNA samples used in the WES pool analyses (n = 120 patients with stroke; n = 104 control patients) to verify the original WES findings. There was very good correlation between the WES genotypes and individual genotypes obtained for the DNA pools used for the discovery phase, with a mean WES genotyping accuracy of >85% for the 22 variants (Table 4). However, there were several genotyping discrepancies, such as the C3R variant in the PRG3 gene. This variant had a genotyping accuracy of only 29% by pooled WES compared with individual genotyping, and this variant was not significantly associated with stroke on verification by individual genotyping. Of the selected 22 candidate variants, 13 maintained their statistically significant associations with stroke in the initial discovery WES cohorts after individual sample verification of the genotypes (P < .05, Table 4).
Individual DNA sample genotyping to validate variants identified by pooled exome sequencing
| Gene symbol . | Protein change . | Variant ID . | WES P value . | WES accuracy (%) . | Individual P value . | Validation P value . |
|---|---|---|---|---|---|---|
| GOLGB1 | Y1212C | rs3732410 | 5.6 × 10−4 | 97.8 | <.0001 | .035 |
| ENPP1 | K173Q | rs1044498 | 7.7 × 10−5 | 96.3 | .0011 | .031 |
| PKD1L3 | S1176R | rs1035543 | 8.7 × 10−7 | 98.1 | .0002 | .109 |
| DUOX2 | P138L | rs2001616 | 5.2 × 10−6 | 82.5 | .0003 | 1.000 |
| HSPB9 | Q2P | rs1122326 | 7.0 × 10−4 | 93.4 | .0018 | .181 |
| TULP2 | A18T | rs7260579 | 3.2 × 10−9 | 93.7 | .0039 | .423 |
| ARGFX | T196I | rs61750878 | 1.4 × 10−4 | 91.5 | .0042 | .309 |
| MS4A14 | I56FS | INDEL_11_60165352 | 1.4 × 10−6 | 94.7 | .0056 | 1.000 |
| CEACAM20 | S369F | rs10414398 | 9.2 × 10−7 | 87.0 | .0074 | .289 |
| DFNA5 | P142T | rs754554 | 1.8 × 10−7 | 93.7 | .0092 | .148 |
| ANKRD33 | Y5F | rs697636 | 9.5 × 10−5 | 76.0 | .0153 | .495 |
| PON1 | Q192R | rs662 | 9.0 × 10−4 | 95.7 | .0252 | .348 |
| PHF14 | K115R | rs218966 | 5.5 × 10−10 | 87.2 | .0261 | .091 |
| ZNF880 | Y150C | rs324125 | 7.6 × 10−9 | 84.6 | .0987 | ND |
| PKD1L2 | P512L | rs7205673 | 5.6 × 10−4 | 96.5 | .1066 | ND |
| SLC7A13 | Q470K | rs9693999 | 5.3 × 10−7 | 77.8 | .1732 | ND |
| DTHD1 | A387C | rs12507599 | 2.8 × 10−5 | 89.9 | .1970 | ND |
| SLC41A3 | L501FS | INDEL_3_125725268 | 1.8 × 10−5 | 63.6 | .2580 | ND |
| C5 | V145I | rs17216529 | 1.5 × 10−5 | 80.5 | .4514 | ND |
| TNFRSF1B | M196R | rs1061622 | 3.0 × 10−4 | 91.6 | .4726 | ND |
| ALG1L | N135D | rs3828357 | 1.3 × 10−4 | 84.1 | .7306 | ND |
| PRG3 | C3R | rs669661 | 7.5 × 10−16 | 29.2 | 1.0000 | ND |
| Gene symbol . | Protein change . | Variant ID . | WES P value . | WES accuracy (%) . | Individual P value . | Validation P value . |
|---|---|---|---|---|---|---|
| GOLGB1 | Y1212C | rs3732410 | 5.6 × 10−4 | 97.8 | <.0001 | .035 |
| ENPP1 | K173Q | rs1044498 | 7.7 × 10−5 | 96.3 | .0011 | .031 |
| PKD1L3 | S1176R | rs1035543 | 8.7 × 10−7 | 98.1 | .0002 | .109 |
| DUOX2 | P138L | rs2001616 | 5.2 × 10−6 | 82.5 | .0003 | 1.000 |
| HSPB9 | Q2P | rs1122326 | 7.0 × 10−4 | 93.4 | .0018 | .181 |
| TULP2 | A18T | rs7260579 | 3.2 × 10−9 | 93.7 | .0039 | .423 |
| ARGFX | T196I | rs61750878 | 1.4 × 10−4 | 91.5 | .0042 | .309 |
| MS4A14 | I56FS | INDEL_11_60165352 | 1.4 × 10−6 | 94.7 | .0056 | 1.000 |
| CEACAM20 | S369F | rs10414398 | 9.2 × 10−7 | 87.0 | .0074 | .289 |
| DFNA5 | P142T | rs754554 | 1.8 × 10−7 | 93.7 | .0092 | .148 |
| ANKRD33 | Y5F | rs697636 | 9.5 × 10−5 | 76.0 | .0153 | .495 |
| PON1 | Q192R | rs662 | 9.0 × 10−4 | 95.7 | .0252 | .348 |
| PHF14 | K115R | rs218966 | 5.5 × 10−10 | 87.2 | .0261 | .091 |
| ZNF880 | Y150C | rs324125 | 7.6 × 10−9 | 84.6 | .0987 | ND |
| PKD1L2 | P512L | rs7205673 | 5.6 × 10−4 | 96.5 | .1066 | ND |
| SLC7A13 | Q470K | rs9693999 | 5.3 × 10−7 | 77.8 | .1732 | ND |
| DTHD1 | A387C | rs12507599 | 2.8 × 10−5 | 89.9 | .1970 | ND |
| SLC41A3 | L501FS | INDEL_3_125725268 | 1.8 × 10−5 | 63.6 | .2580 | ND |
| C5 | V145I | rs17216529 | 1.5 × 10−5 | 80.5 | .4514 | ND |
| TNFRSF1B | M196R | rs1061622 | 3.0 × 10−4 | 91.6 | .4726 | ND |
| ALG1L | N135D | rs3828357 | 1.3 × 10−4 | 84.1 | .7306 | ND |
| PRG3 | C3R | rs669661 | 7.5 × 10−16 | 29.2 | 1.0000 | ND |
Twenty candidate SNV and 2 insertion-deletion variants with the best statistical association with stroke risk (P < .001) were selected for follow-up verification analyses. The DNA samples within each DNA pool were individually genotyped for each of the 22 variants, and the allele frequency was determined. The mean accuracy of the WES genotyping was calculated by comparing the MAF of the DNA pools obtained by individual genotyping to the MAF determined by pooled WES. Variants with no difference between the genotyping methods would have an accuracy of 100%, whereas variants with completely discordant calls would have an accuracy of 0%. The individual genotype calls were used to repeat the statistical association testing between the stroke and control groups. Using the discovery cohorts (stroke n = 120; control n = 104), we verified that 13 of the 22 variants maintained their initial association with stroke in the same direction (increased or decreased stroke risk) as the pooled WES discovery (individual P value). We then tested these 13 variants in an independent validation cohort (stroke n = 57; control n = 231) to validate the association of these variants with risk for stroke (validation P value). All statistical tests were performed using the Fisher exact test and using an allele model.
ND, no analysis performed.
In an attempt to validate the association of these 13 variants discovered by the WES analyses, we next genotyped these 13 variants in an independent validation cohort of control patients (n = 231) and patients with stroke (n = 57), all of whom had not previously undergone exome sequencing. After statistical analyses, 2 of the 13 candidate variants were validated and maintained significant association with the risk for stroke in the validation cohort (Table 4). The 2 mutations were in the GOLGB1 (Y1212C) and ENPP1 (K173Q) genes, and both mutations are near an SNP identified by GWAS, within 127kb and 3kb, respectively (Table 3). When we measured linkage between the WES variants and proximal GWAS SNPs, there was strong linkage between GOLGB1 Y1212C and rs7623901 (D’=0.94), and between ENPP1 K173Q and rs7767502 (D’=0.97).
The GOLGB1 Y1212C mutation is a naturally occurring variant with an MAF of 9.9% in our nonstroke patients with SCA and is associated with protection from stroke, similar to the coinheritance of α-thalassemia (Table 5).15,29,30 This mutation appeared to have a dominant effect: the overall significance of this variant with a reduced risk for stroke in all patients with stroke (MAF, 2.8%; n = 177) vs all control patients (MAF, 9.9%; n = 335) was P < .0001, and the odds ratio was 0.27 (confidence interval [CI], 0.14-0.52). Compared with our nonstroke control patients, the mutation had a similar frequency in a cohort of African American adults without SCA (MAF, 8.8%; n = 94), whereas the mutation was significantly lower in an additional CSSCD cohort of patients with silent infarction (MAF, 3.1%; n = 48) and also in the STOP1 cohort of patients (MAF, 3.9%; n = 117) with abnormal TCD velocities (Table 6). These results highlight that this mutation appears to be protective against ischemic stroke but also may affect the overall development of cerebrovascular disease.
Genotype frequencies of 2 variants associated with risk for stroke in patients with SCA
| . | *GOLGB1 Y1212C mutation (rs3732410) . | †ENPP1 K173Q mutation (rs1044498) . | ||||
|---|---|---|---|---|---|---|
| . | YY . | YC . | CC . | QQ . | QK . | KK . |
| Discovery (pooled WES cohorts) | ||||||
| Control (n = 104) | 76.0% | 24.0% | 0.0% | 54.4% | 39.8% | 5.8% |
| Stroke (n = 120) | 95.0% | 5.0% | 0.0% | 75.8% | 21.7% | 2.5% |
| P value | <.0001 | <.0011 | ||||
| Odds ratio | .17 | 95% CI | 0.06-0.42 | 0.44 | 95% CI | 0.27-0.72 |
| Validation (independent cohorts) | ||||||
| Control (n = 231) | 81.8% | 17.8% | 0.4% | 55.5% | 37.6% | 6.9% |
| Stroke (n = 57) | 93.0% | 7.0% | 0.0% | 67.9% | 30.3% | 1.8% |
| P value | .035 | .031 | ||||
| Odds ratio | 0.37 | 95% CI | 0.12-0.99 | 0.59 | 95% CI | 0.34-0.99 |
| . | *GOLGB1 Y1212C mutation (rs3732410) . | †ENPP1 K173Q mutation (rs1044498) . | ||||
|---|---|---|---|---|---|---|
| . | YY . | YC . | CC . | QQ . | QK . | KK . |
| Discovery (pooled WES cohorts) | ||||||
| Control (n = 104) | 76.0% | 24.0% | 0.0% | 54.4% | 39.8% | 5.8% |
| Stroke (n = 120) | 95.0% | 5.0% | 0.0% | 75.8% | 21.7% | 2.5% |
| P value | <.0001 | <.0011 | ||||
| Odds ratio | .17 | 95% CI | 0.06-0.42 | 0.44 | 95% CI | 0.27-0.72 |
| Validation (independent cohorts) | ||||||
| Control (n = 231) | 81.8% | 17.8% | 0.4% | 55.5% | 37.6% | 6.9% |
| Stroke (n = 57) | 93.0% | 7.0% | 0.0% | 67.9% | 30.3% | 1.8% |
| P value | .035 | .031 | ||||
| Odds ratio | 0.37 | 95% CI | 0.12-0.99 | 0.59 | 95% CI | 0.34-0.99 |
The frequency is given for individuals who are wild type (YY), heterozygous (YC), or homozygous (CC) for the GOLGB1 Y1212C mutation in the discovery cohorts and in the validation cohorts.
The frequency is given for individuals who are wild type (QQ), heterozygous (QK), and/or homozygous (KK) for the ENPP1 K173Q mutation in the same discovery and validation cohorts. The allele frequency of each mutation was used to perform statistical testing as described in “Methods.”
Overall genotype frequencies of the GOLGB1 Y1212C mutation in all tested cohorts
| . | Frequency of the GOLGB1 Y1212C mutation in all cohorts . | ||
|---|---|---|---|
| Cohort . | YY (%) . | YC (%) . | CC (%) . |
| All SCA controls (n = 335) | 80.3 | 19.7 | 0.0 |
| Non-SCA controls (n = 91) | 83.5 | 15.4 | 1.1 |
| All stroke patients (n = 177) | 94.4 | 5.6 | 0.0 |
| Silent infarction cohort (n = 48) | 93.8 | 6.2 | 0.0 |
| Abnormal TCD cohort (n = 117) | 92.3 | 7.7 | 0.0 |
| . | Frequency of the GOLGB1 Y1212C mutation in all cohorts . | ||
|---|---|---|---|
| Cohort . | YY (%) . | YC (%) . | CC (%) . |
| All SCA controls (n = 335) | 80.3 | 19.7 | 0.0 |
| Non-SCA controls (n = 91) | 83.5 | 15.4 | 1.1 |
| All stroke patients (n = 177) | 94.4 | 5.6 | 0.0 |
| Silent infarction cohort (n = 48) | 93.8 | 6.2 | 0.0 |
| Abnormal TCD cohort (n = 117) | 92.3 | 7.7 | 0.0 |
The frequency of individuals' wild-type (YY), heterozygous (YC), and homozygous (CC) for the GOLGB1 Y1212C mutation are given for each cohort.
The ENPP1 K173Q mutation is also associated with protection from stroke and with a predicted dominant effect (Table 5). The overall significance of this K173Q variant with a reduced risk for stroke in all patients with stroke (n = 177; MAF, 14.4%) vs all control patients (n = 335; MAF, 25.8%) was P < .0001, and the odds ratio was 0.49 (CI, 0.35-0.69). In contrast to the Y1212C mutation in GOLGB1, no significant difference was observed in the frequency of the K173Q mutation in the CSSCD cohort of patients with silent infarctions (MAF, 27.1%; n = 48) or in the STOP1 cohort of patients with abnormal TCD velocities (MAF, 20.4%; n = 117), compared with our nonstroke control patients.
As a final analysis of the 13 candidate variants identified by WES (Table 4), the allele frequency of each variant was compared between all patients with stroke (n = 177) and all nonstroke control patients (n = 335). Apart from the GOLGB1 Y1212C and ENPP1 K173Q variants, the only other genetic variant with a significant association with stroke was the Q192R mutation of PON1 (P = .017) with an odds ratio of 1.41 (CI, 1.07-1.86). This PON1 mutation was associated with an increased risk for stroke, especially when comparing the frequency of recessive homozygotes where 13.7% of all patients with stroke were homozygous for Q192R compared with only 3.6% of all control patients (P < .0001). The PON1 Q192R variant was in complete linkage with the rs2057681 SNP identified by the GWAS (D’=1.00).
Discussion
Current paradigms for children with SCA recognize that 10% will go on to have a clinically overt stroke without clinical intervention. The consequent sequelae of stroke in children with SCA are considerable, leading to severe complications such as residual motor deficits, neurologic damage, and cognitive impairment. Because of the limitations of current screening methods to accurately predict stroke risk,31 coupled with evidence for a genetic component to the development of stroke,5 there is a need to define the principal genetic modifiers that determine the risk for stroke. Previous studies using candidate gene approaches have linked more than 35 SNPs with stroke in pediatric patients with SCA.7-14 However, we recently performed an independent validation study and found that the majority of these potential modifiers did not have a replicated association with stroke.15 Only a few genetic modifiers had a confirmed association, with polymorphisms in the ANXA2, TEK, and TGFBR3 genes being associated with increased stroke risk, whereas the α-thalassemia trait and a variant in the ADCY9 gene were protective against stroke. It is unlikely that these associated polymorphisms constitute the entire genetic contribution to the risk for stroke, and only the α-thalassemia trait is a polymorphism with a known functional effect, being a copy-number polymorphism resulting in decreased α-globin production.
In an attempt to better define the principal genetic modifiers affecting stroke risk in patients with SCA, we performed an unbiased GWAS using a combination of polymorphism arrays and WES. Compared with our previous efforts using a limited candidate gene approach, only the TEK SNP (rs489347) had genotype data available from the polymorphism arrays and was associated with an increased risk for stroke (P = .004). Although this significant association was far from the best P value results achieved by the GWAS (P < .00001), this SNP maintained the same direction of association as our previous result.15 None of our results from GWAS showed an association that passed a genome-wide statistical significance or were located in a gene with a known association with stroke risk. However, the results from the GWAS were helpful when we analyzed the results from WES, where we found 300 nonsynonymous variants that are significantly associated with stroke risk in children with SCA. In combination, 11 variants identified by WES were located within 250kb of at least 1 SNP identified by GWAS (Table 3). When we performed validation tests of 22 candidate SNVs that had the best statistical association with stroke risk from the WES results, the 2 variants with validated associations were both located near an SNP with significant association from the GWAS results (Table 3 and 4). These mutations in GOLGB1 (Y1212C) and in ENPP1 (K173Q) were both associated with a significantly decreased risk for stroke.
GOLGB1, also known as Giantin, is a Golgi apparatus–associated protein. The Golgi apparatus is the central biosynthesis hub of the secretory pathway and uses vesicle transport for recycling of its resident enzymes. The best characterized function of GOLGB1 is to tether coat protein 1 (COP1) vesicles to the Golgi apparatus, allowing bidirectional cargo transport through the Golgi stack.32-34 The Y1212C mutation is predicted by bioinformatics to have a deleterious effect on protein function, with a PolyPhen2 score of 0.99 (sensitivity of 0.14 and specificity of 0.99), and the mutation is located in a highly conserved coiled-coiled region near the N-terminus of the protein (Figure 3). Although there is no immediately apparent reason why the Y1212C mutation should protect against stroke, our discovery and validation studies document that this mutation has a significantly lower allele frequency in patients with stroke, patients with silent infarction, and in patients with high TCD velocities compared with nonstroke control patients (Table 6). These results highlight that this mutation appears to be protective against ischemic stroke but also may affect the overall development of cerebrovascular disease. The remaining obstacle to the extension of this discovery will be our ability to characterize the effects of this mutation on GOLGB1 function, specifically in the biological context of stroke such as neuronal, vascular, or endothelial injury.
Conservation of the GOLGB1 protein near the Y1212C mutation. This figure shows the GOLGB1 protein alignments near the Y1212C mutation across human, mouse, rat, and marmoset species. The protein is highly conserved in this region, and PolyPhen2 predicts that this mutation will have a damaging effect on the protein, with a score of 0.99 (sensitivity of 0.14 and specificity of 0.99).
Conservation of the GOLGB1 protein near the Y1212C mutation. This figure shows the GOLGB1 protein alignments near the Y1212C mutation across human, mouse, rat, and marmoset species. The protein is highly conserved in this region, and PolyPhen2 predicts that this mutation will have a damaging effect on the protein, with a score of 0.99 (sensitivity of 0.14 and specificity of 0.99).
ENPP1 is a type 2 transmembrane glycoprotein that cleaves a variety of substrates, including the phosphodiester and pyrophosphate bonds of nucleotides and nucleotide sugars. Mutations in the ENPP1 gene have been documented to cause generalized arterial calcification of infancy ([MIM 208000]). ENPP1 is a potent generator of pyrophosphate (PPi), and loss-of-function mutations in ENPP1 result in depleted extracellular PPi levels.35,36 Lower levels of PPi are the main cause of the arterial calcification in patients with calcification of infancy and in the respective mouse models of defective ENPP1.36-38 In contrast, increased levels of PPi are known to inhibit vascular calcification.39 The ENPP1 K173Q mutation identified in this study has previously been shown to affect intracellular ENPP1 activity and serum ENPP1 enzyme levels.40,41 In both our discovery and validation studies, we found that the ENPP1 K variant was associated with a significantly decreased stroke risk, which suggests that decreased ENPP1 activity may protect against stroke via PPi levels. The K173Q mutation did not have any effect on our cohorts of patients with silent infarctions or elevated TCD velocities and so appears to be associated with more severe cerebrovascular disease. Future work will delineate the role that the ENPP1 gene and PPi levels play in the development of cerebrovascular disease in SCA.
The other genetic variant that displayed a significant association with the risk for stroke was the Q192R mutation in the PON1 gene, which conferred an additional stroke risk. This mutation had been associated with adult ischemic stroke but not previously in the context of SCA or in pediatric stroke.26,27 This mutation showed significant association with an increased risk for stroke when all patients were tested, and there was a marked difference in the number of Q192R homozygotes in all patients with stroke (n = 177; 13.7%) compared with all control patients (n = 335; 3.6%). This variant causes a nonsynonymous change of glutamine to arginine at amino acid position 192 of the encoded enzyme, which normally has both paraoxonase and esterase functions. This mutation warrants further testing in other independent cohorts of patients with SCA to confirm this association of Q192R with increased stroke risk.
Our study describes the first discoveries of functional genetic mutations that may affect susceptibility toward stroke among patients with SCA. Crucially, we have performed an unbiased genome search using both GWAS and WES in a large cohort of patients with known stroke phenotype and appropriate nonstroke control patients, and then corroborated our findings in a subsequent validation step with independent cohorts. Therefore, these genetic mutations represent important candidates for further investigation into the cause of stroke in SCA. Genetic risk factors are not typically considered to be modifiable, but identification of genetic modifiers could provide insights into the pathophysiological pathways of stroke and potential targets for drug therapy. Future studies will focus on how these functional mutations may influence the risk for stroke in the context of SCA.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank all of the patients and their families, as well as the clinical investigators and research staff, for their participation in the Stroke With Transfusions Changing to Hydroxyurea (SWiTCH), Hydroxyurea Study of Long-Term Effects (HUSTLE), Cooperative Study of Sickle Cell Disease (CSSCD), Comprehensive Sickle Cell Centers Collaborative Data Project (C-DATA), and Stroke Prevention in Sickle Cell Anemia (STOP1) studies.
This work was supported by grants from the Doris Duke Charitable Foundation; National Institutes of Health National Heart, Lung, and Blood Institute (R01HL090941-03 and U01HL078787-05); and the American Lebanese Syrian Associated Charities.
Authorship
Contribution: J.M.F. analyzed the data and wrote the manuscript. V.S., H.L., and T.A.H. performed experiments. C.C.H., B.A., and R.J.A. helped design the study and collected samples. Y.-D.W. and G.A.N. performed the Affymetrix SNP6.0 genotyping and WES. R.E.W. designed the study and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jonathan M. Flanagan, Texas Children’s Hematology Center, Department of Pediatrics, Baylor College of Medicine, 1102 Bates, Ste. C1030, Houston, TX 77030; e-mail: jmflanag@bcm.edu.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal