Key Points
Richter syndrome has genomic complexity intermediate between chronic lymphocytic leukemia and diffuse large B-cell lymphoma.
Inactivation of TP53 and of CDKN2A is a main mechanism in the transformation to Richter syndrome.
Abstract
Richter syndrome (RS) occurs in up to 15% of patients with chronic lymphocytic leukemia (CLL). Although RS, usually represented by the histologic transformation to a diffuse large B-cell lymphoma (DLBCL), is associated with a very poor outcome, especially when clonally related to the preexisting CLL, the mechanisms leading to RS have not been clarified. To better understand the pathogenesis of RS, we analyzed a series of cases including 59 RS, 28 CLL phase of RS, 315 CLL, and 127 de novo DLBCL. RS demonstrated a genomic complexity intermediate between CLL and DLBCL. Cell-cycle deregulation via inactivation of TP53 and of CDKN2A was a main mechanism in the histologic transformation from CLL phase, being present in approximately one half of the cases, and affected the outcome of the RS patients. A second major subgroup was characterized by the presence of trisomy 12 and comprised one third of the cases. Although RS shared some of the lesions seen in de novo DLBCL, its genomic profile was clearly separate. The CLL phase preceding RS had not a generalized increase in genomic complexity compared with untransformed CLL, but it presented clear differences in the frequency of specific genetic lesions.
Introduction
Richter syndrome (RS), which is defined by the histologic transformation to a diffuse large B-cell lymphoma (DLBCL), occurs in up to 15% patients with chronic lymphocytic leukemia (CLL).1-3 RS is associated with a very poor outcome, especially when clonally related.1-3
Mechanisms leading to RS have not been clarified, and strong molecular and clinical predictors of transformation at the time of the diagnosis of CLL are needed. In the vast majority of the studies, authors have assessed RS for the genetic events known to occur in CLL and in DLBCL.2-9 Only a few, unbiased, genome-wide studies have been reported, which were performed on a limited number of samples.7,10 So far, the most important events recently elucidated regarding RS pathogenesis are TP53 inactivation and the activation of MYC pathway,3 which also could be mediated by somatic mutations activating NOTCH1.9
Better characterization of the genetics of RS could serve diverse purposes. The assessment of specific copy number (CN) changes underlying the CLL phase of patients with RS could help in an early identification of CLL patients with an enhanced transforming risk. The evaluation of genomic aberrations acquired during the transition from CLL phase to RS eventually would lead to a better understanding of the implicated biologic processes and potentially introduce better-tailored therapeutic approaches.
With these aims, we performed genome-wide DNA profiling on a large series of samples derived from the RS and CLL phases of patients with RS and compared them with untransformed CLL (CLL-U) and de novo DLBCL.
Materials and methods
Tumor panel
All clinical specimens were derived from pathological sites and obtained in the course of routine diagnostic procedures, before the initiation of therapy. CLL-phase and CLL-U samples were collected from peripheral blood at diagnosis. RS and DLBCL samples were collected from nodal or extranodal biopsies. Diagnoses were made following the recommended criteria.1 All RS cases represented DLBCL transformed from a typical CLL. The distinction between germinal center B-cell (GCB) and non-GCB type, on the basis of gene expression profiling, was available for 108 de novo DLBCL samples as previously published.11-13 Cases were selected on the basis of the availability of frozen material, with the fraction of neoplastic cells in the specimen representing >70% of overall cellularity as determined by morphologic and/or immunophenotypic studies. Informed consent was obtained in accordance to the Declaration of Helsinki following the procedures approved by the local ethical committees and institutional review boards of each participating institution. The study was approved by the Bellinzona Ethical Committee.
Genome-wide DNA profiling
Genome-wide DNA profiles were obtained from high-molecular-weight genomic DNA by use of the Genome-Wide Human SNP Array 6.0 (Affymetrix, Santa Clara, CA), as previously reported.14 The raw data reported in this article have been deposited in the Gene Expression Omnibus database (accession number GSE50252). For 73 of 127 DLBCL cases, CEL files were kindly provided by Laura Pasqualucci.11 The raw CN was extracted from CEL files as previously reported.14,15 Genomic profiles were segmented with the Fast first-derivative segmentation algorithm.15 Profiles were considered as poor quality and discarded from further analyses if they showed severe oversegmentation or no aberrations at all as evaluated by two independent investigators (I.K., F.B.). Minimal common regions (MCRs)16 were calculated. MCRs containing genes encoding the immunoglobulin heavy chain (IGHV) genes and the κ and λ light chains were discarded.13 Focal aberrations additionally were identified with the Genomic Identification of Significant Targets in Cancer 2.0 algorithm17 following the default criteria on the filtered and merged segments.
Mutational analysis
Rearrangements of the IGHV-D-J were amplified and sequenced as previously described.3 We documented the clonal relationship between the RS phase and the CLL phase by analyzing the IGHV-D-J rearrangements.3 Mutational analysis of the TP53 and NOTCH1 genes was performed as previously described.3,18 Mutational analysis of the CDKN2A gene was performed by the use of 5′-ACC GGA GGA AGA AAG AGG AG-3′ and 5′-AGA ATC GAA GCG CTA CCT GA-3′ as forward and reverse primers for the p16INK4a isoform (NM_000077) and 5′-TGG GTC CCA GTC TGC AGT TA-3′ and 5′-TAG CCT GGG CTA GAG ACG AA-3′ as forward and reverse primers for the p14ARF isoform (NM_058195).
Analysis of clinical data
Overall survival (OS) was calculated from the time of histologic transformation to the last follow-up or death of any cause. Log-rank test was used to investigate the impact on OS of categorical variables. The cumulative probability of OS was plotted as curve according to the Kaplan-Meier method. The χ2 test was used to compare differences in clinical parameters among subgroups. A P value of < 0.05 was considered as statistically significant. Statistical analyses were performed with Stata/SE v.12.1 (StataCorp, College Station, TX).
Results
RS has a genomic complexity intermediate between CLL and DLBCL
To better understand the pathogenesis of RS, we have analyzed, by using the same high-resolution genome wide-DNA profiling assay, tumor samples derived from: (1) RS, that is, DLBCL in patients with a previous documented history of CLL (n = 59, Table 119 ); (2) CLL phase of RS, that is, CLL samples from patients who have then undergone histologic transformation to DLBCL (n = 28, 15 with paired RS); (3) CLL, from patients who have not undergone histologic transformation to an aggressive lymphoma (n = 315); and (4) de novo DLBCL, that is, from patients without having a previous history of CLL (n = 127).
Clinical characteristics of the RS series
| Characteristics . | Number/available data . | % . | 95% CI . |
|---|---|---|---|
| Clinical features at RS diagnosis | |||
| Age >60 y | 29/41 | 70 | 54-84 |
| Male | 24/43 | 56 | 39-71 |
| ECOG PS > 1 | 10/19 | 52 | 29-76 |
| Ann Arbor stage III-IV | 38/39 | 97 | 86-99 |
| Binet stage | 20 | ||
| A | 4/20 | 20 | 6-44 |
| B | 8/20 | 40 | 19-64 |
| C | 8/20 | 40 | 19-64 |
| B symptoms | 13/28 | 46 | 27-66 |
| Extranodal sites >1 | 8/26 | 31 | 14-52 |
| Tumor size >5 cm | 16/34 | 47 | 30-65 |
| Nodal areas >5 | 11/28 | 39 | 21-59 |
| ALC >5.0 × 109/L | 12/27 | 44 | 25-65 |
| Hb < 10 g/dL | 11/30 | 37 | 20-56 |
| Platelets < 100 × 109/L | 7/29 | 24 | 10-43 |
| LDH > 1.5 ULN | 14/28 | 50 | 31-69 |
| CD38 expression | 12/26 | 46 | 27-67 |
| ZAP70 expression | 11/18 | 61 | 36-83 |
| Previous CLL therapies >1 | 17/22 | 77 | 54-92 |
| Pathologic features at RS diagnosis | |||
| Non-GC phenotype* | 25/25 | 100 | 87-100 |
| CD10 expression | 0/27 | 0 | 0-13 |
| BCL6 expression | 9/24 | 38 | 19-55 |
| MUM1 expression | 21/22 | 95 | 77-100 |
| CD5 expression† | 10/13 | 77 | 46-95 |
| CD23 expression† | 12/15 | 80 | 52-96 |
| MYC translocation‡ | 3/17 | 18 | 4-43 |
| TP53 mutations‡ | 12/20 | 60 | 36-81 |
| Ki-67+>70% | 14/30 | 47 | 28-66 |
| CLL treatment previous to RS | |||
| Fludarabine-based regimens | 14/22 | 64 | 41-83 |
| Previous alkylating agents | 12/22 | 55 | 32-76 |
| RS treatment | |||
| Rituximab chemotherapy | 18/28 | 64 | 44-81 |
| CHOP-based treatment | 19/28 | 68 | 48-84 |
| NHL second-line regimens | 2/28 | 7 | 9-23 |
| Allogeneic stem cell transplant | 2/28 | 7 | 9-23 |
| Characteristics . | Number/available data . | % . | 95% CI . |
|---|---|---|---|
| Clinical features at RS diagnosis | |||
| Age >60 y | 29/41 | 70 | 54-84 |
| Male | 24/43 | 56 | 39-71 |
| ECOG PS > 1 | 10/19 | 52 | 29-76 |
| Ann Arbor stage III-IV | 38/39 | 97 | 86-99 |
| Binet stage | 20 | ||
| A | 4/20 | 20 | 6-44 |
| B | 8/20 | 40 | 19-64 |
| C | 8/20 | 40 | 19-64 |
| B symptoms | 13/28 | 46 | 27-66 |
| Extranodal sites >1 | 8/26 | 31 | 14-52 |
| Tumor size >5 cm | 16/34 | 47 | 30-65 |
| Nodal areas >5 | 11/28 | 39 | 21-59 |
| ALC >5.0 × 109/L | 12/27 | 44 | 25-65 |
| Hb < 10 g/dL | 11/30 | 37 | 20-56 |
| Platelets < 100 × 109/L | 7/29 | 24 | 10-43 |
| LDH > 1.5 ULN | 14/28 | 50 | 31-69 |
| CD38 expression | 12/26 | 46 | 27-67 |
| ZAP70 expression | 11/18 | 61 | 36-83 |
| Previous CLL therapies >1 | 17/22 | 77 | 54-92 |
| Pathologic features at RS diagnosis | |||
| Non-GC phenotype* | 25/25 | 100 | 87-100 |
| CD10 expression | 0/27 | 0 | 0-13 |
| BCL6 expression | 9/24 | 38 | 19-55 |
| MUM1 expression | 21/22 | 95 | 77-100 |
| CD5 expression† | 10/13 | 77 | 46-95 |
| CD23 expression† | 12/15 | 80 | 52-96 |
| MYC translocation‡ | 3/17 | 18 | 4-43 |
| TP53 mutations‡ | 12/20 | 60 | 36-81 |
| Ki-67+>70% | 14/30 | 47 | 28-66 |
| CLL treatment previous to RS | |||
| Fludarabine-based regimens | 14/22 | 64 | 41-83 |
| Previous alkylating agents | 12/22 | 55 | 32-76 |
| RS treatment | |||
| Rituximab chemotherapy | 18/28 | 64 | 44-81 |
| CHOP-based treatment | 19/28 | 68 | 48-84 |
| NHL second-line regimens | 2/28 | 7 | 9-23 |
| Allogeneic stem cell transplant | 2/28 | 7 | 9-23 |
ALC, absolute lymphocyte count; CHOP, cyclophosphamide, doxorubicin, vincristine, prednisone; CI, confidence interval; CR, complete remission; ECOG PS, Eastern Cooperative Oncology Group Performance Status; GC, germinal center; LDH, lactate dehydrogenase; NHL, non-Hodgkin lymphoma; R-CHOP, rituximab, cyclophosphamide, doxorubicin, vincristine, prednisone; ULN, upper limit of normal.
According to Hans et al19 with immunohistochemistry performed locally during the diagnostic procedure.
With immunohistochemistry performed locally during the diagnostic procedure.
From Rossi et al.3
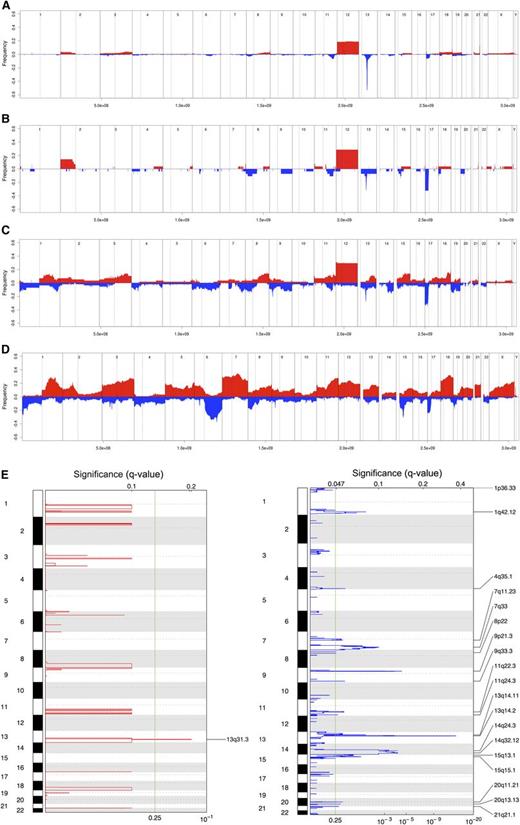
The median number of CN changes was three among CLL-U, 3.5 for CLL-phase, 8.5 for RS, and 16 for DLBCL. The differences were statistically significant between RS and CLL phase (P = .0035) or CLL-U (P < .0001) and between RS and DLBCL (P = .003). Similarly, the frequency of recurrent lesions also showed a progressive increase across the spectrum of the four diseases (Figure 1A-D).
Genomic aberrations in RS. Frequency of DNA gains (red, up) and losses (blue, down) observed in 315 cases of CLL-U (A), 28 cases of CLL-phase of RS (B), 58 cases of RS (C), and 127 cases of de novo DLBCL (D); representation of the focal gains (left) and losses (right) as detected via GISTIC in RS samples (E). (A-D) X-axis represents chromosome localization and physical mapping and y-axis the proportion of cases showing the aberrations. (E) False-discovery rate q values are plotted along the X-axis with chromosomal position along the Y-axis; altered regions with significant levels exceeding the vertical green line (significance threshold) were deemed significant; chromosomal positions are shown for each significant region on the right side of the plots.
Genomic aberrations in RS. Frequency of DNA gains (red, up) and losses (blue, down) observed in 315 cases of CLL-U (A), 28 cases of CLL-phase of RS (B), 58 cases of RS (C), and 127 cases of de novo DLBCL (D); representation of the focal gains (left) and losses (right) as detected via GISTIC in RS samples (E). (A-D) X-axis represents chromosome localization and physical mapping and y-axis the proportion of cases showing the aberrations. (E) False-discovery rate q values are plotted along the X-axis with chromosomal position along the Y-axis; altered regions with significant levels exceeding the vertical green line (significance threshold) were deemed significant; chromosomal positions are shown for each significant region on the right side of the plots.
RS presents a series of recurrent lesions
In RS, the four most prevalent MCRs, likely containing the candidate cancer genes, were deletions at 17p (affecting TP53), at 13q14.3 (DLEU2/MIR15A/MIR16-1), trisomy 12, and losses at 9p21 (CDKN2A; Table 2). Recurrent homozygous losses were detected at 9p21 (7%; CDKN2A) and 13q14.3 (3%; DLEU2/MIR15A/MIR16). The targets of the 17p and 9p deletions also were sequenced. TP53 was found mutated in 12 of 21 (57%) cases, and the mutations were concomitant to 17p loss in 5 of 12. No CDKN2A mutations were found among 16 cases analyzed (12 without 9p21 loss). There were amplifications targeting known oncogenes: JAK2 (two cases, in one also KDM4C/JMJD2C), MIR17HG (one case), MYC (one case), BCL2 (one case).
Minimal common regions observed in >10% of the cases of RS and CLL phase
| Diagnosis . | Lesion . | Cytoband . | Frequency, % . | Start* . | Size, Kb* . | Candidate genes . | No. genes† . |
|---|---|---|---|---|---|---|---|
| RS | Gains | ||||||
| 12 | 27 | 1.00E+00 | 1.34E+05 | >100 | |||
| 3q21.1-q29 | 13 | 1.23E+08 | 7.44E+04 | >100 | |||
| 8q21.2-q24.3 | 13 | 8.57E+07 | 6.06E+04 | MYC | >100 | ||
| 15q21.3-q26.3 | 15 | 5.64E+07 | 4.60E+04 | >100 | |||
| 18q21.2-q23 | 13 | 5.15E+07 | 2.65E+04 | BCL2, NFATC1 | 97 | ||
| 1q32.1 | 15 | 2.02E+08 | 3.53E+03 | MDM4 | 60 | ||
| 1q25.3 | 15 | 1.81E+08 | 1.76E+03 | 11 | |||
| 1q21.1-q32.1 | 15 | 1.46E+08 | 6.05E+04 | >100 | |||
| 11q24.3-q25 | 13 | 1.29E+08 | 6.34E+03 | 32 | |||
| 11q22.1-q22.3 | 13 | 1.02E+08 | 1.98E+03 | 18 | |||
| 13q31.3 | 12 | 9.28E+07 | 1.65E+02 | MIR17HG | 1 | ||
| Losses | |||||||
| 17p13.3-p11.2 | 33 | 5.26E+02 | 2.01E+04 | TP53 | >100 | ||
| 13q14.2-q14.3 | 28 | 4.99E+07 | 1.86E+03 | DLEU2, MIR15A, MIR16 | 20 | ||
| 9p21.3 | 22 | 2.13E+07 | 1.30E+03 | CDKN2A | 17 | ||
| 7q33 | 17 | 1.35E+08 | 1.76E+03 | 9 | |||
| 8p21.2 | 18 | 2.35E+07 | 7.45E+02 | 4 | |||
| 8p23.1 | 18 | 1.26E+07 | 8.14E+01 | 4 | |||
| 9q33.2-q34.3 | 15 | 1.23E+08 | 1.78E+04 | >100 | |||
| 14q32.11-q32.33 | 15 | 9.19E+07 | 1.40E+04 | >100 | |||
| 19p13.2-p13.11 | 15 | 1.29E+07 | 6.17E+03 | >100 | |||
| 9q21.33-q34.3 | 13 | 8.90E+07 | 5.21E+04 | >100 | |||
| 1q42.12-q42.3 | 15 | 2.26E+08 | 1.03E+04 | 90 | |||
| 1p36.33-p36.23 | 15 | 6.17E+04 | 8.75E+03 | TNFRSF14 | >100 | ||
| 4p14 | 13 | 3.87E+07 | 2.23E+03 | 24 | |||
| 7q32.3-q36.3 | 13 | 1.31E+08 | 2.81E+04 | >100 | |||
| 19p | 12 | 9.09E+04 | 2.80E+04 | TNFSF9 | >100 | ||
| 6p25.2-p24.2 | 12 | 3.43E+06 | 8.12E+03 | 49 | |||
| 11q22.3-q23.3 | 12 | 1.04E+08 | 1.23E+04 | ATM, MIR24B, MIR34C | 86 | ||
| CLL phase | Gains | ||||||
| 12 | 29 | 1.00E+00 | 1.34E+05 | >100 | |||
| 2p25.2-p12 | 14 | 1.28E+04 | 8.18E+04 | MYCN, REL | >100 | ||
| 21q22.3 | 11 | 4.73E+07 | 8.26E+02 | 14 | |||
| Losses | |||||||
| 17p13.3-p11.2 | 32 | 5.26E+02 | 1.94E+04 | TP53 | >100 | ||
| 13q14.2-q14.3 | 32 | 4.97E+07 | 2.14E+03 | DLEU2, MIR15A, MIR16 | 83 | ||
| 15q14-q15.3 | 14 | 3.97E+07 | 4.13E+03 | MGA | 83 | ||
| 8p23.3-q13.2 | 11 | 4.64E+04 | 6.81E+04 | TNFRSF10A, TNFRSF10B | >100 | ||
| 11q14.1-q23.3 | 11 | 7.99E+07 | 3.69E+04 | ATM, BIRC3, MIR34B, MIR34C | >100 | ||
| 18p | 11 | 1.15E+04 | 1.50E+04 | >100 | |||
| 20p11.23 | 11 | 1.93E+07 | 5.38E+02 | 8 |
| Diagnosis . | Lesion . | Cytoband . | Frequency, % . | Start* . | Size, Kb* . | Candidate genes . | No. genes† . |
|---|---|---|---|---|---|---|---|
| RS | Gains | ||||||
| 12 | 27 | 1.00E+00 | 1.34E+05 | >100 | |||
| 3q21.1-q29 | 13 | 1.23E+08 | 7.44E+04 | >100 | |||
| 8q21.2-q24.3 | 13 | 8.57E+07 | 6.06E+04 | MYC | >100 | ||
| 15q21.3-q26.3 | 15 | 5.64E+07 | 4.60E+04 | >100 | |||
| 18q21.2-q23 | 13 | 5.15E+07 | 2.65E+04 | BCL2, NFATC1 | 97 | ||
| 1q32.1 | 15 | 2.02E+08 | 3.53E+03 | MDM4 | 60 | ||
| 1q25.3 | 15 | 1.81E+08 | 1.76E+03 | 11 | |||
| 1q21.1-q32.1 | 15 | 1.46E+08 | 6.05E+04 | >100 | |||
| 11q24.3-q25 | 13 | 1.29E+08 | 6.34E+03 | 32 | |||
| 11q22.1-q22.3 | 13 | 1.02E+08 | 1.98E+03 | 18 | |||
| 13q31.3 | 12 | 9.28E+07 | 1.65E+02 | MIR17HG | 1 | ||
| Losses | |||||||
| 17p13.3-p11.2 | 33 | 5.26E+02 | 2.01E+04 | TP53 | >100 | ||
| 13q14.2-q14.3 | 28 | 4.99E+07 | 1.86E+03 | DLEU2, MIR15A, MIR16 | 20 | ||
| 9p21.3 | 22 | 2.13E+07 | 1.30E+03 | CDKN2A | 17 | ||
| 7q33 | 17 | 1.35E+08 | 1.76E+03 | 9 | |||
| 8p21.2 | 18 | 2.35E+07 | 7.45E+02 | 4 | |||
| 8p23.1 | 18 | 1.26E+07 | 8.14E+01 | 4 | |||
| 9q33.2-q34.3 | 15 | 1.23E+08 | 1.78E+04 | >100 | |||
| 14q32.11-q32.33 | 15 | 9.19E+07 | 1.40E+04 | >100 | |||
| 19p13.2-p13.11 | 15 | 1.29E+07 | 6.17E+03 | >100 | |||
| 9q21.33-q34.3 | 13 | 8.90E+07 | 5.21E+04 | >100 | |||
| 1q42.12-q42.3 | 15 | 2.26E+08 | 1.03E+04 | 90 | |||
| 1p36.33-p36.23 | 15 | 6.17E+04 | 8.75E+03 | TNFRSF14 | >100 | ||
| 4p14 | 13 | 3.87E+07 | 2.23E+03 | 24 | |||
| 7q32.3-q36.3 | 13 | 1.31E+08 | 2.81E+04 | >100 | |||
| 19p | 12 | 9.09E+04 | 2.80E+04 | TNFSF9 | >100 | ||
| 6p25.2-p24.2 | 12 | 3.43E+06 | 8.12E+03 | 49 | |||
| 11q22.3-q23.3 | 12 | 1.04E+08 | 1.23E+04 | ATM, MIR24B, MIR34C | 86 | ||
| CLL phase | Gains | ||||||
| 12 | 29 | 1.00E+00 | 1.34E+05 | >100 | |||
| 2p25.2-p12 | 14 | 1.28E+04 | 8.18E+04 | MYCN, REL | >100 | ||
| 21q22.3 | 11 | 4.73E+07 | 8.26E+02 | 14 | |||
| Losses | |||||||
| 17p13.3-p11.2 | 32 | 5.26E+02 | 1.94E+04 | TP53 | >100 | ||
| 13q14.2-q14.3 | 32 | 4.97E+07 | 2.14E+03 | DLEU2, MIR15A, MIR16 | 83 | ||
| 15q14-q15.3 | 14 | 3.97E+07 | 4.13E+03 | MGA | 83 | ||
| 8p23.3-q13.2 | 11 | 4.64E+04 | 6.81E+04 | TNFRSF10A, TNFRSF10B | >100 | ||
| 11q14.1-q23.3 | 11 | 7.99E+07 | 3.69E+04 | ATM, BIRC3, MIR34B, MIR34C | >100 | ||
| 18p | 11 | 1.15E+04 | 1.50E+04 | >100 | |||
| 20p11.23 | 11 | 1.93E+07 | 5.38E+02 | 8 |
Numbering according to Genome Reference Consortium Human Build 37 (GRCh37) (hg19).
According to the NCBI RefSeq database.
To identify significantly recurrently affected regions, we applied the GISTIC algorithm (Figure 1E, Table 3). The MIR17HG locus, coding for the mir17-92 miRNA cluster, at 13q31.3 was the only significant region of gain. On the converse, multiple regions of losses were identified, with, among the most significant, again deletions at 13q14.2-q14.3 (DLEU2/MIR15A/MIR16-1) and 9p21 (CDKN2A), but also at 14q24.2-q32.33.
Significant regions affected by DNA gains and losses in the RS genome, as estimated using the GISTIC algorithm
| Lesion . | Cytoband . | Frequency, % . | Start* . | Size, Kb* . | q value . | Residual q value . | Candidate genes . |
|---|---|---|---|---|---|---|---|
| Gains | |||||||
| 13q31.3 | 12 | 9.02E+07 | 2.40E+06 | 0.13392 | 0.13392 | MIR17HG | |
| Losses | |||||||
| 13q14.2-q14.3 | 29 | 5.07E+07 | 6.16E+05 | 7.17E-15 | 9.14E-15 | DLEU2/MIR15A/MIR16 | |
| 9p21.3 | 22 | 2.16E+07 | 8.94E+05 | 3.13E-05 | 3.13E-05 | CDKN2A | |
| 14q24.2-q32.33 | 16 | 7.11E+07 | 3.53E+07 | 7.71E-05 | 0.0013523 | TRAF3 | |
| 7q32.3-q36.2 | 12 | 1.31E+08 | 2.24E+07 | 0.0025523 | 0.0025523 | ||
| 1q42.12-q43 | 14 | 2.26E+08 | 1.11E+07 | 0.014836 | 0.014836 | ||
| 15q14-q15.1 | 9 | 3.89E+07 | 3.70E+06 | 0.026537 | 0.041218 | MGA | |
| 4q34.1-q35.2 | 10 | 1.74E+08 | 1.67E+07 | 0.11945 | 0.11556 | ||
| 9q33.3 | 16 | 1.28E+08 | 6.22E+05 | 0.12385 | 0.12385 | ||
| 15q11.2-q14 | 9 | 2.24E+07 | 1.24E+07 | 0.057148 | 0.12387 | ||
| 1p36.33-p34.2 | 14 | 1.00E+00 | 4.12E+07 | 0.13304 | 0.13304 | TNFRSF14 | |
| 11q21-q23.3 | 12 | 9.57E+07 | 2.16E+07 | 0.13304 | 0.13304 | ATM, BIRC3, MIR34B, MIR34C | |
| 7q11.1-q11.23 | 9 | 5.55E+07 | 2.17E+07 | 0.14934 | 0.14934 | ||
| 20q11.21-q11.22 | 5 | 2.96E+07 | 4.20E+06 | 0.14804 | 0.14934 | ||
| 20q13.13 | 7 | 4.89E+07 | 2.79E+05 | 0.17164 | 0.16885 | ||
| 11q24.2-q25.3 | 5 | 1.25E+08 | 9.52E+06 | 0.20946 | 0.19129 |
| Lesion . | Cytoband . | Frequency, % . | Start* . | Size, Kb* . | q value . | Residual q value . | Candidate genes . |
|---|---|---|---|---|---|---|---|
| Gains | |||||||
| 13q31.3 | 12 | 9.02E+07 | 2.40E+06 | 0.13392 | 0.13392 | MIR17HG | |
| Losses | |||||||
| 13q14.2-q14.3 | 29 | 5.07E+07 | 6.16E+05 | 7.17E-15 | 9.14E-15 | DLEU2/MIR15A/MIR16 | |
| 9p21.3 | 22 | 2.16E+07 | 8.94E+05 | 3.13E-05 | 3.13E-05 | CDKN2A | |
| 14q24.2-q32.33 | 16 | 7.11E+07 | 3.53E+07 | 7.71E-05 | 0.0013523 | TRAF3 | |
| 7q32.3-q36.2 | 12 | 1.31E+08 | 2.24E+07 | 0.0025523 | 0.0025523 | ||
| 1q42.12-q43 | 14 | 2.26E+08 | 1.11E+07 | 0.014836 | 0.014836 | ||
| 15q14-q15.1 | 9 | 3.89E+07 | 3.70E+06 | 0.026537 | 0.041218 | MGA | |
| 4q34.1-q35.2 | 10 | 1.74E+08 | 1.67E+07 | 0.11945 | 0.11556 | ||
| 9q33.3 | 16 | 1.28E+08 | 6.22E+05 | 0.12385 | 0.12385 | ||
| 15q11.2-q14 | 9 | 2.24E+07 | 1.24E+07 | 0.057148 | 0.12387 | ||
| 1p36.33-p34.2 | 14 | 1.00E+00 | 4.12E+07 | 0.13304 | 0.13304 | TNFRSF14 | |
| 11q21-q23.3 | 12 | 9.57E+07 | 2.16E+07 | 0.13304 | 0.13304 | ATM, BIRC3, MIR34B, MIR34C | |
| 7q11.1-q11.23 | 9 | 5.55E+07 | 2.17E+07 | 0.14934 | 0.14934 | ||
| 20q11.21-q11.22 | 5 | 2.96E+07 | 4.20E+06 | 0.14804 | 0.14934 | ||
| 20q13.13 | 7 | 4.89E+07 | 2.79E+05 | 0.17164 | 0.16885 | ||
| 11q24.2-q25.3 | 5 | 1.25E+08 | 9.52E+06 | 0.20946 | 0.19129 |
Numbering according to Genome Reference Consortium Human Build 37 (GRCh37) (hg19).
CLL-phase presents a limited number of recurrent lesions
In CLL-phase samples, the most common MCRs were deletions at 17p (TP53) and at 13q14.3 (DLEU2/MIR15A/MIR16), followed by trisomy 12, 11q, and 8p losses and 2p gains (Table 2). The only recurrent homozygous deletion targeted the 13q14.3 locus. The GISTIC algorithm did not identify any significant gain and only two losses, affecting the DLEU2/MIR15A/MIR16 and the MGA loci, respectively (data not shown).
CDKN2A loss is the most common acquired lesions at transformation
To identify aberrations acquired at histologic transformation, we compared RS with CLL phase (Table 4). Losses at 9p21 (CDKN2A) were the most statistically significant difference, being absent in CLL phase and detected in up to 20% of RS samples. In three cases with paired CLL phase and RS, the loss was clearly acquired at transformation. As also evident in Figure 1, RS presented a series of additional lesions more commonly than CLL phase, although the differences did not reach the statistical significance: losses at 6q and 9q or gains at 8q, 13q, 15q, and 18q. Conversely, losses at 13q14.3 (DLEU2/MIR15A/MIR16), 17p (TP53) and 11q (ATM), 8p, and also trisomy 12 and gains at 2p (REL) were observed at an overall similar frequency in both CLL phase and RS. Among 15 paired samples, TP53 loss was present in six cases, and in one case the lesion was acquired at the time of transformation. Figure 2 shows the acquisition of multiple genomic aberrations at the transformation in three paired genomic profiles.
Regions showing significantly differences between RS and CLL phase, CLL phase and CLL-U, and RS and de novo DLBCL
| Lesion . | Start* . | Size, Kb* . | RS, % . | % . | Candidate genes . | No. genes† . | P . |
|---|---|---|---|---|---|---|---|
| RS and CLL phase | |||||||
| +1q32.1 | 2.02E+08 | 3.53E+03 | 15 | 0 | MDM4 | 60 | .034 |
| +1q25.3 | 1.81E+08 | 1.76E+03 | 15 | 0 | 11 | .034 | |
| +3q21.1-q29 | 1.23E+08 | 7.44E+04 | 13 | 0 | >100 | .046 | |
| +3q25.2-q26.31 | 1.54E+08 | 2.13E+04 | 13 | 0 | 92 | .046 | |
| −1q42.12-q42.3 | 2.26E+08 | 1.03E+04 | 15 | 0 | 90 | .034 | |
| −1p36.33-p36.23 | 6.17E+04 | 8.75E+03 | 15 | 0 | TNFRSF14, TP73 | >100 | .034 |
| −9p21.3 | 2.13E+07 | 1.30E+03 | 22 | 0 | CDKN2A | 17 | .005 |
| −19p13.2-p13.11 | 1.29E+07 | 6.17E+03 | 15 | 0 | TNFSF9 | >100 | .034 |
| CLL phase and CLL-U | |||||||
| +2p25.3-p13.1 | 1.28E+04 | 7.40E+04 | 14 | 3 | REL, MYCN | >100 | .00432 |
| −8p23.3-p12 | 4.64E+04 | 3.45E+04 | 11 | 3 | >100 | .0302 | |
| −13q14.2-q14.3 | 5.05E+07 | 1.11E+03 | 32 | 53 | DLEU2/MIR15A/MIR16 | 12 | .034 |
| −15q15.1 | 4.01E+07 | 2.29E+03 | 14 | 3 | MGA | 55 | .0023 |
| −17p13.3-p11.2 | 5.26E+02 | 2.02E+04 | 32 | 6 | TP53 | >100 | <.00001 |
| −15q15.1 (homozygous loss) | 5.26E+02 | 2.02E+04 | 4 | 0.3 | MGA | 17 | .03025 |
| RS and de novo DLBCL | |||||||
| +1q32.1 | 2.02E+08 | 3.37E+03 | 15 | 28 | MDM4 | 57 | .0311 |
| −1p36.33-p36.31 | 6.17E+04 | 6.56E+03 | 15 | 28 | TNFRSF14, TP73 | >100 | .0196 |
| +2p16.1-p15 | 6.07E+07 | 2.38E+03 | 10 | 22 | REL | 19 | .0269 |
| +3q27-q29 | 1.88E+08 | 1.03E+04 | 13 | 28 | 81 | .0087 | |
| +5p15-q15 | 1.55E+04 | 9.55E+04 | 2 | 18 | >100 | .002 | |
| +6p25.3-p12.1 | 1.50E+05 | 5.38E+04 | 3 | 16 | >100 | .016 | |
| −6q23.3 | 1.37E+08 | 1.73E+03 | 9 | 38 | TNFAIP3 | 12 | .00003 |
| −6q16.3-q21 | 1.01E+08 | 6.50E+03 | 10 | 35 | PRDM1 | 17 | .00049 |
| +7 | 4.33E+04 | 1.59E+05 | 5 | 34 | >100 | .00002 | |
| −7q31.31-q36.3 | 1.18E+08 | 4.10E+04 | 13 | 3 | >100 | .017 | |
| +9q31.1-q34.11 | 1.08E+08 | 2.29E+04 | 4 | 12 | >100 | .023 | |
| −11q22.3 | 1.08E+08 | 4.97E+03 | 13 | 2 | ATM | 1 | .0046 |
| +12 | 1.00E+00 | 1.34E+08 | 27 | 17 | >100 | .046 | |
| −13q14.3 | 5.05E+07 | 1.10E+06 | 28 | 12 | DLEU2/MIR15A/MIR16 | 12 | .0075 |
| −14q24.1-q32.33 | 6.93E+07 | 3.69E+04 | 15 | 3 | TRAF3 | >100 | .0062 |
| −15q21.1-q21.1 | 4.11E+07 | 3.93E+03 | 7 | 31 | B2M, MGA | 78 | .0017 |
| +18 | 1.15E+04 | 7.80E+07 | 14 | 32 | BCL2, MALT1, NFATC1 | >100 | .011 |
| +20 | 6.13E+04 | 6.29E+04 | 0 | 13 | >100 | .0033 | |
| +21p11.2-q22.3 | 1.07E+07 | 3.74E+07 | 3 | 17 | >100 | .0090 |
| Lesion . | Start* . | Size, Kb* . | RS, % . | % . | Candidate genes . | No. genes† . | P . |
|---|---|---|---|---|---|---|---|
| RS and CLL phase | |||||||
| +1q32.1 | 2.02E+08 | 3.53E+03 | 15 | 0 | MDM4 | 60 | .034 |
| +1q25.3 | 1.81E+08 | 1.76E+03 | 15 | 0 | 11 | .034 | |
| +3q21.1-q29 | 1.23E+08 | 7.44E+04 | 13 | 0 | >100 | .046 | |
| +3q25.2-q26.31 | 1.54E+08 | 2.13E+04 | 13 | 0 | 92 | .046 | |
| −1q42.12-q42.3 | 2.26E+08 | 1.03E+04 | 15 | 0 | 90 | .034 | |
| −1p36.33-p36.23 | 6.17E+04 | 8.75E+03 | 15 | 0 | TNFRSF14, TP73 | >100 | .034 |
| −9p21.3 | 2.13E+07 | 1.30E+03 | 22 | 0 | CDKN2A | 17 | .005 |
| −19p13.2-p13.11 | 1.29E+07 | 6.17E+03 | 15 | 0 | TNFSF9 | >100 | .034 |
| CLL phase and CLL-U | |||||||
| +2p25.3-p13.1 | 1.28E+04 | 7.40E+04 | 14 | 3 | REL, MYCN | >100 | .00432 |
| −8p23.3-p12 | 4.64E+04 | 3.45E+04 | 11 | 3 | >100 | .0302 | |
| −13q14.2-q14.3 | 5.05E+07 | 1.11E+03 | 32 | 53 | DLEU2/MIR15A/MIR16 | 12 | .034 |
| −15q15.1 | 4.01E+07 | 2.29E+03 | 14 | 3 | MGA | 55 | .0023 |
| −17p13.3-p11.2 | 5.26E+02 | 2.02E+04 | 32 | 6 | TP53 | >100 | <.00001 |
| −15q15.1 (homozygous loss) | 5.26E+02 | 2.02E+04 | 4 | 0.3 | MGA | 17 | .03025 |
| RS and de novo DLBCL | |||||||
| +1q32.1 | 2.02E+08 | 3.37E+03 | 15 | 28 | MDM4 | 57 | .0311 |
| −1p36.33-p36.31 | 6.17E+04 | 6.56E+03 | 15 | 28 | TNFRSF14, TP73 | >100 | .0196 |
| +2p16.1-p15 | 6.07E+07 | 2.38E+03 | 10 | 22 | REL | 19 | .0269 |
| +3q27-q29 | 1.88E+08 | 1.03E+04 | 13 | 28 | 81 | .0087 | |
| +5p15-q15 | 1.55E+04 | 9.55E+04 | 2 | 18 | >100 | .002 | |
| +6p25.3-p12.1 | 1.50E+05 | 5.38E+04 | 3 | 16 | >100 | .016 | |
| −6q23.3 | 1.37E+08 | 1.73E+03 | 9 | 38 | TNFAIP3 | 12 | .00003 |
| −6q16.3-q21 | 1.01E+08 | 6.50E+03 | 10 | 35 | PRDM1 | 17 | .00049 |
| +7 | 4.33E+04 | 1.59E+05 | 5 | 34 | >100 | .00002 | |
| −7q31.31-q36.3 | 1.18E+08 | 4.10E+04 | 13 | 3 | >100 | .017 | |
| +9q31.1-q34.11 | 1.08E+08 | 2.29E+04 | 4 | 12 | >100 | .023 | |
| −11q22.3 | 1.08E+08 | 4.97E+03 | 13 | 2 | ATM | 1 | .0046 |
| +12 | 1.00E+00 | 1.34E+08 | 27 | 17 | >100 | .046 | |
| −13q14.3 | 5.05E+07 | 1.10E+06 | 28 | 12 | DLEU2/MIR15A/MIR16 | 12 | .0075 |
| −14q24.1-q32.33 | 6.93E+07 | 3.69E+04 | 15 | 3 | TRAF3 | >100 | .0062 |
| −15q21.1-q21.1 | 4.11E+07 | 3.93E+03 | 7 | 31 | B2M, MGA | 78 | .0017 |
| +18 | 1.15E+04 | 7.80E+07 | 14 | 32 | BCL2, MALT1, NFATC1 | >100 | .011 |
| +20 | 6.13E+04 | 6.29E+04 | 0 | 13 | >100 | .0033 | |
| +21p11.2-q22.3 | 1.07E+07 | 3.74E+07 | 3 | 17 | >100 | .0090 |
Numbering according to Genome Reference Consortium Human Build 37 (GRCh37) (hg19).
According to the NCBI RefSeq database.
Examples of unbalanced genomic aberration acquired at the transformation in three paired CLL-phase and RS samples. Black arrows highlight losses at 9p (CDKN2A; pt#97, pt#203) and 17p (TP53; pt#203, pt#72) and an amplification at 9p (JAK2; pt#72). Black dots, raw CN values; red line, smoothed CN values. X-axis, genomic mapping; Y-axis, log2 copy-number values.
Examples of unbalanced genomic aberration acquired at the transformation in three paired CLL-phase and RS samples. Black arrows highlight losses at 9p (CDKN2A; pt#97, pt#203) and 17p (TP53; pt#203, pt#72) and an amplification at 9p (JAK2; pt#72). Black dots, raw CN values; red line, smoothed CN values. X-axis, genomic mapping; Y-axis, log2 copy-number values.
CLL in patients who develop RS exhibits a greater frequency of certain genomic aberrations compared with untransformed CLL
To identify lesions capable of identifying CLL patients at greater risk of RS transformation, we then compared the genomic profiles observed in the CLL-phase samples in patients who developed RS with the samples of CLL cases who with a median follow-up of 6.25 years have not transformed to RS (Table 4). The most significant differences between the two groups concerned a greater frequency of losses at 17p (TP53), 15q (MGA, including homozygous losses), and gains at 2p (MYCN, REL) in samples derived from CLL phase, which, conversely, presented less deletions targeting MIR15/MIR16 (13q14.3). Although not reaching statistical significance, also +12 and ATM losses were observed more frequently in CLL phase of RS-transformed cases than in CLL-U: 29% vs 18% and 11% vs 7%, respectively.
Because CLL genomic lesions show a differential distribution on the basis of the IGHV gene mutational status20 and the CLL phase of RS patients is mostly associated with unmutated IGHV,2,8 the genomic profiles of CLL phase were compared with CLL-U both with unmutated IGHV (uCLL-U, n = 111) and with mutated IGHV genes (mCLL-U, n = 199). Even when compared with the uCLL-U samples, CLL-phase samples more commonly displayed the loss of TP53 (32% vs 11%; P = .0046), but other lesions (−8p, RS 11% vs uCLL-U 4%; +2p, 14% vs 8%; −13q14.3, 32% vs 44%), although differentially affected, did not reach statistical significance. Similar frequencies were observed for 15q15 (MGA, RS 11% vs uCLL-U 7%; homozygous losses, 4% vs 1%) and 11q21-q23 (ATM, 11% vs 14%) losses, and for trisomy 12 (29% vs 24%).
Differences in distribution of genomic aberrations were largely more evident when CLL phase was compared with 199 mCLL-U cases. CLL phase presented more commonly 2p25.3-p12 gains (REL/MYCN, 14%% vs 1%; P < .00001), losses affecting 8p (11% vs 2%; P = .028), 11q21-q23.3 (ATM; 11% vs 2%; P = .028), 15q15 (MGA, 14% vs 2%, P = .0002), and 17p (TP53, 32% vs 4%; P < .00001); in contrast, they exhibited less frequently 13q14.3 deletions (DLEU2/MIR15A/MIR16B, 32% vs 58%; P = .0077).
In CLL, different patterns of 13q14.3 deletions have been described, mainly determined by concomitant loss or otherwise of both the MIR15/MIR16 and of the more centromeric RB1 locus.14,21-23 Here, we observed that the 13q14 losses also encompassed the RB1 locus more commonly in CLL phase and RS than in CLL-U: 56%, 63%, and 42%, respectively, but the differences were not statistically significant (P = .064).
RS shares some of the DLBCL lesions, but its profile remains separate
Finally, we compared RS with de novo DLBCL (Table 4). A large series of statistically significant differences were observed, largely attributable to a lower frequency in RS of frequently associated lesions of de novo DLBCL. The only lesions that appeared more commonly in RS were lesions typical for CLL (13q14.3 and 11q22.3 losses and +12) and the 7q31-q36 deletion. Lesions that were the most significantly overrepresented in RS vs CLL-phase or in the CLL-phase vs CLL-U, namely losses of CDKN2A and TP53, showed similar frequency in both RS and DLBCL: 21% vs 17% and 33% vs 24%, respectively.
Because two main DLBCL subtypes are recognized on the basis of gene expression profiling, GCB, and non-GCB DLBCL, segregating unique recurrent genomic lesions,16 we compared the RS vs the two individual DLBCL subgroups (supplemental Table 1; see the Blood Web site). The levels of statistical significance of the observed differences were largely maintained, showing differences with both GCB- and non−GCB-DLBCL.
Two main genetic pathways lead to RS
Considering the most frequent lesions, we found that the 59 RS cases could be separated in three main subgroups that were mutually exclusive (Figure 3A). A first group, accounting for one half of the RS cases (30/60), was characterized by TP53 inactivation (by loss or by somatic mutations) and/or CDKN2A loss, alongside MYC activation (five gains and two translocations), 13q14.3 loss (n = 9) and additional lesions. A second group presented almost exclusively +12 (17/60, 28%). A third group was heterogeneous in terms of genomic lesions (13/60, 22%). Seven had genomic aberrations other than TP53/CDKN2A inactivation or +12, often including some of the most recurrent lesions; six cases did not apparently present any aberration.
Heatmap with the relative distribution of the most common lesions detected in RS samples (A), Kaplan-Meier graph showing OS in 24 RS patients according to the presence of TP53 inactivation and/or CDKN2A loss (B). (A) Red, presence; gray, absence; white, not available; cell proliferation (as measured by assessment of the percentage of Ki-67+ cells at immunohistochemistry) is also visualized. (B) X-axis, months; Y-axis, percentage of alive patients. The spikes on the survival curves indicate the censored patients.
Heatmap with the relative distribution of the most common lesions detected in RS samples (A), Kaplan-Meier graph showing OS in 24 RS patients according to the presence of TP53 inactivation and/or CDKN2A loss (B). (A) Red, presence; gray, absence; white, not available; cell proliferation (as measured by assessment of the percentage of Ki-67+ cells at immunohistochemistry) is also visualized. (B) X-axis, months; Y-axis, percentage of alive patients. The spikes on the survival curves indicate the censored patients.
NOTCH1, which can be activated by somatic mutations in CLL and RS,9,24-27 was sequenced in 28 cases: seven mutations were detected: 5 of 7 (71%) mutations were detected in cases bearing +12 and were mutually exclusive with MYC activation.
The vast majority of patients with a high proliferation rate (Ki-67+ cells > 70%) presented TP53 and/or CDKN2 inactivation (79%, 11/14), and the latter group showed a trend for a greater proliferation rate than the remaining cases (P = .057). A high proliferation rate was observed in 61% of the cases (11/18) with inactivation of TP53 and/or CDKN2, in 33% (2/6) with +12 and in 17% (1/6) of the genetically heterogeneous group. Among the 24 cases with available follow-up data, the cases with TP53 and/or CDKN2A inactivation had poorer OS than the remaining patients (P = .0105; hazard ratio; 3.6, 95% confidence interval, 1.3-10.1; Figure 3B). No associations were found for any of the three RS subgroups with IGHV gene mutational status, B-cell receptor stereotypy, expression of BCL6, MUM1, CD5, CD23, or the clonal relatedness of the CLL phase with the RS phase.
Discussion
We analyzed a large series of RS and its preexisting CLL phase, untransformed CLL, and de novo DLBCL cases by high-density genome-wide DNA microarray to better define the genetic aberrations underlying RS and to understand its relationship with the other three conditions. Our results are indicating that: (1) RS has a genomic complexity intermediate between CLL and DLBCL, as suggested by the CN aberration rates; (2) being present in approximately one half of the cases, inactivation of TP53 and of CDKN2A is a major mechanism in the histologic transformation from CLL phase and can affect the outcome of patients with RS; (3) a second major subgroup defined by genetic lesions is characterized by the presence of trisomy 12 and comprises approximately one third of the cases; (4) RS shares some of the DLBCL lesions, but its profile remains clearly separate; (5) the CLL phase preceding RS does not have an overall increase in genomic complexity compared with CLL-U, but it presents clear differences in the frequency of specific genetic lesions.
In RS cases in the present study, both the median number of lesions per sample and the recurrent lesions were intermediate between CLL and DLBCL, whereas the profiles of the latter two diseases demonstrated findings similar to what is reported in the literature.9,11 RS bore more lesions than CLL-U or even CLL phase, which is in accordance with the greater biologic and clinical aggressiveness of the disease. However, the observation that RS showed less CN changes than DLBCL did not seem in keeping with the more aggressive clinical course in RS. In the immunodeficiency-related DLBCL, the presence of Epstein-Barr virus infection is associated with a lower number of genomic aberrations.28,29 Although RS arises in the preexisting immunodeficiency associated with CLL,1 Epstein-Barr virus infection does not play a major role in RS2,3 and is an unlikely explanation for our data. Epigenetic changes, such as specific methylation patterns,30 or still-unidentified somatic mutations, might explain the observed relative low genomic complexity in RS vs DLBCL.
The most common lesions in RS concerned TP53 losses, trisomy 12, 13q14.3 losses (DLEU2/MIR15A/MIR16B), and CDKN2A losses. All these lesions occurred in >20% of the cases and were accompanied by a series of additional aberrations, such as gains of the MIR17HG locus, losses at 14q32 (including TRAF3), at 15q15 (MGA), at 1p (including TNFRSF14), 11q (including ATM, BIRC3, MIR34B, MIR34C), and on the long arm of chromosome 7.
Apart from TP53 inactivation, trisomy 12 and 13q14.3 losses, already present in the CLL phase, the vast majority of the RS genomic aberrations are acquired at the time of transformation. The most frequent acquired lesions were deletions targeting CDKN2A, a known tumor suppressor gene involved in cell-cycle regulation. CDKN2A inactivation, by DNA losses and not by somatic mutations, was exclusively observed in RS and in DLBCL at similar frequencies, but neither in CLL-U nor in CLL phase. Importantly, in DLBCL, CDKN2A losses have been associated with a poor outcome and with a non−GCB-phenotype.16,31,32 In addition, there are previous suggestions of the importance of cell-cycle deregulation in RS.30,33-35 In our RS series, CDKN2A losses also were associated with poor outcome and occurred mostly concomitantly with TP53 inactivation.
The inactivation of either CDKN2A or TP53 was accompanied by additional genomic lesions (RB1 losses, MYC activation, gains of MIR17HG) and defined a subgroup of patients characterized by high proliferation rate and deregulation of cell cycle, apoptosis, senescence, and cellular metabolism. Of clinical relevance, these patients had a poorer outcome. Similarly to what has been very recently suggested for a subset of de novo DLBCL,35 it might be speculated that these patients might benefit by treatment with compounds targeting the dysregulated apoptotic machinery, inhibiting the cyclin-dependent kinases or reactivating TP53 activity.
TP53 inactivation and/or CDKN2A loss were mutually exclusive with trisomy 12, indicating that RS might develop via two main genetic pathways. The most common mechanism, used by at least one half of the cases, entails the accumulation of a series of genomic lesions, resulting in TP53 inactivation, MYC activation, and, finally, loss of CDKN2A. A second mechanism, occurring in one third of the patients, is driven by the early acquisition of trisomy 12. This lesion is followed by the acquisition of NOTCH1 mutations, in agreement with recent publications,9,24,25 and/or of a limited number of additional genomic aberrations not exclusively observed in this group of patients.
Approximately one fifth of the cases had neither TP53/CDKN2A inactivation nor trisomy 12 and exhibited a heterogeneous pattern of genomic aberrations, with individual patients bearing lesions (11q losses, 14q losses, MYC activation, NOTCH1 mutation, MIR17HG gain) also detected in the two major RS subgroups defined by TP53/CDKN2A losses or trisomy 12.
In agreement with the literature,2,3 the histology of all our cases of RS was that of DLBCL, and the vast majority of them presented a non-GCB phenotype. Overall, the RS genomic profile appeared statistically distant from both non−GCB-DLBCL and GCB-DLBCL but RS did not fully recapitulate the genomic lesions observed in DLBCL, neither in the GCB- nor the non−GCB-subgroups. RS shared some of the lesions commonly observed in DLBCL (such as gains of 1q, 3q, 13q, and 18q and losses at 9p and 17p), but other lesions, recurrent in DLBCL (such as gains of chromosomes 5, 7, or 6q losses) were clearly underrepresented in RS. Indeed, we had also previously reported that RS has an extremely low rate of deletions and mutations inactivating the two known tumor suppressor genes mapped at 6q, coding for PRDM1/BLIMP1 (6q21) and TNFAIP3/A20 (6q23),3 which prevent plasmacytic differentiation and activate the nuclear factor κB pathway, respectively.36
The lesions specifically detected in RS and not, or at lower frequency, in DLBCL comprised mainly the aberrations typically observed in CLL (+12, and 13q14.3 and 11q23 losses).20 The deletions targeting chromosome 13 often encompassed the RB1 gene, alongside DLEU2/MIR15A/MIR16B. This pattern is similar to the one recently described in DLBCL, in which the lesion seems to contribute to cell-cycle deregulation and also to the tumor immune escape,36 and compatible with the notion that that a wider 13q14 lesion, comprising also RB1, is associated with a more aggressive form of disease.14,21,23,38 Losses affecting the long arm of 7 losses were observed exclusively in RS; these losses were more telomeric than the deletions present in splenic marginal zone lymphomas,39 and their extension did not allow the identification of putative tumor suppressor genes.
RS cells usually carry unmutated IGHV genes; thus, it was not surprising that the genomic profiles of the paired CLL-phase samples were similar to that seen in uCLL-U. CLL phase presented an elevated frequency of specific genomic aberrations (such as 2p gains, losses at 8p, 15q, and TP53 inactivation) but not a general increase of genomic complexity. In our previous study of 323 unselected CLL cases, gains occurring at 2p, possibly affecting REL and/or MYCN, were associated with a poor outcome and also with a high risk of RS transformation20 : this observation is herein confirmed on a larger number of CLL-phase samples. Losses at 8p have been reported to be associated with TP53 inactivation and with a poorer outcome in lymphoid tumors, including CLL and DLBCL, often together with TP53 inactivation.20,40 Deletions at 15q affected MGA, a negative MYC regulator, which, thus, might be directly involved in CLL and RS pathogenesis.41,42
Finally, indications on new therapeutic targets for RS patients come from the observed genomic amplifications of 9p24.1 (JAK2, JMJD2C), 13q31.3 (MIR17HG), and 18q21.33 (BCL2), which are all directly involved in the positive regulation of MYC expression and activity,43,44 and, notably, amplifications of 8q24.21 (MYC) also were observed underlining the importance of the gene in RS pathogenesis. Moreover, MIR17HG also contributes to the activation of the PI3K/AKT/mTOR pathway.45 Our data provide the rational for the evaluation of new targeted therapies such JAK2 inhibitors, MYC inhibitors,46 BCL2 inhibitors, phosphatidylinositol 3-kinase inhibitors, AKT inhibitors, and mammalian target of rapamycin inhibitors.47
In conclusion, our data establish specific patterns of genomic lesions in RS patients with prognostic and potentially therapeutic relevance. Also, lesions differentiating RS from DLBCL and the RS CLL phase from untransformed CLL have been identified.
Partially presented as an oral communication at the 53rd annual meeting of the American Society of Hematology, San Diego, CA, December 10-13, 2011.
The data reported in this article have been deposited in the Gene Expression Omnibus database (accession number GSE50252).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by Oncosuisse OCS 02296-08-2008 (to F. Bertoni), the Helmut Horten Foundation (to F. Bertoni), the San Salvatore Foundation (to F. Bertoni), the Nelia et Amadeo Barletta Foundation (to F. Bertoni), Computational Life Sciences/Ticino in Rete (to F. Bertoni), and DFG-SFB1074, subproject B2 (to S.S.). E.C. was a recipient of an European Society for Medical Oncology (ESMO) Fellowship Grant.
Authorship
Contribution: E.C. interpreted data and cowrote the manuscript; A.R. performed genomic profiling; I.K. performed statistical analysis and interpreted data; P.M.V.R. performed statistical analysis; D.R. and G.G. designed research, collected and characterized tumor samples, and provided advice; J.C.S, D.O., K.S., T.P., F. Berger, K.H.Y., F.M., R.R., T.C.G., W.C.C., E.M.O., M. Lucioni, R.M., G.I., M. Ladetto, F.F., S.C., H.V., S.H.S., S.S., M.A.P, A.M., D.S., E.N., A.Z., V.G., G.B., and G.O. collected and characterized tumor samples; E.Z. provided advice and cowrote the manuscript; F. Bertoni designed research, performed statistical analysis, interpreted data, and cowrote the manuscript; and all authors approved the final manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Francesco Bertoni, MD, Lymphoma and Genomics Research Program, Institute of Oncology Research, via Vincenzo Vela 6, 6500 Bellinzona, Switzerland; e-mail: frbertoni@me.com.
References
Author notes
E.C. and A.R contributed equally to this study.