Key Points
Novel crosstalk between SMO and NF-κB representing additional level of NF-κB regulation independent of genetic constitutive activation.
SMO activates NF-κB by recruiting Gαi and Gα12 to activate PKCβ/CARMA1 and assembling CARMA1/BCL10/MALT1/TRAF6 to SMO.
Abstract
Diffuse large B-cell lymphoma (DLBCL) is the most common lymphoid malignancy in adults. Aberrant activation of Hedgehog (Hh) and nuclear factor (NF)-κB pathways is ubiquitously observed and known to mediate tumor growth, survival, and chemoresistance in DLBCL. Here, we find that activation of Hh signaling is positively correlated with NF-κB pathway in DLBCL tumors, and that smoothened (SMO), the signal transducer subunit of Hh pathway, contributes to NF-κB activation through recruiting G protein subunits Gαi and Gα12 to activate PKCβ/CARMA1/TRAF6/NEMO signaling axis followed by assembling of the CARMA1/BCL10/MALT1/TRAF6 complex to SMO. Moreover, functional inhibition of SMO enhances the cytotoxic effects of NF-κB inhibitor. Altogether, our study reveals a noncanonical Hh signaling pathway in which SMO activates trimeric G proteins and CARMA1-associated signaling complex, leading to NF-κB activation. This signaling cascade contributes to the survival of DLBCL and may serve as a potential target for combination therapies in DLBCL.
Introduction
Nuclear factor (NF)-κB pathway plays a critical role in B-cell physiology and contributes to cell survival, proliferation, and chemoresistance of diffuse large B-cell lymphoma (DLBCL), the most common B-cell non-Hodgkin lymphoma in adults.1,2 The NF-κB transcription factor family consists of 5 proteins, 3 canonical (p65, p50 and c-Rel) and 2 alternative (p52 and RelB) that form various homo- and heterodimers.3 Among them, the heterodimeric p65/p50 complex is the most abundant and responsible for regulating inflammatory responses.4 When NF-κB pathway is inactive, the p65/p50 complex binds to IκBα and is retained in the cytoplasm. When NF-κB pathway is activated, IκBα is phosphorylated by IκB kinase complex (IKK) and degradated in the proteosome. Subsequently, p65/p50 translocates to the nucleus to bind NF-κB target genes. Nuclear detection of NF-κB components, direct evidence of NF-κB activation, has been found in ∼90% of DLBCL of activated B cell (ABC) type and in ∼30% of DLBCL of germinal center (GC) type.5 Genetic alterations and mutations that explain the activation of NF-κB signaling have been found in ∼63% of ABC type and in ∼30% of GC type.5,6 However, many DLBCLs have constitutive activation of NF-κB but do not carry genetic lesions justifying NF-κB activation.
G protein–coupled receptors (GPCR) are a large family of 7 transmembrane domain proteins linking extracellular inputs with diverse cellular responses. GPCRs play important roles in regulating cell migration, differentiation, proliferation, and survival.7 GPCRs are integrated by a receptor that binds the soluble signal and a heterotrimeric (αβγ) G protein, which is able to exchange guanosine diphosphate for guanosine triphosphate (GTP), resulting in activation of the Gα subunit and dissociation of the Gβγ subunits followed by a biological response.8 The Gα subunit contains several subgroups, including Gi, Gs, Gq, G16, and G12/13, which independently activate several downstream signaling cascades including NF-κB pathway through a CARMA-Bcl10-MALT1 (CBM) complex-dependent mechanism.9
Hedgehog (Hh) signaling is an evolutionarily conserved pathway involved in organogenesis, embryogenesis, and homeostasis of adult tissues.10 Hh signaling is deregulated in several cancers, including DLBCL.11-14 Sonic Hh (SHh), Indian Hh, and Desert Hh are the ligands. Patched 1 (PTCH1) and smoothened (SMO) are the receptors. PTCH1 is the ligand receptor subunit and, in the absence of Hh ligands, inhibits SMO.15 In the presence of Hh ligands, the inhibition of PTCH1 over SMO is abrogated, resulting in SMO activation. Upon activation, SMO transduces the signal to the cytoplasm using glioma-associated oncogene homolog (GLI) proteins as major transcriptional effectors (canonical Hh signaling).16,17
We find that Hh and NF-kB pathways are positively correlated and that Hh ligands contribute to NF-κB activation in DLBCL. SMO has 7 transmembrane domains and has been established as a GPCR-like protein after the identification of its binding with Gαi.18,19 Because of the similarities between SMO with GPCR proteins, we sought to determine whether SMO contributed to NF-κB activation in DLBCL. To the best of our knowledge, the contribution of SMO in NF-κB activation has not been previously determined. Here, we demonstrate that SMO contributes to NF-κB activation through GPCR signaling mechanisms and that inhibition of SMO enhances the cytotoxic effects of NF-κB inhibitors in DLBCL.
Material and methods
Cell lines
We used 5 DLBCL GC type (DOHH2, SUDHL4, OCI-Ly19, OCI-Ly7, and BJAB), 1 DLBCL ABC type (HBL1) human bone marrow stromal cell line HS-5 and HEK293T cells. BJAB is an immortalized Epstein-Barr virus B-cell lymphoma cell line considered to have a GC phenotype.20,21 DOHH2, SUDHL4, and OCI-Ly19 were purchased from DSMZ (Braunschweig, Germany); HS-5 and HEK293T were obtained from ATCC (Manassas, VA). HBL1 and BJAB were a gift from Dr Samaniego (Department of Lymphoma and Myeloma, MD Anderson Cancer Center, Houston, TX). OCI-Ly7 was provided by Dr Lin (Molecular and Cellular Oncology; MD Anderson Cancer Center). The tissue samples were used under the approved and active institutional review board laboratory protocol LAB07-0920, and the study was conducted in accordance with the Declaration of Helsinki. All DLBCL cell lines were maintained at 37°C in RPMI1640 (ATCC) supplemented with 10% to 20% heat-inactivated fetal bovine serum (FBS) (Sigma, Ronkonkoma, NY) and 0.2% MycoZapTM Plus-CL (Lonza, Walkersville, MD) in 5% CO2 humidified atmosphere. HEK293T and HS-5 were cultured in Dulbecco’s modified Eagle medium (Mediatech, Manassas, VA) supplemented with 10% FBS.
Coculture
HS-5 cells were plated in 6-well plates in Dulbecco’s modified Eagle medium with 2% FBS and allowed to attach and grow for 48 h followed with or without 5E1 anti-Hh blocking antibody (Hybridoma Bank, University of Iowa, Iowa City, Iowa) to block secreted Hh ligands. Then, cell culture trans-wells (BD Falcon, San Jose, CA) with a 0.4-μM membrane size were inserted and DLBCL cells were plated in the inserts and cocultured with HS-5 for 1 hour.
DLBCL tissue microarray and immunohistochemistry
We used a tissue microarray with 54 DLBCL tumors collected from formalin fixed, paraffin-embedded archival tumor specimens. According to the expression of CD10, BCL-6, and MUM-1 (30% cutoff) cases were classified as either GC or non-GC type using the system of Hans et al.22 Immunohistochemical studies were performed as previously described using primary antibodies for GLI1 (Santa Cruz, Dallas, TX) and p-p65 (Cell Signaling, Boston, MA).1,14 The percentage of tumor cells positive for each marker was quantified in representative fields counting up to 1000 cells.
Electrophoretic mobility shift assay (EMSA)
Nuclear extracts (4 µg) were extracted as described23 and incubated for 15 min at room temperature with a 32P-labeled double-stranded NF-κB–specific or Oct-1 oligonucleotide probes (Promega, Madison, WI). Samples were run on nondenaturing polyacrylamide gels and visualized by autoradiography.
Nucleotransfection and stable cell line establishment
Transfections were done by electroporation using the Nucleofector transfection kit (Amaxa Biosystems, Gaithersburg, MD) per the manufacturer’s instructions. Small interfering RNAs (siRNAs) were purchased from Dharmacon (Lafayette, CO) and nonspecific scrambled siRNA was used as a control. Whole-cell lysates were prepared 48 hours after transfection. To generate stable SMO and GLI1 silenced cell lines, HEK293T and OCI-LY19 were infected with lentiviral transduction particles containing control shRNA, SMO, or GLI1 shRNA plasmids from Sigma. Cells were grown in the presence of puromycin for clone selection.
Western blot analysis
Total cell lysates were processed for immunoblotting as previously described.24 Primary antibodies used were SMO,T642p-PKCβ1, PKCβ1, S660p-PKCβ2, PKCβ2, anti-HA (Abcam, Burlingame, CA), PTCH1, TRAF6, BCL10, IKBα, Gαs, Gαi, Gαq, Gα12 (Santa Cruz), Gα16 (Thermo Scientific, Waltham, MA), S536p-p65, p65, S32/36p-IκBα, GLI1, S652p-CARMA1, CARMA1, MALT1 (Cell Signaling), and T759p-PLCγ2, PLCγ2, anti-Flag, actin (Sigma). Reactions were visualized with suitable secondary antibodies (Thermo Scientific) conjugated with horseradish peroxidase using enhanced chemiluminescence reagents (Millipore, Billerica, MA).
DNA binding enzyme-linked immunosorbent assay
The TransAM NF-κB Transcription factor assay kit (Active Motif, Carlsbad, CA) was used to detect and quantify binding of NF-κB transcription factors (p65 and p50 subunits) to its DNA consensus site (5′-GGGACTTTCC-3′).
Luciferase assays
A luciferase reporter plasmid PGL3-NF-κB-6× and SMO/Control siRNA or GFP/SMO-GFP were cotransfected into DLBCL cells using the Nucleofector transfection kit (Amaxa Biosystems). Plasmid for Renilla expression was also cotransfected for transfection efficiency normalization. Luciferase and Renilla activities were measured using the Luciferase assay kit (Promega).
Cytokines and receptors PCR arrays and PKC kinase assay
The Human Inflammatory Cytokines & Receptors polymerase chain reaction (PCR) Array (Qiagen, Valencia, CA) was used to detect expression of downstream cytokines of NF-κB. Expression levels for each gene were normalized to glyceraldehyde-3-phosphate dehydrogenase and data were analyzed using Qiagen website-based RT2 Profilter PCR Array Data Analysis software. Protein kinase C (PKC) kinase activity was detected using PepTag Assay for Non-Radioactive Detection of PKC (Promega). The relative PKC kinase activity was quantified by spectrophotometry (absorbance 570 nm).
Immunoprecipitation and in vivo ubiquitination assays
Total cell lysates and indicated antibodies were incubated for 1 hour at 4°C followed by protein agarose beads (Santa Cruz) with overnight immunoprecipitation and immunoblotting. HEK293T cells were transiently transfected with the indicated plasmids to detect exogenous ubiquitination of TRAF6 and NEMO. Cells were harvested and lysed 48 hours posttransient transfection. Ubiquitinated TRAF6 and NEMO were immunoprecipitated by M2 beds (Sigma) and immunoblotted by anti-HA.
GTPase assay
GTPase activity was measured with the colorimetric GTPase assay kit from Innova Biosciences (Babraham, UK) following the manufacturer’s protocol. Briefly, DLBCL cells treated with vehicle or recombinant SHh for 10 minutes were solubilized under nondenaturing conditions and PiBindTM resin was added to remove π. Gα-subunits were then immunoprecipitated, incubated for 1 hour with selective Gα antibodies followed by immunoprecipitation for 1 hour at 4°C using agarose beads. GTP was then added to the well-washed protein agarose beads and incubated at 30°C for 30 minutes. Gold mix and stabilizer were further added to the mixture. Optical density was determined at 595 nm.
RNA extraction and quantitative real time PCR analysis
Total messenger RNA (mRNA) was extracted using RNeasy Minikit (Qiagen) as per the manufacturer’s protocol. Quantitative real-time PCR analysis was performed according to the described protocol.13 The primers and probes for GLI1, BCL2, BCL2L1, SMO, and 18S were obtained from Applied Biosystems (Carlsbad, CA). Each target was amplified in duplicate and data analyses were done using the 2-(ΔΔCT) method.25
Cell viability and apoptosis assays
DLBCL cells were plated in triplicate with desired concentrations of SMO and NF-kB inhibitors for 48 hours. A cell proliferation kit Cell Titer 96 Aqueous One Solution (MTS) (Promega) was used to determine cell viability and fluorescein isothiocyanate-annexin V and propidium iodide (PI) staining kit (BD Biosciences) was used to detect cell apoptosis followed by FlowJo7.6.5 software analysis.
Statistical analysis
Gene expression data were retrieved from public repositories (ArrayExpress E-GEOD-11318; Gene Expression Omnibus GSE 11318). The raw intensities were log(2) transformed and robust multi-array analysis normalized. log(2) transformed and median centered data from Lenz et al26 was used to compare GLI1 expression (y-axis). The Student t test was used to evaluate the significant difference of 2 groups of data in all the pertinent experiments. The Spearman test was used to analyze the correlation between nuclear protein expression of GLI1 and p-p65. A P value < .05 was thought to be significantly different.
Results
Hh and NF-κB pathways are positively correlated and Hh contributes to NF-κB activation in DLBCL
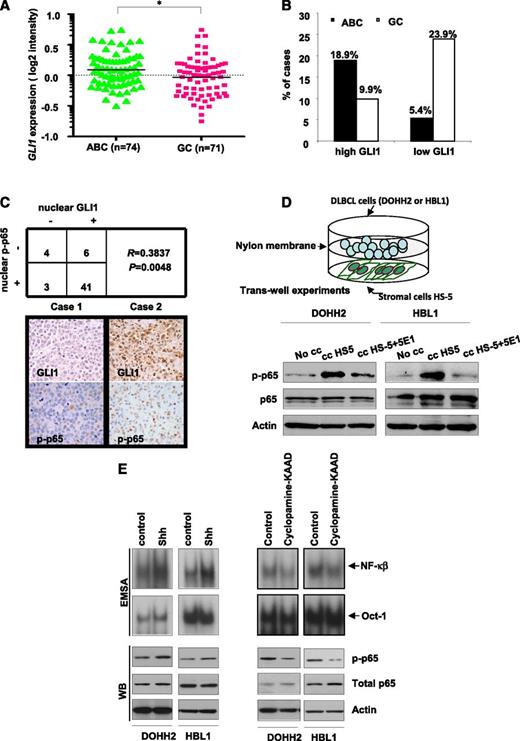
Because NF-κB pathway is more frequently activated in DLBCL of ABC type than in GC,5 we compared GLI1 expression levels (readout of Hh signaling activation) between 145 DLBCL of ABC and GC type. We found that cases of ABC type showed significantly higher GLI1 levels than those of GC (*P < .0, Figure 1A). As shown in Figure 1B, high GLI1 expression (normalized signal cutoff of 0.3) was more frequent in cases of ABC (14 of 74 [18.9%]) than in GC (7 of 71 [9.9%]) and low GLI1 expression was more frequent in cases of GC (17 of 71 [23.9%]) than in ABC (4 of 74 [5.4%]). We then performed immunohistochemical studies to evaluate expression of nuclear GLI1 and nuclear p-p65 (readout of NF-κB signaling activation) in a series of 54 DLBCL, including 23 cases of ABC, 29 of GC, and 2 undetermined. We quantified the percentage of positive cells for each immunomarker (Figure 1C and supplemental Figure 1A). We found that both Hh and NF-κB pathways were frequently activated in DLBCL tumors of either ABC or GC type. Nuclear GLI1 was detected in 47 of 54 (87%) cases, and nuclear p-p65 was detected in 44 of 54 (81.4%). More important, there was a significant positive correlation between expression of nuclear GLI1 and nuclear p-p65 (R = 0.3837, P = .0048). These data raise the possibility that Hh signaling contributes to NF-kB activation in DLBCL. To test our hypothesis, we cocultured DLBCL cells (DOHH2 and HBL-1 cell lines) with human stromal cells (HS-5), known to secrete Hh ligands that activate Hh signaling.13 Indeed, we observed an enhancement of p65 phosphorylation in the presence of HS-5 cells in both cell lines, and this effect was abrogated in the presence of the anti-Hh blocking antibody 5E1 (2.5 µg/mL) (Figure 1D). Because phosphorylation of p65 at S536 has been related with p65 nuclear localization,27,28 our results indicate that Hh signaling contributes to NF-kB activation.
Hh and NF-κB pathways are positively correlated in DLBCL, and Hh pathway contributes to NF-κB activation in DLBCL. (A) GLI1 expression levels in 145 DLBCL tumor biopsies. (B) High GLI1 expression was more frequent in DLBCL of ABC type than in GC type; in addition, low GLI1 expression was more frequent in cases of GC type than in those of ABC type. *P < .05. (C) A significant positive correlation between nuclear GLI1 and nuclear p-p65 (R = 0.3837, P = .0048) in a series of 54 DLBCL analyzed by immunohistochemistry (IHC). Representative IHC staining results for nuclear GLI1 and p-p65 in 2 DLBCL cases. Left (case 1), the tumor cells were negative for GLI1 and p-p65. Right (case 2), the tumor cells expressed both GLI1 and p-p65 in the nuclei. A subset of lymphoma-associated macrophages was also frequently and strongly positive for p-p65, which was confirmed by double immunohistochemical studies using p-p65 and the macrophage marker CD68 (supplemental Figure 1A). (D) Coculture of DLBCL cells (DOHH2 and HBL-1) with the stromal cell HS-5 in trans-well experiments resulted in increased p-p65. This effect was abrogated, at least in part, in the presence of the anti-Hh blocking antibody 5E1 (2.5 µg/mL). (E) DOHH2 and HBL1 cells treated with or without recombinant Shh (250 ng/mL) for 30 minutes or cyclopamine-KAAD (4.8 µM) for 1 hour were subjected to EMSA by using 32P-labeled NF-κB or Oct-1 probes (Upper panel) as well as western blot to detect p-p65 (Lower panel). Dimethylsulfoxide (DMSO; vehicle for cyclopamine-KAAD) and phosphate-buffered saline (for recombinant Shh) were used as controls for drugs treatments.
Hh and NF-κB pathways are positively correlated in DLBCL, and Hh pathway contributes to NF-κB activation in DLBCL. (A) GLI1 expression levels in 145 DLBCL tumor biopsies. (B) High GLI1 expression was more frequent in DLBCL of ABC type than in GC type; in addition, low GLI1 expression was more frequent in cases of GC type than in those of ABC type. *P < .05. (C) A significant positive correlation between nuclear GLI1 and nuclear p-p65 (R = 0.3837, P = .0048) in a series of 54 DLBCL analyzed by immunohistochemistry (IHC). Representative IHC staining results for nuclear GLI1 and p-p65 in 2 DLBCL cases. Left (case 1), the tumor cells were negative for GLI1 and p-p65. Right (case 2), the tumor cells expressed both GLI1 and p-p65 in the nuclei. A subset of lymphoma-associated macrophages was also frequently and strongly positive for p-p65, which was confirmed by double immunohistochemical studies using p-p65 and the macrophage marker CD68 (supplemental Figure 1A). (D) Coculture of DLBCL cells (DOHH2 and HBL-1) with the stromal cell HS-5 in trans-well experiments resulted in increased p-p65. This effect was abrogated, at least in part, in the presence of the anti-Hh blocking antibody 5E1 (2.5 µg/mL). (E) DOHH2 and HBL1 cells treated with or without recombinant Shh (250 ng/mL) for 30 minutes or cyclopamine-KAAD (4.8 µM) for 1 hour were subjected to EMSA by using 32P-labeled NF-κB or Oct-1 probes (Upper panel) as well as western blot to detect p-p65 (Lower panel). Dimethylsulfoxide (DMSO; vehicle for cyclopamine-KAAD) and phosphate-buffered saline (for recombinant Shh) were used as controls for drugs treatments.
To further confirm whether modulating Hh signaling affects NF-κB activation, we performed an EMSA and supershift with antibodies specific to p65, p50, c-Rel, RelB, and p52. Using 3 different DLBCL cell lines, we confirmed that NF-κB binding activity was induced (Ly7 cells) or enhanced (DOHH2 and HBL1 cells) after treatment with recombinant Shh peptide (250 ng/mL) for 30 minutes and decreased after cyclopamine-KAAD (4.8 μM) for 1 hour (Figure 1E and supplemental Figure 1B). Supershift analysis has revealed that recombinant Shh increases activity of classical NF-kB members including p65, p50, and c-Rel in all examined cell lines (supplemental Figure 1B). Next, we checked the phosphorylation status of p65 after modulating the functional status of SMO with short treatments with cyclopamine-KAAD and recombinant Shh. The phosphorylation of p65 at S536 was increased after treatment with Shh and decreased by cyclopamine-KAAD in DOHH2 and HBL1 cell (Figure 1E). Similar results were seen in BJAB and SUDHL4 cells (supplemental Figure 1C). Consistently, treatment with the same doses of cyclopamine-KAAD or recombinant Shh for 24 h resulted in decrease or increase of the nuclear levels of p-p65 (supplemental Figure 1E). The effect of cyclopamine-KAAD and recombinant Shh on Hh pathway was confirmed by measuring GLI1 mRNA levels (supplemental Figure 2).
These data suggest that Hh and NF-κB pathways are positively correlated and Hh signaling contributes to NF-κB activation in DLBCL.
SMO activates NF-κB signaling independently of GLI1
To uncover which component of Hh signaling contributed to NF-κB signaling activation in DLBCL, we silenced SMO by siRNA and then examined the phosphorylation of p65 at S536. SMO silencing in DLBCL cell lines resulted in decrease of p-p65 associated with decrease of the nuclear binding of p65 and p50 (2 major components of the canonical NF-κB pathway) (Figures 2A-B and supplemental Figure 1D) and NF-κB activity measured by NF-κB–dependent luciferase reporter assays (Figure 2C). SMO silencing also resulted in a significant decrease expression of 16 cytokines—CCL15, CXCL10, CXCL9, CCL7, CCL1, CXCL6, C3, IL8, CXCL5, CCR5, CXCL11, IL9, CXCR2, IL1B, LTB, and CCL2—that are direct downstream targets of NF-κB pathway (Figure 2D and supplemental Figure 3).
SMO activates NF-κB pathway independent of GLI1. (A) Silencing SMO in DOHH2 and HBL1 resulted in decrease of p-p65, (B) decrease of nuclear DNA binding activity of p65 and p50, and (C) decrease of total NF-κB luciferase activity. (D) Silencing SMO in HBL1 cells also resulted in decrease of 16 known direct NF-κB downstream target genes. The heat map was generated using TreeView software v1.1 (normalized, centered values). (E) Silencing PTCH in DOHH2 and HBL1 cells resulted in increase of p-p65. (F) No changes in p-p65 were seen after transiently silencing GLI1 by siRNA in DOHH2 and HBL1 or stably silencing GLI1 by shRNA in OCI-Ly19 cells. Data are presented as mean ± standard deviation of at least 2 independent experiments. P < .05.
SMO activates NF-κB pathway independent of GLI1. (A) Silencing SMO in DOHH2 and HBL1 resulted in decrease of p-p65, (B) decrease of nuclear DNA binding activity of p65 and p50, and (C) decrease of total NF-κB luciferase activity. (D) Silencing SMO in HBL1 cells also resulted in decrease of 16 known direct NF-κB downstream target genes. The heat map was generated using TreeView software v1.1 (normalized, centered values). (E) Silencing PTCH in DOHH2 and HBL1 cells resulted in increase of p-p65. (F) No changes in p-p65 were seen after transiently silencing GLI1 by siRNA in DOHH2 and HBL1 or stably silencing GLI1 by shRNA in OCI-Ly19 cells. Data are presented as mean ± standard deviation of at least 2 independent experiments. P < .05.
Because PTCH1 is a physiological inhibitor of SMO, we also silenced PTCH1 and examined the effect on p-p65 (Figure 2E). Silencing PTCH1 by siRNA resulted in increase of p-p65. To address whether GLI1 was also involved in the modulation of NF-κB by SMO, we silenced GLI1 using both siRNA and short hairpin RNA (shRNA) approaches. Transient or stable silencing of GLI1 did not affect p-p65 compared with the controls (Figure 2F).
Altogether, these data further support that Hh pathway contributes to NF-κB activation in DLBCL cells; this contribution is dependent of the functional status of SMO but independent of GLI1.
SMO activates NF-κB pathway through recruiting Gαi and Gα12 followed by activating PKCβ-CARMA1-MALT1-BCL10-TRAF6-NEMO axis
We hypothesized that a GPCR-related mechanism such as PKC-CARMA1-BCL10/MALT1/TRAF6 axis may be involved in SMO-dependent NF-κB activation. This hypothesis is supported by earlier studies that PKC and CARMA3 (CARMA1 homolog in nonhematopoietic cells) mediate NF-κB activation induced by several GPCR ligands29 and that Shh ligands increase activity of the PKC isoforms α, δ, and z in mouse stem cells.9,29-31
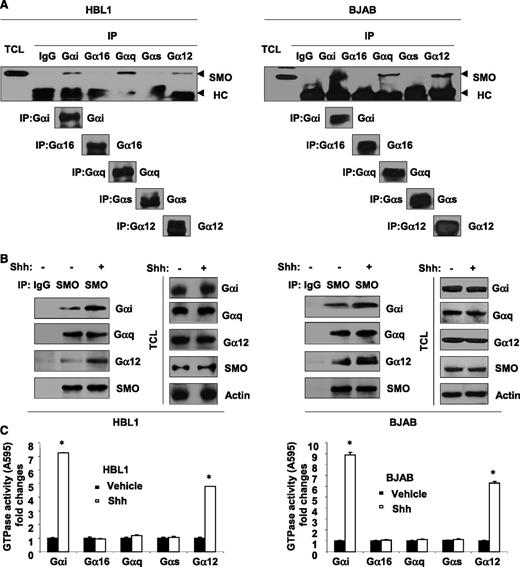
We first examined whether Gα proteins are associated with SMO by immunoprecipitating representative members of the Gα subfamilies (Gαi, Gα16, Gαq, Gαs, and Gα12) and screening for SMO. We found that SMO was associated with Gαi, Gαq, and Gα12 in DLBCL cell lines (Figure 3A). Stimulation of SMO with recombinant Shh increased the recruitment of Gαi and Gα12 to SMO (Figure 3B) and also increased the GTPase activity of Gαi and Gα12 (Figure 3C). These findings suggest that SMO may form a signaling complex with Gαi and Gα12 and activation of SMO results in the activation of these Gα proteins in DLBCL cells.
SMO recruits and activates Gαi and Gα12 in DLBCL. (A) Immunoprecipitation (IP) assays showed that SMO coupled to Gαi, Gαq, and Gα12, but not to Gαs and Gα16 in HBL1 and BJAB. TC, total cell lysate; HC, heavy chain. Immunoprecipitation (B) and GTPase assays (C) showed that stimulation of SMO with recombinant Shh (250 ng/mL) for 10 minutes was associated with increased recruitment of Gαi and Gα12 to SMO as well as increased GTPase activity of Gαi and Gα12 in HBL1 and BJAB cells. Data are presented as mean ± standard deviation of 3 experiments. *P < .05.
SMO recruits and activates Gαi and Gα12 in DLBCL. (A) Immunoprecipitation (IP) assays showed that SMO coupled to Gαi, Gαq, and Gα12, but not to Gαs and Gα16 in HBL1 and BJAB. TC, total cell lysate; HC, heavy chain. Immunoprecipitation (B) and GTPase assays (C) showed that stimulation of SMO with recombinant Shh (250 ng/mL) for 10 minutes was associated with increased recruitment of Gαi and Gα12 to SMO as well as increased GTPase activity of Gαi and Gα12 in HBL1 and BJAB cells. Data are presented as mean ± standard deviation of 3 experiments. *P < .05.
Next, we determined the contribution of SMO to the activity of total PKC in HEK293T and DLBCL cells. Transient or stable silencing SMO resulted in decreased total PKC activity and overexpressing SMO resulted in increased PKC activity (Figure 4A). Treatments with cyclopamine-KAAD or recombinant Shh resulted in decrease or increase of total PKC activity, respectively. Because PKC isoforms β-1 and β-2 are the major isoforms expressed in B-lymphocytes32,33 and PKCβ mediates NF-κB activation induced by B-cell receptor,34 we assessed the activation status of these 2 β isoforms in response to changes of SMO activity. Inhibiting SMO with cyclopamine-KAAD or silencing SMO by siRNA decreased the phosphorylation of PKCβ-1 and PKCβ-2 (Figure 4B). In contrast, activating SMO with recombinant Shh increased the phosphorylation of PKCβ-1 and PKCβ-2 (Figure 4B). Then, we assessed if SMO could modulate the activity of CARMA1. CARMA1 is a scaffold protein that integrates the upstream signal of PKCs with downstream effectors in hematopoietic cells.35 We found that inhibiting or silencing SMO decreased the phosphorylation of CARMA1 at S652 and that activation of SMO with recombinant Shh increased CARMA1 phosphorylation at this site (Figure 4B). These changes in the activation status of PKCβ isoforms and CARMA1 were associated with reverse changes of the total levels of IKBα, thus further indicating that the activation status of SMO correlates with the activation of NF-κB (Figure 4B). Phosphorylation of CARMA1 at S652 (S652 is the human site and is equivalent to S657 in mouse and S668 in chicken) is mediated by PKC and is associated with recruitment of BCL10 and MALT1 proteins to CARMA1.36
SMO activates NF-κB through PKCβ-CARMA1-MALT1-BCL10-TRAF6-NEMO axis. (A) Stably knocking down SMO or overexpressing SMO showed a decrease or increase in total PKC activity in HEK293T cells, respectively. Inhibition of SMO by cyclopamine-KAAD-KAAD in BJAB or silencing SMO in HBL1 also resulted in decrease of total PKC activity, whereas activating SMO by recombinant Shh in BJAB resulted in increase of total PKC activity. (B) Inhibition of SMO by cyclopamine-KAAD or silencing of SMO in DLBCL cell lines resulted in decrease of p-PKCβ-1 and p-PKCβ-2 and a decrease of p-CARMA1. In contrast, activating SMO by recombinant Shh resulted in increase of p-PKCβ-1 and p-PKCβ-2 followed by increased of p-CARMA1. (C) Activation of SMO with recombinant Shh increased the binding between SMO and CARMAL1/BCL10/MALT1/TRAF6 complex (central lane; +SMO). TCL, total cell lysates. (D) Transient overexpression of SMO resulted in increased TRAF6 and NEMO ubiquitination on K63 lysine indicating stabilization of these proteins and supporting NF-κB activation.
SMO activates NF-κB through PKCβ-CARMA1-MALT1-BCL10-TRAF6-NEMO axis. (A) Stably knocking down SMO or overexpressing SMO showed a decrease or increase in total PKC activity in HEK293T cells, respectively. Inhibition of SMO by cyclopamine-KAAD-KAAD in BJAB or silencing SMO in HBL1 also resulted in decrease of total PKC activity, whereas activating SMO by recombinant Shh in BJAB resulted in increase of total PKC activity. (B) Inhibition of SMO by cyclopamine-KAAD or silencing of SMO in DLBCL cell lines resulted in decrease of p-PKCβ-1 and p-PKCβ-2 and a decrease of p-CARMA1. In contrast, activating SMO by recombinant Shh resulted in increase of p-PKCβ-1 and p-PKCβ-2 followed by increased of p-CARMA1. (C) Activation of SMO with recombinant Shh increased the binding between SMO and CARMAL1/BCL10/MALT1/TRAF6 complex (central lane; +SMO). TCL, total cell lysates. (D) Transient overexpression of SMO resulted in increased TRAF6 and NEMO ubiquitination on K63 lysine indicating stabilization of these proteins and supporting NF-κB activation.
Because PKC-CARMA-BCL10-MALT1 complex constitutes a common axis to transduce signals from PKC to NF-κB, we explored if SMO activation was associated with increased recruitment of this protein complex to SMO. Once activated at the plasma membrane, the CARMA1-BCL10-MALT1 complex facilitates the activation of the IKK (I κ B kinase) complex, which phosphorylates IκBα targeting IκBα for proteasomal degradation, thereby allowing NF-κB transcription factors to enter the nucleus to drive the expression of NF-κB target genes.4 We found that activation of SMO with recombinant Shh was associated with increased recruitment of CARMA1-MALT1-BCL10-TRAF6 to SMO (Figure 4C) supporting that this signaling axis is involved in the transmission of the signal between SMO with NF-κB pathway.
Finally, because the polyubiquitination of TRAF6 and NEMO (IKKγ) at lysine 63 (K63) are also important events in propagating NF-κB signaling, we examined the effect of SMO overexpression on K63-linked polyubiquitination of TRAF6 and NEMO. Transient overexpression of SMO resulted in increased TRAF6 and NEMO polyubiquitination (Figure 4D), supporting activation of both proteins by SMO and thereby leading to NF-κB pathway activation.
These findings suggest that 1 mechanism by which SMO contributes to the activation of NF-κB signaling pathway is through recruiting Gαi and Gα12 to activate PKCβ/CARMA1/TRAF6/NEMO signaling axis followed by assembling of the CARMA1/BCL10/MALT1/TRAF6 complex to SMO.
SMO inhibitor enhances the cytotoxic effects of NF-κB inhibitor in DLBCL
Because SMO activation influences NF-κB activity, we investigated whether inhibiting SMO enhanced the cytotoxic effect of NF-κB inhibitor in DLBCL. As measured by MTS assays, combined treatments of cyclopamine-KAAD with BAY-11-7082 (NF-κB inhibitor) resulted in a higher decrease of cell viability than that observed by BAY-11-7082 treatment alone (Figure 5A). Annexin V and PI staining assays also indicated that combining cyclopamine-KAAD with BAY-11-7082 resulted in increase of apoptosis (Figure 5B). The increased apoptotic rate and decreased cell viability with the combined treatment was further confirmed by the increased cleavage of poly (adenosine 5′-diphosphate-ribose) polymerase 1 and decrease in p-p65 expression seen with the drug combination in comparison with that with BAY-11-7082 alone (Figure 5C). The combination treatment also significantly reduced mRNA levels of GLI1, as well as anti-apoptotic genes BCL2 and BCL-xL (Figure 5D).
SMO inhibitor enhanced the cytotoxic effect of NF-κB inhibitor on DLBCL cells. (A) BJAB and DOHH2 were treated with cyclopamine-KAAD and/or BAY11-7082. Combined treatments resulted in enhanced decrease of cell viability, as determined by MTS assays (at 48 hours), compared with treatment with the BAY11-7082 alone. (B) Annexin V and PI plots showed the percentage of apoptosis at 48 hours baseline (dimethylsulfoxide [DMSO], a), after treating with cyclopamine-KAAD or BAY11-7082 alone (b-c), and after combination of both inhibitors (d). The percentage of apoptosis was measured 48 hours after treatments and determined by Annexin V and PI assays. Data are presented as mean ± standard deviation (SD) of 3 experiments. *P < .05. (C) Western blot analysis showed that combining cyclopamine-KAAD with BAY11-7082 resulted in an increase of cleaved poly (adenosine 5′-diphosphate-ribose) polymerase 1 and a decrease of p-p65 compared with that treated with BAY11-7082 alone. (D) Quantitative real-time PCR showed that combined treatments resulted in marked decrease in the mRNA levels of GLI1, BCL2, and BCL-xL compared with BAY11-7082 treatment alone. These results were normalized to 18S mRNA level and expressed as fold changes in mRNA expression compared with control. Data are presented as mean ± SD of 3 experiments. *P < .05. (E) Proposed working model for SMO-mediated NF-κB activation using a GPCR signal transduction pathway. In the presence of Hh ligands, SMO is released from PTCH1 inhibition and gets activated. Activated SMO transduces signals through a canonical Hh pathway (GLI transcription factors) as well as through noncanonical a Hh pathway by recruiting and activating Gαi and Gα12. As a consequence, the Gβγ subunit is released and may contribute to phosphorylate and activate PKC-β. Gα12 may also contribute to the activation of PKC-β through the ρ family of small GTPases. Activated PKC-β phosphorylates and activates CARMA1 leading to the assembly of the CBM signalosome, polyubiquitination of TRAF6 and NEMO at K63, and followed by degradation of IκBα and activation of NF-κB pathway.
SMO inhibitor enhanced the cytotoxic effect of NF-κB inhibitor on DLBCL cells. (A) BJAB and DOHH2 were treated with cyclopamine-KAAD and/or BAY11-7082. Combined treatments resulted in enhanced decrease of cell viability, as determined by MTS assays (at 48 hours), compared with treatment with the BAY11-7082 alone. (B) Annexin V and PI plots showed the percentage of apoptosis at 48 hours baseline (dimethylsulfoxide [DMSO], a), after treating with cyclopamine-KAAD or BAY11-7082 alone (b-c), and after combination of both inhibitors (d). The percentage of apoptosis was measured 48 hours after treatments and determined by Annexin V and PI assays. Data are presented as mean ± standard deviation (SD) of 3 experiments. *P < .05. (C) Western blot analysis showed that combining cyclopamine-KAAD with BAY11-7082 resulted in an increase of cleaved poly (adenosine 5′-diphosphate-ribose) polymerase 1 and a decrease of p-p65 compared with that treated with BAY11-7082 alone. (D) Quantitative real-time PCR showed that combined treatments resulted in marked decrease in the mRNA levels of GLI1, BCL2, and BCL-xL compared with BAY11-7082 treatment alone. These results were normalized to 18S mRNA level and expressed as fold changes in mRNA expression compared with control. Data are presented as mean ± SD of 3 experiments. *P < .05. (E) Proposed working model for SMO-mediated NF-κB activation using a GPCR signal transduction pathway. In the presence of Hh ligands, SMO is released from PTCH1 inhibition and gets activated. Activated SMO transduces signals through a canonical Hh pathway (GLI transcription factors) as well as through noncanonical a Hh pathway by recruiting and activating Gαi and Gα12. As a consequence, the Gβγ subunit is released and may contribute to phosphorylate and activate PKC-β. Gα12 may also contribute to the activation of PKC-β through the ρ family of small GTPases. Activated PKC-β phosphorylates and activates CARMA1 leading to the assembly of the CBM signalosome, polyubiquitination of TRAF6 and NEMO at K63, and followed by degradation of IκBα and activation of NF-κB pathway.
Discussion
We previously reported that Hh signaling was activated in DLBCL and involved in cell survival, proliferation, and stromal-induced chemotolerance of this neoplasm.12-14 In this study, we reveal a novel noncanonical (GLI1-independent) Hh pathway in DLBCL, in which SMO recruits Gα12 and Gαi followed by activation of PKC-CARMA1–dependent signaling cascade contributing to NF-κB activation.
To link SMO with PKC activity, we investigate which Gα subunit binds to SMO and gets activated by SMO in DLBCL. A previous study showed that SMO can couple and activate members of Gαi family in sf9 cells and HEK293 cells,37 whereas another group reported that SMO can activate Gα12 in HEK293 cells.19 Using 2 different lymphoma cell lines, we found that SMO binds to Gαi, Gαq, and Gα12, but only that Gαi and Gα12 showed increased association with SMO following stimulation with recombinant Shh. These findings are of interest because it has been shown that Gα12 uses ρ family of small GTPases to activate serum response factor and NF-κB.38,39 In addition, it has been shown that Gα12 enhances PKC phosphorylation by promoting the expression of an E3 ubiquitin ligase rififylin and subsequent ubiquitination and degradation of PRR5L that is a suppressor of mTORC2-mediated phosphorylation of PKC.40 Activated Gαi subunits modulate the activity of the cyclic adenosine monophosphate–dependent kinase PKA but not directly PLC-β. Although the activation of Gαi has not been linked to PKC activation,8 the released Gβγ subunits from Gαi may activate kinases such as PKC and PI3K providing an additional link between SMO and PKC.37
In addition, our data indicate that the mechanistic link between SMO and NF-κB involves activation of PKCβ-1 and PKCβ-2 and the recruitment of CARMA1, BCL10, and MALT1 (CBM signalosome) to the SMO receptor complex. However, it remains to be determined if the CBM complex binds directly to SMO or if other scaffold proteins are required for its assembly.
The main PKC isoforms expressed in B cells are PKCβ-1 and PKCβ-232,33 and the contribution of SMO on the activation of these 2 PKC isoforms has not been previously studied. Previous studies have showed that activation of different PKC isoforms results in NF-κB activation.41-43 It has been reported that, in embryonic mouse stem cells, Shh enhances the translocation of PKCα, δ, and z isoforms from the cytosol to the cytoplasmic membrane supporting Hh-induced activation of these PKC isoforms.31 Earlier studies have found that CARMA3 and TRAF6 are required for GPCR-induced NF-κB activation in Mef cells.29 CARMA1 is the CARMA3-equivalent scaffold protein expressed in hematopoietic cells.9 It has not been investigated whether CARMA1 is required for GPCR-induced NF-κB activation in hematopoietic cells. In the T-cell receptor signaling pathway, stimulation of T-cell receptor results in phosphorylation of PKCθ followed by phosphorylation and activation of CARMA144 that recruits BCL10, MALT1, and TRAF6.45,46 TRAF6 functions as an E3 ligase inducing K63-linked polyubiquitination of NEMO (IKKγ) followed by activation of the IKK complex and NF-κB.45
DLBCL of ABC type has upregulated NF-κB pathway and is oncogenically “addicted” to high NF-κB activity.47 Our data show that SMO contributes to the activation NF-κB in cell lines of both GC and ABC type. These findings suggest that the crosstalk between SMO and NF-κB represents an additional level of NF-κB activity regulation that seems to be independent of the constitutive NF-κB activation induced by genetic abnormalities. Thus, in addition to genetic abnormalities, external signals such as Hh ligands secreted by stromal cells as well as by the own tumor cells may contribute to augment the activation of NF-κB.
Additional support of the functional crosslink between SMO and NF-κB was also obtained from the demonstration of enhanced effect on apoptosis after combining cyclopamine-KAAD and BAY-11-7082. The treatment with both inhibitors also resulted in a synergistic decrease in the mRNA levels of GLI1 as well as BCL2 and BCL-xL. BCL2 and BCL-xL genes are downstream targets of both Hh and NF-κB pathways and GLI1 is a direct downstream target of Hh signaling per se. These findings provide a molecular explanation for the beneficial effect on cell apoptosis of combining SMO and NF-κB inhibitors.
Finally, in addition to the link between SMO with NF-κB documented here, there is experimental evidence that NF-κB pathway can also modulate the activity of Hh signaling.48,49 Recently, Kasperczyk and colleagues50 have demonstrated that SHH is a direct downstream target gene of NF-κB as p65 binds to the SHH promoter. These experimental data and our findings support the existence of complex bidirectional crosstalk between NF-κB and Hh pathways that likely contributes to promote tumor growth and apoptosis resistance.
In summary, we describe a novel noncanonical role of SMO that contributes to NF-κB activation in DLBCL. The propagation of the SMO signal is mediated by a GPCR-like mechanism with activation of PKCβ and assembling the CBM signalosome (Figure 5E). These findings are of clinical interest and provide a rationale for the combination of SMO and NF-κB inhibitors as potential therapeutic approaches in DLBCL as well as in other malignancies.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Drs Jeremy Reiter, Hui-Kuan Lin, and Lan Pham for providing the following plasmids: Smo-GFP, eGFP, HA-K63, Flag-TRAF6, and PGL3-NF-κB-6X.
This study was supported by funds from the Translational Grant of The Leukemia & Lymphoma Society (R.R.S., F.V.), National Institutes of Health K08 Physician-Scientist Award 1 K08 CA143151-01 (F.V.), and The University Cancer Foundation via the Institutional Research Grant program at the University of Texas Anderson Cancer Center (F.V.). The primary tumor samples were provided by the Hematopathology Tissue Bank of the UT Anderson Cancer Center (supported by the National Cancer Institute/National Institutes of Health Grant CA16672).
Authorship
Contribution: C.Q., R.R.S., X.L., and F.V. designed research; C.Q., Y.L., and K.K. performed research; C.Q., M.B., N.K.A., and F.V. analyzed data; and C.Q. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Francisco Vega, Department of Hematopathology, Unit 72, The University of Texas MD Anderson Cancer Center, 1515 Holcombe Blvd, Houston, TX 77030; e-mail: fvegava@mdanderson.org; and Nitin Kumar Agarwal, Department of Hematopathology, Unit 72, The University of Texas MD Anderson Cancer Center, 1515 Holcombe Blvd, Houston, TX 77030; email: nkagarwa@mdanderson.org.
References
Author notes
N.K.A. and F.V. contributed equally to the study.



![Figure 5. SMO inhibitor enhanced the cytotoxic effect of NF-κB inhibitor on DLBCL cells. (A) BJAB and DOHH2 were treated with cyclopamine-KAAD and/or BAY11-7082. Combined treatments resulted in enhanced decrease of cell viability, as determined by MTS assays (at 48 hours), compared with treatment with the BAY11-7082 alone. (B) Annexin V and PI plots showed the percentage of apoptosis at 48 hours baseline (dimethylsulfoxide [DMSO], a), after treating with cyclopamine-KAAD or BAY11-7082 alone (b-c), and after combination of both inhibitors (d). The percentage of apoptosis was measured 48 hours after treatments and determined by Annexin V and PI assays. Data are presented as mean ± standard deviation (SD) of 3 experiments. *P < .05. (C) Western blot analysis showed that combining cyclopamine-KAAD with BAY11-7082 resulted in an increase of cleaved poly (adenosine 5′-diphosphate-ribose) polymerase 1 and a decrease of p-p65 compared with that treated with BAY11-7082 alone. (D) Quantitative real-time PCR showed that combined treatments resulted in marked decrease in the mRNA levels of GLI1, BCL2, and BCL-xL compared with BAY11-7082 treatment alone. These results were normalized to 18S mRNA level and expressed as fold changes in mRNA expression compared with control. Data are presented as mean ± SD of 3 experiments. *P < .05. (E) Proposed working model for SMO-mediated NF-κB activation using a GPCR signal transduction pathway. In the presence of Hh ligands, SMO is released from PTCH1 inhibition and gets activated. Activated SMO transduces signals through a canonical Hh pathway (GLI transcription factors) as well as through noncanonical a Hh pathway by recruiting and activating Gαi and Gα12. As a consequence, the Gβγ subunit is released and may contribute to phosphorylate and activate PKC-β. Gα12 may also contribute to the activation of PKC-β through the ρ family of small GTPases. Activated PKC-β phosphorylates and activates CARMA1 leading to the assembly of the CBM signalosome, polyubiquitination of TRAF6 and NEMO at K63, and followed by degradation of IκBα and activation of NF-κB pathway.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/121/23/10.1182_blood-2012-12-470153/4/m_4718f5.jpeg?Expires=1769155482&Signature=Zvb~dM3jugY8xr987xJ921Y2-GKYX1j~5xmmQIwm0L43nFPnZujJkw1vbftq7DhJ5IbvsZPcVhz5XWLLw5LsrFs1B7hFwCVJxLTc6dqBqIXnxnRCoK71Z13EAXiejm9ygp6XF4oxYnX~vFx-yqmf8IajbaOXruRLymP-SnEEFZLQO-1oDhFjnssxpzwm-a95jE2ujLqfHdiq7mIHxEUdAs4Oi5EhxXQLhNN22SByL48ChP8oW-aOSJ55zSUtSfc0LfERjW2-A7rogJPiaGy0Oa2EBqXWPeBVOEAoiv-aQ3KtNJ406yIzctZhZbkvjvuVkDbUTnlMHUARUelTqFjApQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal